Abstract
The localization of the adenovirus E1B-55K-E4orf6 protein complex is critical for its function. Prior studies demonstrated that E4orf6 directs the nuclear localization of E1B-55K in human cells and in rodent cells that contain part of human chromosome 21. We show here that the relevant activity on chromosome 21 maps to RUNX1. RUNX1 proteins are transcription factors that serve as scaffolds for the assembly of proteins that regulate transcription and RNA processing. After transfection, the RUNX1a, RUNX1b, and RUNX1-ΔN variants allowed E4orf6-directed E1B-55K nuclear localization. The failure of RUNX1c to allow nuclear colocalization was relieved by the deletion of amino-terminal residues of this protein. In the adenovirus-infected mouse cell, RUNX1 proteins were localized to discrete structures about the periphery of viral replication centers. These sites are enriched in viral RNA and RNA-processing factors. RUNX1b and RUNX1a proteins displaced E4orf6 from these sites. The association of E1B-55K at viral replication centers was enhanced by the RUNX1a and RUNX1b proteins, but only in the absence of E4orf6. In the presence of E4orf6, E1B-55K occurred in a perinuclear cytoplasmic body resembling the aggresome and was excluded from the nucleus of the infected mouse cell. We interpret these findings to mean that a dynamic relationship exists between the E4orf6, E1B-55K, and RUNX1 proteins. In cooperation with E4orf6, RUNX1 proteins are able to modulate the localization of E1B-55K and even remodel virus-specific structures that form at late times of infection. Subsequent studies will need to determine a functional consequence of the interaction between E4orf6, E1B-55K, and RUNX1.
Human adenoviruses express genes early in a productive infection to create an environment suited for viral DNA replication, late gene expression, and virus assembly. The early region 1B 55-kDa (E1B-55K) and early region 4 open reading frame 6 (E4orf6) proteins are two such viral products that neutralize cellular antiviral responses (82) and promote late viral gene expression (7, 20, 24, 83). The E1B-55K and E4orf6 proteins form a physical complex in the nucleus of infected cells at late times of infection. This complex has been implicated in regulating mRNA transport by promoting the export of late viral messages while preventing the export of cellular messages (reviewed in reference 20). In addition, the complex directs the proteasome-dependent degradation of p53 (77), MRE11 (78), and DNA ligase IV (4) by reconfiguring a cellular ubiquitin ligase composed of Cul5, Rbx1, and elongins B and C (30, 67). Notably, mutant viruses that fail to express the E1B-55K or E4orf6 genes are restricted for replication. This is due in part to the inability to promote the efficient transport of late viral RNA, which leads to reduced late viral gene expression (24). Because mRNA transport in adenovirus-infected cells is dependent on the proteasomal degradation machinery, the regulation of cellular and viral gene expression appears to depend on the activity of the E1B-55K-E4orf6 protein complex as a viral ubiquitin ligase (14, 83). The localization of the E1B-55K and E4orf6 proteins at late times of infection suggests that this activity is found in the nucleus of the infected cell (25, 60).
The E1B-55K and E4orf6 proteins shuttle between the nucleus and cytoplasm (18, 41) and accumulate in the nucleus of infected cells. The E4orf6 protein is evenly distributed throughout the nucleoplasm and is excluded from nucleoli (16). E1B-55K exhibits a complex distribution throughout the nucleus and cytoplasm. Within the nucleus at late times of infection, E1B-55K is distributed throughout the nucleus, is found in small filamentous spicules, and is concentrated about the periphery of the viral replication centers (25, 40, 60). These centers, sometimes called viral factories, have long been recognized as sites of viral DNA replication (49) and late viral RNA synthesis (48). Interestingly, E1B-55K fails to associate with the viral replication centers in cells infected with mutant viruses unable to express the E4orf6 gene (60). The phenotype of the E4orf6 mutant virus is similar to that of the E1B-55K mutant virus. Cells infected with E4orf6 mutant viruses export late viral messages to the cytoplasm poorly compared to that by wild-type virus-infected cells (29, 32, 71) and fail to degrade cellular proteins such as p53, MRE11, and DNA ligase IV (4, 68, 77, 78). The ability of the E4orf6 protein to direct the association of E1B-55K to viral replication centers in the nucleus may be a critical element in the function of the E1B-55K-E4orf6 protein complex (25, 60).
When expressed by transfection, E1B-55K is restricted to the cytoplasm (26, 57). The E4orf6 protein is able to direct the nuclear localization of E1B-55K in primate cells but not in most rodent cells (13, 26). Because heterokaryons of human cells and rodent cells enabled the E4orf6 protein to retain E1B-55K in the rodent cell nucleus, we proposed that a primate-specific activity or factor promotes an interaction between the E1B-55K and E4orf6 proteins (26). Further studies mapped this activity to the distal region of human chromosome 21 (13). We report here that this activity mapped to human chromosome 21 is RUNX1, a gene previously identified as AML-1.
The RUNX1 proteins are so named because of their similarity to the Runt protein of Drosophila melanogaster, which was the first member of this family of proteins to be described (34). The RUNX1 proteins bind DNA and also serve as molecular scaffolds to promote protein-protein interaction at appropriate sites within the nucleus (31, 34, 87). At least four RUNX1 proteins have been described: RUNX1a, RUNX1b, RUNX1c, and RUNX1ΔN. These proteins are translated from alternatively spliced transcripts derived from two promoters (22, 44, 54). RUNX1 proteins are closely related to RUNX2 and RUNX3 proteins by their shared genomic architecture and through highly similar sequences in the amino terminus, called the Runt domain. The Runt domain is responsible for DNA binding (34, 56). RUNX1 proteins form the heterodimeric transcription factor identified as core binding factor (CBF), also known as the polyoma enhancer binding protein 2 (PEBP2), when associated with core binding factor beta (CBFβ) (56). RUNX1 members are involved in both transcriptional activation and repression during hematopoietic differentiation (34, 75). Consistently with their dual nature as scaffold proteins and DNA-binding transcription factors, RUNX1 proteins have a complex and dynamic physical structure composed of activation, inhibition, and negative regulatory domains (34). Negative regulatory regions for DNA binding and heterodimerization are present at both the amino and carboxyl termini. Signals within the cell of an unidentified nature cause these regions to mask the Runt domain and prevent it from associating with CBFβ or DNA (33). The RUNX1 proteins interact with many proteins within the nucleus, some of which act as corepressors of transcription while others act as coactivators (62). Proteins such as TLE1 and mSin3A are corepressors that contribute to RUNX1-mediated repression (47). RUNX1-associated coactivators include Myb and ALY/REF, which activate the T-cell receptor alpha enhancer; C/EBP, which promotes the coactivation of the macrophage colony-stimulating factor receptor promoter; and p300/CBP, which promotes the transcription of many genes required for myeloid cell differentiation (12, 36, 45). Interestingly, ALY/REF is deposited on newly synthesized mRNA in a sequence-independent manner and promotes efficient mRNA transport from the nucleus (61, 90). Therefore, the RUNX1 proteins may indirectly regulate mRNA transport by recruiting proteins such as ALY/REF to appropriate sites in the nucleus.
In this study, we demonstrate that human RUNX1 variants a, b, and ΔN promote the E4orf6-directed nuclear localization of E1B-55K in mouse cells, suggesting that it is the nuclear scaffold nature and not the DNA-binding ability of the RUNX1 protein that affects the apparent interaction between the E1B-55K and E4orf6 proteins. In addition, both endogenous RUNX1 protein and the RUNX1b variant protein localize at the periphery of virus replication centers at late times in adenovirus infection. RUNX1 localization at the virus replication centers appears to disrupt the normal localization of E4orf6 at these sites. In the absence of E4orf6, the RUNX1 proteins are able to retain E1B-55K at the periphery of virus replication centers. Therefore, E4orf6 and RUNX1 differentially affect the localization of E1B-55K during infection. Strikingly, viral replication centers were less developed in the absence of E1B-55K. However, the RUNX1b protein appeared to compensate for the absence of E1B-55K with respect to the development of viral replication centers. Taken together, these results suggest that the RUNX1b protein significantly affects the architecture of the virus replication center by mislocalizing the E4orf6 protein, ultimately leading to the inefficient trafficking of E1B-55K to the cytoplasm. The disruption of E4orf6-E1B-55K interactions at sites of virus replication within the nucleus could lead to defects in the virus replication cycle.
MATERIALS AND METHODS
Cells, plasmids, and viruses.
Cell culture media, cell culture supplements, and sera were obtained from Invitrogen Life Technologies (Gaithersburg, MD) or HyClone (Logan, UT). Jurkat cells and Raji cells were maintained in HYQ RPMI-1640 (HyClone) medium supplemented with 10% fetal bovine serum (FBS). SaOS-2 cells were maintained in McCoy's 5a medium supplemented with 15% FBS. Mouse A9 cells were maintained in DMEM supplemented with 10% FBS. Mouse A9-21 cells were maintained in DMEM supplemented with 10% FBS and 400 U of hygromycin B per ml (15). The GM11130 cell line (Coriell Institute, Camden, NJ) was maintained in DMEM supplemented with 0.1 mM sodium hypoxanthine, 0.4 μM aminopterin, 16 μM thymidine, and 10% FBS. The GM10063 cell line (Coriell Institute) was maintained in DMEM supplemented with 0.1 mM sodium hypoxanthine, 0.4 μM aminopterin, 16 μM thymidine, and 15% FBS. The Chinese hamster ovary (CHO) cell lines bearing portions of human chromosome 21 (21q+, MRC-2G, 6918, R2-10W, Raj-5, 643C-13, 72532x6, and E7b) were generated at the Eleanor Roosevelt Institute (Denver, CO) and were previously described (27). CHO cell lines were maintained in Ham's F12 medium supplemented with 10% FBS.
The plasmids encoding E1B-55K and E4orf6 under the control of both the T7 promoter and cytomegalovirus (CMV) immediate-early enhancer and promoter were described previously (26). The plasmids encoding human RUNX2, human RUNX3, mouse Runx1 (previously described as PEBP2αC, PEBP2αA, and PEBP2αB1, respectively), and human RUNXΔN under the control of the human EF-1α promoter were described previously (89). The plasmids encoding the influenza virus hemagglutinin (HA) epitope-tagged RUNX1a and RUNX1b under the control of the simian virus 40 immediate-early promoter were described previously (39). These plasmids also confer resistance to G418 by directing the expression of the Tn5 aminoglycoside 3′-phosphotransferase. The plasmid encoding human RUNX1c under the control of the CMV enhancer/promoter was kindly provided by Scott Hiebert (Vanderbilt University, Nashville, TN) and was described previously (53). The plasmid encoding an amino-terminally deleted RUNX1c under the control of the CMV enhancer/promoter was created by Scott Hiebert and kindly provided by Gary Stein (University of Massachusetts Medical School, Worcester). This RUNX1c plasmid originally was designated AML1b (or RUNX1b) but now has been correctly identified as RUNX1c. Herring sperm DNA was used as the carrier DNA in transfection experiments.
The adenovirus type 5 strain dl309 served as the wild-type adenovirus used in these studies. This virus is deleted of part of the E3 region that has been shown to be dispensable for growth in tissue culture (37). The E1B-55K mutant virus dl1520 contains a large deletion of the E1B-55K gene and is unable to express small splice variants of the larger E1B-55K protein. This virus has been previously described (5). The E1B-55K mutant virus dl338 contains a smaller deletion of the E1B-55K region and can direct the expression of E1B proteins that are not expressed by dl1520 (63). The E4orf6/E4orf7 mutant virus dl356, which expresses the E4orf6 gene but not the E4orf6/E4orf7 fusion product, was described previously (29). The E4orf6 mutant virus used for this study was dl355*, which includes the wild-type E3 gene, and was described previously (32). Viruses were grown in 293 cells, and concentrated virus stock was prepared by sequential centrifugation through CsCl gradients as previously described (26).
Antibodies.
Adenovirus-specific antibodies included the mouse monoclonal antibody Rsa#3 (50) for the E4orf6 protein used as undiluted hybridoma culture supernatant fluid, the rat monoclonal antibody 9C10 (EMD Calbiochem, San Diego, CA) for the E1B-55K protein used at 1 μg per ml, the mouse monoclonal antibody 2A6 (72) for the E1B-55K protein used as undiluted hybridoma culture supernatant fluid, and the mouse monoclonal antibody B6-8 (70) for the E2A-DNA-binding protein (E2A-DBP) used as a fivefold diluted hybridoma culture supernatant fluid. Rabbit polyclonal antibodies against RUNX1 (Sigma, St. Louis, MO), RUNX2 (Active Motif, Carlsbad, CA), and RUNX3 (Active Motif) were used at 1:500 dilutions. HA-tagged RUNX1 proteins were visualized with the high-affinity rat monoclonal antibody 3F10 (Roche Diagnostics, Indianapolis, IN), which was used at 200 ng per ml. The mouse monoclonal antibody RmcB (CRL-2379; ATCC, Manassas, VA) against the coxsackie and adenovirus receptor (CAR) was used for cell sorting as the undiluted hybridoma culture supernatant fluid. Ubiquitin-specific antibody (clone P4D1; Cell Signaling Technology, Danvers, MA) was used at a 1:1,000 dilution. Fluorescently labeled secondary antibodies, qualified for multiple labeling experiments, were used at 1 to 4 μg per ml. These antibodies included Alexa Fluor 488 conjugated to goat anti-mouse immunoglobulin G (IgG) (Molecular Probes/Invitrogen), Alexa Fluor 568 conjugated to goat anti-rat IgG (Molecular Probes/Invitrogen), and rhodamine red X-conjugated goat anti-rat IgG (Jackson ImmunoResearch, West Grove, PA).
Stable HA-RUNX1a and HA-RUNX1b mouse A9 cell lines.
Mouse A9 cells expressing HA-tagged human RUNX1a and RUNX1b cDNAs were generated by transfection and clonal selection. Briefly, 106 mouse A9 cells were transfected with 3 μg of HA-RUNX1a or HA-RUNX1b plasmid with Lipofectamine plus (Invitrogen Life Technologies) according to the manufacturer's recommendation. Two days after transfection, growth medium was replaced with medium supplemented with 600 μg of G418 per ml. Once all nontransfected cells died, single-transfected cell clones were obtained by serial dilution. Uniform HA-RUNX1 expression was confirmed by immunofluorescence, and the appropriate protein size was confirmed by immunoblotting. Two separate clones were maintained for each RUNX1 protein by propagation in G418-containing medium.
Stable mouse A9 cell lines expressing the hCAR gene.
Mouse A9 cells, HA-RUNX1a A9 cells, and HA-RUNX1b A9 cells were transduced with the pLXSN-based retrovirus expressing a carboxyl-terminally truncated variant of the human CAR (hCAR) (59). The virus was generously provided by James DeGregori (University of Colorado, Denver) through Linda Gooding (Emory University, Atlanta, GA). Working stocks of the retrovirus were produced as previously described (51). Because the HA-RUNX1 cells were resistant to G418, the virally transduced cells were selected by two rounds of staining for hCAR followed by cell sorting. Briefly, transduced cells were harvested in enzyme-free cell dissociation buffer (Invitrogen) and stained with the hCAR antibody RmcB for 1 h on ice, followed by being stained with Alexa Fluor 488-conjugated goat anti-mouse Ig for 30 min on ice. Cells were sorted using a FACSAria cell-sorting instrument (BD Biosciences, San Jose, CA). Sorted cells were maintained in medium containing 50 μg of G418 per ml to prevent microbial contamination. An additional round of cell sorting based on extracellular hCAR was performed to enrich for hCAR-positive cells.
Indirect immunofluorescence.
Indirect immunofluorescence with intact cells was conducted as previously described (13). Briefly, cells were fixed with 2% freshly prepared formaldehyde and then permeabilized with 0.2% Triton X-100 before indirect antibody labeling. Infected Jurkat, Raji, SaOS-2, and RUNX1-expressing mouse A9 cells were extracted prior to fixation as described previously (60). Extraction with Triton X-100 enhances the visualization of viral replication centers by removing the diffuse nuclear component of E2A-DBP while sparing the replication center-associated protein (60, 81). Briefly, cells were washed twice in ice-cold phosphate-buffered saline with 1.5 mM MgCl2 and then extracted for 5 min on ice with CSK buffer [100 mM KCl, 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 6.8), 300 mM sucrose, 3 mM MgCl2, 1 mM ethylene glycol tetraacetic acid, 1 mM phenylmethylsulfonyl fluoride] containing 0.5% Triton X-100. Cells then were fixed with 2% formaldehyde in CSK buffer lacking Triton X-100 for 20 min at room temperature, followed by indirect immunofluorescence with the appropriate antibodies. Samples were mounted with a glycerol- or polyvinyl alcohol-based mounting medium containing the DNA dye 4′,6-diamidino-2-phenylindole (DAPI) and were analyzed by epifluorescence microscopy using a Nikon TE300 inverted microscope fitted with filters appropriate for DAPI, Alexa Fluor 488, and Alexa Fluor 568 excitation. Images were acquired using a Retiga EX 1350 digital camera (QImaging Corp., Burnaby, British Columbia, Canada) with a ×60 magnification/1.4-numerical aperture or ×100 magnification/1.4-numerical aperture oil immersion objective. The relative brightness and contrast of the digital images within each figure were adjusted to the same extent based on exposures obtained from control samples stained with secondary antibody alone. Figures were assembled with the open-source image-processing software ImageJ (69) and Canvas 10 (ACD Systems, Miami, FL) using either a Macintosh or Dell microcomputer. Color is used in some micrographs in which the adenovirus protein is shown in green and the RUNX protein in magenta so that colocalization is seen as white.
Quantitative evaluation of E4orf6-mediated nuclear E1B-55K localization.
For the quantitative measurements reported in Fig. 1 and 4, cells were infected with recombinant vaccinia virus vTF7.3 (21) to express the T7 RNA polymerase and were transfected with cDNA for E1B-55K and E4orf6 under the control of the T7 promoter. At 12 to 15 h postinfection (hpi), cells were fixed with formaldehyde before double-label immunofluorescence was performed to visualize the E4orf6 and E1B-55K proteins using the monoclonal antibodies Rsa#3 and 9C10, respectively. Cells in which at least a portion of E1B-55K was coincident in the nucleus with the E4orf6 protein and in which E4orf6 was excluded from the nucleoli were scored as nuclear. Approximately 6 to 10 randomly selected fields that were distributed uniformly across the culture surface were photographed in order to evaluate at least 200 to 400 cells in each experiment. Results from two to six independent experiments were pooled, and the 95% confidence intervals were estimated using the exact binomial test.
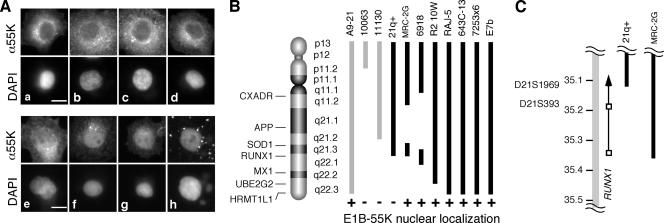
FIG. 1.
E1B-55K and E4orf6 proteins colocalize in the nuclei of rodent cells containing the RUNX1 locus of human chromosome 21. (A) Mouse A9 cells containing human chromosome 21 were infected with the recombinant vaccinia virus vTF7.3 to express the T7 RNA polymerase. The cells were transfected with cDNA for E1B-55K and E4orf6 under the control of the T7 promoter. At 12 hpi, double-label immunofluorescence was used to identify cells expressing both E4orf6 and E1B-55K. (A) Representative micrographs showing the localization of the E1B-55K protein, in which the localization was identified as predominantly cytoplasmic (a to d) or predominantly nuclear (e to h). The bar represents 5 μm. (B) Hamster (black) and mouse (gray) cells containing portions of human chromosome 21 were infected and transfected as described for panel A. At least three independent experiments were scored for the nuclear localization of E1B-55K. Cell lines that demonstrated the nuclear localization of E1B-55K in the presence of E4orf6 are indicated by a plus sign. (C) The portion of human chromosome 21 DNA in the 21q+ and MRC-2G hamster cells at the distal breakpoint is represented by black bars. Chromosome 21 is represented by the gray bar; the scale is indicate in megabase pairs. The approximate locations of STS markers are indicated. The RUNX1 gene is represented by the arrow; open boxes identify the two RUNX1 promoters.
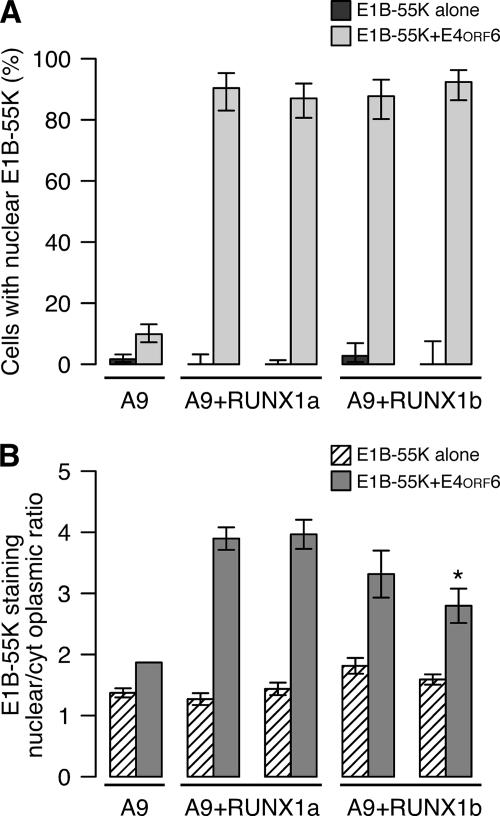
FIG. 4.
E1B-55K and E4orf6 proteins efficiently colocalize in the nuclei of mouse cells that express human RUNX1a and RUNX1b. Two independent stable mouse A9 cell lines expressing human RUNX1a or RUNX1b were established, and the ability of E4orf6 to direct the nuclear localization of E1B-55K was evaluated as described for Fig. 1. (A) Cells were scored as negative or positive for nuclear E1B-55K staining, and the percentages of cells with nuclear E1B-55K protein are indicated along with the standard errors of the means. (B) The ratio of nuclear to cytoplasmic staining for E1B-55K was quantified as described in Materials and Methods and is plotted as the means and standard deviations.
E4orf6-mediated E1B-55K nuclear localization was measured after the transient expression of selected RUNX1 constructs shown in Table 1 by first transfecting 5 × 104 cells with 0.1 μg of RUNX plasmid using Lipofectamine plus (Invitrogen Life Technologies). Approximately 30 h after transfection, E1B-55K and E4orf6 plasmids were expressed using the recombinant vaccinia virus vTF7.3, and the localization of the E1B-55K protein was determined as described above.
TABLE 1.
Human RUNX1 proteins permit E4orf6-directed nuclear localization of E1B-55K in mouse cells
| Transfected construct | Cells with nuclear E1B-55K (%) | 95% Confidence interval |
|---|---|---|
| Controls | ||
| No E4orf6, no RUNX | 1.3 | 0.36-4.8 |
| No RUNX | 6.2 | 3.0-1.3 |
| Mouse Runx1 | ||
| Mouse Runx1b | 3.5 | 0.5-25 |
| Human RUNX related | ||
| RUNX2 (6p21) | 10.0 | 3.7-29 |
| RUNX3 (1p36) | 7.3 | 2.2-24 |
| Human RUNX1 | ||
| RUNX1a | 62.0 | 51-75 |
| RUNX1b | 63.0 | 36-100 |
| RUNX1ΔN | 50.0 | 49-52 |
| RUNX1c | 9.6 | 3.3-28 |
| N terminus-deleted RUNX1c | 50.0 | 49-52 |
Functional mapping of human chromosome 21.
The E1B-55K cDNA alone or the E1B-55K and E4orf6 cDNAs were expressed in rodent cell lines containing various fragments of human chromosome 21 using the recombinant vaccinia virus vTF7.3, and the localization of the proteins was determined by double-label immunofluorescence as described above. Cell lines for which the frequency of E1B-55K-E4orf6 nuclear colocalization was greater than that of the parental (mouse or hamster) cell line were scored as positive.
Quantitative evaluation of the relative intensity of E1B-55K staining in the nucleus.
The macro facility of the open-source software ImageJ (69) was used to quantify the ratio of nuclear to cytoplasmic staining for E1B-55K. Code and additional details will be provided upon request. Briefly, the expression of the E1B-55K and E4orf6 cDNAs were established with the recombinant vaccinia virus vTF7.3 as described above. The cells were processed for immunofluorescence, and the localization of E1B-55K was recorded. A corresponding high-contrast image of the DAPI-stained nucleus was recorded and used to generate a binary image of the nuclear border that then was superimposed on the image of E1B-55K protein localization. A transect line was placed across each cell to be analyzed. Additional pseudorandomly distributed transect lines were generated automatically. The fluorescent intensity across the transect lines was measured, and the borders of the cell and the nucleus were recorded on each line. The mean fluorescent intensity outside of the cell was measured to serve as the local background. This value was subtracted from the mean fluorescent intensity for the nucleus and the cytoplasm before determining the ratio of nuclear to cytoplasmic fluorescence. Approximately 10 to 20 arbitrarily selected cells were analyzed, with four transect lines for each cell in each experiment.
RESULTS
Expressed by transfection, the E1B-55K and E4orf6 proteins colocalize in the nuclei of mouse cells containing the RUNX1 locus of human chromosome 21.
Mouse A9 fibroblast cells containing a copy of human chromosome 21 (15) were infected with a recombinant vaccinia virus to express the bacteriophage T7 polymerase. The infected cells were transfected with cDNA for the adenovirus E1B-55K protein or both E1B-55K and E4orf6 proteins under the direction of a T7 promoter, and the localization of the E4orf6 and E1B-55K proteins was determined by indirect immunofluorescence. Representative cells that received only the E1B-55K cDNA show predominantly cytoplasmic staining for E1B-55K (Fig. 1A, images a to d). Cells transfected with cDNAs for both viral genes typically showed predominantly nuclear staining for E1B-55K (Fig. 1A, images e to h), although the relative intensity of nuclear staining varied among cells and not all cells contained predominantly nuclear E1B-55K protein. These results confirm previously reported findings (13) and illustrate the method used to score cells as containing predominantly nuclear E1B-55K protein.
Additional mouse and hamster cell lines containing portions of human chromosome 21 were evaluated as described above. At least three independent experiments were scored for the nuclear localization of E1B-55K. Cell lines that contained the nuclear localization of E1B-55K in the presence of the E4orf6 protein are indicated in Fig. 1B. E1B-55K remained cytoplasmic in all cell lines in the absence of the E4orf6 protein (data not shown). Both mouse (A9-21) and hamster (E7b, 72532x6, 643C-13, and Raj-5) cell lines with an intact copy of chromosome 21 as well as three hamster cell lines containing fragments of the chromosome (MRC-2G, 6918, and R2-10W) allowed the E4orf6-directed nuclear localization of E1B-55K (Fig. 1B).
The difference in the localization of E1B-55K between the 21q+ and MRC-2G cell lines was especially informative. By cytological criteria, the distal breakpoints of chromosome 21 in the MRC-2G and 21q+ cell lines appear identical. However, the 21q+ cell line, which does not permit E1B-55K protein nuclear localization, contains human chromosome 21 from the p terminus to sequence-tagged site (STS) marker D21S1969 and lacks STS marker D21S1950. By contrast, the distal fragment of chromosome 21 in the MRC-2G cell line, which permits E1B-55K protein nuclear localization, extends from the SOD1 marker to STS marker D21S393 (27). Therefore, this localizes the activity previously mapped to the q terminus of chromosome 21 that allows the E4orf6-directed nuclear localization of E1B-55K to a region of approximately 100 kb on chromosome 21. The only features identified in this region of chromosome 21 include CLIC6, a gene for an intracellular channel protein, and RUNX1, formerly known as AML-1. Because the RUNX1 proteins are nuclear proteins that mediate protein-protein interactions, we chose to investigate further the potential role of RUNX1 in permitting E4orf6-directed nuclear localization.
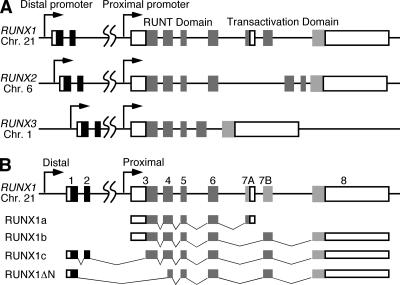
A representation of the 260-kb RUNX1 gene is shown in Fig. 2. The gene contains nine identified exons that are transcribed from telomere to centromere. RUNX genes express multiple isoforms from two promoters. RUNX1 encodes at least four isoforms in humans that arise from two promoters and alternative splicing (Fig. 2B). RUNX1 transcripts originating from the proximal promoter are translated in a cap-independent manner under the direction of the large 5′-untranslated sequence that serves as an internal ribosome entry site (65). The distal RUNX1 promoter directs the synthesis of mRNA translated by canonical cap-dependent means (65).
FIG. 2.
Schematic representation of the human RUNX1, RUNX2, and RUNX3 genes. (A) Boxes in the RUNX genes represent exons, open boxes represent noncoding exons, and shaded boxes indicate coding exons. The dual promoters, Runt domain, and transcriptional transactivation domains are highlighted. (B) The four major splice variants of RUNX1 are indicated.
Human RUNX proteins accumulate at viral replication centers at late times during adenovirus infection.
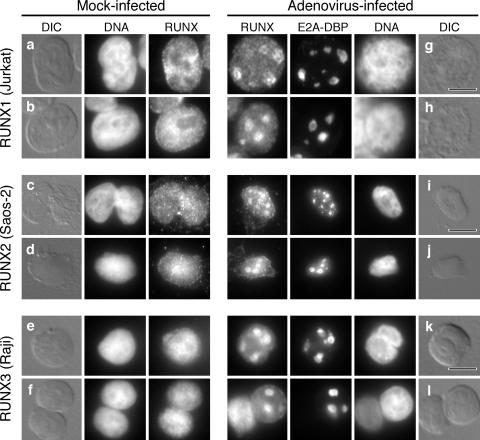
In human cells, the E4orf6 protein directs a portion of E1B-55K to the periphery of viral replication centers at late times of infection (60). These sites are thought to be a key site of E1B-55K-E4orf6 protein interaction within the nucleus (25, 40, 60). To determine if RUNX1 proteins also accumulate at viral replication centers, T-cell lymphoma-derived Jurkat cells, which contain abundant levels of RUNX1 proteins, were infected, and the localization of the endogenous RUNX1 protein was determined with respect to the E2A-DBP-stained replication centers. For comparison, the localization of the RUNX2 protein was determined in the osteosarcoma cell line SaOS-2, and the localization of the RUNX3 protein was determined in the Burkitt's B-cell lymphoma cell line Raji. In mock-infected cells, each RUNX protein exhibited a uniform granular staining pattern throughout the nucleus with exclusion from the nucleoli (Fig. 3a to f). The staining pattern for each RUNX protein changed after infection, such that a portion of the RUNX proteins relocalized to nuclear viral inclusions in the cells. Because these inclusions contained the E2A-DBP, we believe that these are sites of viral DNA replication and are the same as the viral replication centers that have been described previously for adenovirus-infected epithelial cells.
FIG. 3.
Human RUNX proteins accumulate at viral replication centers at late times during adenovirus infection. Jurkat cells, SaOS-2 cells, and Raji cells were either mock infected (a to f) or infected with the wild-type virus dl309 (g to l). After 24 h, the cells were extracted with Triton X-100 as described in Materials and Methods. Viral replication centers were visualized with the mouse monoclonal antibody B6-8 against the E2A-DNA-binding protein, and the RUNX protein was simultaneously visualized with rabbit polyclonal antibodies specific for RUNX1 (a, b, g, and h), RUNX2 (c, d, i, and j), or RUNX3 (e, f, k, and l). DNA was visualized by being stained with DAPI. The bar in each differential interference contrast (DIC) image represents 10 μm.
The localization of the RUNX protein with respect to the E2A-DBP differed among the different cell lines. The RUNX1 protein in the infected Jurkat cells was concentrated in discrete granules under 1 μm in diameter that formed a ring about the periphery of the E2A-DBP-containing structures (Fig. 3g and h). This localization resembled that of E1B-55K (see Fig. 2 of reference 60). A similar staining pattern was described by Bridge and associates for the snRNP proteins identified by the Y12 antibody in adenovirus-infected HeLa cells (11). These investigators suggested that this pattern was characteristic of cells at the onset of late transcription. By contrast, staining for both RUNX2 and RUNX3 was coincident with staining for the E2A-DBP, suggesting that the RUNX2 (Fig. 3i and j) and RUNX3 (Fig. 3k and l) proteins were found throughout the viral replication centers and perhaps were more closely associated with the viral DNA than with nascent viral RNA. Nonetheless, the superficially similar distribution of RUNX1, RUNX2, and RUNX3 in late-infected cells led us to test the possibility that the RUNX2 and RUNX3 proteins enhance the E4orf6-directed nuclear localization of E1B-55K in mouse cells.
A subset of RUNX1 proteins permits E4orf6-directed nuclear localization of E1B-55K in transfected mouse cells.
Mouse A9 cells were transfected with cDNAs for each human RUNX1 isoform or the predominant forms of human RUNX2, RUNX3, and mouse Runx1. The expression of the E1B-55K and E4orf6 cDNAs was established, and the localization of E1B-55K in the presence of the E4orf6 protein was scored as cytoplasmic or nuclear as described for Fig. 1. Consistently with previous findings, whether expressed alone or coexpressed with E4orf6 in mouse A9 cells, E1B-55K was restricted to the cytoplasm and perinuclear aggregates in over 90% of these cells (Table 1). The mouse Runx1b or human RUNX2 and RUNX3 proteins had no significant effect on the ability of E4orf6 to direct nuclear E1B-55K protein localization. By contrast, RUNX1a, RUNX1b, and RUNX1ΔN proteins allowed E4orf6-directed nuclear E1B-55K protein localization in over 60% of the cells expressing the viral genes. Interestingly, RUNX1c was unable to promote E1B-55K nuclear localization. The RUNX1c protein contains a unique sequence in its amino-terminal portion (Fig. 2B) with the potential to prevent DNA binding and association with CBFβ (33). It is possible that this region, termed the negative regulatory region for heterodimerization and DNA binding, also prevents RUNX1c from affecting E4orf6-mediated E1B-55K protein nuclear localization. Evidence in support of this notion was derived from the property of an amino-terminally truncated RUNX1c variant that permitted E1B-55K nuclear localization. This variant contains an HA epitope tag in place of the first 26 amino acids of the RUNX1c amino terminus and therefore contains only 3 amino acids of the original RUNX1c negative regulatory region. This N-terminally deleted RUNX1c variant allowed the E4orf6-directed nuclear localization of E1B-55K in 50% of the mouse A9 cells (Table 1). This result suggests that sequences in the amino-terminal portion of RUNX1c can prevent RUNX1 from promoting E4or6-directed E1B-55K nuclear localization in mouse cells. These results also suggest that the ability to direct E1B-55K nuclear localization is not uniformly shared by RUNX1 variants. To overcome limitations associated with the transient expression required for these experiments, we established mouse A9 cell lines expressing RUNX1a and RUNX1b for further studies.
Expressed by transfection, the E1B-55K and E4orf6 proteins efficiently colocalize in the nuclei of mouse cells that express human RUNX1a and RUNX1b.
Mouse A9 cells were transfected with plasmids encoding epitope-tagged human RUNX1a and RUNX1b cDNAs linked to a neomycin-selectable marker. Independent single-cell clones expressing HA-RUNX1a and HA-RUNX1b were isolated and designated HA-RUNX1a-A9 (clones 1 and 2) and HA-RUNX1b-A9 (clones 1 and 5). Uniform RUNX1 expression was confirmed by immunofluorescence, and the predicted size of the protein was confirmed by immunoblotting (data not shown). It should be noted that repeated attempts to establish mouse cells expressing either epitope-tagged or native forms of RUNX1c and RUNX1ΔN using a variety of different expression constructs failed. The consistent failure to establish the expression of these two constructs leads us to suggest that mouse A9 cells cannot tolerate the stable expression of these particular forms of human RUNX1 (data not shown). Curiously, the long-term expression of the RUNX1b construct also changed the growth properties of the A9 cells. The HA-RUNX1b cells grew more slowly and became difficult to detach from the culture vessel. The basis for these changes and their significance are not understood.
The nuclear localization assay for E1B-55K was performed as described for Fig. 1. As before, E1B-55K was restricted to the cytoplasm and perinuclear aggregates in the absence of the E4orf6 protein; this distribution was not affected by the RUNX1 protein. However, when coexpressed with E4orf6, E1B-55K was found in the nucleus of over 90% of the HA-RUNX1a-A9 and HA-RUNX1b-A9 cells (Fig. 4A). This result, which represents a substantial increase above the size of the fraction measured after transient transfection, confirms that both the RUNX1a and RUNX1b proteins permit the E4orf6-directed nuclear localization of E1B-55K.
Although both RUNX1a and RUNX1b enabled E4orf6 to the direct the nuclear localization of E1B-55K in mouse cells, we observed differences in the staining intensity for the nuclear E1B-55K protein between the HA-RUNX1a and HA-RUNX1b cell lines. To determine if these differences were significant, the ratio of nuclear to cytoplasmic staining intensity for E1B-55K was quantified. This ratio was between 1 and 2 in the absence of E4orf6 (Fig. 4B). The coexpression of the E4orf6 cDNA with the E1B-55K cDNA led to a substantial and statistically significant increase in the nuclear staining-to-cytoplasmic staining ratio in both the HA-RUNX1a and HA-RUNX1b A9 cell lines (Fig. 4B). No significant change in this ratio was measured for the vector-transfected A9 cell line (Fig. 4B and data not shown). However, both HA-RUNX1b cell lines showed greater cell-to-cell variability in the nuclear staining-to-cytoplasmic staining ratio. This difference was confirmed by an analysis of variance in which the variance for the HA-RUNX1b cells transfected with both E4orf6 and E1B-55K was 1.5-fold greater than that of RUNX1a cells (P = 0.007). The HA-RUNX1b cells also appeared to contain relatively less nuclear E1B-55K protein than the HA-RUNX1a cells. However, this decrease was statistically significant for only one of the two HA-RUNX1b-A9 cell lines (Fig. 4B) (P < 0.002 by Tukey's test for multiple comparisons). These results lead us to suggest that although RUNX1a and RUNX1b are able to permit the E4orf6-directed nuclear localization of E1B-55K, the RUNX1 isoforms are not identical in this regard.
RUNX1a and RUNX1b proteins differ in their association with viral replication centers in mouse cells.
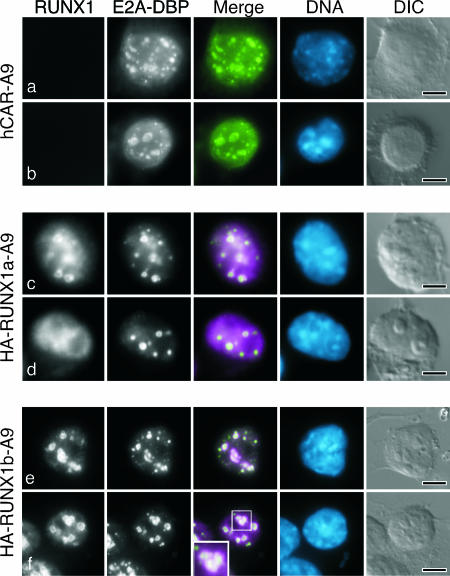
To determine if the RUNX1a and RUNX1b proteins associated with virus-specific structures in the infected mouse cell, the parental and HA-RUNX1 A9 mouse cell lines were transduced with a retrovirus to express the hCAR. After two rounds of enrichment for CAR expression, the cells were infected with adenovirus at an multiplicity of infection of ∼1, corresponding to 300 to 1,000 viral particles. As noted by others, human adenovirus replicates more slowly in mouse cells than in human epithelial cells. Maximal early gene expression was observed at approximately 24 hpi, with peak virus production measured at 72 hpi (data not shown). Therefore, the infected cells were extracted with Triton X-100 at 24 hpi as described previously (60), and viral replication centers were visualized by staining for the E2A-DBP with a mouse monoclonal antibody. Extraction with Triton X-100 enhances the visualization of viral replication centers by removing the diffuse nuclear component of E2A-DBP while sparing the replication center-associated protein (81). The localization of the RUNX1 proteins was determined simultaneously using a rat monoclonal antibody for the HA epitope tag. Both RUNX1a and RUNX1b proteins exhibited a uniform granular staining pattern throughout the nucleus of mock-infected cells (data not shown). Both RUNX1a and RUNX1b proteins associated with the viral replication centers at late times of infection. However, the extent of this association differed, as seen in the representative cells shown in Fig. 5. In the majority of the cells, RUNX1a protein was observed at the periphery of the nuclear viral replication centers (Fig. 5c) or more coincident with E2A-DBP staining (data not shown). In the remaining infected cells, very little RUNX1a protein was associated with viral replication centers (Fig. 5d). In all infected HA-RUNX1a cells, a significant portion of the RUNX1a protein remained diffusely distributed throughout the nucleus. By contrast, over 90% of the HA-RUNX1b cells contain distinct viral replication centers surrounded by staining for the RUNX1b protein. The RUNX1b protein was not coincident with E2A-DBP. Rather, the RUNX1b protein was primarily at the periphery of the replication centers (Fig. 5f). These results demonstrate that the RUNX1a and RUNX1b proteins associate with centers of viral DNA replication in the infected mouse cell but that the nature of this association differs between the two proteins.
FIG. 5.
RUNX1a and RUNX1b proteins vary in the extent of association with viral replication centers. Mouse A9 cells that express hCAR were transduced with vector DNA (a and b) or cDNA of HA-tagged human RUNX1a (c and d) or HA-tagged human RUNX1b (e and f) and were infected with the wild-type virus dl309. After 24 h, the cells were extracted with Triton X-100 as described in Materials and Methods, and double-label immunofluorescence was performed to visualize viral replication centers with the monoclonal antibody B6-8 against the E2A-DBP and a rat monoclonal antibody against the HA epitope. In the merged image, E2A-DBP is shown in green and RUNX1 is shown in magenta. (f) The inset in the merge image shows RUNX1 protein (magenta) surrounding the E2A-DBP (green). DNA was visualized by being stained with DAPI and is shown in blue. Representative micrographs are shown; the bar in each differential interference contrast (DIC) image represents 5 μm.
RUNX1 displaces E4orf6 from viral replication centers in adenovirus-infected mouse cells.
A portion of the E4orf6 protein localizes to the periphery of viral replication centers in HeLa cells (60). To determine if the E4orf6 protein showed a similar localization in mouse cells, A9-hCAR and HA-RUNX1 A9 cells were infected and stained for the E4orf6 and HA-tagged RUNX1 proteins. For these experiments, the RUNX1 protein was used as a marker for viral replication centers, because both E2A-DBP and E4orf6 antibodies are mouse antibodies, precluding their use for double labeling. At 24 hpi, approximately 25% of the infected parental A9-hCAR cells displayed a strong E4orf6-specific staining pattern resembling that of viral replication centers (Fig. 6a and b). This distribution and its frequency were similar to those observed in infected human cells. Remarkably, the E4orf6 protein was no longer concentrated about the viral replication centers in A9 cells expressing RUNX1a or RUNX1b. Although viral replication centers were readily visualized by staining for the appropriate RUNX1 protein, the E4orf6 protein was uniformly distributed through the nucleus and was excluded from nucleoli (Fig. 6c, e, and f). In a representative experiment, only 13 of 75 (17%) HA-RUNX1a cells with distinct viral replication centers showed any indication of increased E4orf6 staining at these centers. Furthermore, the intensity of E4orf6 staining at the replication centers in these few cells was substantially less than that observed in A9-hCAR cells (compare Fig. 6b to Fig. 6d). In the same experiment, none of 85 HA-RUNX1b cells with distinct replication centers showed increased E4orf6 staining at these centers (Fig. 6e and f). These results indicate that the RUNX1 proteins displace or preclude E4orf6 from concentrating at the viral replication centers in the infected mouse cell, with the RUNX1b protein showing a stronger ability to perturb E4orf6 localization at the viral replication centers compared to that of the RUNX1a protein.
FIG. 6.
RUNX1b precludes E4orf6 localization at the periphery of viral replication centers. The hCAR-transduced mouse A9 cells analyzed in Fig. 5 were infected with the E4orf6/E4orf7 mutant virus dl356. After 24 h, the cells were extracted with Triton X-100 as described in Materials and Methods and processed for double-label immunofluorescence using the E4orf6-specific antibody Rsa#3 and the HA-specific rat antibody. The phenotypically wild-type virus dl356 was used for this experiment, because the Rsa#3 antibody recognizes both E4orf6 protein and the 17-kDa E4orf6/E4orf7 fusion protein. In the absence of staining for the E2A-DBP, viral replication centers can be recognized by differential interference contrast (DIC) illumination or by the absence of DAPI staining in the DNA image. In the merged image, the E4orf6 protein is shown in green and RUNX1 is shown in magenta. DNA was visualized by being stained with DAPI and is shown in blue. Representative micrographs are shown, except for that for the HA-RUNX1a cell in panel d. This cell represents a rare cell displaying (<5%) E4orf6 protein concentrated about the viral replication centers defined by RUNX1 staining. The bar in each DIC image represents 5 μm.
E4orf6 and RUNX1 exert different effects on the localization of E1B-55K in adenovirus-infected mouse cells.
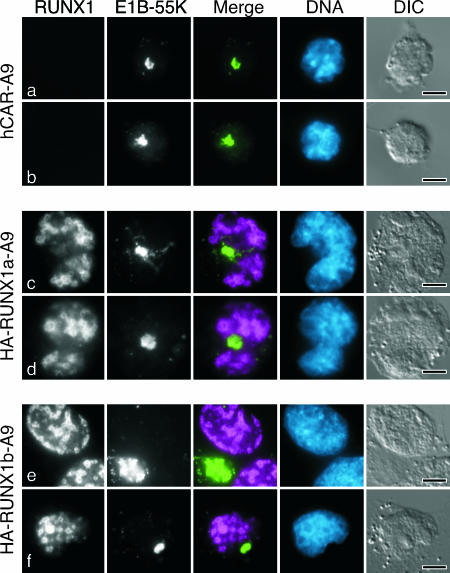
In contrast to the ability of the RUNX1 proteins to perturb E4orf6 protein localization, the RUNX1 proteins had no apparent impact on the localization of E1B-55K in mouse cells infected with the wild-type virus. In these cells, E1B-55K occurred primarily in a perinuclear cytoplasmic inclusion body (Fig. 7). Virtually identical staining patterns for E1B-55K were observed for A9-hCAR, HA-RUNX1a, and HA-RUNX1b A9 cells. Similar perinuclear inclusions of E1B-55K were observed in adenovirus-transformed rodent cells (86). Recently, inclusion bodies similar in appearance to these were identified as aggresomes and are associated with the terminal stages of E1B-55K-E4orf6-mediated protein degradation (1, 46). An antibody to ubiquitin revealed pronounced staining of cytoplasmic bodies of the same frequency and relative position to the nucleus as the perinuclear E1B-55K bodies in the infected mouse cells. This observation is consistent with the possibility that this structure is an aggresome (data not shown). These results distinctly differ from findings from human cells in which a portion of the E1B-55K and E4orf6 proteins colocalized at the periphery of viral replication centers. Therefore, although the RUNX1 proteins promote the nuclear colocalization of the E4orf6 and E1B-55K proteins after transfection, the RUNX1 proteins fail to exert a similar influence following infection.
FIG. 7.
E1B-55K accumulates at cytoplasmic structures resembling the aggresome in wild-type virus-infected mouse A9 cells. The hCAR-transduced mouse A9 cells analyzed in Fig. 5 were infected with the wild-type virus dl309. After 24 h, the cells were extracted with Triton X-100 as described in Materials and Methods and processed for double-label immunofluorescence using the E1B-55K-specific mouse monoclonal antibody 2A6 and the HA-specific rat antibody. In the merged image, E1B-55K is shown in green and RUNX1 is shown in magenta. DNA was visualized by being stained with DAPI and is shown in blue. The bar represents 5 μm. DIC, differential interference contrast.
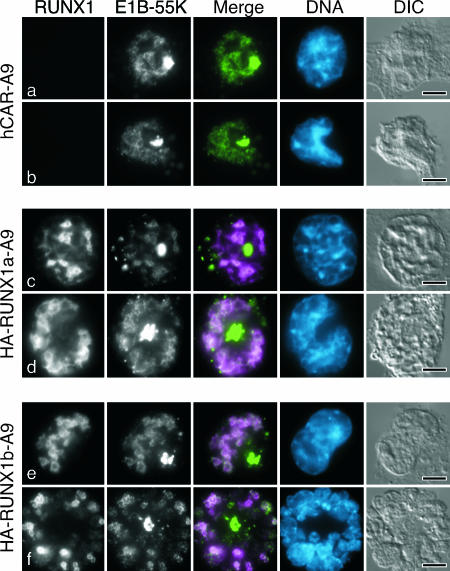
To determine if the E4orf6 protein had any impact on E1B-55K localization in the infected mouse cell, cells were infected with the E4orf6 mutant virus dl355* and E1B-55K was visualized. As in wild-type virus-infected cells, some of the E1B-55K protein accumulated in the aggresome. Surprisingly, a significant portion of E1B-55K remained in the infected hCAR-A9 mouse cell nucleus in the absence of the E4orf6 protein (Fig. 8a and b). Furthermore, some cells displayed a limited association between E1B-55K and viral replication centers (Fig. 8a). The localization of E1B-55K at the periphery of virus replication centers increased in both HA-RUNX1a (Fig. 8c and d) and HA-RUNX1b (Fig. 8e and f) cells. In HA-RUNX1b cells with prominent viral replication centers, most of the nuclear E1B-55K protein was coincident with staining for the RUNX1b protein. Taken together with the results shown in Fig. 7, these findings indicate that E4orf6 acts in a dominant manner to exclude E1B-55K from the nucleus of infected mouse cells. In the absence of E4orf6, the RUNX1a and RUNX1b proteins promote the retention of E1B-55K at viral replication centers. Furthermore, the similarity of HA-RUNX1 staining in cells infected with the wild-type virus (Fig. 7) and E4orf6 mutant virus (Fig. 8) reveals that E4orf6 has little impact on the localization of the RUNX1 proteins.
FIG. 8.
E4orf6 and RUNX1 differentially affect the localization of E1B-55K in adenovirus-infected mouse cells. The hCAR-transduced mouse A9 cells analyzed in Fig. 5 were infected with the E4orf6 mutant virus dl355*. After 24 h, the cells were extracted with Triton X-100 as described in Materials and Methods and processed for double-label immunofluorescence after 24 h using the E1B-55K-specific mouse monoclonal antibody 2A6 and the HA-specific rat antibody. In the merged image, E1B-55K is shown in green and RUNX1 is shown in magenta. DNA was visualized by being stained with DAPI and is shown in blue. The bar represents 5 μm. DIC, differential interference contrast.
E1B-55K promotes the formation of viral replication centers, and RUNX1b can compensate for the loss of E1B-55K.
Cells were infected with the E1B-55K mutant virus dl1520 in order to determine if E1B-55K affected the localization of the RUNX1 proteins. The infected cells were extracted with Triton X-100 after 24 h, and the RUNX1 protein and E2A-DBP were visualized by double-label immunofluorescence as before. Surprisingly, no E2A-DBP staining was evident in the Triton X-100-extracted HA-RUNX1a cells. However, nonextracted cells were uniformly stained for E2A-DBP throughout the nucleus. Identical results were obtained following infection with the E1B mutant virus dl338 (63), which also bears a deletion in the E1B-55K coding region but is able to direct the expression of minor E1B-55K-related proteins that are not expressed by dl1520 (data not shown). Thus, the failure to see staining in the extracted cells was due to the unexpected discovery that E1B-55K is required for well-developed viral replication centers to form in hCAR-A9 and HA-RUNX1a A9 cells (Fig. 9a to d). In HA-RUNX1a cells infected with the E1B-55K mutant virus, the RUNX1a protein was diffusely distributed throughout the nucleus and excluded from the nucleoli (Fig. 9c and d). These results show that the absence of E1B-55K affects the localization of RUNX1a, which fails to accumulate at viral replication centers. However, it seems likely that this is a secondary effect due to the failure to form viral replication centers in the mutant virus-infected RUNX1a-expressing cells.
FIG. 9.
E1B-55K supports the formation of virus replication centers, and RUNX1b can compensate for the loss of E1B-55K in adenovirus-infected mouse cells. The hCAR-transduced mouse A9 cells analyzed in Fig. 5 were infected with the E1B-55K mutant virus dl1520 and processed for double-label immunofluorescence as intact cells after 24 h using the E2A-DBP-specific mouse monoclonal antibody B6-8 and the HA-specific rat antibody. In the merged image, E2A-DBP is shown in green and RUNX1 is shown in magenta. DNA was visualized by being stained with DAPI and is shown in blue. The bar represents 5 μm. DIC, differential interference contrast.
In sharp contrast to dl1520-infected HA-RUNX1a cells, viral replication centers were prominent and abundant in HA-RUNX1b cells that were infected with the E1B-55K mutant virus. The staining pattern for E2A-DBP in the mutant virus-infected HA-RUNX1b cells (Fig. 9e and f) was similar to that of wild-type virus-infected cells (Fig. 5e and f). Again, the RUNX1b protein was found at the periphery of the viral replication centers, often forming concentric rings about the E2A-DBP-containing structures (Fig. 9f). These results indicate that in mouse A9 cells, E1B-55K is necessary for the efficient development of viral replication centers. Furthermore, these results reveal a striking functional difference between the RUNX1a and RUNX1b proteins, wherein only RUNX1b can compensate for the absence of E1B-55K expression by promoting the development of viral replication centers.
DISCUSSION
In this study, we identify the previously described activity on human chromosome 21 (13) that allows E4orf6 to direct the nuclear localization of E1B-55K following transfection in rodent cells. This activity maps to the RUNX1 gene at 21q22.3. The human RUNX1 protein variants a, b, and ΔN, but not RUNX1c, permit the E4orf6-directed nuclear localization of E1B-55K. Human RUNX1b and RUNX1c mRNAs were detected by reverse transcription followed by PCR, and RUNX1-related proteins of the appropriate size were detected by immunoblotting the A9-21 mouse cell line that carries human chromosome 21 (data not shown). Because we show here that the RUNX1a, RUNX1b, and RUNX1ΔN variants, but not the RUNX1c variant, permit E4orf6-directed E1B-55K nuclear localization in mouse cells, we suggest that the relevant activity detected in the A9-21 cells was that of RUNX1b.
RUNX1 proteins are canonical DNA-binding transcription factors that serve as a scaffold for the assembly of a multiprotein complex. This complex can include both transcriptional coactivators and corepressors (62). The RUNX1 isoforms differ in their ability to bind DNA and transcriptional modulators (34). We show here that three of the four major RUNX1 variants (a, b, and ΔN) permitted the E4orf6-directed nuclear localization of E1B-55K in mouse cells. It seems reasonable that this property resides in the sequence common to these three proteins, which is limited to part of the Runt domain encoded by exons 4, 5, and 6. However, this sequence is not sufficient for this activity, because RUNX1c, which also contains this sequence, does not permit E1B-55K nuclear localization. RUNX1c contains 31 more amino-terminal residues than RUNX1b. This domain can interfere with binding to both DNA and the heterodimerization partner CBFβ (33). The results reported here indicate that the negative regulatory amino-terminal domain also precludes RUNX1c from promoting the E4orf6-mediated nuclear localization of E1B-55K. Further support for this idea derives from a mutant form of RUNX1c in which all but three amino acids of the unique N terminus were replaced with an epitope tag. This variant permitted the E4orf6-directed nuclear localization of E1B-55K as effectively as the RUNX1a and RUNX1b proteins. RUNX1ΔN is missing 12 of the 31 amino acids in the amino terminus and half of the DNA-binding Runt domain. This variant is unable to bind DNA (88). However, RUNX1ΔN allowed E4orf6-directed E1B-55K nuclear localization, indicating that the ability to bind DNA is dispensable for the RUNX1 proteins to affect E1B-55K-E4orf6 colocalization. These results lead us to suggest that the scaffolding nature of the RUNX1 proteins is important for their ability to affect the E4orf6-directed nuclear localization of E1B-55K.
In contrast to the relatively simple relationship between E1B-55K, E4orf6, and the RUNX1 proteins observed in transfected cells, the behavior of these proteins in adenovirus-infected cells is complex. Endogenous RUNX1 protein present in Jurkat cells and human RUNX1a and RUNX1b proteins expressed in mouse cells accumulate about the periphery of viral replication centers at late times of infection. This localization, which is close to sites of late viral RNA biogenesis, does not depend on either E1B-55K or E4orf6. Also, because viral DNA replication proceeded with similar kinetics among RUNX1-expressing cell lines (data not shown), it seems unlikely that different rates of viral DNA synthesis affected protein localization about viral replication centers. Rather, the RUNX1 proteins appear to expel most of the E4orf6 protein from these sites. Furthermore, instead of retaining E1B-55K in the nucleus of adenovirus-infected mouse cells, E4orf6 expression excludes E1B-55K from the nucleus of infected mouse cells. In the absence of E4orf6 expression, E1B-55K accumulates in the nucleus and at viral replication centers; this localization is enhanced by the RUNX1b protein. Although RUNX1 may increase the retention of E1B-55K at viral replication centers by expelling a portion of E4orf6 from these sites, we predict that RUNX1 disrupts functions of the E1B-55K-E4orf6 protein complex. Because E1B-55K rapidly shuttles between the nucleus and cytoplasm of transfected (41) and infected cells (18), we interpret these findings to suggest that the E4orf6 protein accelerates the trafficking of E1B-55K through the nucleus, which ultimately results in its accumulation in structures resembling the aggresome in the infected mouse cell. Recently, the trafficking of the E1B-55K protein from the nucleus to the aggresome has been linked to the sequestration of the candidate tumor suppressor protein, sequence-specific single-stranded DNA-binding protein 2 (19).
The E1B-55K protein is a multifunctional protein with a complex distribution throughout the nucleus and cytoplasm of the adenovirus-infected cell. The intracellular trafficking of E1B-55K is affected by interactions with viral factors such as the E4orf3 (42, 43) and E4orf6 proteins (17, 58, 60) as well as cellular factors such as elongins B and C, Cullin5, and RING Box protein-1 (30, 67). With some of these binding partners, the E1B-55K and E4orf6 proteins form a virus-specific E3 ubiquitin ligase that, among other activities, regulates mRNA transport (see especially references 7, 20, and 83). A portion of the E1B-55K protein localizes to nodules or crenulations about the periphery of viral replication centers; this localization is reduced during infection with E4orf6 mutant viruses (60). These nodules and crenulations are enriched in splicing factors and viral RNA (9, 11, 64) and are the initial sites of late viral transcription and RNA processing (2, 3, 64). The localization of RUNX1 to these sites is consistent with the possibility that the RUNX1 proteins alter viral RNA biogenesis through the E1B-55K-E4orf6 protein complex. In the infected mouse cells, RUNX1b is found at the viral replication centers at a much higher frequency than RUNX1a. Staining for RUNX1b at the periphery of these structures resembles staining for small nuclear ribonucleoproteins in ring cells described by Aspegren and Bridge (2, 3). Because the localization of RUNX1b at these sites expels some of the E4orf6 protein, RUNX1b may disrupt the function of the E1B-55K-E4orf6 protein complex. We previously reported that a greater fraction of rodent cells with the q terminus of human chromosome 21 expressed late viral genes than cells without that fragment of human DNA (13). However, a comparable effect on virus yield was not observed in these studies. Consequently, the results reported here will guide additional studies to elucidate the impact of specific RUNX1 variants on adenovirus replication.
Centers of viral DNA replication and late viral transcription, termed viral factories, viral inclusion bodies, viral centers, or viral replication centers, are complex structures that evolve during the course of a virus infection (see, for example, references 3, 6, 10, and 66). Specific localizations within this structure during the late phase of an infection can indicate function. The RUNX1b protein is found in the peripheral replicative zone (66), which has been shown to be the site of late viral RNA processing. Staining for the RUNX1 protein in the T-cell lymphoma Jurkat cell line most closely resembled staining for the RUNX1b protein. By contrast, RUNX1a was most often coincident with the E2A-DBP, which is linked to accumulations of single-stranded viral DNA in the virus replication centers. These E2A-DBP-rich sites are devoid of factors involved in RNA processing (64). This localization was most similar to that of the RUNX2 and RUNX3 proteins in SaOS-2 and Raji cells, respectively. It is possible that common properties of the RUNX proteins, such as the Runt DNA-binding domain, enable the RUNX proteins to associate with the viral replication centers while specific protein-protein interactions target this protein to substructures at these sites. The molecular basis for the association between the RUNX1 proteins and the adenovirus proteins as well as virus-specific structures in the infected cell remains to be elucidated.
The RUNX1 proteins are important regulators of transcription and replication for several viruses. As the DNA-binding component of the core binding factor, the larger RUNX1 proteins regulate transcription from viral enhancers in murine leukemia viruses (73, 84, 85), maedi visna virus (79), and polyomavirus (reviewed in reference 76). It has been suggested that RUNX1 proteins contribute to the E2-mediated repression of bovine and human papillomavirus transcription (8, 74). Mouse Runx1 was identified as a protein that binds polyomavirus DNA and activates viral DNA replication (38). The inability of polyomavirus to replicate in mouse embryonal carcinoma cells has been attributed to the absence of the Runx1 protein (35). Recently, Runx1 was shown to be recruited to virus replication factories during polyomavirus infection. At these sites, Runx1 was proposed to couple the viral DNA to the nuclear matrix while recruiting T antigen to the origin of replication (55). Although the contribution of RUNX1 to the outcome of an adenovirus infection has yet to be determined, the findings we report here suggest that the DNA-binding ability of RUNX1 is not important for adenovirus. Rather, because of the similar distribution for RUNX1b and viral RNA and RNA processing factors, we suggest that properties of RUNX1 that link it to RNA processing, such as the ability to bind ALY/REF (12), will be important to the outcome of an adenovirus infection.
In summary, the results presented here reveal a dynamic relationship between the E4orf6, E1B-55K, and RUNX1 proteins. In cooperation with the E4orf6 protein, RUNX1 proteins are able to affect the localization of E1B-55K. The localization of RUNX1b in the infected cell is consistent with a possible role for this cellular protein in modulating viral RNA metabolism. Additional studies are required to determine the molecular mechanism by which these proteins interact or influence the localization of each other. It also will be important to determine if the activity of the E1B-55K-E4orf6 protein complex is altered in human cells with high levels of the RUNX1 proteins. Lymphocytic cells express high levels of RUNX1 at various stages in development. Interestingly, some of these cells are able to harbor adenovirus in a quiescent or possibly latent state (23, 52). Perhaps by targeting the E1B-55K-E4orf6 protein complex, the RUNX1 proteins can suppress adenovirus gene expression as part of a program to establish the quiescent state. We show here that RUNX1a and RUNX1b exert significant differences on the development of virus-specific structures in the infected cell. Recently, human RUNX1a and RUNX1b were shown to differ in their ability to promote hematopoietic stem cell expansion and differentiation (80). In human cord blood lymphocytes, RUNX1a expression was restricted to the CD34+ progenitor cells and was highest in the most primitive CD34+ compartment (80). The recently identified association between adenovirus and childhood acute lymphocytic leukemia (28) makes it important to evaluate the impact of RUNX1 expression on the outcome of an adenovirus infection in the developing hematopoietic compartment.
Acknowledgments
We thank Scott Hiebert (Vanderbilt University) and Gary Stein (University of Massachusetts, Worcester) for generously providing RUNX1 plasmids, Tom Shenk (Princeton University), Arnie Berk (UCLA), and Pat Hearing (SUNY Stony Brook) for mutant adenoviruses, and James DeGregori for the hCAR retrovirus. We especially thank Isabelle Berquin, Nathan Iyer, and James Woods (WFU Health Sciences) for their help in creating hCAR-transduced cells. We thank Linda Gooding (Emory University) for valuable discussions during this work.
Cell culture reagents and services were provided by the Cell and Virus Vector Core Laboratory, a service of the Comprehensive Cancer Center of Wake Forest University, which is supported in part by the National Cancer Institute grant CA 12197. This work was supported by the Towne Foundation (D.P.) and Public Health Service grant CA 77342 from the National Cancer Institute to D.A.O.
Footnotes
Published ahead of print on 16 April 2008.
REFERENCES
- 1.Araujo, F. D., T. H. Stracker, C. T. Carson, D. V. Lee, and M. D. Weitzman. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 7911382-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aspegren, A., and E. Bridge. 2002. Release of snRNP and RNA from transcription sites in adenovirus-infected cells. Exp. Cell Res. 276273-283. [DOI] [PubMed] [Google Scholar]
- 3.Aspegren, A., C. Rabino, and E. Bridge. 1998. Organization of splicing factors in adenovirus-infected cells reflects changes in gene expression during the early to late phase transition. Exp. Cell Res. 245203-213. [DOI] [PubMed] [Google Scholar]
- 4.Baker, A., K. J. Rohleder, L. A. Hanakahi, and G. Ketner. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 817034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156107-121. [DOI] [PubMed] [Google Scholar]
- 6.Besse, S., and F. Puvion-Dutilleul. 1994. Compartmentalization of cellular and viral DNAs in adenovirus type 5 infection as revealed by ultrastructural in situ hybridization. Chromosome Res. 2123-135. [DOI] [PubMed] [Google Scholar]
- 7.Blanchette, P., C. Y. Cheng, Q. Yan, G. Ketner, D. A. Ornelles, T. Dobner, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 249619-9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeckle, S., H. Pfister, and G. Steger. 2002. A new cellular factor recognizes E2 binding sites of papillomaviruses which mediate transcriptional repression by E2. Virology 293103-117. [DOI] [PubMed] [Google Scholar]
- 9.Bridge, E., M. Carmo-Fonseca, A. Lamond, and U. Pettersson. 1993. Nuclear organization of splicing small nuclear ribonucleoproteins in adenovirus-infected cells. J. Virol. 675792-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bridge, E., and U. Pettersson. 1996. Nuclear organization of adenovirus RNA biogenesis. Exp. Cell Res. 229233-239. [DOI] [PubMed] [Google Scholar]
- 11.Bridge, E., D. X. Xia, M. Carmo-Fonseca, B. Cardinali, A. I. Lamond, and U. Pettersson. 1995. Dynamic organization of splicing factors in adenovirus-infected cells. J. Virol. 69281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruhn, L., A. Munnerlyn, and R. Grosschedl. 1997. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev. 11640-653. [DOI] [PubMed] [Google Scholar]
- 13.Chastain-Moore, A. M., T. Roberts, D. A. Trott, R. F. Newbold, and D. A. Ornelles. 2003. An activity associated with human chromosome 21 permits nuclear colocalization of the adenovirus E1B-55K and E4orf6 proteins and promotes viral late gene expression. J. Virol. 778087-8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbin-Lickfett, K. A., and E. Bridge. 2003. Adenovirus E4-34kDa requires active proteasomes to promote late gene expression. Virology 315234-244. [DOI] [PubMed] [Google Scholar]
- 15.Cuthbert, A. P., D. A. Trott, R. M. Ekong, S. Jezzard, N. L. England, M. Themis, C. M. Todd, and R. F. Newbold. 1995. Construction and characterization of a highly stable human: rodent monochromosomal hybrid panel for genetic complementation and genome mapping studies. Cytogenet. Cell Genet. 7168-76. [DOI] [PubMed] [Google Scholar]
- 16.Cutt, J. R., T. Shenk, and P. Hearing. 1987. Analysis of adenovirus early region 4-encoded polypeptides synthesized in productively infected cells. J. Virol. 61543-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 164276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dosch, T., F. Horn, G. Schneider, F. Kratzer, T. Dobner, J. Hauber, and R. H. Stauber. 2001. The adenovirus type 5 E1B-55K oncoprotein actively shuttles in virus-infected cells, whereas transport of E4orf6 is mediated by a CRM1-independent mechanism. J. Virol. 755677-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleisig, H. B., N. I. Orazio, H. Liang, A. F. Tyler, H. P. Adams, M. D. Weitzman, and L. Nagarajan. 2007. Adenoviral E1B55K oncoprotein sequesters candidate leukemia suppressor sequence-specific single-stranded DNA-binding protein 2 into aggresomes. Oncogene 264797-4805. [DOI] [PubMed] [Google Scholar]
- 20.Flint, S. J., and R. A. Gonzalez. 2003. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 272287-330. [DOI] [PubMed] [Google Scholar]
- 21.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 838122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita, Y., M. Nishimura, M. Taniwaki, T. Abe, and T. Okuda. 2001. Identification of an alternatively spliced form of the mouse AML1/RUNX1 gene transcript AML1c and its expression in early hematopoietic development. Biochem. Biophys. Res. Commun. 2811248-1255. [DOI] [PubMed] [Google Scholar]
- 23.Garnett, C. T., D. Erdman, W. Xu, and L. R. Gooding. 2002. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J. Virol. 7610608-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez, R., W. Huang, R. Finnen, C. Bragg, and S. J. Flint. 2006. Adenovirus E1B 55-kilodalton protein is required for both regulation of mRNA export and efficient entry into the late phase of infection in normal human fibroblasts. J. Virol. 80964-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez, R. A., and S. J. Flint. 2002. Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism. J. Virol. 764507-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodrum, F. D., T. Shenk, and D. A. Ornelles. 1996. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J. Virol. 706323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graw, S. L., K. Gardiner, K. Hall-Johnson, I. Hart, A. Joetham, K. Walton, D. Donaldson, and D. Patterson. 1995. Molecular analysis and breakpoint definition of a set of human chromosome 21 somatic cell hybrids. Somat. Cell Mol. Genet. 21415-428. [DOI] [PubMed] [Google Scholar]
- 28.Gustafsson, B., W. Huang, G. Bogdanovic, F. Gauffin, A. Nordgren, G. Talekar, D. A. Ornelles, and L. R. Gooding. 2007. Adenovirus DNA is detected at increased frequency in Guthrie cards from children who develop acute lymphoblastic leukaemia. Br. J. Cancer 97992-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harada, J. N., A. Shevchenko, A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 769194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrington, K. S., A. Javed, H. Drissi, S. McNeil, J. B. Lian, J. L. Stein, A. J. Van Wijnen, Y. L. Wang, and G. S. Stein. 2002. Transcription factors RUNX1/AML1 and RUNX2/Cbfa1 dynamically associate with stationary subnuclear domains. J. Cell Sci. 1154167-4176. [DOI] [PubMed] [Google Scholar]
- 32.Huang, M. M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 632605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito, Y. 1999. Molecular basis of tissue-specific gene expression mediated by the runt domain transcription factor PEBP2/CBF. Genes Cells 4685-696. [DOI] [PubMed] [Google Scholar]
- 34.Ito, Y. 2004. Oncogenic potential of the RUNX gene family: overview. Oncogene 234198-4208. [DOI] [PubMed] [Google Scholar]
- 35.Ito, Y. 2008. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv. Cancer Res. 99C33-76. [DOI] [PubMed] [Google Scholar]
- 36.Javed, A., B. Guo, S. Hiebert, J. Y. Choi, J. Green, S. C. Zhao, M. A. Osborne, S. Stifani, J. L. Stein, J. B. Lian, A. J. van Wijnen, and G. S. Stein. 2000. Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBFα/AML/PEBP2α) dependent activation of tissue-specific gene transcription. J. Cell Sci. 1132221-2231. [DOI] [PubMed] [Google Scholar]
- 37.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17683-689. [DOI] [PubMed] [Google Scholar]
- 38.Kamachi, Y., E. Ogawa, M. Asano, S. Ishida, Y. Murakami, M. Satake, Y. Ito, and K. Shigesada. 1990. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J. Virol. 644808-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitabayashi, I., A. Yokoyama, K. Shimizu, and M. Ohki. 1998. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 172994-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.König, C., J. Roth, and M. Dobbelstein. 1999. Adenovirus type 5 E4orf3 protein relieves p53 inhibition by E1B-55-kilodalton protein. J. Virol. 732253-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krätzer, F., O. Rosorius, P. Heger, N. Hirschmann, T. Dobner, J. Hauber, and R. H. Stauber. 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 19850-857. [DOI] [PubMed] [Google Scholar]
- 42.Leppard, K. N., and R. D. Everett. 1999. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J. Gen. Virol. 80997-1008. [DOI] [PubMed] [Google Scholar]
- 43.Lethbridge, K. J., G. E. Scott, and K. N. Leppard. 2003. Nuclear matrix localization and SUMO-1 modification of adenovirus type 5 E1b 55K protein are controlled by E4 Orf6 protein. J. Gen. Virol. 84259-268. [DOI] [PubMed] [Google Scholar]
- 44.Levanon, D., G. Glusman, T. Bangsow, E. Ben-Asher, D. A. Male, N. Avidan, C. Bangsow, M. Hattori, T. D. Taylor, S. Taudien, K. Blechschmidt, N. Shimizu, A. Rosenthal, Y. Sakaki, D. Lancet, and Y. Groner. 2001. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene 26223-33. [DOI] [PubMed] [Google Scholar]
- 45.Levanon, D., R. E. Goldstein, Y. Bernstein, H. Tang, D. Goldenberg, S. Stifani, Z. Paroush, and Y. Groner. 1998. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. USA 9511590-11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, Y., A. Shevchenko, A. Shevchenko, and A. J. Berk. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 7914004-14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lutterbach, B., J. J. Westendorf, B. Linggi, S. Isaac, E. Seto, and S. W. Hiebert. 2000. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 275651-656. [DOI] [PubMed] [Google Scholar]
- 48.Martinez-Palomo, A. 1968. Ultrastructural study of the replication of human adenovirus type 12 in cultured cells. Pathol. Microbiol. 31147-164. [DOI] [PubMed] [Google Scholar]
- 49.Martinez-Palomo, A., and N. Granboulan. 1967. Electron microscopy of adenovirus 12 replication. II. High-resolution autoradiography of infected KB cells labeled with tritiated thymidine. J. Virol. 11010-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marton, M. J., S. B. Baim, D. A. Ornelles, and T. Shenk. 1990. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J. Virol. 642345-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNees, A. L., C. T. Garnett, and L. R. Gooding. 2002. The adenovirus E3 RID complex protects some cultured human T and B lymphocytes from Fas-induced apoptosis. J. Virol. 769716-9723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McNees, A. L., J. A. Mahr, D. Ornelles, and L. R. Gooding. 2004. Postinternalization inhibition of adenovirus gene expression and infectious virus production in human T-cell lines. J. Virol. 786955-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyers, S., and S. W. Hiebert. 1995. Indirect and direct disruption of transcriptional regulation in cancer: E2F and AML-1. Crit. Rev. Eukaryot. Gene Expr. 5365-383. [DOI] [PubMed] [Google Scholar]
- 54.Miyoshi, H., M. Ohira, K. Shimizu, K. Mitani, H. Hirai, T. Imai, K. Yokoyama, E. Soeda, and M. Ohki. 1995. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 232762-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murakami, Y., L. F. Chen, N. Sanechika, H. Kohzaki, and Y. Ito. 2007. Transcription factor Runx1 recruits the polyomavirus replication origin to replication factories. J. Cell Biochem. 1001313-1323. [DOI] [PubMed] [Google Scholar]
- 56.Ogawa, E., M. Maruyama, H. Kagoshima, M. Inuzuka, J. Lu, M. Satake, K. Shigesada, and Y. Ito. 1993. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc. Natl. Acad. Sci. USA 906859-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orlando, J. S., and D. A. Ornelles. 1999. An arginine-faced amphipathic alpha helix is required for adenovirus type 5 e4orf6 protein function. J. Virol. 734600-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Orlando, J. S., and D. A. Ornelles. 2002. E4orf6 variants with separate abilities to augment adenovirus replication and direct nuclear localization of the E1B 55-kilodalton protein. J. Virol. 761475-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orlicky, D. J., J. DeGregori, and J. Schaack. 2001. Construction of stable coxsackievirus and adenovirus receptor-expressing 3T3-L1 cells. J. Lipid Res. 42910-915. [PubMed] [Google Scholar]
- 60.Ornelles, D. A., and T. Shenk. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pérez-Alvarado, G. C., M. Martinez-Yamout, M. M. Allen, R. Grosschedl, H. J. Dyson, and P. E. Wright. 2003. Structure of the nuclear factor ALY: insights into post-transcriptional regulatory and mRNA nuclear export processes. Biochemistry 427348-7357. [DOI] [PubMed] [Google Scholar]
- 62.Perry, C., A. Eldor, and H. Soreq. 2002. Runx1/AML1 in leukemia: disrupted association with diverse protein partners. Leuk. Res. 26221-228. [DOI] [PubMed] [Google Scholar]
- 63.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pombo, A., J. Ferreira, E. Bridge, and M. Carmo-Fonseca. 1994. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 135075-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pozner, A., D. Goldenberg, V. Negreanu, S. Y. Le, O. Elroy-Stein, D. Levanon, and Y. Groner. 2000. Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol. Cell. Biol. 202297-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Puvion-Dutilleul, F., and E. Puvion. 1990. Analysis by in situ hybridization and autoradiography of sites of replication and storage of single- and double-stranded adenovirus type 5 DNA in lytically infected HeLa cells. J. Struct. Biol. 103280-289. [DOI] [PubMed] [Google Scholar]
- 67.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 153104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 713788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rasband, W. S. 2007. ImageJ, version 1.38q ed. National Institutes of Health, Bethesda, MD.
- 70.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128480-484. [DOI] [PubMed] [Google Scholar]
- 71.Sandler, A. B., and G. Ketner. 1989. Adenovirus early region 4 is essential for normal stability of late nuclear RNAs. J. Virol. 63624-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sarnow, P., P. Hearing, C. W. Anderson, D. N. Halbert, T. Shenk, and A. J. Levine. 1984. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J. Virol. 49692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Satake, M., M. Inuzuka, K. Shigesada, T. Oikawa, and Y. Ito. 1992. Differential expression of subspecies of polyomavirus and murine leukemia virus enhancer core binding protein, PEBP2, in various hematopoietic cells. Jpn. J. Cancer Res. 83714-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmidt, H. M., G. Steger, and H. Pfister. 1997. Competitive binding of viral E2 protein and mammalian core-binding factor to transcriptional control sequences of human papillomavirus type 8 and bovine papillomavirus type 1. J. Virol. 718029-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Speck, N. A., T. Stacy, Q. Wang, T. North, T. L. Gu, J. Miller, M. Binder, and M. Marin-Padilla. 1999. Core-binding factor: a central player in hematopoiesis and leukemia. Cancer Res. 591789s-1793s. [PubMed] [Google Scholar]
- 76.Speck, N. A., and S. Terryl. 1995. A new transcription factor family associated with human leukemias. Crit. Rev. Eukaryot. Gene Expr. 5337-364. [DOI] [PubMed] [Google Scholar]
- 77.Steegenga, W. T., N. Riteco, A. G. Jochemsen, F. J. Fallaux, and J. L. Bos. 1998. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 16349-357. [DOI] [PubMed] [Google Scholar]
- 78.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418348-352. [DOI] [PubMed] [Google Scholar]
- 79.Sutton, K. A., C. T. Lin, G. D. Harkiss, I. McConnell, and D. R. Sargan. 1997. Regulation of the long terminal repeat in visna virus by a transcription factor related to the AML/PEBP2/CBF superfamily. Virology 229240-250. [DOI] [PubMed] [Google Scholar]
- 80.Tsuzuki, S., D. Hong, R. Gupta, K. Matsuo, M. Seto, and T. Enver. 2007. Isoform-specific potentiation of stem and progenitor cell engraftment by AML1/RUNX1. PLoS Med. 4e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Voelkerding, K., and D. F. Klessig. 1986. Identification of two nuclear subclasses of the adenovirus type 5-encoded DNA-binding protein. J. Virol. 60353-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weitzman, M. D., and D. A. Ornelles. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 247686-7696. [DOI] [PubMed] [Google Scholar]
- 83.Woo, J. L., and A. J. Berk. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zaiman, A. L., and J. Lenz. 1996. Transcriptional activation of a retrovirus enhancer by CBF (AML1) requires a second factor: evidence for cooperativity with c-Myb. J. Virol. 705618-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zaiman, A. L., A. F. Lewis, B. E. Crute, N. A. Speck, and J. Lenz. 1995. Transcriptional activity of core binding factor-alpha (AML1) and beta subunits on murine leukemia virus enhancer cores. J. Virol. 692898-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zantema, A., J. A. Fransen, A. Davis-Olivier, F. C. Ramaekers, G. P. Vooijs, B. DeLeys, and A. J. Van der Eb. 1985. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology 14244-58. [DOI] [PubMed] [Google Scholar]
- 87.Zeng, C., S. McNeil, S. Pockwinse, J. Nickerson, L. Shopland, J. B. Lawrence, S. Penman, S. Hiebert, J. B. Lian, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1998. Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proc. Natl. Acad. Sci. USA 951585-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang, Y. W., S. C. Bae, G. Huang, Y. X. Fu, J. Lu, M. Y. Ahn, Y. Kanno, T. Kanno, and Y. Ito. 1997. A novel transcript encoding an N-terminally truncated AML1/PEBP2 αB protein interferes with transactivation and blocks granulocytic differentiation of 32Dcl3 myeloid cells. Mol. Cell. Biol. 174133-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang, Y. W., N. Yasui, K. Ito, G. Huang, M. Fujii, J. Hanai, H. Nogami, T. Ochi, K. Miyazono, and Y. Ito. 2000. A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc. Natl. Acad. Sci. USA 9710549-10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou, Z., M. J. Luo, K. Straesser, J. Katahira, E. Hurt, and R. Reed. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407401-405. [DOI] [PubMed] [Google Scholar]