Abstract
The glycan shield of the human immunodeficiency virus type 1 (HIV-1) envelope (Env) protein serves as a barrier to antibody-mediated neutralization and plays a critical role in transmission and infection. One of the few broadly neutralizing HIV-1 antibodies, 2G12, binds to a carbohydrate epitope consisting of an array of high-mannose glycans exposed on the surface of the gp120 subunit of the Env protein. To produce proteins with exclusively high-mannose carbohydrates, we generated a mutant strain of Saccharomyces cerevisiae by deleting three genes in the N-glycosylation pathway, Och1, Mnn1, and Mnn4. Glycan profiling revealed that N-glycans produced by this mutant were almost exclusively Man8GlcNAc2, and four endogenous glycoproteins that were efficiently recognized by the 2G12 antibody were identified. These yeast proteins, like HIV-1 gp120, contain a large number and high density of N-linked glycans, with glycosidase digestion abrogating 2G12 cross-reactivity. Immunization of rabbits with whole Δoch1 Δmnn1 Δmnn4 yeast cells produced sera that recognized a broad range of HIV-1 and simian immunodeficiency virus (SIV) Env glycoproteins, despite no HIV/SIV-related proteins being used in the immunization procedure. Analyses of one of these sera on a glycan array showed strong binding to glycans with terminal Manα1,2Man residues, and binding to gp120 was abrogated by glycosidase removal of high-mannose glycans and terminal Manα1,2Man residues, similar to 2G12. Since S. cerevisiae is genetically pliable and can be grown easily and inexpensively, it will be possible to produce new immunogens that recapitulate the 2G12 epitope and may make the glycan shield of HIV Env a practical target for vaccine development.
The development of a human immunodeficiency virus (HIV) vaccine able to induce neutralizing antibodies against a broad spectrum of primary isolates is complicated by the large diversity of HIV type 1 (HIV-1) strains, the continual mutation of the envelope (Env) glycoprotein in the face of immune selective pressure, and the presence of numerous N-linked glycans that mask polypeptide epitopes (7). Indeed, genetic deletion of N-linked carbohydrate sites can greatly increase the sensitivity of HIV-1 to antibody-mediated neutralization (3, 12, 25, 26, 34, 35). One of the few broadly neutralizing monoclonal antibodies (MAbs) isolated from HIV-1-infected patients, 2G12, circumvents these obstacles by binding to relatively conserved high-mannose-type oligosaccharides exposed on the glycan shield of the gp120 subunit of Env (47, 49, 54). The 2G12 epitope consists of an array of at least three such glycans presented as a dense cluster of terminal mannose sugars (49, 54). Crystal structures of the 2G12 Fab in complex with carbohydrates reveal a specificity toward Manα1,2Man disaccharides, alone or terminally exposed on the D1 and D3 arms of Man9GlcNAc2 (Man9) and Man8GlcNAc2 (Man8) structures, without recognizing other mannose disaccharides, including Manα1,3Man and Manα1,6Man (8, 9). The relatively conserved nature of the 2G12 epitope and the role of N-linked carbohydrates in protecting HIV-1 from antibodies make the glycan shield of Env a viable vaccine target (46, 48).
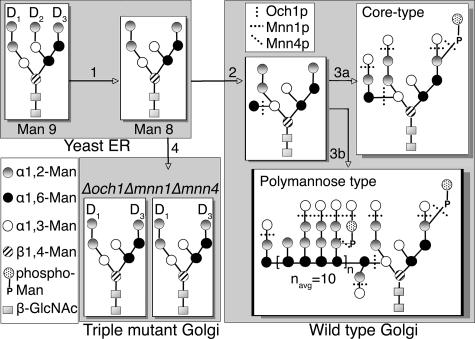
The gp120 subunit of the HIV-1 Env protein contains an average of 25 N-linked glycosylation sites, approximately half of which are composed of high-mannose or hybrid-type glycans (13, 31, 59). To mimic the 2G12 epitope, glycoantigens have been constructed by several laboratories through chemical synthesis of mannose oligosaccharides and chemoenzymatic conjugation to different scaffolds (8, 9, 27, 29, 41, 56). However, to our knowledge, these approaches have yet to elicit antibodies that cross-react with gp120 or neutralize the virus. An alternative approach is to identify and produce other proteins that contain carbohydrate structures similar to those comprising the 2G12 epitope on HIV-1 Env. Analysis of the Saccharomyces cerevisiae genome reveals the presence of numerous proteins that contain a large number of potential N-linked glycosylation sites, making yeast a possible source of proteins with closely arrayed N-linked glycans with the potential to cross-react with the 2G12 antibody. However, while essentially identical high-mannose core structures are added to the N-linked glycosylation sites on both yeast and mammalian cell proteins in the endoplasmic reticulum (ER) (4, 14, 18, 23), subsequent carbohydrate processing pathways in the Golgi apparatus diverge significantly. In yeast cells, numerous mannose residues are added to the core structure in the Golgi apparatus, forming polymannose-type glycans (14). Over a dozen proteins in the Golgi apparatus of S. cerevisiae are involved in processing N-glycans (20), three of which are vital for the modification of the core Man8 structure that is exported from the ER (Fig. 1). In the cis-Golgi, Och1p initiates the first mannose residue necessary for the extended α1,6-linked mannose branch, a key component of polymannose-type glycans (30). Deletion of the Och1 gene alone leads to the lack of the outer chain, resulting in a majority of Man9 and Man10 structures, which represent core Man8 capped at the D1 and/or D3 arm with α1,3-linked mannose residues (40). In the trans-Golgi, Mnn1p is the sole α1,3-mannosytransferase responsible for adding these α1,3-mannose caps to the core glycans (11, 39, 58), while Mnn4p is a positive regulator involved in adding phosphomannose residues to both the core and outer chain (43, 44).
FIG. 1.
N-linked glycosylation pathway in wild-type and Δoch1 Δmnn1 Δmnn4 S. cerevisiae. In the ER, after en bloc transfer of Glc3Man9GlcNAc2 to nascent polyproteins, the three glucose residues are cleaved. In wild-type yeast, N-linked glycans are then processed as follows. In step 1, Man9GlcNAc2 is trimmed by Mns1p to Man8GlcNAc2 in the ER. In step 2, the first α1,6-linked-mannose backbone residue is added by Och1p in the cis-Golgi. In step 3a, for core-type glycans, a few mannose residues are added by Mnn1p and Mnn4p (through GDP-mannose-dependent mannosyltransferase protein) in the Golgi apparatus. In step 3b, for polymannose type, numerous mannosyltransferases in the Golgi apparatus (including Mnn5p to Mnn9p, Ktr1p to Ktr3p, Kre2p, and Mnn1p) lead to glycans with an average of 160 mannose residues (14). Mannose sugars added by Och1p, Mnn1p, and Mnn4p in the Golgi apparatus of wild-type yeast are depicted with dotted vertical, horizontal, and diagonal lines, respectively. In step 4, the absence of these proteins in Δoch1 Δmnn1 Δmnn4 yeast cells is expected to produce a majority of Man8GlcNAc2, with minor amounts of untrimmed Man9GlcNAc2, on proteins exported from the Golgi apparatus.
In this study, we sought to mimic the 2G12 epitope on the HIV-1 Env protein by producing a yeast strain that would express exclusively unprocessed, high-mannose glycans. We achieved this by generating a triple mutant of S. cerevisiae, a Δoch1 Δmnn1 Δmnn4 mutant (that we named TM [for triple mutant]) that produced almost homogenous Man8 glycans. The MAb 2G12 bound efficiently to the TM mutant, but not to the wild-type S. cerevisiae (that we named WT [for wild type]). At least four heavily glycosylated S. cerevisiae proteins supported 2G12 binding, with each of these possessing numerous N-linked glycosylation sites at a high density. Importantly, immune sera raised with whole cells of this mutant yeast, but not WT, cross-reacted in a carbohydrate-dependent manner with a broad array of mammalian cell-expressed Env glycoproteins from HIV-1 and simian immunodeficiency virus (SIV) strains, suggesting that genetically modified yeast proteins may serve as molecular scaffolds that recapitulate carbohydrate-dependent epitopes on the surface of the HIV-1 Env protein.
MATERIALS AND METHODS
Materials.
The 2G12 antibody was purchased from Polymum Scientific (Vienna, Austria). The AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH supplied the following reagents: SF162 gp120 (catalog no. 7363), 96ZM651 gp120 (catalog no. 10080), SIVmac1A11 gp140 (catalog no. 2209), and SIVmac239 gp130 (catalog no. 2322). Horseradish peroxidase (HRP)-labeled goat anti-rabbit immunoglobulin G (IgG) and goat anti-human IgG were purchased from Jackson ImmunoResearch (West Grove, PA).
S. cerevisiae strains and mating.
All S. cerevisiae strains used in this study are listed in Table 1, with the WT strain from Invitrogen (Carlsbad, CA), mating testers from the American Type Culture Collection (ATCC) (Manassas, VA), and yeast knockouts from Open Biosystems (West Grove, PA). Yeast strains were mated and sporulated according to standard molecular biology techniques (53). Knockout of indicated genes was verified by colony PCR using synthesized primers specific to Mnn1, Och1, and Mnn4 and to the kanMX4 insertion cassette (19). After mating, all yeast cultures used for analysis and immunization were grown in yeast peptone dextrose (YPD) medium (1% Bacto yeast extract, 2% Bacto peptone, and 2% glucose) or YPDK (YPD supplemented with 0.3 M KCl) as indicated.
TABLE 1.
S. cerevisiae strains used in this study
| S. cerevisiae strain | Genotype | Source |
|---|---|---|
| INVSc1 (WT) | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 trp1-289/trp1-289 ura3-52/ura3-52 | Invitrogen |
| BY4710 | MATatrp1Δ63 | ATCC |
| BY4711 | MATα trp1Δ63 | ATCC |
| Δmnn1 mutant | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 mnn1::kanMX4 | Open Biosystems |
| Δmnn4 mutant | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mnn4::kanMX4 | Open Biosystems |
| Δoch1 mutant | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 och1::kanMX4 | Open Biosystems |
| Δmnn1 Δmnn4 mutant | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mnn1::kanMX4 mnn4::kanMX4 | This study |
| Δoch1 Δmnn1 mutant | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 mnn1::kanMX4 och1::kanMX4 | This study |
| Δoch1 Δmnn1 Δmnn4 mutant | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mnn1::kanMX4 mnn4::kanMX4 och1::kanMX4 | This study |
| Δoch1 Δmnn1 Δmnn4 diploid mutant (TM) | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 LYS2/lys2Δ0ura3Δ0/ura3Δ0 mnn1::kanMX4/mnn1::kanMX4 mnn4::kanMX4/mnn4::kanMX4 och1::kanMX4/och1::kanMX4 | This study |
Production of antibodies.
Rabbit polyclonal antibody against zymosan was generated using the yeast cell wall extract, zymosan A from Sigma (St. Louis, MO). Two rabbits were immunized with 200 μg of zymosan A in complete Freund's adjuvant (Sigma) at week 0 and boosted with 100 μg of antigen in incomplete Freund's adjuvant (Sigma) at weeks 2 and 4 and thereafter every 4 weeks. The resulting sera, collected 1 week after each boost, were pooled, and the IgG was purified with protein A-agarose (Invitrogen). Rabbit antibodies against yeast glycoproteins were produced using synthetic peptides coupled to keyhole limpet hemocyanin from Pierce Biotechnology (Rockford, IL) and using the immunization procedure described above. The peptides used and their sequences were as follows: Ecm33 (DSIKKITGDLNMQE), Gp38 (AVNGVTSKSALESIFP), Gas1 (TPKEQLSFVMNLYYEKSGGSKSD), and YJL171c (EVGDRVWFSGKNAPLADY). Antibodies were purified by immunoaffinity chromatography using the antigenic peptide coupled to SulfoLink gel (Pierce).
Glycan profiling.
N-linked glycans on log-phase TM yeast cells or purified Gp38 glycoprotein were analyzed by the Glycotechnology Core Resource, University of California, San Diego. Peptide-N-glycosidase F (PNGase F) digestion was used to release N-linked oligosaccharides. Glycans were permethylated for analysis by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) using a QSTAR hybrid mass spectrometer (Applied Biosystems, Foster City, CA) and derivatized with 2-aminobenzamide (2AB) for normal-phase high-performance liquid chromatography (NP-HPLC) analysis on a Dionex DX-600 HPLC system (Sunnyvale, CA). 2AB-derivatized N-linked glycans released from RNase B were used as a standard for HPLC.
Immunofluorescence.
Log-phase TM and WT yeast cells grown in YPDK were fixed with 4% paraformaldehyde for 1 h. The cells were washed with phosphate-buffered saline (PBS) and transferred to polylysine-coated slides. After the cells were allowed to air dry, they were incubated with 0.5% sodium dodecyl sulfate (SDS) for 15 min. Antizymosan and 2G12 antibodies were diluted to 10 μg/ml and 20 μg/ml, respectively, and incubated with the cells for 1 h. The slides were washed as described above and probed using Alexa Fluor 488-conjugated goat anti-rabbit IgG and Alexa Fluor 568-conjugated goat anti-human IgG (Invitrogen). Images were obtained using a Zeiss Axioskop fluorescence microscope (McBain Instruments, Chatsworth, CA).
Western blotting.
All yeast strains were grown to an optical density at 600 nm (OD600) of 3.0 in YPDK medium, and the collected cell pellets were lysed with acid-washed glass beads in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 10 mM Tris [pH 7.4], 1 mM EDTA [pH 8.0], 1% Triton X-100, 1% deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, and 1 μg/ml leupeptin). Cell debris was removed by centrifugation, and the lysates were diluted to 0.24 mg/ml for SDS-polyacrylamide gel electrophoresis. Western blotting was conducted using 2G12 antibody, followed by HRP-labeled anti-human IgG secondary antibody. Chemiluminescence detection was employed using an ECL (enhanced chemiluminescence) kit from GE Biosciences (Piscataway, NJ).
ELISA.
For whole-cell enzyme-linked immunosorbent assay (ELISA), log-phase yeast cells were diluted to 5.0 × 107 cells/ml in PBS to coat plates. Antigens (100 μl) were incubated in each well overnight at room temperature. The wells were blocked for 2 h with blocking buffer (PBS, 0.1% bovine serum albumin [BSA], 0.02% thimerosal), rinsed, and incubated for 2 h with 2G12 antibody at 10 μg/ml with a threefold serial dilution. The wells were washed with wash buffer (PBS, 0.05% Tween 20, 0.02% thimerosal) and incubated 1 h with HRP-labeled secondary antibodies. After the wells were washed, they were incubated with 3,3′,5,5′-tetramethylbenzidine for color development, which was stopped with 1 M HCl. Absorbance at an optical density at 450 nm (OD450) was read using an EMax microplate reader (Molecular Devices, Sunnyvale, CA). For gp120 ELISA, HIV-1 gp120 glycoproteins from strains JR-FL, ADA, R2, Du179, Q1769, 98IN012, HxB, Yu2, Tybe, and R3A were expressed and purified by concanavalin A (ConA)-lectin affinity chromatography from 293T cells using recombinant vaccinia virus vectors (21). Antigens were diluted to 3 μg/ml in 50 mM carbonate buffer, pH 9.5, with 100 μl incubated in each well overnight at room temperature with wells blocked and rinsed as described above. Immune sera against TM yeast cells were added at a 1:500 dilution in blocking buffer, along with 2G12 antibody as a control at 0.2 μg/ml, and incubated for 2 h. ELISA plates were processed as described above.
Protein identification.
Partially purified proteins detected by antibody 2G12 were excised from either analytic two-dimensional (2D) gels or full-size SDS-polyacrylamide gels and identified by Proteomic Research Services, Inc. (Ann Arbor, MI) using trypsin digestion followed by nanoliter liquid chromatography coupled to tandem MS (nano-LC-MS-MS) on a Micromass Q-ToF 2. Peptide masses were searched using MASCOT (Matrix Science, Boston, MA). Only proteins with at least two corroborating peptides with quality MS-MS profiles were considered positive identifications.
Immunoprecipitation.
Log-phase TM (haploid) yeast cells were grown in YPDK and separated from the culture supernatant by centrifugation. Cells were lysed in RIPA buffer as described above. Cell lysate and culture supernatant samples were precleared by incubation with protein A-agarose (GE Biosciences) for 2 h. Then, 10 μg of 2G12 antibody was added and incubated on a shaker for 1 h. After incubation, 20 μl of protein A-agarose conjugate was added and incubated for 1 h. Beads were pelleted and washed twice with 800 μl of PBS. Proteins were eluted by boiling with 40 μl of 1× Laemmli sample buffer and loaded onto a 4 to 20% gradient SDS-polyacrylamide gel. Proteins were blotted onto nitrocellulose for Western blot analysis using anti-Ecm33, anti-Gp38, anti-YJL171c, and anti-Gas1.
Glycosidase digestion.
The endogenous yeast protein Gp38 was purified using 95% saturated ammonium sulfate precipitation followed by preparative 2D separation to >95% purity using the Rotofor Cell and model 491 Prep Cell from Bio-Rad Laboratories (Hercules, CA). Purified protein samples (1 μg each) were denatured and reduced by boiling in 1% SDS and 50 mM dithiothreitol. Samples were incubated at 37°C with 50 mU endoglycosidase H (Endo H) or without Endo H (mock) for 24 h in 20 μl of the manufacturer's recommended buffer (EMD Chemicals, San Diego, CA).
For digestion by glycosidases, 30 μg of R2 or JR-FL gp120 (30 μl) was denatured in 50 μl of 50 mM NaH2PO4 (pH 7.5), 0.1% SDS, and 50 mM dithiothreitol for 5 min at 100°C. In total, 10 μg of each gp120 was digested with 10 U of PNGase F (EMD Chemicals) and 10 mU of Endo H and mock digested by adding 2 μl of 50 mM NaH2PO4 (pH 7.5) for 18 h at 37°C. For digestion by mannosidases, 10 μg of R2 or JR-FL gp120 was digested with 0.5 U of jack bean mannosidase for 3 h at 37°C, or 1 mU of Aspergillus saitoi mannosidase for 24 h at 37°C according to the manufacturer's protocols (Prozyme, San Leandro, CA).
Whole-cell immunization.
Two groups of 10 rabbits were immunized with heat-killed, diploid WT or TM yeast cells using a modified protocol as described previously (4). Briefly, log-phase cells were suspended in 0.9% NaCl at 3.0 × 107 cells/ml and heat killed at 70°C for 90 min. Rabbits were injected with 0.25 ml, 0.5 ml, 0.75 ml, and 1.0 ml of this suspension in the marginal ear vein three times a week at week 1, week 2, week 3, and week 4, respectively. Three days after the last injection in weeks 3, 4, and 5, the rabbits were bled. After 4 weeks of rest, the rabbits were injected with 0.5 ml, 0.75 ml, and 1.0 ml of the suspension in the marginal ear vein three times a week at week 9, week 10, and week 11, respectively. A fourth, final bleed at week 12 was collected 3 days after the final injection (week 12).
After analysis of the primary immunization, a second group of four rabbits was immunized with heat-killed whole yeast cells at 1.0 × 108 cells/ml. Rabbits were immunized continuously three times a week for 9 weeks with a 1-ml suspension of cells in sterile 0.9% NaCl. Rabbits were bled at weeks 4, 6, 8, and 10.
Titration ELISA.
Rabbit serum titers were evaluated by an ELISA using purified HIV-1 gp120 as the coating antigen. Purified gp120 at 150 ng per well was adsorbed on Immulon 2HB 96-well plates (Thermo Labsystems, Waltham, MA) in ELISA capture buffer (0.15 M sodium carbonate and 0.35 M sodium bicarbonate in PBS [pH 9.6]) overnight. Wells were washed three times with 200 μl wash buffer (PBS with 0.05% Tween 20) and blocked with 200 μl blocking solution (PBS with 2% BSA) for 1 h. Rabbit sera were diluted in blocking solution, added to the plate at 100 μl/well, and allowed to bind for 2 h with slight rocking. After the wells were washed three times, HRP-conjugated goat anti-rabbit IgG was added at 1:10,000 dilution in blocking buffer, 100 μl/well, and allowed to incubate for 1 h. Following three final washes, 100 μl of 3,3′,5,5′-tetramethylbenzidine solution was added to each well. After a 5-min incubation at 37°C, the reaction was stopped with 100 μl of 1 M phosphoric acid per well, and OD450 was measured using a MRX Revelation microplate reader (Dynex Technologies, Chantilly, VA).
Glycan microarray screening.
Preimmune (week 0) and postimmune (week 10) sera from rabbit 1141 were purified using affinity chromatography with protein A-agarose (Invitrogen). These IgG antibodies were tested for binding to 320 synthetic and natural glycans (printed array version 3.0, glycans listed at http://www.functionalglycomics.org/static/consortium/resources/resourcecoreh8.shtml) on glass microscope slides as described previously (1, 5). This work was done by Core H, Consortium for Functional Glycomics (CFG), Emory University. Briefly, 70 μl of rabbit 1141 antibody samples, diluted to 1:50 (20 μg/ml) in standard PBS binding buffer with 1% BSA and 0.05% Tween 20, was applied to the printed surface of the microarray and incubated in a humidified chamber for 1 h. The slide was rinsed four times in PBS buffer. Binding was detected using a fluorescence (Alexa Fluor 488)-labeled anti-rabbit IgG at 5 μg/ml and incubated for 1 h. The slide was rinsed four times in PBS buffer. The binding image was read in a Perkin-Elmer microarray XL4000 (Waltham, MA) scanner and analyzed using Imagene (V.6) image analysis software (BioDiscovery, El Segundo, CA). 2G12 antibody was tested similarly at 30 μg/ml, with detection using fluorescence-labeled (Alexa Fluor 488) anti-human IgG.
RESULTS
Creation of mutant yeast strains to produce Man8 glycans.
To produce glycoproteins with carbohydrate structures that are potentially able to mimic the 2G12 epitope, double and triple mutant strains of S. cerevisiae were generated through successive mating and sporulation of three single open reading frame knockout strains, the Δoch1, Δmnn1, and Δmnn4 strains. The final genotypes of the resulting strains, Δmnn1 Δmnn4, Δoch1 Δmnn1, and Δoch1 Δmnn1 Δmnn4 (TM), were verified by PCR using primers specific to each gene and the KanMX4 insertion cassette (data not shown). TM yeast had growth characteristics similar to those of the Δoch1 strain, such as a loss of viability at nonpermissive temperatures (above 37°C) and slow growth (38); the cells took more than three times as long as WT to reach similar cell densities, a phenotype that was partially rescued in medium with osmotic support (YPDK).
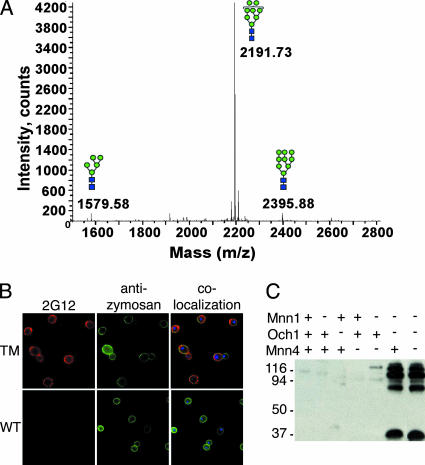
To assay the oligosaccharides present on glycoproteins from TM yeast, MALDI-TOF and NP-HPLC glycan profiling were conducted on samples from whole yeast cells. MALDI-TOF results for N-linked glycans released from an extract of whole TM cells showed that Man8 was the predominant oligosaccharide, with minor amounts of Man9 and Man5 detected (Fig. 2A). NP-HPLC results were consistent with the MALDI-TOF data, with the major peak eluting at 25.4 min, the same as the 2AB-labeled Man8 standard (data not shown). Thus, the N-linked glycans derived from TM yeast contained mostly Man8, the glycan structure that comprises the epitope for MAb 2G12 on the surface of HIV-1 gp120.
FIG. 2.
Cross-reactivity of 2G12 with TM yeast expressing mostly Man8 glycans. (A) MALDI-TOF MS was performed on permethylated glycans, released from homogenized TM yeast with PNGase F. The major mass peak at a mass-to-charge ratio (m/z) of 2191.73 is representative of over 90% of the total glycans and corresponds to permethylated Man8GlcNAc2. (B) Immunofluorescence staining of whole TM (top) and WT (bottom) yeast cells using MAb 2G12 (left) and a rabbit polyclonal antibody specific to the yeast cell wall particle zymosan (middle), with colocalization of staining depicted (right). Note that TM yeast cells are noticeably larger than WT yeast cells. (C) Western blot with whole-cell lysates from a panel of yeast strains probed with the HIV-1 MAb 2G12. Yeast strains contain functional (+) or deleted (−) Och1, Mnn1, and Mnn4 genes as indicated, with WT and TM yeast in the first and last lanes, respectively. The migration positions of molecular mass markers (in kilodaltons) are indicated to the left of the blot.
Cross-reaction of TM yeast proteins with MAb 2G12.
To determine whether MAb 2G12 specifically recognized TM yeast, we tested for 2G12 binding using various techniques. First, we conducted immunofluorescence using MAb 2G12 and a polyclonal antibody specific to zymosan, a yeast cell wall component consisting of α-mannan and β-glucan (16). MAb 2G12 strongly stained TM yeast and colocalized with antizymosan on the cell wall (Fig. 2B, top). WT yeast showed staining only with antizymosan, with no staining seen for 2G12 (Fig. 2B, bottom). Second, a whole-cell ELISA was conducted to measure binding of 2G12 to native proteins on the yeast cell surface. MAb 2G12 failed to show any binding to WT cells even at a high concentration of 10 μg/ml, while showing obvious binding to TM yeast at a concentration as low as 1 μg/ml (data not shown). Finally, we performed Western blot analysis on cell lysates derived from a panel of yeast glycosylation mutants, including WT, single, double, and triple mutant haploid yeast. In the lysates of the Δoch1 Δmnn1 and TM yeast strains, 2G12 detected multiple bands, several of which showed strong signals (Fig. 2C). By contrast, cell lysates from other yeast strains showed no or very faint bands recognized by 2G12. Five additional Δoch1 Δmnn1 Δmnn4 haploid yeast strains generated in our laboratory showed similar 2G12 Western blot results, with comparable protein sizes and signal strengths, indicating that there were no genes complementary to those deleted (data not shown). Together, these results show that 2G12 not only cross-reacts with native proteins on the surfaces of TM yeast cells but also recognizes multiple denatured proteins by Western blotting.
Identification of yeast proteins that cross-react with MAb 2G12.
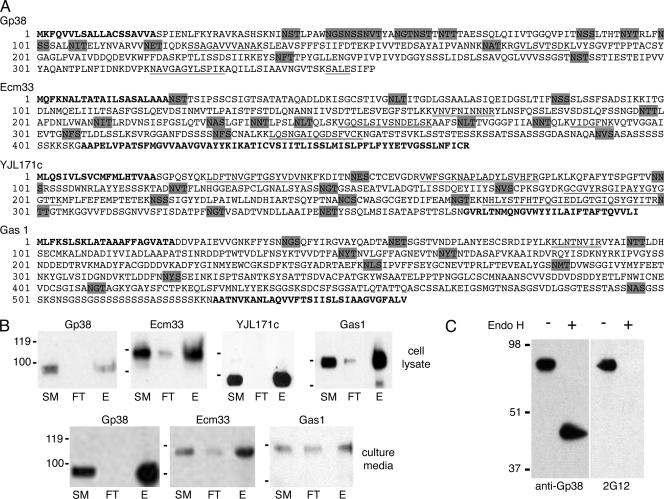
The endogenous yeast glycoproteins that bound to MAb 2G12 (Fig. 2C) were isolated and purified by employing differential centrifugation to separate membrane-associated proteins from cytoplasmic proteins in TM cell lysates, followed by ConA-lectin affinity chromatography. The resulting ConA elutes were separated by two 2D gels in parallel. Spots from the Spyro Ruby-stained gel corresponding to MAb 2G12-positive signals analyzed by Western blotting were excised for nano-LC-MS-MS identification. Two 2G12-reactive proteins with a molecular mass of approximately 110 to 115 kDa were identified as Ecm33 and Gas1, while one protein at approximately 95 kDa was identified as YJL171c. In addition, 2G12-reactive proteins present in the culture supernatant of TM yeast cells were identified using a combination of ConA chromatography, full-size SDS-polyacrylamide gel separation, and nano-LC-MS-MS identification. Ecm33 was again identified at approximately 110 kDa, and a new protein, Gp38, was identified at 90 kDa. The sequences of these proteins are shown in Fig. 3A. Like gp120, these proteins have numerous potential N-linked glycosylation sites, ranging from 10 to 15 (Table 2). The majority of these sites are likely to be glycosylated, since all four proteins migrated in SDS-polyacrylamide gels to positions that were approximately twice their expected molecular masses calculated from the mature polypeptides. All of these proteins are related to the cell wall of yeast, with Ecm33, YJL171c, and Gas1 being glycosylphosphatidylinositol (GPI)-anchored proteins important for maintaining the architecture of the cell wall and responding to damage (15, 28, 45, 52).
FIG. 3.
Identification and verification of yeast glycoproteins that cross-react with MAb 2G12. (A) The amino acid sequences of glycoproteins from TM yeast that cross-reacted with MAb 2G12 by immunoblotting are shown. Underlined sequences represent peptides identified by nano-LC-MS-MS, with double underlines representing peptides identified two or more times. Bold letters at the N and C termini depict predicted signal peptides and GPI anchor signal sequences, respectively. Sequences highlighted by gray shading are potential N-linked glycosylation sites. The amino acid sequences were obtained from GenBank under accession numbers P38616 for Gp38 (15), P38248 for Ecm33 (55), P46992 for YJL171c (17), and P22146 for Gas1 (42). (B) The glycoproteins from TM cells were immunoprecipitated with MAb 2G12 and blotted with polyclonal antibodies specific to the identified yeast glycoproteins. TM cell lysate (top panels) or culture media (bottom panels) was used as the starting material (SM) for 2G12 binding. Antibody-protein complexes were precipitated with protein A-agarose, unbound proteins were removed as flowthrough (FT) followed by washes, and bound proteins were eluted with Laemmli sample buffer (E). (C) Immunoblot of purified Gp38 after either mock digestion (−) or Endo H digestion (+) and probed with anti-Gp38 (left) or MAb 2G12 (right). The migration positions of molecular mass markers (in kilodaltons) are indicated to the left of the blots.
TABLE 2.
Features of MAb 2G12-reactive glycoproteins in TM yeast
| GenBank accession no. | Protein name | Mature protein
|
GPIe | ||||
|---|---|---|---|---|---|---|---|
| No. of aaa | N-glycan sites
|
Molecular mass (kDa)
|
|||||
| No. | Density (%)b | aac | Exp.d | ||||
| P38616 | Gp38 | 335 | 15 | 4.8 | 35 | 90 | No |
| P38248 | Ecm33 | 386 | 13 | 3.4 | 40 | 110 | Yes |
| P46992 | YJL171c | 349 | 11 | 3.2 | 38 | 95 | Yes |
| P22146 | Gas1 | 506 | 10 | 2 | 54 | 115 | Yes |
No. of aa, number of amino acids in mature protein.
N-glycan density [(number of N-linked sites/number of amino acids in mature protein) × 100)].
Calculated molecular mass based on the number of amino acids (aa).
Experimental (Exp.) molecular mass from TM cells based on SDS-polyacrylamide gel electrophoresis.
Predicted GPI-anchored protein.
We performed immunoprecipitations to confirm that these identified yeast glycoproteins were indeed MAb 2G12 reactive. Proteins in the cell lysates and culture media of TM cells were immunoprecipitated with MAb 2G12 under nondenaturing conditions, and the blots were probed with peptide-generated antibodies specific to each yeast protein. Gp38, Ecm33, YJL171c, and Gas1 were each immunoprecipitated from TM cell lysate by MAb 2G12 (Fig. 3B, top), and Gp38, Ecm33, and Gas1 were also immunoprecipitated from TM culture supernatant (Fig. 3B, bottom). These results confirm the identity of the 2G12-reactive yeast glycoproteins from TM yeast and demonstrate that each glycoprotein can individually bind to 2G12 under native conditions.
The specificity of MAb 2G12 to mannose glycans on these endogenous yeast proteins was verified for Gp38. This 2G12-reactive protein was purified to greater than 95% from TM culture supernatant. MALDI-TOF analysis of N-linked glycans released from Gp38 showed that Man8 is the predominant form (data not shown), similar to the glycan profile for whole TM yeast cells (Fig. 2A). We removed high-mannose-type, N-linked carbohydrates from Gp38 by Endo H digestion (47). After digestion, the protein exhibited a size shift from approximately 90 kDa to 45 kDa (Fig. 3C) as judged by Western blotting with an anti-Gp38 antibody. However, 2G12 cross-reactivity was lost after Endo H digestion (Fig. 3C). These results confirm that approximately half of the molecular mass of the mature Gp38 protein from TM is from mannose glycans and that 2G12 cross-reactivity is dependent upon the carbohydrate moieties of the protein.
Induction of antibodies that cross-react with HIV-1 Env glycoproteins.
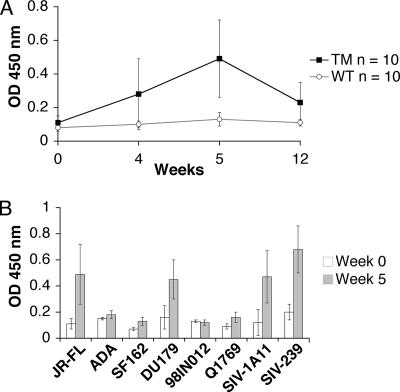
To determine whether TM yeast could induce 2G12-like antibodies in animals, two groups of 10 rabbits were immunized intravenously with heat-killed, whole yeast cells from WT and TM strains. Serum samples from 0, 4, 5, and 12 weeks were tested by ELISA using microwell plates containing immobilized, purified gp120 protein from HIV-1JR-FL. Sera from rabbits immunized with WT yeast cells failed to bind HIV-1JR-FL gp120 (Fig. 4A). By contrast, the sera from rabbits immunized with TM cells showed time-dependent binding activity, with a significant increase from week 0 to week 5 (P < 0.001). All 10 rabbits immunized with TM cells produced antibodies that cross-reacted with JR-FL gp120. The immune sera raised with TM cells were further tested against a variety of Env glycoproteins from HIV-1 and SIV to determine the breadth of binding activity. All of the tested Env glycoproteins were produced in mammalian cells and purified under native conditions. The postimmune sera at week 5 showed variable binding activities to the Env proteins from clade B (JR-FL) and clade C (DU179) of HIV-1 and also recognized two SIV Env proteins from SIVmac1A11 and SIVmac239 strains, while the preimmune sera showed no binding (Fig. 4B).
FIG. 4.
Induction of antibodies that cross-react with Env proteins from HIV-1 and SIV. (A) Sera at weeks 0, 4, 5, and 12 from all TM- and WT-immunized rabbits were tested for binding to 293T-expressed gp120 from HIV-1JR-FL coated on microwell plates. Each sample was tested at a 1:500 dilution in duplicate, and the means of all 10 TM and WT rabbits are presented with standard deviations (SDs) (error bars). (B) Multiple Env glycoproteins were immobilized on microwell plates and incubated with pre- and postimmune sera from TM-immunized rabbits at a 1:500 dilution. The means ± SDs (error bars) for all 10 rabbits are presented. The Env glycoproteins from JR-FL, ADA, DU179, 98IN012 and Q1769 were produced in 293T cells, SF162 and SIVmac239 (SIV-239) were produced in CHO cells, and SIVmac1A11 (SIV-1A11) was produced in CV-1 cells. All Env proteins have a purity of greater than 90%. OD, optical density.
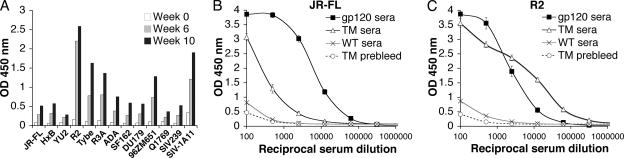
On the basis of the results from TM-immunized rabbits, we attempted to increase the titer of antibodies cross-reactive with HIV-1 gp120 by immunizing a second group of four rabbits at a higher dose (1.0 × 108 cells) and a longer continuous time period (9 weeks). However, the TM yeast cells, which are larger than WT cells, aggregated at this high concentration, and only one of the animals received the full immunization schedule. We analyzed sera from weeks 0, 6, and 10 of rabbit 1141 and found that the immune serum from week 10 exhibited broad reactivity to 11 out of 13 Env proteins from HIV-1 and SIV when using an OD450 of ≤0.5 as a cutoff (Fig. 5A). The preimmune serum had no reactivity to any tested Env proteins using the same cutoff. These data are in contrast to the results with MAb 2G12, which showed higher titers against gp120 proteins from clade B viruses but no binding to Env proteins from SIV and some clade C isolates (data not shown). When the antibody concentrations in the ELISA are considered, our immune serum has a 50- to 100-fold lower Env binding activity than MAb 2G12 does against clade B gp120s.
FIG. 5.
Induction of antisera with stronger and broader binding to a range of Env proteins from HIV-1 and SIV. (A) Sera at weeks 0, 6, and 10 from rabbit 1141 were tested by ELISA at 1:500 dilution against the indicated Env gp120 proteins. Proteins from JR-FL, HxB, YU2, R2, Tybe, R3A, ADA, DU179, 96ZM651, and Q1769 were produced in 293-T cells, SF162 and SIV239 were produced in CHO cells, and SIVmac1A11 (SIV-1A11) was produced in CV-1 cells. (B) gp120-specific IgG antibody titers in rabbit serum samples were measured using 150 ng purified JR-FL in ELISA format. Sera tested include TM-immunized rabbit 1141 at week 0 (TM prebleed) and week 10 (TM sera), the average of four postimmune serum samples from WT-immunized rabbits (WT sera), and serum from a rabbit immunized with JRFL gp120 (gp120 sera). The values are the averages ± standard errors of the means (error bars) of three independent experiments. (C) gp120-specific IgG antibody titers in rabbit serum samples were measured using 150 ng purified R2 Env proteins in ELISA format. The values are the averages ± standard errors of the means (error bars) of three independent experiments. OD, optical density.
Titration of rabbit 1141 postimmune serum (week 10) against purified gp120 proteins derived from the HIV-1 strains JR-FL and R2 Env showed significantly higher binding compared to rabbit 1141 prebleed and WT-immunized yeast sera (Fig. 5B and C). Serum from a rabbit immunized with JR-FL Env was tested in parallel for comparison. These results show that whole TM yeast cells can induce antibodies that cross-react with a broad range of gp120 glycoproteins produced in mammalian cells.
Specific binding of the antibodies to terminal α1,2-linked mannose residues on high-mannose, N-linked glycans.
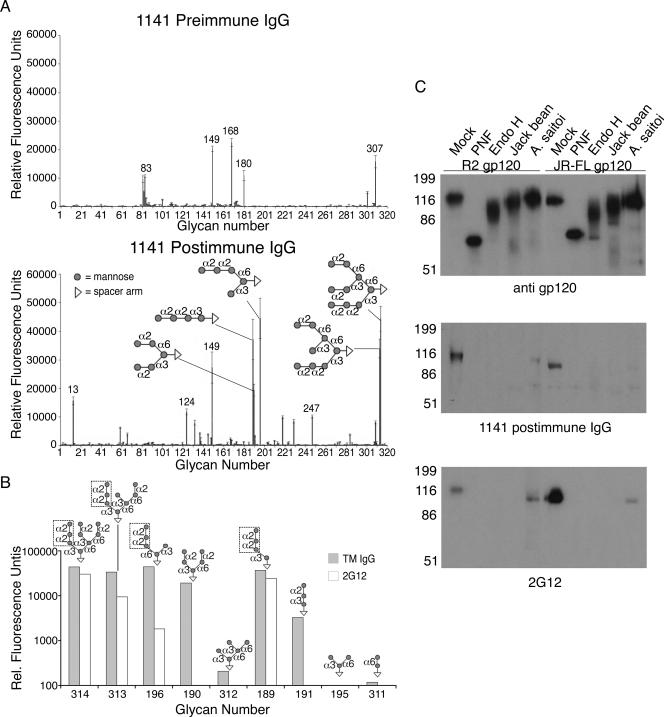
To characterize the glycan specificity recognized by serum from TM-immunized rabbit 1141, we purified the IgG from the pre- and postimmune samples and analyzed them for binding to an array of 320 different glycans. The antibody sample from preimmune serum (week 0) showed relatively low levels of preexisting antibodies to five glycan structures on the array with relative fluorescence units (RFU) above 10,000. None of these oligosaccharides contained mannose residues (Fig. 6A, top). By contrast, the IgG antibody from postimmune serum (week 10) showed strong binding (RFU from 33,973 to 44,545) to the synthetic oligomannoses that contain a terminal Manα1,2Manα1,2Man trisaccharide, including branched Man9, Man8, and Man5, and linear Man4 with glycan numbers 314, 313, 196, and 189, respectively (Fig. 6A, bottom). This antibody also bound with less strength (19,408 RFU) to a different form of Man5 (glycan number 190), which contains two branches with terminal α1,2-linked mannoses, Manα1,2Manα1,3Man and Manα1,2Manα1,6Man. Three of the remaining synthetic oligomannoses, glycan numbers 195, 311, and 312, contain only terminal α1,3-linked and/or α1,6-linked mannose structures and were undetected. Glycan number 149 with a high RFU (>20,000) is a lactosamine unit commonly found on polylactosamines and was detected by both pre- and postimmune sera. These results suggest that the antibodies induced by TM yeast cells specifically bind to mannose glycans with preference to Manα1,2Manα1,2Man trimannose structures.
FIG. 6.
Analysis of TM-induced antibodies reveals specificity to high-mannose-type glycans and terminal Manα1,2Man structures. (A) Purified IgG from rabbit 1141 sera at week 0 (preimmune) and week 10 (postimmune) was used to probe a microarray of 320 glycans. All hits to high-mannose glycans above 10,000 RFU are shown with the indicated linkages (α2 is α1-2 linkage, etc.), and each is conjugated to a spacer arm (CH2)5NH2. All hits to other glycans above 10,000 RFU are indicated with their corresponding glycan number. Glycan 13 is α-l-Rhα, glycan 124 is Galβ1-3(Neu5Acα2-6)GlcNAcβ1-4Galβ1-4Glcβ, glycan 149 is Galβ1-4GlcNAcβ1-3Galβ1-4Glcβ, and glycan 247 is Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAcβ. (B) Glycan binding by MAb 2G12 was tested on the microarray, and a comparison with purified IgG from rabbit 1141 at week 10 is shown for all synthetic mannose glycans. Manα1,2Manα1,2Man trisaccharides are boxed. Glycan microarray analyses were conducted by the Consortium for Functional Glycomics, Core H facility, Emory University. Rel., Relative. (C) Purified 293T-expressed gp120 was digested with PNGase F (PNF), Endo H, jack bean mannosidase, or A. saitoi mannosidase, with mock digestion using the same denatured conditions as in PNGase F digestion but without enzyme. Samples were loaded onto SDS-polyacrylamide gels at 1.25 μg/lane for purified rabbit 1141 IgG (middle) and at 100 ng/lane for 2G12 (bottom) and anti-gp120 (top) Western blots.
To directly compare the glycan specificity between MAb 2G12 and IgG from rabbit 1141 serum, MAb 2G12 was tested on the glycan microarray assay, and a side-by-side comparison of the results is shown (Fig. 6B). There were different binding profiles observed for the two samples, such that the TM-induced antibody exhibited broader and stronger binding to synthetic mannose glycans than 2G12, despite similar concentrations tested. MAb 2G12 showed binding only to glycans exhibiting a Manα1,2Manα1,2Man trisaccharide structure, normally found on the D1 arm of natural Man8 and Man9. In contrast, while our polyclonal IgG bound most strongly to these D1 arm-containing glycans, it also showed reactivity to glycans with Manα1,2Man disaccharides. This is consistent with the broader Env binding activity exhibited by the antisera to both HIV and SIV Env proteins.
To characterize the glycan specificity of the immune serum in the context of HIV-1 Env, native gp120 glycoproteins derived from two primary clade B strains were digested by a variety of glycosidases. PNGase F cleaves all N-linked glycans, Endo H cleaves high-mannose and hybrid-type glycans, jack bean mannosidase cleaves terminal α1,2-, 1,3-, and, 1,6-linked mannose residues, and A. saitoi mannosidase specifically cleaves terminal α1,2-linked mannose residues (22, 47). Digestion of the gp120 proteins with PNGase F, Endo H, jack bean mannosidase, and A. saitoi mannosidase resulted in masses of approximately 70 to 80, 85 to 105, 115, and 120 kDa, respectively, as detected by anti-gp120 (Fig. 6C, top). Mock-digested gp120 continued to migrate at 120 kDa. Probing the same digested proteins with rabbit 1141 IgG from postimmune serum (week 10) showed that the antibody detected only the mock-digested gp120 from R2 and JR-FL (Fig. 6C, middle), implicating terminal α1,2-linked mannose residues on high-mannose N-linked glycans as being vital epitopes for the antibody binding. This result is similar to that from MAb 2G12 (Fig. 6C, bottom), although there was some residual binding to A. saitoi mannosidase-digested gp120, perhaps due to incomplete digestion. These results confirm that TM yeast cells can induce the production of gp120 cross-reactive antibodies with specificity to terminal Manα1,2Man residues on high-mannose-type glycans, similar to 2G12.
DISCUSSION
Of the few epitopes on the HIV-1 Env protein to which broadly neutralizing antibodies can bind, only that recognized by MAb 2G12 is composed of carbohydrates. More specifically, the 2G12 epitope is composed of several closely spaced, high-mannose carbohydrate structures that present terminally exposed Manα1,2Man moieties on their D1 or D3 arms to which this neutralizing antibody binds (8, 49, 54). The very dense packing of this carbohydrate array on the outer domain of the gp120 subunit of Env may limit MAb 2G12 autoreactivity: while high-mannose carbohydrate structures are sometimes found on cell surface proteins, a dense cluster of such oligosaccharides is unusual if not entirely absent on mammalian cell surfaces, as most N-linked carbohydrate chains are processed to more complex forms in the Golgi apparatus. This idea is corroborated by the facts that passive immunization with high doses of MAb 2G12 was well tolerated in patients (2, 24) and that 2G12-like antibodies were found in long-term nonprogressors with broader neutralization capacity (6). In addition, the dense clustering of high-mannose structures on HIV-1 Env coupled with the unique Vh domain-exchanged configuration of 2G12 may limit carbohydrate flexibility, reducing the entropic cost and enabling 2G12 to bind to Env with high affinity (9). This unique aspect of the HIV Env glycan shield raises the possibility that it can be targeted for vaccine development (48). However, despite the fact that the epitope recognized by MAb 2G12 has been well characterized by structural studies, attempts to elicit 2G12-like antibodies have not been successful. Immunization with gp120 or other forms of HIV Env have failed to produce neutralizing antibodies to the dense glycan array that covers much of the gp120 subunit (7, 32). As a result, attempts have been made to reproduce the 2G12 epitope in different contexts in hopes of improving its immunogenicity. Indeed, several synthetic carbohydrate antigens, as well as peptide carbohydrate mimotopes, have been produced that can interact with 2G12 (8, 33, 36, 46, 56, 57). In a different approach, Scanlan et al. treated human 293T cells with kifunensine, a plant alkaloid that inhibits alpha-mannosidases, resulting in the expression of high-mannose oligosaccharides, particularly Man9GlcNAc2 (50). MAb 2G12 bound to these cells and also bound efficiently to a human protein (CEACAM1) expressed in 293T cells treated with kifunensine, suggesting that hypermannosylation of heterologous proteins may yield antigens that can be used for immunization trials.
While high-mannose structures that support MAb 2G12 binding can be produced on the surfaces of cells or synthetically, immunization studies with such glycoantigens have not, to the best of our knowledge, elicited antibodies capable of recognizing the HIV Env protein, let alone neutralize the virus. We took a genetic approach to recapitulate the epitope recognized by MAb 2G12, taking advantage of the power of yeast genetics and the fact that S. cerevisiae encodes a large number of proteins that contain numerous N-linked glycosylation sites. We found that deletion of the genes encoding two key carbohydrate processing enzymes, Och1 and Mnn1, resulted in efficient recognition of yeast by 2G12; deletion of Mnn4 does not appear to have an effect on 2G12 binding when comparing TM and Δoch1 Δmnn1 cells (Fig. 2C), but mannosylphosphate residues are strong immunogenic determinants making them undesirable for immunization studies (4). As expected, analysis of the glycans released from TM yeast cells showed that they contained exclusively high-mannose-type N-glycans, with the majority being Man8GlcNAc2, a result consistent with previous studies which sought to produce Man5GlcNAc2 and hybrid-type glycans in yeast (10, 51). Enzymatic treatment confirmed that the specificity of 2G12 to TM yeast glycoproteins was due to the presence of these high-mannose oligosaccharides, verified as mostly Man8GlcNAc2 by MALDI-TOF profiling. Additionally, the lack of 2G12 reactivity in Δoch1 and Δoch1 Δmnn4 mutants by Western blotting indicates that the specificity was due to the exposure of terminal Manα1,2Man upon Mnn1 deletion.
In part because of the ease with which yeast can be grown and manipulated, we were able to identify the major proteins to which MAb 2G12 bound. As shown in Fig. 3 and Table 2, these proteins contain a high density of potential N-linked glycosylation sites, similar to gp120 with an N-glycan density of greater than 4%. All these proteins are associated with the yeast cell wall, and most are important for maintaining cell wall architecture and responding to cell wall damage. Ecm33 is a cell surface protein important for maintaining proper cell wall structure and in anchoring mannoproteins to the cell wall (45), Gas1 is a β-1-3-glucanosyltransferase required for cell wall structure and remodeling (52), Gp38 is a cell wall-related secretory glycoprotein induced by nutrient deprivation (15), and YJL171c is a GPI-anchored cell wall protein of unknown function induced in response to cell wall damage (28). In addition to these four proteins, other strong 2G12-reactive bands in TM yeast remain to be identified, including a protein band at 35 kDa, additional bands in the range of 80 to 120 kDa (Fig. 2C), as well as a few bands greater than 150 kDa (data not shown).
Due to the efficient binding of MAb 2G12 to TM yeast, we initiated preliminary immunization trials using whole yeast cells to test the validity of our approach. We chose to use heat-killed yeast because this method was proven to successfully induce antimannose antibodies with α1,3- and α1,6-linked specificity using WT yeast and Δmnn1 Δmnn2 yeast, respectively (4), and because whole, heat-killed S. cerevisiae cells have proven safe in phase I clinical trials (37). We consistently found that immune sera from rabbits immunized with TM (but not WT) yeast contained antibodies that could cross-react with HIV-1 gp120. Importantly, sera from animals immunized with TM yeast recognized a variety of clade B and clade C gp120 proteins as well as gp120 proteins derived from SIV, exceeding the breadth of reactivity exhibited by 2G12. This is likely due to the broader recognition of mannose-containing epitopes seen by our sera in the glycan microarray analysis. Like MAb 2G12, the binding of these antibodies to Env proteins was abrogated by removal of N-linked, high-mannose glycans, as well as by selective removal of terminal Manα1,2Man structures (47, 49); reactivity of TM postimmune sera with deglycosylated gp120 was never observed. The TM-induced antisera exhibited significant differences in the recognition of Env proteins, such that some isolates (R2 and SIV-1A11) were bound more efficiently than others (YU2). This can be explained by the fact that the number of high-mannose glycans (defined as Man5-Man9) on a given gp120 protein can vary widely, from 33% to 60% of the total glycan, based on the isolate and source of the protein (46). Since TM-induced sera showed a broad range of specificity for Manα1,2Man-containing glycans, the differential binding may be a consequence of this heterogeneity of glycans displayed by different gp120 proteins. Regardless, the immunization of rabbits with TM yeast resulted in the elicitation of antibodies with at least some 2G12-like properties.
We have begun to test sera from all rabbits immunized with TM yeast for neutralization activity. Our preliminary tests showed that 50% neutralization titers were rarely observed and only at serum dilutions of <1:60. Additionally, the sera did not show enhanced neutralization against two of the strains whose gp120 proteins were most efficiently recognized by ELISA. Thus, it appears that our first generation immunization strategy with whole TM yeast cells can induce antibodies that cross-react with a variety of Env proteins of HIV and SIV, without consistent, reproducible neutralizing activity. This is likely a consequence of the low levels of antibodies, as demonstrated by 50- to 100-fold lower gp120 binding activity than that of MAb 2G12 and the fact that the antiserum recognized a larger variety of mannose-dependent epitopes.
Our results represent a promising step toward generating an immunogen that can elicit antibodies to the glycan shield of HIV-1. Our immunization platform consists of a genetically modified yeast strain that contains epitopes on at least four cell wall-associated proteins to which MAb 2G12 binds. These highly glycosylated yeast proteins recapitulate facets of the glycan shield sufficiently well for immunization with whole, fixed TM yeast cells to result in antibodies that bind to HIV glycans. While good titers of antibodies that cross-react with gp120 glycans were obtained, convincing neutralizing activity was not observed, indicating that we have either elicited antibodies to carbohydrates on Env other than those composing the 2G12 epitope and/or the antibodies produced to the 2G12 epitope are too dilute in the complex polyclonal array elicited by whole yeast cells to neutralize the virus efficiently. To resolve this, a number of steps can be taken to focus the immune response and hopefully improve neutralization activity. For example, it will be important to identify additional yeast proteins to which MAb 2G12 binds, to identify the proteins to which 2G12 binds with highest affinity, and to initiate mutagenesis studies in attempts to further increase the affinity of 2G12 binding in hopes of developing more useful immunogens. Once proteins that bind to 2G12 most efficiently are identified, they can be purified and used as immunogens. We can also investigate new immunization methods, adjuvants, formulations, or prime-boost strategies to enhance the neutralizing immune response. The facts that S. cerevisiae can be grown easily and inexpensively and that it is genetically pliable will make it possible to further refine its 2G12-binding characteristics and to produce varied, new immunogens in large amounts that will hopefully make targeting the glycan shield of HIV Env a practical reality.
Acknowledgments
We thank Regina Guo for her excellent technical help and Anup K. Datta at Glycotechnology Core Resource, University of California, San Diego, for his glycan analyses.
This work was supported by the Neutralizing Antibody Consortium of the International AIDS Vaccine Initiative and by National Institutes of Health grants AI058724 to Y.G., AI040880 to R.W.D., and T32007632 to C.A.
Yu Geng is President and Chief Executive Officer of ProSci Incorporated and acknowledges a potential conflict of interest.
R.J.L. and Y.G. designed the research; R.J.L., J.L., H.F., K.K.C., C.A., I.M., F.-H.L., D.M., and D.F.S. performed the research; and R.J.L., R.W.D., and Y.G. wrote the paper.
Footnotes
Published ahead of print on 23 April 2008.
REFERENCES
- 1.Amonsen, M., D. F. Smith, R. D. Cummings, and G. M. Air. 2007. Human parainfluenza viruses hPIV1 and hPIV3 bind oligosaccharides with α2-3-linked sialic acids that are distinct from those bound by H5 avian influenza virus hemagglutinin. J. Virol. 818341-8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armbruster, C., G. M. Stiegler, B. A. Vcelar, W. Jager, U. Koller, R. Jilch, C. G. Ammann, M. Pruenster, H. Stoiber, and H. W. Katinger. 2004. Passive immunization with the anti-HIV-1 human monoclonal antibody (hMAb) 4E10 and the hMAb combination 4E10/2F5/2G12. J. Antimicrob. Chemother. 54915-920. [DOI] [PubMed] [Google Scholar]
- 3.Back, N. K., L. Smit, J. J. De Jong, W. Keulen, M. Schutten, J. Goudsmit, and M. Tersmette. 1994. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199431-438. [DOI] [PubMed] [Google Scholar]
- 4.Ballou, C. E. 1990. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 185440-470. [DOI] [PubMed] [Google Scholar]
- 5.Blixt, O., S. Head, T. Mondala, C. Scanlan, M. E. Huflejt, R. Alvarez, M. C. Bryan, F. Fazio, D. Calarese, J. Stevens, N. Razi, D. J. Stevens, J. J. Skehel, I. van Die, D. R. Burton, I. A. Wilson, R. Cummings, N. Bovin, C. H. Wong, and J. C. Paulson. 2004. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. USA 10117033-17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braibant, M., S. Brunet, D. Costagliola, C. Rouzioux, H. Agut, H. Katinger, B. Autran, and F. Barin. 2006. Antibodies to conserved epitopes of the HIV-1 envelope in sera from long-term non-progressors: prevalence and association with neutralizing activity. AIDS 201923-1930. [DOI] [PubMed] [Google Scholar]
- 7.Burton, D. R., R. C. Desrosiers, R. W. Doms, W. C. Koff, P. D. Kwong, J. P. Moore, G. J. Nabel, J. Sodroski, I. A. Wilson, and R. T. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5233-236. [DOI] [PubMed] [Google Scholar]
- 8.Calarese, D. A., H. K. Lee, C. Y. Huang, M. D. Best, R. D. Astronomo, R. L. Stanfield, H. Katinger, D. R. Burton, C. H. Wong, and I. A. Wilson. 2005. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc. Natl. Acad. Sci. USA 10213372-13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 3002065-2071. [DOI] [PubMed] [Google Scholar]
- 10.Chiba, Y., M. Suzuki, S. Yoshida, A. Yoshida, H. Ikenaga, M. Takeuchi, Y. Jigami, and E. Ichishima. 1998. Production of human compatible high mannose-type (Man5GlcNAc2) sugar chains in Saccharomyces cerevisiae. J. Biol. Chem. 27326298-26304. [DOI] [PubMed] [Google Scholar]
- 11.Cohen, R. E., W. Zhang, and C. E. Ballou. 1982. Effects of mannoprotein mutations on Saccharomyces cerevisiae core oligosaccharide structure. J. Biol. Chem. 2575730-5737. [PubMed] [Google Scholar]
- 12.Cole, K. S., J. D. Steckbeck, J. L. Rowles, R. C. Desrosiers, and R. C. Montelaro. 2004. Removal of N-linked glycosylation sites in the V1 region of simian immunodeficiency virus gp120 results in redirection of B-cell responses to V3. J. Virol. 781525-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cutalo, J. M., L. J. Deterding, and K. B. Tomer. 2004. Characterization of glycopeptides from HIV-I(SF2) gp120 by liquid chromatography mass spectrometry. J. Am. Soc. Mass Spectrom. 151545-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean, N. 1999. Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta 1426309-322. [DOI] [PubMed] [Google Scholar]
- 15.Destruelle, M., H. Holzer, and D. J. Klionsky. 1994. Identification and characterization of a novel yeast gene: the YGP1 gene product is a highly glycosylated secreted protein that is synthesized in response to nutrient limitation. Mol. Cell. Biol. 142740-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Carlo, F. J., and J. V. Fiore. 1958. On the composition of zymosan. Science 127756-757. [DOI] [PubMed] [Google Scholar]
- 17.Galibert, F., D. Alexandraki, A. Baur, E. Boles, N. Chalwatzis, J. C. Chuat, F. Coster, C. Cziepluch, M. De Haan, H. Domdey, P. Durand, K. D. Entian, M. Gatius, A. Goffeau, L. A. Grivell, A. Hennemann, C. J. Herbert, K. Heumann, F. Hilger, C. P. Hollenberg, M. E. Huang, C. Jacq, J. C. Jauniaux, C. Katsoulou, L. Kirchrath, K. Kleine, E. Kordes, P. Koetter, S. Liebl, E. J. Louis, V. Manus, H.-W. Mewes, T. Miosga, B. Obermaier, J. Perea, T. M. Pohl, D. Portetelle, A. Pujol, B. Purnelle, M. Ramezani Rad, S. W. Rasmussen, M. Rose, R. Rossau, I. Schaaff-Gerstenschlaeger, P. H. M. Smits, T. Scarcez, N. Soriano, D. Tovan, M. Tzermia, A. Van Broekhoven, M. Vandenbol, H. Wedler, D. von Wettstein, R. Wambutt, M. Zagulski, A. Zollner, and L. Karpfinger-Hartl. 1996. Complete nucleotide sequence of Saccharomyces cerevisiae chromosome X. EMBO J. 152031-2049. [PMC free article] [PubMed] [Google Scholar]
- 18.Gemmill, T. R., and R. B. Trimble. 1999. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim. Biophys. Acta 1426227-237. [DOI] [PubMed] [Google Scholar]
- 19.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418387-391. [DOI] [PubMed] [Google Scholar]
- 20.Herscovics, A. 1999. Processing glycosidases of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1426275-285. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman, T. L., C. C. LaBranche, W. Zhang, G. Canziani, J. Robinson, I. Chaiken, J. A. Hoxie, and R. W. Doms. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc. Natl. Acad. Sci. USA 966359-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichishima, E., M. Arai, Y. Shigematsu, H. Kumagai, and R. Sumida-Tanaka. 1981. Purification of an acidic alpha-D-mannosidase from Aspergillus saitoi and specific cleavage of 1,2-alpha-D-mannosidic linkage in yeast mannan. Biochim. Biophys. Acta 65845-53. [DOI] [PubMed] [Google Scholar]
- 23.Jigami, Y., and T. Odani. 1999. Mannosylphosphate transfer to yeast mannan. Biochim. Biophys. Acta 1426335-345. [DOI] [PubMed] [Google Scholar]
- 24.Joos, B., A. Trkola, H. Kuster, L. Aceto, M. Fischer, G. Stiegler, C. Armbruster, B. Vcelar, H. Katinger, and H. F. Gunthard. 2006. Long-term multiple-dose pharmacokinetics of human monoclonal antibodies (MAbs) against human immunodeficiency virus type 1 envelope gp120 (MAb 2G12) and gp41 (MAbs 4E10 and 2F5). Antimicrob. Agents Chemother. 501773-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang, S. M., F. S. Quan, C. Huang, L. Guo, L. Ye, C. Yang, and R. W. Compans. 2005. Modified HIV envelope proteins with enhanced binding to neutralizing monoclonal antibodies. Virology 33120-32. [DOI] [PubMed] [Google Scholar]
- 26.Koch, M., M. Pancera, P. D. Kwong, P. Kolchinsky, C. Grundner, L. Wang, W. A. Hendrickson, J. Sodroski, and R. Wyatt. 2003. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313387-400. [DOI] [PubMed] [Google Scholar]
- 27.Krauss, I. J., J. G. Joyce, A. C. Finnefrock, H. C. Song, V. Y. Dudkin, X. Geng, J. D. Warren, M. Chastain, J. W. Shiver, and S. J. Danishefsky. 2007. Fully synthetic carbohydrate HIV antigens designed on the logic of the 2G12 antibody. J. Am. Chem. Soc. 12911042-11044. [DOI] [PubMed] [Google Scholar]
- 28.Lagorce, A., N. C. Hauser, D. Labourdette, C. Rodriguez, H. Martin-Yken, J. Arroyo, J. D. Hoheisel, and J. Francois. 2003. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 27820345-20357. [DOI] [PubMed] [Google Scholar]
- 29.Lee, H. K., C. N. Scanlan, C. Y. Huang, A. Y. Chang, D. A. Calarese, R. A. Dwek, P. M. Rudd, D. R. Burton, I. A. Wilson, and C. H. Wong. 2004. Reactivity-based one-pot synthesis of oligomannoses: defining antigens recognized by 2G12, a broadly neutralizing anti-HIV-1 antibody. Angew. Chem. Int. Ed. Engl. 431000-1003. [DOI] [PubMed] [Google Scholar]
- 30.Lehle, L., A. Eiden, K. Lehnert, A. Haselbeck, and E. Kopetzki. 1995. Glycoprotein biosynthesis in Saccharomyces cerevisiae: ngd29, an N-glycosylation mutant allelic to och1 having a defect in the initiation of outer chain formation. FEBS Lett. 37041-45. [DOI] [PubMed] [Google Scholar]
- 31.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 26510373-10382. [PubMed] [Google Scholar]
- 32.Letvin, N. L. 2006. Progress and obstacles in the development of an AIDS vaccine. Nat. Rev. Immunol. 6930-939. [DOI] [PubMed] [Google Scholar]
- 33.Li, H., and L. X. Wang. 2004. Design and synthesis of a template-assembled oligomannose cluster as an epitope mimic for human HIV-neutralizing antibody 2G12. Org. Biomol. Chem. 2483-488. [DOI] [PubMed] [Google Scholar]
- 34.Li, Y., B. Cleveland, I. Klots, B. Travis, B. A. Richardson, D. Anderson, D. Montefiori, P. Polacino, and S. L. Hu. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J. Virol. 82638-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCaffrey, R. A., C. Saunders, M. Hensel, and L. Stamatatos. 2004. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J. Virol. 783279-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menendez, A., D. A. Calarese, R. L. Stanfield, K. C. Chow, C. N. Scanlan, R. Kunert, H. Katinger, D. R. Burton, I. A. Wilson, and J. K. Scott. 15 January 2008, posting date. A peptide inhibitor of HIV-1 neutralizing antibody 2G12 is not a structural mimic of the natural carbohydrate epitope on gp120. FASEB J. 221380-1392. doi: 10.1096/fj.07-8983com. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munson, S., J. Parker, T. H. King, Y. Lu, V. Kelly, Z. Guo, V. Borges, and A. Franzusoff. 2008. Coupling innate and adaptive immunity with yeast-based cancer immunotherapy, p. 131-149. In R. Orentas, J. W. Hodge, and B. D. Johnson (ed.), Cancer vaccines and tumor immunity. John Wiley & Sons, Hoboken, NJ.
- 38.Nagasu, T., Y. Shimma, Y. Nakanishi, J. Kuromitsu, K. Iwama, K. Nakayama, K. Suzuki, and Y. Jigami. 1992. Isolation of new temperature-sensitive mutants of Saccharomyces cerevisiae deficient in mannose outer chain elongation. Yeast 8535-547. [DOI] [PubMed] [Google Scholar]
- 39.Nakajima, T., and C. E. Ballou. 1975. Yeast manno-protein biosynthesis: solubilization and selective assay of four mannosyltransferases. Proc. Natl. Acad. Sci. USA 723912-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakanishi-Shindo, Y., K. Nakayama, A. Tanaka, Y. Toda, and Y. Jigami. 1993. Structure of the N-linked oligosaccharides that show the complete loss of alpha-1,6-polymannose outer chain from och1, och1 mnn1, and och1 mnn1 alg3 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 26826338-26345. [PubMed] [Google Scholar]
- 41.Ni, J., H. Song, Y. Wang, N. M. Stamatos, and L. X. Wang. 2006. Toward a carbohydrate-based HIV-1 vaccine: synthesis and immunological studies of oligomannose-containing glycoconjugates. Bioconjug. Chem. 17493-500. [DOI] [PubMed] [Google Scholar]
- 42.Nuoffer, C., P. Jeno, A. Conzelmann, and H. Riezman. 1991. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol. Cell. Biol. 1127-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Odani, T., Y. Shimma, A. Tanaka, and Y. Jigami. 1996. Cloning and analysis of the MNN4 gene required for phosphorylation of N-linked oligosaccharides in Saccharomyces cerevisiae. Glycobiology 6805-810. [DOI] [PubMed] [Google Scholar]
- 44.Odani, T., Y. Shimma, X. H. Wang, and Y. Jigami. 1997. Mannosylphosphate transfer to cell wall mannan is regulated by the transcriptional level of the MNN4 gene in Saccharomyces cerevisiae. FEBS Lett. 420186-190. [DOI] [PubMed] [Google Scholar]
- 45.Pardo, M., L. Monteoliva, P. Vazquez, R. Martinez, G. Molero, C. Nombela, and C. Gil. 2004. PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology 1504157-4170. [DOI] [PubMed] [Google Scholar]
- 46.Pashov, A., M. Perry, M. Dyar, M. Chow, and T. Kieber-Emmons. 2007. Defining carbohydrate antigens as HIV vaccine candidates. Curr. Pharm. Des. 13185-201. [DOI] [PubMed] [Google Scholar]
- 47.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 767293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scanlan, C. N., J. Offer, N. Zitzmann, and R. A. Dwek. 2007. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature 4461038-1045. [DOI] [PubMed] [Google Scholar]
- 49.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 767306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scanlan, C. N., G. E. Ritchie, K. Baruah, M. Crispin, D. J. Harvey, B. B. Singer, L. Lucka, M. R. Wormald, P. Wentworth, Jr., N. Zitzmann, P. M. Rudd, D. R. Burton, and R. A. Dwek. 2007. Inhibition of mammalian glycan biosynthesis produces non-self antigens for a broadly neutralising, HIV-1 specific antibody. J. Mol. Biol. 37216-22. [DOI] [PubMed] [Google Scholar]
- 51.Takamatsu, S., Y. Chiba, T. Ishii, K. Nakayama, T. Yokomatsu-Kubota, T. Makino, Y. Fujibayashi, and Y. Jigami. 2004. Monitoring of the tissue distribution of fibroblast growth factor containing a high mannose-type sugar chain produced in mutant yeast. Glycoconj. J. 20385-397. [DOI] [PubMed] [Google Scholar]
- 52.Tomishige, N., Y. Noda, H. Adachi, H. Shimoi, A. Takatsuki, and K. Yoda. 2003. Mutations that are synthetically lethal with a gas1Delta allele cause defects in the cell wall of Saccharomyces cerevisiae. Mol. Genet. Genomics 269562-573. [DOI] [PubMed] [Google Scholar]
- 53.Treco, D. A. 1988. Growth and manipulation of yeast, p. 13.2.1-13.2.11. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. Greene Publishing Associates and Wiley-Interscience, New York, NY. [Google Scholar]
- 54.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 701100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van der Aart, Q. J., C. Barthe, F. Doignon, M. Aigle, M. Crouzet, and H. Y. Steensma. 1994. Sequence analysis of a 31 kb DNA fragment from the right arm of Saccharomyces cerevisiae chromosome II. Yeast 10959-964. [DOI] [PubMed] [Google Scholar]
- 56.Wang, J., H. Li, G. Zou, and L. X. Wang. 2007. Novel template-assembled oligosaccharide clusters as epitope mimics for HIV-neutralizing antibody 2G12. Design, synthesis, and antibody binding study. Org. Biomol. Chem. 51529-1540. [DOI] [PubMed] [Google Scholar]
- 57.Wang, L. X. 2006. Toward oligosaccharide- and glycopeptide-based HIV vaccines. Curr. Opin. Drug Discov. Dev. 9194-206. [PubMed] [Google Scholar]
- 58.Yip, C. L., S. K. Welch, F. Klebl, T. Gilbert, P. Seidel, F. J. Grant, P. J. O'Hara, and V. L. MacKay. 1994. Cloning and analysis of the Saccharomyces cerevisiae MNN9 and MNN1 genes required for complex glycosylation of secreted proteins. Proc. Natl. Acad. Sci. USA 912723-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, X., C. Borchers, R. J. Bienstock, and K. B. Tomer. 2000. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 3911194-11204. [DOI] [PubMed] [Google Scholar]