Abstract
Influenza viruses are classified into three types: A, B, and C. The genomes of A- and B-type influenza viruses consist of eight RNA segments, whereas influenza C viruses only have seven RNAs. Both A and B influenza viruses contain two major surface glycoproteins: the hemagglutinin (HA) and the neuraminidase (NA). Influenza C viruses have only one major surface glycoprotein, HEF (hemagglutinin-esterase fusion). By using reverse genetics, we generated two seven-segmented chimeric influenza viruses. Each possesses six RNA segments from influenza virus A/Puerto Rico/8/34 (PB2, PB1, PA, NP, M, and NS); the seventh RNA segment encodes either the influenza virus C/Johannesburg/1/66 HEF full-length protein or a chimeric protein HEF-Ecto, which consists of the HEF ectodomain and the HA transmembrane and cytoplasmic regions. To facilitate packaging of the heterologous segment, both the HEF and HEF-Ecto coding regions are flanked by HA packaging sequences. When introduced as an eighth segment with the NA packaging sequences, both viruses are able to stably express a green fluorescent protein (GFP) gene, indicating a potential use for these viruses as vaccine vectors to carry foreign antigens. Finally, we show that incorporation of a GFP RNA segment enhances the growth of seven-segmented viruses, indicating that efficient influenza A viral RNA packaging requires the presence of eight RNA segments. These results support a selective mechanism of viral RNA recruitment to the budding site.
Influenza viruses, which belong to the family of Orthomyxoviridae, are classified into three types: A, B, and C (31). The genomes of A- and B-type influenza viruses consist of eight single-stranded, negative-sense RNA segments, whereas influenza C viruses have only seven RNAs (31). Both A and B influenza viruses contain two major surface glycoproteins: the hemagglutinin (HA), possessing the receptor-binding and fusion activities; and the neuraminidase (NA), which destroys the receptor by cleaving sialic acid from host cell membranes, thereby releasing newly formed virus particles (31). Influenza C viruses contain only one major surface glycoprotein, HEF (hemagglutinin-esterase fusion), which has the receptor-binding fusion as well as receptor-destroying/esterase activities (13, 31). HEF forms a homotrimer and binds to 9-O-acetylneuraminic acid on the cell surface, while the HAs of influenza A and B viruses, which also form a homotrimer, instead recognize N-acetylneuraminic acid as their receptors (31). The fusion activity of both HEF and HA requires proteolytic cleavage of the full-length precursor into two disulfide bond-linked subunits: HEF0 into HEF1 and HEF2 and HA0 into HA1 and HA2. After cleavage, the fusion peptide on the HEF2 or HA2 subunit is able to mediate membrane fusion upon the low pH activation in the endosome (12, 31).
A late step of influenza virus replication in cells involves the packaging of each individual viral RNA (vRNA) segment into budding virions, in an equimolar manner. The mechanism of vRNA packaging is not fully understood. Two models have been proposed for influenza A virus: the random incorporation model and the selective packaging model (31). The random incorporation model argues that all vRNA segments possess a common packaging feature and are incorporated indiscriminately, with the result that full genomes are assembled only by chance (1, 6). The selective packaging model argues that each RNA segment has a specific feature—the unique packaging sequence—which allows the selective incorporation of eight segments into a budding virion. Recently, evidence supporting the specific packaging model has accumulated. Several groups have found that the 5′ and 3′ noncoding sequences as well as sequences at both ends of the coding region are important for vRNA packaging (9, 10, 18, 19, 21, 22, 25, 29, 30, 43). Utilizing these packaging sequences, foreign transgenes can be carried by the influenza virus either transiently, only for several passages, or stably, for multiple passages (20, 21, 36, 43).
Reverse genetics, the generation of negative-sense RNA virus from cloned cDNA, has revolutionized research on influenza virus (8, 27). Methods to rescue all three types of influenza viruses, A, B, and C, have been successfully established (5, 8, 14, 15, 24, 27). Reverse genetics has been utilized to study the influenza viral replication cycle, to characterize the function of influenza viral proteins, and to understand the high pathogenicity of the 1918 and H5N1 viruses (4, 11, 16, 39, 44). Reverse genetics can also be used to design live attenuated vaccines, such as influenza A and B viruses expressing altered type I interferon antagonist (NS1) proteins, live in ovo vaccine against avian influenza and Newcastle disease viruses, and H5N1 influenza A virus carrying an M2 protein with deletions in the cytoplasmic tail (37, 38, 42). Because the replication cycle of influenza virus lacks a DNA phase, the viral genome cannot integrate into the host genome. This property makes the influenza virus a promising, safe candidate for a vaccine or gene delivery vector (28). With reverse genetics, foreign RNAs that are flanked by influenza virus packaging sequences can be incorporated into influenza virions, allowing the expression of foreign proteins in infected host cells (21, 36, 43).
In this study, we substituted the two major glycoproteins of influenza virus A/Puerto Rico/8/34 (A/PR/8/34) virus, HA and NA, with the single major glycoprotein of influenza virus C/Johannesburg/1/66 (C/JHB/1/66) virus, HEF, and successfully generated two seven-segmented chimeric influenza viruses. The two viruses each possess six influenza A virus RNA segments (PB1, PB2, PA, NP, M, and NS) and one chimeric RNA segment encoding either the influenza C virus HEF full-length protein or the chimeric protein HEF-Ecto, which consists of HEF ectodomain and HA transmembrane and cytoplasmic regions. Both HEF and HEF-Ecto RNA segments carry sequences from A/WSN/33 HA RNA at the two ends to allow packaging into virions. Furthermore, both viruses are able to stably incorporate one extra green fluorescent protein (GFP) RNA segment carrying the NA packaging sequences, demonstrating a new approach to developing a vaccine or gene delivery vector.
MATERIALS AND METHODS
Cells and viruses.
293T cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum. Madin-Darby canine kidney (MDCK) cells were grown in Eagle's minimal essential medium with 10% fetal calf serum. Both cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The viruses were grown in 8- or 10-day-old specific-pathogen-free chicken embryos (Charles River Laboratories, SPAFAS, Preston, CT).
Plasmid construction. (i) Generation of pPOL I-HEF construct.
The open reading frame (ORF) of the HEF protein, which is 1,965 bp long, was amplified by reverse transcription-PCR (RT-PCR) from influenza virus C/JHB/1/66 vRNA (Fig. 1A). Two restriction sites, NheI and XhoI, were introduced to flank the HEF ORF. The pPOL I-HA (A/WSN/33) plasmid carrying two restriction sites, NheI (nucleotides 78 to 83) and XhoI (nucleotides 1,645 to 1,650), was generated previously (21), and the original HA translation start codon ATG (nucleotides 33 to 35) was mutated to CTG. The HEF ORF PCR fragment was digested with NheI and XhoI and ligated to the NheI and XhoI sites of pPOL I-HA with 45 and 80 nucleotides of packaging sequences from the HA ORF. The two restriction sites used for ligation, NheI and XhoI, were removed by site-directed mutagenesis after ligation, generating the final pPOL I-HEF construct (Fig. 1A).
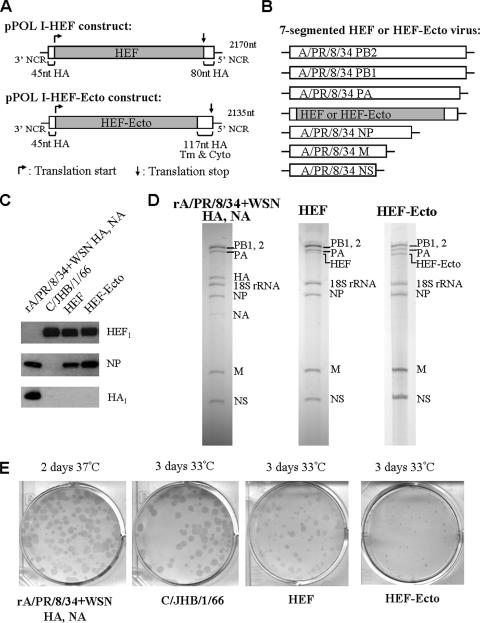
FIG. 1.
Generation of recombinant seven-segmented influenza viruses HEF and HEF-Ecto. (A) Construction of pPOL I-HEF and pPOL I-HEF-Ecto plasmids. The HEF ORF or HEF ectodomain is flanked by A/WSN/33 HA 3′ and 5′ NCRs and packaging sequences from the HA ORF at the two ends. Both chimeric genes are driven by a human RNA polymerase I promoter to produce vRNAs. “Tm” and “Cyto” designate the transmembrane domain and cytoplasmic region of HA, respectively. The light gray portion in each construct designates the ORF of C/JHB/1/66 HEF. The translation start and stop codons of each construct are indicated by arrows. The length of each RNA segment in nucleotides (nt) is labeled on the right of the diagram. (B) Genome structure of seven-segmented HEF or HEF-Ecto virus. Six A/PR/8/34 ambisense plasmids (pDZ-PB2, pDZ-PB1, pDZ-PA, pDZ-NP, pDZ-M, and pDZ-NS) and one pPOL I plasmid (pPOL I-HEF or pPOL I-HEF-Ecto) were used to generate the recombinant HEF or HEF-Ecto virus by using reverse genetics (8, 33). (C) Incorporation of HEF full-length or HEF-Ecto chimeric glycoprotein into the recombinant seven-segmented virions. The rA/PR/8/34 virus expressing the A/WSN/33 HA and NA (rA/PR/8/34+WSN HA, NA), which was grown at 37°C, and three viruses (C/JHB/1/66, HEF, and HEF-Ecto) which were grown at 33°C were inoculated into 8- or 10-day-old embryonated chicken eggs. Two to three days later, allantoic fluids were harvested and virions were purified by centrifugation through a 30% sucrose cushion. A Western blot was performed to detect the presence of HEF, NP, and HA proteins using specific monoclonal antibodies. (D) The genome of HEF or HEF-Ecto virus contains seven RNA segments. RNA was isolated from purified rA/PR/8/34 expressing the A/WSN/33 HA and NA, HEF, and HEF-Ecto viruses, run on a 2.8% acrylamide gel, and visualized by silver staining. RNA (0.5 to 1 μg) was loaded for each virus. Each RNA segment is labeled to the right of the gel. (E) Immunostaining of viruses grown in MDCK cells. Confluent MDCK cells were infected with rA/PR/8/34 expressing the A/WSN/33 HA and NA and the C/JHB/1/66, HEF, and HEF-Ecto viruses at a multiplicity of infection of 0.0001, covered with agar overlay containing 1 μg/ml TPCK-trypsin, and incubated at either 37°C or 33°C. Two to three days postinfection, the agar overlay was removed after formaldehyde fixation. The cells were incubated at 4°C overnight with rabbit polyclonal antiserum against A/PR/8/34 for the rA/PR/8/34 virus expressing the A/WSN/33 HA and NA or with mouse monoclonal antibody against HEF for the C/JHB/1/66, HEF, and HEF-Ecto viruses. The second day, the cells were incubated with horseradish peroxidase-linked anti-rabbit or anti-mouse immunoglobulin G for 1 h at room temperature, and the “plaques” were visualized by using TrueBlue peroxidase substrate.
(ii) Generation of pPOL I-HEF-Ecto construct.
By using a similar strategy, the pPOL I-HA (A/WSN/33) plasmid carrying restriction sites NheI (nucleotides 78 to 83) and XhoI (nucleotides 1,605 to 1,610) was generated, and the original HA translation start codon was also mutated to CTG (Fig. 1A). The ORF encoding the HEF ectodomain, which is 1,893 bp long and flanked by NheI and XhoI restriction sites at the two ends, was amplified by RT-PCR from C/JHB/1/66 vRNA and ligated to the NheI and XhoI sites of a pPOL I-HA plasmid backbone with 45 and 117 nucleotides of packaging signals from the HA ORF. The NheI and XhoI sites were also removed by site-directed mutagenesis after ligation, generating the final pPOL I-HEF-Ecto construct.
(iii) Generation of pDZ-GFP-1 and pDZ-GFP-2 constructs.
The previously constructed pDZ-NA (A/PR/8/34) (21, 33) was digested with SphI and BmtI, treated with a DNA polymerase I Klenow fragment, and then self-ligated using T4 DNA ligase in order to remove the NheI and XhoI sites on the vector backbone, generating the plasmid pDZ-NA (−NheI, −XhoI) (Fig. 2A). The 2.2-kb EcoRI-HindIII fragment from the pDZ-NA plasmid, which contains the A/PR/8/34 NA gene, was subcloned into the EcoRI and HindIII sites of pUC18 vector and subjected to site-directed mutagenesis to generate NheI (nucleotides 132 to 137) and XhoI (nucleotides 1,223 to 1,228) sites on the NA ORF. The two ATG codons before the NheI site were mutated to TTG. Finally, the 2.2-kb EcoRI-Hind III fragment was subcloned back into the EcoRI and HindIII sites of the pDZ-NA (−NheI, −XhoI) plasmid, replacing the original 2.2-kb fragment. The resulting plasmid was digested with NheI and XhoI and ligated with the GFP fragment digested with the same enzymes, generating the construct pDZ-GFP-1, in which the GFP ORF is flanked by 111 nucleotides and 157 nucleotides of packaging sequences from the NA ORF. The GFP ORF was derived from the previously constructed pPOL I HA(45)GFP(80) plasmid (21). The plasmid pDZ-GFP-2, with 153 and 181 nucleotides of NA ORF packaging sequences at the 3′ and 5′ ends of GFP, was constructed by using similar methods. The three ATG codons before the NheI site were also mutated to TTG by site-directed mutagenesis. Sequences of primers used for cloning and site-directed mutagenesis are available from the authors upon request.
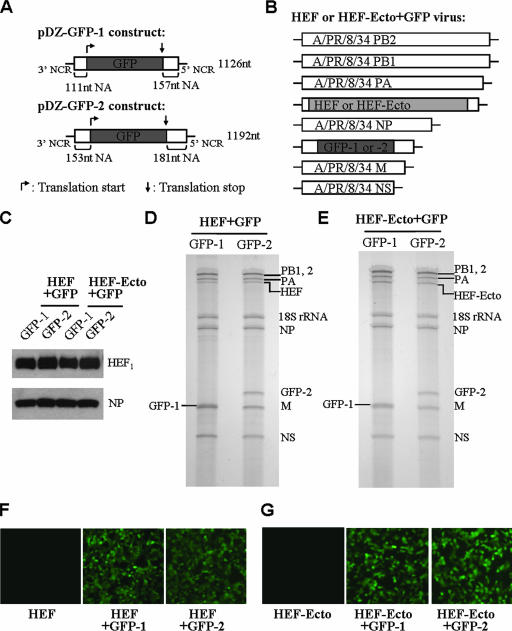
FIG. 2.
Generation of HEF- or HEF-Ecto-containing viruses carrying a GFP RNA segment. (A) Generation of pDZ-GFP-1 and pDZ-GFP-2 constructs. The GFP ORF is flanked by the A/PR/8/34 NA 3′ and 5′ NCRs and packaging sequences from the NA ORF. For the pDZ-GFP-1 construct, GFP is flanked by 111 and 157 nucleotides of the NA ORF; and for the pDZ-GFP-2 construct, 153 and 181 nucleotides of packaging sequences are used. Both GFP constructs are in the ambisense pDZ vector backbone which was described previously (33). The dark gray designates the GFP ORF. The translation start and stop codons are indicated by the arrows. The length of each RNA segment in nucleotides (nt) is labeled on the right of the diagram. (B) Genome structure of HEF+GFP and HEF-Ecto+GFP viruses. The rescue procedure was the same as that for HEF and HEF-Ecto viruses (Fig. 1B), except that one additional construct, pDZ-GFP-1 or pDZ-GFP-2, was also included in the rescue transfection. (C) Incorporation of HEF full-length or HEF-Ecto chimeric glycoprotein into the recombinant HEF+GFP or HEF-Ecto+GFP virions. The experiment was performed together with that shown in Fig. 1C. (D, E) GFP-1 or GFP-2 RNA is incorporated into HEF+GFP (D) and HEF-Ecto+GFP (E) virions. The method was the same as that described in the legend for Fig. 1D. (F, G) Expression of GFP in 293T cells by the recombinant HEF+GFP (F) and HEF-Ecto+GFP (G) viruses. 293T cells were infected with recombinant HEF, HEF+GFP-1, or HEF+GFP-2 (F) viruses or with HEF-Ecto, HEF-Ecto+GFP-1, or HEF-Ecto+GFP-2 (G) viruses at a multiplicity of infection of 0.5. The expression of GFP was observed 8 h postinfection.
Reverse genetics for recombinant viruses.
The method for generating recombinant influenza viruses was modified from previously described protocols (8, 33). Briefly, for the generation of HEF- or HEF-Ecto-containing viruses, 293T cells were transfected with six A/PR/8/34 plasmids (pDZ-PB2, pDZ-PB1, pDZ-PA, pDZ-NP, pDZ-M, and pDZ-NS), one pPOL I-HEF or pPOL I-HEF-Ecto plasmid, three A/WSN/33 protein expression plasmids (pCAGGS-HA, pCAGGS-NA, and pCAGGS-NS1), plus or minus the GFP plasmid (pDZ-GFP-1 or pDZ-GFP-2). For the generation of recombinant A/PR/8/34 (rA/PR/8/34) virus expressing the A/WSN/33 HA and NA, six A/PR/8/34 plasmids (pDZ-PB2, pDZ-PB1, pDZ-PA, pDZ-NP, pDZ-M, and pDZ-NS) and two A/WSN/33 plasmids (pPOL I-HA and pPOL I-NA) were used to transfect 293T cells. The plasmids (0.5 μg each) were added in one well of a six-well plate. Twenty-four hours posttransfection, the supernatants and cells were harvested and inoculated into 8- or 10-day-old embryonated chicken eggs, and the eggs were incubated at 33°C for three days. The allantoic fluids were harvested and subjected to hemagglutination assay to determine the presence of viruses.
Western blotting.
Viruses were grown in 8- or 10-day-old chicken embryos at 33°C or 37°C for two to three days. The allantoic fluids were harvested, and the viruses were purified by centrifugation through a 30% sucrose cushion. Purified viruses were resuspended in 1× phosphate-buffered saline (PBS) and lysed in 2× protein loading buffer (100 mM Tris-HCl [pH 6.8], 4% sodium dodecyl sulfate, 20% glycerol, 5% β-mercaptoethanol, and 0.2% bromophenol blue). The protein lysates were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to a nitrocellulose membrane (Whatman, Inc.). The membrane was then probed with mouse monoclonal antibody against A/PR/8/34 NP protein (HT103, 1 μg/ml), A/WSN/33 HA protein (2G9, 1 μg/ml), or C/JHB/1/66 HEF protein (8D6D3, 1 μg/ml).
Acrylamide gel electrophoresis of purified vRNA.
The viruses were grown in 8- to 10-day-old embryonated chicken eggs at either 33°C for HEF- or HEF-Ecto-containing viruses or 37°C for rA/PR/8/34 virus expressing the WSN HA and NA. Two to three days later, the embryos were killed by transferring them to 4°C and were left overnight. The allantoic fluids were harvested and clarified by centrifugation at 6,000 rpm at 4°C for 15 min using a Beckman rotor SW28. Clarified supernatant was then layered on a 30% sucrose cushion and further centrifuged at 25,000 rpm for 2.5 h. Pelleted virus was resuspended in 1× PBS buffer, and vRNA was extracted by using TRIzol reagent (Invitrogen). Precipitated vRNA was resuspended in a final volume of 15 μl DNase- and RNase-free H2O and stored at −80°C. RNA (0.5 to 1 μg) was separated on a 2.8% denaturing polyacrylamide gel containing 7 M urea. The gel was stained with a SilverXpress silver staining kit (Invitrogen) to visualize the RNA bands.
Immunostaining of plaques.
The protocol was modified from a previously reported method (23). Briefly, a MDCK cell monolayer in six-well plates was infected with viruses in serial 10-fold dilutions for 1 h and covered with agar overlay containing 1 μg/ml l-1-tosylamide-2-phenyl-ethyl chloromethyl ketone (TPCK)-trypsin (Sigma). Two to three days later, the cells were fixed with 4% formaldehyde containing 0.01% Triton X-100 for 3 h. The agar overlay was removed and blocked with 1× PBS containing 5% dried nonfat milk and 0.05% Tween 20 for 1 h at room temperature. Then, the cells were incubated overnight with the first antibody: rabbit anti-A/PR/8/34 polyclonal antibody for rA/PR/8/34 virus expressing the A/WSN/33 HA and NA (1:2,000 dilution); or mouse anti-HEF monoclonal antibody (8D6D3, 1 μg/ml) for HEF- and HEF-Ecto-containing viruses and influenza C/JHB/1/66 virus. The cell monolayer was washed with PBS and incubated for 1 h at room temperature with Amersham ECL-HRP linked anti-rabbit or anti-mouse secondary antibody (GE Healthcare). The cells were washed three times with PBS and stained with TrueBlue peroxidase substrate (KPL).
Viral growth kinetics.
Eight-day-old embryonated chicken eggs were inoculated with influenza viruses (100 PFU/egg) and incubated at 33°C. At 24, 48, and 72 h postinoculation, the eggs were transferred to 4°C and left overnight, after which the allantoic fluids were harvested. The titer of the viruses was determined by plaque assay or immunostaining of the plaques in MDCK cells.
RESULTS
Generation of recombinant seven-segmented influenza viruses.
In this study, we attempted to generate an influenza A virus containing the influenza virus C/JHB/1/66 glycoprotein HEF instead of HA and NA. The 1,965-bp full-length HEF ORF was amplified by RT-PCR from the C/JHB/1/66 vRNA. In order for the HEF segment to be packaged into influenza A virions, the ORF was flanked by the previously identified packaging sequences of A/WSN/33 HA (the 3′ and 5′ noncoding regions [NCRs] plus the 45 nucleotides proximal to the start codon and the 80 nucleotides proximal to the stop codon of the HA ORF [21, 43]). The resultant cDNA was then inserted into the pPOL I plasmid to generate the pPOL I-HEF construct (Fig. 1A). The original HA translation start codon ATG was mutated to CTG, such that the HEF protein is translated from its own initiation codon (Fig. 1A).
Although the overall structures are similar, HA and HEF only share about 12% sequence identity (35). Compared to HA, C/JHB/1/66 HEF has a shorter transmembrane domain and cytoplasmic tail, which are 20 and 3 amino acids long, respectively (26, 32). In addition, for HEF, the cysteine residue next to the cytoplasmic domain is acylated with stearic acid, while for HA, the cysteine residues of C-termini undergo palmitoylation (40, 41). Considering these differences and the possibility that the C-terminal portion of the HA protein may be required for the assembly of virions (3, 45, 46), we also generated an HEF-Ecto chimeric gene, in which the 1,893-bp ORF encoding the HEF ectodomain was fused to the sequences encoding the HA transmembrane and cytoplasmic domains (Fig. 1A). The HEF-Ecto chimeric gene is also flanked by HA NCRs and 45 nucleotides of packaging sequence from the HA ORF that are proximal to the start codon. Expression of both HEF and HEF-Ecto RNAs is driven by a human RNA polymerase I promoter and terminated with the hepatitis delta virus ribozyme sequence.
By using previously established methods (see Materials and Methods), two chimeric HEF- and HEF-Ecto-containing viruses (hereafter called the HEF and HEF-Ecto viruses) were successfully rescued (Fig. 1B) and were found to be stable for at least five passages in embryonated chicken eggs. HEF virus contains six RNA segments (PB2, PB1, PA, NP, M, and NS) from A/PR/8/34 and one HEF segment flanked by the influenza A virus packaging sequences. HEF-Ecto virus contains the same six backbone segments from A/PR/8/34 and one HEF-Ecto chimeric segment (Fig. 1B). In order to characterize the protein compositions, the viruses were grown in the allantoic cavities of embryonated chicken eggs and purified by centrifugation through a 30% sucrose cushion. Western blotting was performed to detect the presence of individual proteins using specific monoclonal antibodies. The results showed that both HEF and HEF-Ecto viral particles contain the A/PR/8/34 NP and the C/JHB/1/66 glycoprotein HEF1, which is the proteolytic cleavage product of full-length precursor HEF0. Both viruses were also shown not to contain influenza A virus HA (Fig. 1C), since HA1, the cleavage product of full-length HA0, is only detected in rA/PR/8/34 virus expressing the A/WSN/33 HA and NA. In order to show that the newly obtained HEF or HEF-Ecto chimeric virus indeed contains seven RNA segments, RNA was purified and run on an acrylamide gel to visualize the genome compositions by silver staining (Fig. 1D). The results showed that both HEF and HEF-Ecto viruses contain seven RNA segments: PB2, PB1, PA, NP, M, NS, and one chimeric segment, HEF or HEF-Ecto. The chicken 18S rRNA derived from ribosomes, which were copurified with virions, was also visible on the gel. Both viruses lack the influenza A virus RNA segments HA and NA, which are present in rA/PR/8/34 virus expressing the A/WSN/33 HA and NA (Fig. 1D). It should be noted that the abundance of A/WSN/33 NA vRNA in the A/PR/8/34 background is low possibly because of inefficient packaging (Fig. 1D). Interestingly, the intensity of the HEF segment RNA from the HEF virus is about 50% less than that of the corresponding PA segment, while the HEF-Ecto RNA segment of the HEF-Ecto virus is comparable to that of PA (Fig. 1D). This indicates that the packaging efficiency of the HEF-Ecto RNA segment is higher than that of HEF RNA, which may be due to the presence of longer HA packaging sequences at the 5′ end (Fig. 1A).
When traditional plaque assay methods are used, the influenza C/JHB/1/66 virus does not lyse cells and forms clear plaques in MDCK cells (34). In this study, we found that the newly rescued HEF and HEF-Ecto chimeric viruses grew very poorly in MDCK cells and were also unable to form visible plaques by regular plaque assay methods. Thus, in order to titrate the C/JHB/1/66, HEF, and HEF-Ecto viruses, we performed immunostaining of plaque assays, using a method modified from a previous report (23). Briefly, the viruses were subjected to regular plaque assay using MDCK cells. After fixation with formaldehyde, instead of staining the cell monolayer with crystal violet, we used specific monoclonal antibody against HEF protein and peroxidase substrate to visualize the “plaques” (Fig. 1E). At 33°C, C/JHB/1/66 forms bigger “plaques” than both the HEF and HEF-Ecto viruses, and the “plaques” formed by HEF-Ecto are the smallest (Fig. 1E).
Generation of HEF or HEF-Ecto viruses carrying a GFP RNA segment.
The selective packaging model argues that eight segments are required for efficient packaging of influenza A virus genomes (9, 10, 18, 19, 21, 22, 25, 29, 30, 43). If this model is correct, the HEF or HEF-Ecto chimeric viruses could be modified to carry one additional RNA segment that contains the NA packaging sequences. To determine whether this is the case, we generated GFP constructs in which the GFP ORF is flanked by the A/PR/8/34 NA 3′ and 5′ packaging sequences: pDZ-GFP-1 and pDZ-GFP-2 (Fig. 2A). GFP-1 carries 111 and 157 nucleotides of the NA coding region flanking the 3′ and 5′ ends, respectively, and GFP-2 carries slightly longer sequences from the NA ORF, 153 and 181 nucleotides at the 3′ and 5′ ends, respectively. Because the packaging sequences of the A/PR/8/34 NA segment have not been characterized, we tested shorter NA terminal sequences in pDZ-GFP-1 and longer sequences in pDZ-GFP-2. All ATG codons before the GFP translation initiation codon in both constructs were mutated to TTG; thus, the translation of GFP utilized its own start codon. Unpublished data from our lab showed that mutating these ATGs did not interfere with vRNA packaging. GFP-1 and GFP-2 chimeric genes are expressed from the ambisense pDZ plasmids, which were described previously (33). By using reverse genetics techniques, we successfully rescued the GFP-carrying HEF and HEF-Ecto viruses (HEF+GFP and HEF-Ecto+GFP), each of which contains the six A/PR/8/34 backbone segments (PB2, PB1, PA, NP, M, and NS), one chimeric segment HEF or HEF-Ecto, and one extra segment GFP-1 or GFP-2 (Fig. 2B). Western blot results showed that both HEF+GFP and HEF-Ecto+GFP virions contain A/PR/8/34 NP and C/JHB/1/66 glycoprotein HEF1 but not HA (Fig. 2C). Furthermore, to show that both viruses do contain the GFP segment, the RNA was isolated from purified viruses and run on an acrylamide gel to visualize the viral genome compositions by silver staining (Fig. 2D and E). GFP-1 and GFP-2 RNA segments can be seen in both HEF+GFP (Fig. 2D) and HEF-Ecto+GFP (Fig. 2E) virions, and with similar incorporation efficiencies. Consistent with this result, GFP protein was expressed in 293T cells infected with viruses HEF+GFP or HEF-Ecto+GFP, with either GFP-1 or GFP-2, but not in cells infected with seven-segmented HEF or HEF-Ecto virus (Fig. 2F and G).
Incorporation of an extra GFP segment increases the growth rate of the seven-segmented viruses.
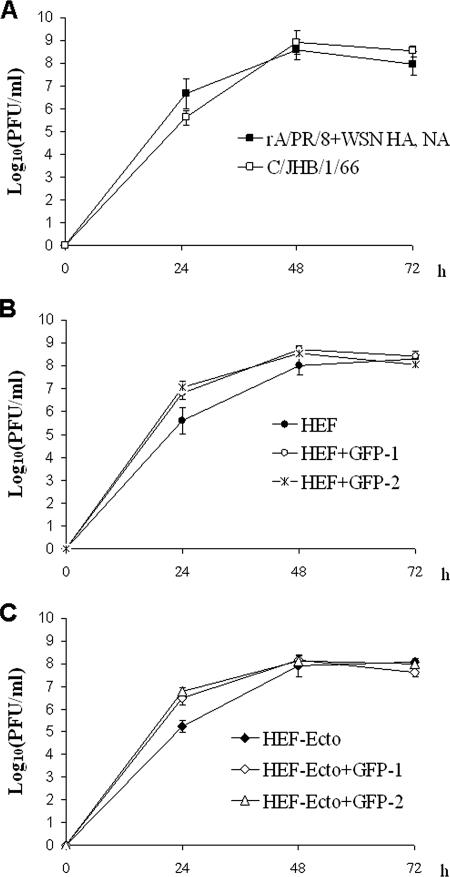
We compared the growth kinetics of HEF and HEF-Ecto viruses with or without a GFP segment, as well as those of the rA/PR/8/34 virus expressing the A/WSN/33 HA and NA and the C/JHB/1/66 virus, in 8-day-old embryonated chicken eggs at 33°C (Fig. 3). For each virus at every time point, the allantoic fluids from three eggs were analyzed. The results showed that incorporation of a GFP-1 or GFP-2 RNA segment significantly increased the growth rate of HEF and HEF-Ecto viruses (Fig. 3B and C). At 24 h postinoculation, the titers of both HEF+GFP (HEF+GFP-1 and HEF+GFP-2) and HEF-Ecto+GFP (HEF-Ecto+GFP-1 and HEF-Ecto+GFP-2) viruses were about 10 times higher than those of the seven-segmented viruses (Fig. 3B and C). At 48 and 72 h postinoculation, there were no significant differences in the titers (Fig. 3B and C). These results indicate that the presence of eight RNA segments enhances viral replication, possibly through more-efficient vRNA packaging. Although HEF-Ecto virus forms much smaller “plaques” than HEF virus in MDCK cells (Fig. 1E), no significant differences were observed for the growth rates of HEF and HEF-Ecto viruses in embryonated chicken eggs (Fig. 3B and C). The mechanism underlying this phenomenon remains to be determined.
FIG. 3.
Incorporation of a GFP RNA segment enhances the growth of HEF or HEF-Ecto seven-segmented virus. Eight-day-old embryonated chicken eggs were inoculated with 100 PFU of (A) rA/PR/8/34 expressing the A/WSN/33 HA and NA or C/JHB/1/66, (B) HEF, HEF+GFP-1, or HEF+GFP-2, or the (C) HEF-Ecto, HEF-Ecto+GFP-1, or HEF-Ecto+GFP-2 viruses and incubated at 33°C. At 24, 48, and 72 h postinoculation, the eggs were transferred to 4°C overnight and allantoic fluids were harvested. The titer of the virus at each time point was determined by immunostaining of the plaques in MDCK cells. The growth analyses shown in panels A, B, and C were performed in the same experiment.
Stability of HEF or HEF-Ecto viruses carrying a GFP segment.
The rescued HEF+GFP-1, HEF+GFP-2, HEF-Ecto+GFP-1, and HEF-Ecto+GFP-2 viruses were passaged in 8-day-old embryonated chicken eggs to determine whether their genomes are stable over multiple passages. As shown in Fig. 4A, similar levels of GFP-2 RNA were observed for HEF+GFP-2 virus at passage 1 and passage 7. Although the GFP-1 vRNA of HEF+GFP-1 virus is not separable from M vRNA in the gel (Fig. 4A), we were still able to assess its similar levels of presence at passage 1 and passage 7 by observing the GFP expression in infected cells (data not shown). The same phenomenon was also observed for HEF-Ecto+GFP-1 and HEF-Ecto+GFP-2 viruses (Fig. 4B). At passage 1 and passage 7, overall titers of the viruses were similar, and comparable levels of expression of GFP in 293T or MDCK cells were also observed. In addition, no mutations were observed for either HEF or HEF-Ecto vRNA segments after seven passages.
FIG. 4.
Stable incorporation of a GFP RNA segment by the HEF or HEF-Ecto virus over multiple passages. (A) HEF+GFP-1 and HEF+GFP-2 viruses were passaged in 8-day-old embryonated chicken eggs seven times. The viruses at passage 1 (P1) and P7 were purified by centrifugation through a sucrose cushion, and RNA was isolated using TRIzol reagent. The RNA (0.5 μg/lane) was separated on a 2.8% acrylamide gel and visualized by silver staining. (B) The method was the same as that described for panel A, except that HEF-Ecto+GFP-1 and HEF-Ecto+GFP-2 viruses were analyzed.
DISCUSSION
RNA viruses with segmented genomes can undergo reassortment when two viruses coinfect the same cell, allowing them to exchange genetic information and generate new strains which may be more adapted to the environment (31). Although influenza A, B, and C viruses have a common ancestor, reassortment between different types has never been observed (31). This could be due to the incompatibility of either viral proteins or vRNAs. Previously, Flandorfer et al. generated influenza A viruses carrying the full length of either the NA or HA ectodomain from influenza B virus, demonstrating a certain degree of compatibility between influenza A and B virus RNAs and proteins (7). During the past several years, the packaging signals have been identified in each RNA segment of influenza A virus. These packaging signals not only include 3′ and 5′ NCRs but also the termini of each ORF, which vary in length for each segment (9, 10, 18, 19, 21, 22, 25, 29, 43). By using the packaging sequences, foreign RNA segments can be incorporated into virions of influenza A virus (21, 36). In this study, using the HA packaging sequences, we generated two influenza A viruses that express the influenza C virus glycoprotein HEF instead of HA and NA. Indeed, both viruses contain seven RNA segments (Fig. 1D).
The successful rescue and the high titers of these viruses show that the influenza C virus HEF glycoprotein is fully compatible with the other influenza A viral components. This is surprising given the low (12%) amino acid sequence identity shared between HEF and HA (32, 35). In addition, the transmembrane and cytoplasmic regions of HEF are shorter than those of HA, and the cysteine residues undergo different modifications (26, 32, 40, 41). Despite these differences, HEF can fully replace the two glycoproteins, HA and NA, in the seven-segmented viruses generated in this study, indicating that the relatively long cytoplasmic region and palmitoylation of HA are not specifically required for influenza A virus. This conclusion is further supported by the data derived from HEF-Ecto chimeric viruses, in which the HEF ectodomain is fused to the HA transmembrane and cytoplasmic regions. Although the HEF-Ecto RNA segment has a higher packaging efficiency than that of the HEF RNA segment (Fig. 1D), the growth rate of HEF-Ecto viruses is similar to that of HEF viruses (Fig. 3B and C). It is also possible that the HEF ectodomain and HA transmembrane and cytoplasmic regions used in this study may not be fully compatible with each other. Exactly how many amino acids each domain contains and especially what the optimal junction site is still need to be demonstrated.
Influenza virus is a promising candidate for vaccine vector development because its replication does not have a DNA phase and it is highly immunogenic. Previous work by Luytjes et al. first described that a chloramphenicol acetyltransferase gene segment can be maintained for several passages by influenza virus (20). Other strategies, including inserting foreign peptides into the HA antigenic sites or the NA stalk region, have also been shown to be successful (2, 17). In the past several years, the sequences that are required for packaging of vRNA into virions have been identified by several groups. The characterization of these packaging sequences supports a specific packaging theory, which argues that for efficient vRNA packaging of influenza A virus, eight RNA segments are required (9, 10, 18, 19, 21, 22, 25, 29, 30, 43). How these packaging sequences function during the virion assembly process is still unclear. Foreign genes that are flanked by these sequences can be packaged into virions, which can then serve as vaccine or gene delivery vectors. In this study, the generation of HEF or HEF-Ecto viruses carrying an extra GFP segment further supports the potential of influenza viruses to act as gene vectors (Fig. 2). Despite the fact that the HEF or HEF-Ecto viruses carrying seven RNA segments can be rescued and are stable over multiple passages, the addition of an extra segment flanked by the NA packaging sequences enhances the viral growth in embryonated chicken eggs (Fig. 3). The fact that eight-segmented virus has a higher growth rate than the seven-segmented virus indicates that the HEF or HEF-Ecto virus is a suitable candidate to develop a vaccine or gene delivery vector. Previous work by Watanabe et al. also showed that influenza virus can carry an extra GFP-encoding segment after substituting the HA and NA segments with a segment expressing the glycoprotein of vesicular stomatitis virus (43). The use of influenza C virus HEF protein in this study provides another example of how influenza virus can be exploited as a vaccine vector.
In conclusion, in this study we generated two seven-segmented influenza A viruses expressing the C glycoprotein HEF instead of HA and NA. Both of the viruses are able to stably carry an additional RNA segment expressing a foreign gene, indicating that the seven-segmented viruses can be used as vaccine or gene delivery vectors carrying transgenes. This study also supports the specific packaging theory of influenza vRNA incorporation into virions.
Acknowledgments
We thank John Steel for helping with the immunostaining and RNA gel experiments and Anice C. Lowen for helpful comments on the manuscript.
This work was partially supported by NIH grants UO1AI070469 (Live Attenuated Vaccines for Epidemic and Pandemic Flu), HHSN2662000700010C (Center for Research on Influenza Pathogenesis), U54 AI057158-04 (Northeast Biodefense Center), and 1 UC19 AI062623-023 (Center For Investigating Viral Immunity and Antagonism).
Footnotes
Published ahead of print on 30 April 2008.
REFERENCES
- 1.Bancroft, C. T., and T. G. Parslow. 2002. Evidence for segment-nonspecific packaging of the influenza A virus genome. J. Virol. 767133-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castrucci, M. R., S. Hou, P. C. Doherty, and Y. Kawaoka. 1994. Protection against lethal lymphocytic choriomeningitis virus (LCMV) infection by immunization of mice with an influenza virus containing an LCMV epitope recognized by cytotoxic T lymphocytes. J. Virol. 683486-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, B. J., G. P. Leser, E. Morita, and R. A. Lamb. 2007. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 817111-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conenello, G. M., D. Zamarin, L. A. Perrone, T. Tumpey, and P. Palese. 2007. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 31414-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crescenzo-Chaigne, B., and S. van der Werf. 2007. Rescue of influenza C virus from recombinant DNA. J. Virol. 8111282-11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enami, M., G. Sharma, C. Benham, and P. Palese. 1991. An influenza virus containing nine different RNA segments. Virology 185291-298. [DOI] [PubMed] [Google Scholar]
- 7.Flandorfer, A., A. Garcia-Sastre, C. F. Basler, and P. Palese. 2003. Chimeric influenza A viruses with a functional influenza B virus neuraminidase or hemagglutinin. J. Virol. 779116-9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 739679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii, K., Y. Fujii, T. Noda, Y. Muramoto, T. Watanabe, A. Takada, H. Goto, T. Horimoto, and Y. Kawaoka. 2005. Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J. Virol. 793766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujii, Y., H. Goto, T. Watanabe, T. Yoshida, and Y. Kawaoka. 2003. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl. Acad. Sci. USA 1002002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glaser, L., J. Stevens, D. Zamarin, I. A. Wilson, A. Garcia-Sastre, T. M. Tumpey, C. F. Basler, J. K. Taubenberger, and P. Palese. 2005. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J. Virol. 7911533-11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrler, G., R. W. Compans, and H. Meier-Ewert. 1979. A precursor glycoprotein in influenza C virus. Virology 9949-56. [DOI] [PubMed] [Google Scholar]
- 13.Herrler, G., I. Durkop, H. Becht, and H. D. Klenk. 1988. The glycoprotein of influenza C virus is the haemagglutinin, esterase and fusion factor. J. Gen. Virol. 69(Pt. 4)839-846. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann, E., K. Mahmood, C. F. Yang, R. G. Webster, H. B. Greenberg, and G. Kemble. 2002. Rescue of influenza B virus from eight plasmids. Proc. Natl. Acad. Sci. USA 9911411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson, D., A. Cadman, T. Zurcher, and W. S. Barclay. 2002. A reverse genetics approach for recovery of recombinant influenza B viruses entirely from cDNA. J. Virol. 7611744-11747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobasa, D., S. M. Jones, K. Shinya, J. C. Kash, J. Copps, H. Ebihara, Y. Hatta, J. H. Kim, P. Halfmann, M. Hatta, F. Feldmann, J. B. Alimonti, L. Fernando, Y. Li, M. G. Katze, H. Feldmann, and Y. Kawaoka. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445319-323. [DOI] [PubMed] [Google Scholar]
- 17.Li, S. Q., J. L. Schulman, T. Moran, C. Bona, and P. Palese. 1992. Influenza A virus transfectants with chimeric hemagglutinins containing epitopes from different subtypes. J. Virol. 66399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang, Y., Y. Hong, and T. G. Parslow. 2005. cis-acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 7910348-10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang, Y., T. Huang, H. Ly, T. G. Parslow, and Y. Liang. 2008. Mutational analyses of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments. J. Virol. 82229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luytjes, W., M. Krystal, M. Enami, J. D. Parvin, and P. Palese. 1989. Amplification, expression, and packaging of foreign gene by influenza virus. Cell 591107-1113. [DOI] [PubMed] [Google Scholar]
- 21.Marsh, G. A., R. Hatami, and P. Palese. 2007. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 819727-9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh, G. A., R. Rabadan, A. J. Levine, and P. Palese. 2008. Highly conserved regions of influenza A virus polymerase gene segments are critical for efficient viral RNA packaging. J. Virol. 822295-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matrosovich, M., T. Matrosovich, W. Garten, and H. D. Klenk. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muraki, Y., T. Murata, E. Takashita, Y. Matsuzaki, K. Sugawara, and S. Hongo. 2007. A mutation on influenza C virus M1 protein affects virion morphology by altering the membrane affinity of the protein. J. Virol. 818766-8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muramoto, Y., A. Takada, K. Fujii, T. Noda, K. Iwatsuki-Horimoto, S. Watanabe, T. Horimoto, H. Kida, and Y. Kawaoka. 2006. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J. Virol. 802318-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakada, S., R. S. Creager, M. Krystal, R. P. Aaronson, and P. Palese. 1984. Influenza C virus hemagglutinin: comparison with influenza A and B virus hemagglutinins. J. Virol. 50118-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 969345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann, G., M. A. Whitt, and Y. Kawaoka. 2002. A decade after the generation of a negative-sense RNA virus from cloned cDNA—what have we learned? J. Gen. Virol. 832635-2662. [DOI] [PubMed] [Google Scholar]
- 29.Noda, T., H. Sagara, A. Yen, A. Takada, H. Kida, R. H. Cheng, and Y. Kawaoka. 2006. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439490-492. [DOI] [PubMed] [Google Scholar]
- 30.Ozawa, M., K. Fujii, Y. Muramoto, S. Yamada, S. Yamayoshi, A. Takada, H. Goto, T. Horimoto, and Y. Kawaoka. 2007. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J. Virol. 8130-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palese, P., and M. L. Shaw. 2007. Orthomyxoviridae: the viruses and their replication, p. 1647-1689. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 32.Pfeifer, J. B., and R. W. Compans. 1984. Structure of the influenza C glycoprotein gene as determined from cloned DNA. Virus Res. 1281-296. [DOI] [PubMed] [Google Scholar]
- 33.Quinlivan, M., D. Zamarin, A. Garcia-Sastre, A. Cullinane, T. Chambers, and P. Palese. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 798431-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Racaniello, V. R., and P. Palese. 1979. Isolation of influenza C virus recombinants. J. Virol. 321006-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenthal, P. B., X. Zhang, F. Formanowski, W. Fitz, C. H. Wong, H. Meier-Ewert, J. J. Skehel, and D. C. Wiley. 1998. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature 39692-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinya, K., Y. Fujii, H. Ito, T. Ito, and Y. Kawaoka. 2004. Characterization of a neuraminidase-deficient influenza A virus as a potential gene delivery vector and a live vaccine. J. Virol. 783083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steel, J., S. V. Burmakina, C. Thomas, E. Spackman, A. Garcia-Sastre, D. E. Swayne, and P. Palese. 2008. A combination in-ovo vaccine for avian influenza virus and Newcastle disease virus. Vaccine 26522-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talon, J., M. Salvatore, R. E. O'Neill, Y. Nakaya, H. Zheng, T. Muster, A. Garcia-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 974309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tumpey, T. M., T. R. Maines, N. Van Hoeven, L. Glaser, A. Solorzano, C. Pappas, N. J. Cox, D. E. Swayne, P. Palese, J. M. Katz, and A. Garcia-Sastre. 2007. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 315655-659. [DOI] [PubMed] [Google Scholar]
- 40.Veit, M., G. Herrler, M. F. Schmidt, R. Rott, and H. D. Klenk. 1990. The hemagglutinating glycoproteins of influenza B and C viruses are acylated with different fatty acids. Virology 177807-811. [DOI] [PubMed] [Google Scholar]
- 41.Veit, M., H. Reverey, and M. F. Schmidt. 1996. Cytoplasmic tail length influences fatty acid selection for acylation of viral glycoproteins. Biochem. J. 318(Pt. 1)163-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe, T., S. Watanabe, J. H. Kim, M. Hatta, and Y. Kawaoka. 2008. Novel approach to the development of effective H5N1 influenza A virus vaccines: use of M2 cytoplasmic tail mutants. J. Virol. 822486-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe, T., S. Watanabe, T. Noda, Y. Fujii, and Y. Kawaoka. 2003. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J. Virol. 7710575-10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada, S., Y. Suzuki, T. Suzuki, M. Q. Le, C. A. Nidom, Y. Sakai-Tagawa, Y. Muramoto, M. Ito, M. Kiso, T. Horimoto, K. Shinya, T. Sawada, M. Kiso, T. Usui, T. Murata, Y. Lin, A. Hay, L. F. Haire, D. J. Stevens, R. J. Russell, S. J. Gamblin, J. J. Skehel, and Y. Kawaoka. 2006. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature 444378-382. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, J., G. P. Leser, A. Pekosz, and R. A. Lamb. 2000. The cytoplasmic tails of the influenza virus spike glycoproteins are required for normal genome packaging. Virology 269325-334. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 744634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]