Abstract
Chromatin structure is strictly regulated during the cell cycle. DNA viruses occasionally disturb the spatial organization of the host cell chromatin due to formation of the viral DNA replication compartment. To examine chromatin behavior in baculovirus-infected cells, we constructed recombinant plasmids expressing fluorescent protein-tagged histone H4 molecules and visualized the intracellular localization of chromatin by their transient expression in live infected cells. Similar to other DNA viruses, the baculovirus Bombyx mori nucleopolyhedrovirus induced marginal relocation of chromatin within the nuclei of BmN cells, simultaneously with expansion of the viral DNA replication compartment, the virogenic stroma (VS). In the late stage of infection, however, the peristromal region (PR), another virus-induced subnuclear compartment, was also excluded from the chromatin-localizing area. Provided that late-gene products such as PR proteins (e.g., envelope proteins of the occlusion-derived virus) were expressed, blockage of viral DNA synthesis failed to inhibit chromatin relocation, despite abrogation of VS expansion. Instead, chromatin became marginalized concomitantly with PR expansion, suggesting that the PR contributes directly to chromatin replacement. In addition, chromatin was excluded from relatively large subnuclear structures that were induced in uninfected cells by cotransfection with four baculovirus genes, ie1, lef3, p143, and hr. Omission of any of the four genes, however, failed to result in formation of the large structures or chromatin exclusion. This correlation between compartmentalization and chromatin exclusion suggests the possibility that a chromatin-exclusive property of viral molecules, at least in part, supports nuclear compartmentalization of virus-infected cells.
Baculoviruses are arthropod-specific large, rod-shaped, enveloped viruses containing double-stranded circular DNA genomes of 100 to 180 kb. Similar to the case for most other DNA viruses, many steps in baculovirus replication proceed within the cell nucleus, and to facilitate their replication baculoviruses reorganize the nuclear architecture by the induction of subnuclear compartments. One such virus-induced compartment is the virogenic stroma (VS), in which progeny nucleocapsids are assembled. Previously, Okano et al. (17) demonstrated that DNA replication of the baculovirus Bombyx mori nucleopolyhedrovirus (BmNPV) proceeds within a discrete subnuclear compartment in BmN cells. We have subsequently shown that the major capsid protein VP39 (reviewed in reference 6), as well as the viral DNA replication factors IE1 (a multifunctional transactivator [reviewed in reference 5]), LEF3 (a single-stranded DNA binding protein [8]), and P143 (a DNA helicase [13]), localizes to this compartment and that the compartment has a high DNA content (12). We therefore proposed that this DNA replication compartment is the VS, implying that the VS is the site for not only nucleocapsid assembly but also viral DNA replication (12). Previous observations of baculovirus-infected cells by electron microscopy have shown that cellular heterochromatin becomes progressively marginalized concomitantly with VS development (22), suggesting that this DNA replication compartment affects the chromatin organization.
Besides the VS, baculoviruses generate another subnuclear compartment that is functionally distinct from the DNA replication compartment, the peristromal region (PR). In addition to the general processes of DNA virus infection such as DNA replication, the baculovirus life cycle has an unusual process, the intranuclear envelopment of nucleocapsids to produce one type of virion, an occlusion-derived virus (ODV). To accomplish this unusual envelopment and subsequent occlusion body formation, baculoviruses create this second subnuclear compartment. As expected from its function, a number of ODV envelope proteins and ODV-associated proteins (e.g., ODV-E25, P91, or P74) localize to this compartment (2, 18, 19, 21). Since most of these proteins are late-expression gene products, if viral DNA synthesis is blocked, these genes cannot be expressed, resulting in a lack of PR formation (see below). Although ODV is enveloped within the PR, the other type of virion, termed a budded virus (BV), acquires its envelope by budding from the cytoplasmic membrane. The two virions are functionally differentiated; i.e., BV is required for systemic infection of an individual host, whereas ODV mediates interhost transmission. While both nucleocapsids of the two are assembled within the same compartment (VS), the mechanism of determination of the following destination (i.e., intranuclear envelopment versus nuclear egress) or involvement of the PR in the destination is still unknown.
In baculovirus-infected cells, the two compartments, VS and PR, are tightly associated but never overlap, thus creating a sharp boundary between the two that seems to establish a part of their shapes or outlines (12). One possible origin of the boundary might be inherent to its property of mutual exclusion. On the other hand, the overall structure of these compartments that prevents diffuse distribution of their respective components might require a mechanism(s) other than mutual exclusion. In the replication process of herpes simplex virus 1 (a mammalian DNA virus), nuclear marginalization of host chromatin that correlates with expansion of the viral replication compartment is evident (14). It is possible that the exclusion of chromatin partially supports the establishment of the viral replication compartment within the nuclei of cells infected with this virus, similar to how oil droplets fail to diffuse in water. Whereas electron microscopy suggests that baculovirus infection induces cellular heterochromatin marginalization (22), little is known about the details of chromatin dynamics or, in particular, the spatial relationships of chromatin with the VS or PR. By inference from mammalian virus research, however, we would expect that expansion of these compartments may also lead to chromatin marginalization and that chromatin exclusion may function in the organization of the compartments in baculovirus infection.
In this study, we examined the spatial relationship between host cell chromatin and virus-induced subnuclear compartments in baculovirus-infected cells. Our results indicate that expansion of the VS and PR drives chromatin marginalization. Furthermore, we found that cellular chromatin was excluded from subnuclear structures induced by cotransfection of uninfected cells with four baculovirus genes, ie1, lef3, p143, and hr (a DNA sequence element on viral genome [reviewed in reference 5]), suggesting that chromatin exclusion serves to gather the viral proteins and thus supports nuclear compartmentalization in the transfected cells and probably in virus-infected cells.
MATERIALS AND METHODS
Plasmids.
Genomic DNA from BmN cells was used to clone the Bombyx mori histone H4 gene. Cells (1 × 107) were harvested in 1 ml of phosphate-buffered saline (PBS), lysed in 500 μl of cell lysis buffer (10 mM Tris [pH 8.0], 100 mM EDTA, 20 μg/ml RNase A, 0.5% sodium dodecyl sulfate), and incubated for 30 min at 37°C before the addition of 80 μg/ml of proteinase K and incubation overnight at 65°C. Total DNA was phenol extracted, isopropanol precipitated, and suspended in 100 μl of water. Using this DNA sample as a template, with primers 5′-ATGACCGGTCGCGGTAAAGG-3′ and 5′-ACCGCCGAAACCGTACAGGG-3′ (these primers were designed from a Bombyx mori cDNA clone encoding histone H4 [GenBank accession no. BP118365]) and LA Taq (Takara), PCR was performed under the following conditions: 94°C for 3 min and 35 cycles of 98°C for 20 s, 47°C for 1 min, and 72°C for 1 min. The resulting PCR product was cloned into the pIB/V5-His-TOPO vector (Invitrogen) for production of pIB-BmH4. The SalI-XbaI fragment of pEGFP-1 (Clontech) containing the coding sequence for green fluorescent protein (GFP) and the SalI-XbaI fragment of pDsRed2-1 (Clontech) containing the coding sequence for DsRed were inserted into XhoI-XbaI sites of pIB-BmH4 to construct plasmids expressing histone H4-GFP and histone H4-DsRed (pIB-BmH4-GFP and pIB-BmH4-DsR), respectively. A plasmid expressing P74-GFP under the control of its own promoter (pHCS-p74-GFP) was created from a plasmid carrying a BmNPV genomic DNA fragment (the HindIII-SphI subfragment in the HindIII C fragment; map units 84.7 to 88.5) that contained p74, pHCSph. The 12-bp sequence containing the termination codon (underlined) of the p74 gene (TTAAAAAAAAAC) in pHCSph was replaced with the BglII-SpeI cutting sequence by site-directed mutagenesis. The mutagenized plasmid was digested with BglII and SpeI and ligated with the BglII-XbaI fragment of pEGFP-1 (Clontech) containing the GFP gene sequence. Plasmids expressing IE1-DsRed, VP39-GFP, and ODV-E25-GFP under the control of their own promoters were constructed as described previously (12). To visualize nuclear structures associated with IE1, LEF3, and P143, plasmids expressing IE1-GFP, LEF3-GFP, and GFP-P143 (in which P143 was tagged with GFP at the N terminus, in contrast to the other GFP-tagged proteins used) under the control of their own promoters, which were constructed as described previously (12, 16), and the plasmids containing BmNPV genomic fragments, pEcoG (ie1)/90.5-96.4 (map units), pPDSal (lef3)/39.2-42.7, pPEXba (p143)/55.2-60.2, and pBS-hr3 (hr) (15) were employed.
Cells and viruses.
BmN cells were maintained in TC100 medium (Funakoshi Co., Tokyo, Japan) supplemented with 10% fetal bovine serum (12). BmNPV wild-type isolate T3 was propagated in BmN cells (12). Recombinant viruses in which the ie1 and p143 genes were replaced with IE1-GFP and GFP-P143 genes, respectively (vIE1-GFP and vGFP-P143), were constructed as described previously (16). Other recombinant viruses in which p91 and p74 genes were replaced with P91-GFP and P74-GFP genes, respectively (vP91-GFP and vP74-GFP), were generated by homologous recombination of T3 genomic DNA with pPK-p91-GFP (12) or pHCS-p74-GFP and plaque purified three times. To confirm the expected recombination, genomic DNA samples of the obtained viruses were characterized by restriction enzyme digestions and PCR as described previously (16) (data not shown).
Transfection and infection.
Plasmid transfections were performed as described previously (12). Briefly, BmN cells were transfected with 0.5 μg of each plasmid DNA sample by using Lipofectin reagent (Invitrogen). The transfected cells were incubated at 28°C for 24 h and were directly analyzed with a confocal microscope or infected with viruses at a multiplicity of infection of 5 or 10. In infection experiments, time zero was defined as the time point at which fresh medium was added following a 1-hour virus adsorption period. For treatment with aphidicolin, the infected cells were cultured in medium containing the reagent (20 μΜ) from appropriate time points. To detect total DNA, cells were fixed in prechilled methanol at −20°C for 6 min, postfixed in 3.7% paraformaldehyde in PBS for 10 min, and stained with propidium iodide (PI) (1 μg/ml) as described previously (12).
FISH.
Fluorescence in situ hybridization (FISH) techniques were performed as described by Ishov and Maul (10) and Greber et al. (7). The hybridization probes were prepared with biotin-nick translation mix (Roche) using the wild-type virus DNA genome as the template in accordance with the manufacturer's instructions. Following virus infection, cells were fixed as described above and quenched in 25 mM ammonium chloride in PBS at room temperature for 5 min. Cellular and probe DNAs were denatured simultaneously for 3 min at 92°C and hybridized for 2 h at 37°C in a hybridization mixture containing 50% formamide, 10% dextran sulfate, 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate), 1 ng/μl biotinylated DNA, and 100 ng/μl salmon sperm DNA. After hybridization, cells were washed three times in 50% formamide-2× SSC prewarmed to 42°C for 10 min, followed by three washes for 10 min each in 0.1× SSC prewarmed to 60°C. Washed cells were then incubated with 1% blocking reagent (Molecular Probes) in PBS overnight, stained with fluorescein-treated avidin and PI, and analyzed with a confocal microscope.
Microscopy.
Confocal images were obtained with a Leica TCS NT instrument using a 488-nm laser line for GFP and fluorescein and a 568-nm laser line for DsRed and PI and were processed as described previously (12).
RESULTS
Host cell chromatin is repositioned during baculovirus infection.
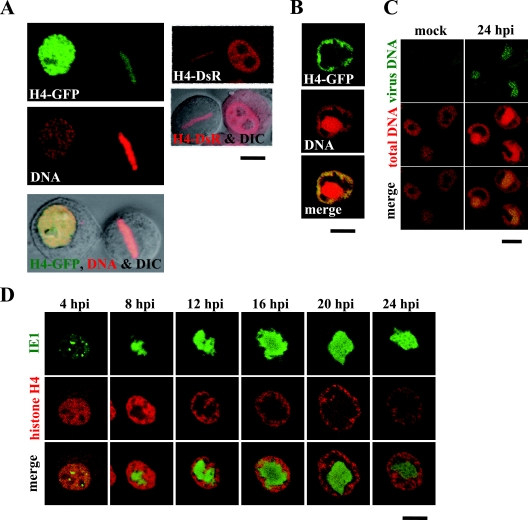
To visualize chromatin dynamics in BmN cells infected with the baculovirus BmNPV, we amplified the histone H4 gene from the genomic DNA of BmN cells by PCR and constructed recombinant plasmids to express the histone H4 gene as fusion proteins with fluorescent proteins GFP and DsRed (H4-GFP and H4-DsR) under control of the baculovirus Op-ie2 promoter. When the plasmid expressing H4-GFP was transfected into uninfected BmN cells, H4-GFP was observed throughout the nuclei of interphase cells, colocalizing with the nuclear DNA (Fig. 1A). In addition, in mitotic-phase cells, where concentrated DNA was aligned at the metaphase plate, H4-GFP was also concentrated at the same place (Fig. 1A). This result indicates that H4-GFP is associated with chromatin and can be used as a marker for chromatin distribution. The distribution of H4-DsR was similar to that of H4-GFP (i.e., nucleoplasmic distribution in interphase cells and localization to the metaphase plate in mitotic-phase cells) and can thus also be used as a chromatin marker in this study (Fig. 1A).
FIG. 1.
Chromatin marginalization is coupled with VS expansion. (A) BmN cells were transfected with plasmids expressing H4-GFP and H4-DsR, and the cells during interphase and mitotic phase were analyzed by confocal microscopy. H4-GFP-expressing cells were fixed at 24 h posttransfection and stained with a DNA-specific dye (PI) before the microscopic analysis. (B) Following transfection with a plasmid expressing H4-GFP, BmN cells were infected with a wild-type virus, fixed at 24 hpi, stained with a DNA-specific dye (PI), and analyzed by confocal microscopy. (C) BmN cells were mock infected or infected with a wild-type virus and fixed at 24 hpi. The fixed cells were hybridized with biotinylated virus DNA, stained with fluorescein-conjugated avidin and a DNA-specific dye (PI), and analyzed by confocal microscopy. (D) Following transfection with a plasmid expressing H4-DsR, BmN cells were infected with a recombinant virus expressing IE1-GFP and analyzed by confocal microscopy at the indicated time points. Bars, 10 μm.
Next, we infected H4-GFP-expressing cells with wild-type BmNPV and analyzed the infected cells at 24 h postinfection (hpi). DNA staining of the cells showed that while highly concentrated DNA was present at the center of the nucleus, a lower concentration of DNA was distributed at the nuclear periphery (Fig. 1B). In those cells, H4-GFP was associated with the peripheral DNA rather than the central DNA, suggesting that chromatin localizes to the nuclear periphery in BmNPV-infected cells (Fig. 1B). We have previously demonstrated that the central DNA accumulation corresponds to the IE1-associated structure, i.e., the VS, and therefore we speculated that the DNA accumulation consists of progeny virus DNA but not cellular DNA (12). To verify this speculation, we performed FISH experiments using the viral DNA genome as a template. Although nonspecific staining was seen in the cytosol of mock-infected cells, no nuclear staining was detected (Fig. 1C). In contrast, the central DNA within the nucleus, but not the peripheral DNA, was clearly stained with the viral DNA probes in virus-infected cells, indicating that the central DNA accumulation contains the viral DNA genome whereas peripheral DNA contains cellular chromatin (Fig. 1C). Taking this together with previous studies in which cellular DNA content remained unchanged during infection (3, 9), we conclude that cellular chromatin becomes localized to the nuclear periphery during baculovirus infection.
To trace the chromatin marginalization in more detail, we next infected H4-DsR-expressing cells with a recombinant BmNPV in which the ie1 gene was replaced with an IE1-GFP gene (16) and analyzed the infected cells at 4-h intervals for 24 h. IE1 is a good marker for VS distribution (12). The chromatin distribution observed at 4 hpi was unchanged in comparison with that in uninfected cells, and we were unable to detect any evidence of chromatin exclusion by IE1 foci that were expected to become VS (12). At 8 hpi, however, chromatin began to marginalize in association with initiation of the VS expansion that was driven by viral DNA synthesis (Fig. 1D). During the VS expansion period (8 to 16 hpi), chromatin became gradually marginalized, with most of the nuclear space occupied by VS. From 16 hpi, however, separation between VS and chromatin became apparent and the VS no longer expanded, possibly because viral DNA synthesis was not as robust (Fig. 1D). Thus, chromatin marginalization is coupled with VS expansion during the period when viral DNA synthesis is highly active, while expansion of the gap between VS and chromatin initiated from 16 hpi occurs after VS expansion. Although we have not performed quantitative measurements, it is likely that the gap is able to expand due to nuclear enlargement at around 20 hpi. On the other hand, it could expand due to shrinkage of the VS at around 24 hpi (Fig. 1D; see Fig. 2B).
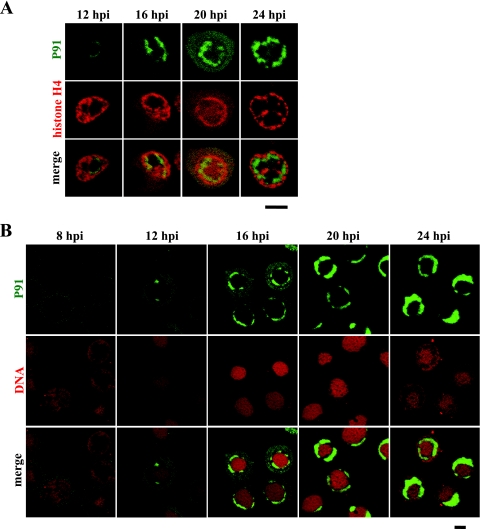
FIG. 2.
PR excludes host cell chromatin. (A) Following transfection with a plasmid expressing H4-DsR, BmN cells were infected with a recombinant virus expressing P91-GFP and analyzed by confocal microscopy at the indicated time points. (B) BmN cells were infected with a recombinant virus expressing P91-GFP, fixed at the indicated time points, stained with a DNA-specific dye (PI), and analyzed by confocal microscopy. Bars, 10 μm.
PR, as well as VS, excludes host cell chromatin.
While we observed chromatin marginalization coupled to VS expansion from 8 to 16 hpi, we also observed the formation of a gap between the VS and chromatin from 16 to 24 hpi. Since we had previously determined that the VS is surrounded by the PR at the late stage of infection (12), we suspected that this gap corresponds to the PR. To test this possibility, we infected H4-DsR-expressing cells with a recombinant BmNPV in which the p91 gene was replaced with a P91-GFP gene (see Materials and Methods). We have already demonstrated that P91-GFP localizes to the VS periphery (12). Whereas we failed to detect nuclear accumulations of P91-GFP at 8 hpi (Fig. 2B), small accumulations were visible in some cells at 12 hpi (Fig. 2A). At 16 hpi the P91-GFP accumulations were evident in many cells, and by 24 hpi these accumulations progressively formed circular structures that would enclose the VS (Fig. 2A). As predicted, the P91-GFP accumulations bordered the chromatin-localizing area, suggesting that the gap between the VS and chromatin corresponds to PR. To make the gap, in conjunction with P91-GFP, more clearly visible, cells infected with the P91-GFP-expressing virus were stained with a DNA-specific dye, PI. In PI-stained cells, the gap is evident as a DNA-poor region between the central heavily staining region (VS) and the peripheral lightly staining region (chromatin) (Fig. 1B). As expected, from the onset of gap formation, P91-GFP consistently filled the gap (Fig. 2B). These results strongly suggest that this gap is the PR; in other words, the PR does not overlap the VS or chromatin. Furthermore, to verify this conclusion, we carried out similar experiments using a recombinant virus in which the p74 gene was replaced with a P74-GFP gene (vP74-GFP) (see Materials and Methods). P74 is an ODV membrane protein that localizes to PR (4, 21). In these experiments, we obtained results similar to those for P91 (see Fig. 4c1 to c6, and data not shown), further supporting our conclusion that the gap is the PR and that the PR excludes both the VS and chromatin.
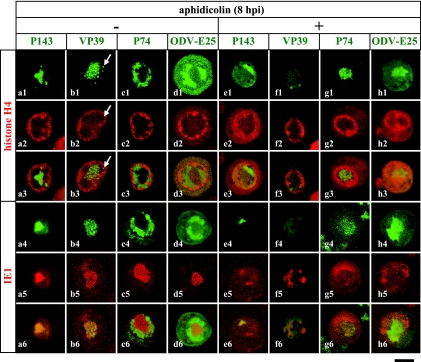
FIG. 4.
Inhibition of viral DNA accumulations causes alteration in VS and PR distributions. Following transfection with plasmids expressing H4-DsR (top panels) and IE1-DsR (bottom panels) in conjunction with plasmids expressing VP39-GFP (b1 to b6 and f1 to f6) and ODV-E25-GFP (d1 to d6 and h1 to h6), BmN cells were infected with recombinant viruses expressing GFP-P143 (a1 to a6 and e1 to e6) and P74-GFP (c1 to c6 and g1 to g6) and with a wild-type virus (b1 to b6, d1 to d6, f1 to f6, and h1 to h6). The infected cells were not treated (left panels) or treated with a DNA synthesis inhibitor (aphidicolin) at 8 hpi (right panels) and analyzed by confocal microscopy at 24 hpi. Arrows indicate VP39 accumulations in the PR. Bars, 10 μm.
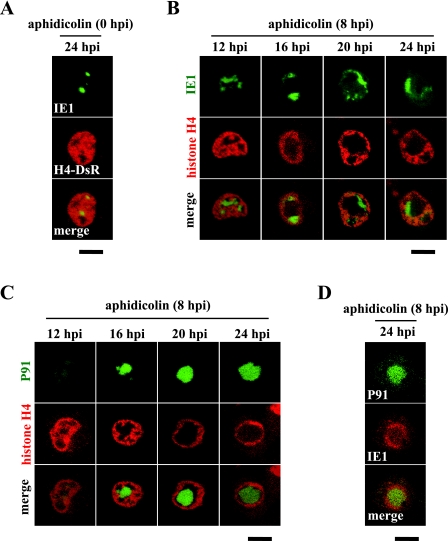
Chromatin marginalization occurs even in the absence of VS expansion.
Although we have shown that the PR excludes chromatin as well as the VS, because the chromatin was fully compressed by the VS before PR formation (Fig. 2A and B), we were unsure whether PR expansion directly caused the chromatin relocation. To assess this issue, we performed DNA synthesis inhibition experiments by using the DNA synthesis inhibitor aphidicolin for prevention of VS expansion. When applied from 0 hpi, this reagent inhibited viral DNA synthesis and consequently blocked VS formation (Fig. 3A). Under this condition, no chromatin movement was observed, indicating that early events such as viral attachment are insufficient for chromatin marginalization (Fig. 3A). In this case, however, we were unable to examine the effect of PR expansion on chromatin relocation because none of the late genes, including the PR protein genes, were expressed and therefore PR formation could not occur (data not shown). To express the late genes, we applied this reagent from 8 hpi, when viral DNA synthesis had already been initiated but had not been completed. As expected, IE1-GFP accumulations failed to expand under this condition even at 24 hpi (Fig. 3B). Nevertheless, chromatin was marginalized and fully compressed, resulting in the formation of a large open space (Fig. 3B). To determine whether this open space was occupied by PR proteins, we next compared the distribution of chromatin with that of a PR protein, P91, within cells that were treated with aphidicolin from 8 hpi. As shown in Fig. 3C, P91-GFP occupied a large portion of the open space (i.e., the chromatin-exclusive region), suggesting that PR expansion plays a direct role in chromatin relocation. To investigate whether the mutual exclusion between the VS and PR was maintained in the presence of the DNA synthesis inhibitor, we analyzed the spatial relationship between IE1 and P91 in cells treated with aphidicolin from 8 hpi. Although reverse images (i.e., IE1 surrounded P91) were observed, the two proteins were tightly associated but did not overlap (Fig. 3D), suggesting that the VS and PR preserve the mutual exclusivity even when VS expansion is suppressed.
FIG. 3.
PR expansion drives chromatin marginalization. Following transfection with plasmids expressing H4-DsR (A to C) and IE1-DsR (D), BmN cells were infected with recombinant viruses expressing IE1-GFP (A and B) and P91-GFP (C and D), treated with a DNA synthesis inhibitor (aphidicolin) at 0 (A) and 8 (B to D) hpi, and analyzed by confocal microscopy at the indicated time points. Bars, 10 μm.
VS and PR are able to recruit their constituents even in the absence of VS expansion.
By treating infected cells with aphidicolin from 8 hpi, we demonstrated chromatin marginalization in association with expansion of the P91-localizaing area rather than the IE1-localizaing area, suggesting that PR expansion plays a direct role in chromatin relocation. This unusual localization pattern, however, may not reflect alterations in the distribution of the compartments, because only two marker proteins were examined. To eliminate the possibility that the two marker proteins mislocalized abnormally in the presence of aphidicolin, we examined the subnuclear localization of other viral proteins that could be used as indicators of the virus-induced compartments. We had previously demonstrated that P143 molecules tagged with GFP at the N terminus (GFP-P143) localize to the VS together with IE1 (16) (Fig. 4a4 to a6). Likewise, in cells infected with a recombinant virus expressing GFP-P143 and subsequently treated with aphidicolin from 8 hpi, colocalization of GFP-P143 and IE1-DsRed was also observed at 24 hpi despite no expansion of their localizing area (Fig. 4e1 to e6). This result strongly suggests that aphidicolin suppresses VS expansion but does not inhibit localization of IE1 to the VS. In the absence of aphidicolin, VP39-GFP predominantly localizes to the VS, although small accumulations of VP39-GFP were sometimes observed in the PR (12) (Fig. 4b1 to b3). When those cells were treated with aphidicolin from 8 hpi, specific accumulations of VP39-GFP were barely detected in the nucleus, even within the IE1-localizing area (Fig. 4f1 to f6). It is possible that VP39 accumulation requires viral DNA accumulation rather than initiation of viral DNA synthesis, and this may be the reason why we could not detect VP39-GFP accumulations. We next examined the PR markers P74 and ODV-E25 to verify PR expansion in the presence of aphidicolin. As described above, we employed the recombinant virus vP74-GFP for P74-GFP expression. For ODV-E25-GFP expression, however, we performed transfection of an ODV-E25-GFP gene (12) and subsequent infection of the wild-type virus because we failed to produce recombinant viruses in which the odv-e25 gene was replaced with an ODV-E25-GFP gene. As shown in Fig. 4g1 to g6 and h1 to h6, both proteins occupied a large space of the nucleus in association with nuclear marginalization of chromatin, suggesting PR expansion in the presence of aphidicolin. Taking the results together, we concluded that aphidicolin treatment from 8 hpi leads to suppression of VS expansion rather than mislocalization of specific viral proteins and that PR expansion, as well as VS expansion, is involved in chromatin relocation.
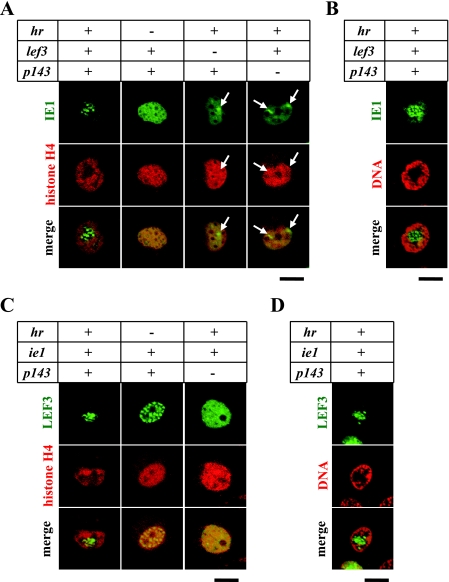
Chromatin is excluded from subnuclear structures induced by four baculovirus genes, ie1, lef3, p143, and hr.
Nuclear compartmentalization of baculovirus-infected cells may depend partially on the chromatin-exclusive properties of viral factors, since viral factors characterized by chromatin exclusion must gather within the cell nucleus, which fills mainly with chromatin. We previously demonstrated that cotransfection of uninfected cells with four baculovirus genes, ie1, lef3, p143, and hr, induces formation of subnuclear structures containing the three proteins; in other words, these viral factors induce nuclear compartmentalization (16). We therefore suspected that the four viral factors might exhibit a chromatin-exclusive property to stabilize the IE1/LEF3/P143-associated structures. To determine whether the IE1/LEF3/P143-associated structures exclude chromatin and, if so, which of the four factors are essential for the chromatin exclusion, uninfected cells were cotransfected with the H4-DsRed gene in various combination with the four genes. IE1-GFP (Fig, 5A and B), LEF3-GFP (Fig. 5C and D), and GFP-P143 (data not shown) were used as markers to visualize the IE1/LEF3/P143-associated structures. Microscopy revealed that H4-DsRed was excluded from the IE1/LEF3/P143-associated structures that formed following transfection with the four genes (Fig. 5A and C). When these cells were stained with PI, the nuclear structures were located within the DNA-poor regions (Fig. 5B and D). These results indicate that the IE1/LEF3/P143-associated structures exclude cellular chromatin. As reported previously (16), omission of any of the four genes failed to result in formation of the subnuclear structures. Under this condition, chromatin was distributed normally, that is, throughout the nucleoplasm but not at specific sites such as the nuclear periphery (Fig. 5A and C). This result indicates a correlation between formation of the IE1/LEF3/P143-associated structures and chromatin exclusion. Omission of LEF3 or P143 induced formation of IE1 foci instead of the IE1/LEF3/P143-associated structures because only IE1 and hr, and not all four genes, were present in the transfected cells (15, 16). In the transfected cells chromatin was not excluded from the IE1 foci (Fig. 5A), which were similar to another type of IE1 foci formed in virus-infected cells at 4 hpi (Fig. 1D). This indicates that virus-induced subnuclear structures do not always exclude cellular chromatin and that chromatin exclusion is not always required for formation of virus-induced subnuclear structures, i.e., nuclear compartmentalization. In addition, omission of hr led to an uneven distribution of LEF3-GFP throughout the nucleoplasm (Fig. 5C), suggesting that the interaction among IE1, LEF3, and P143 affects their spatial distribution in the absence of chromatin exclusion. In this case, however, their interaction merely achieved incomplete, partial compartmentalization (i.e., uneven distribution of LEF3). Thus, it is likely that the chromatin exclusion properties of the viral proteins facilitate nuclear compartmentalization by gathering the viral proteins but are not essential for the compartmentalization (see Discussion).
FIG. 5.
IE1/LEF3/P143-associated structures exclude cellular chromatin. BmN cells were transfected with plasmids expressing IE1-GFP (A and B), LEF3-GFP (C and D), and H4-DsR (A and C), in conjunction with plasmids (+) or no plasmids (−) containing the indicated genes. The transfected cells were directly analyzed by confocal microscopy at 24 h posttransfection (A and C) or fixed at 24 h posttransfection and stained with a DNA-specific dye (PI) before the microscopic analysis (B and D). Lack of the ie1 gene caused a failure to express LEF3-GFP (data not shown). Arrows indicate IE1 foci. Bars, 10 μm.
DISCUSSION
Here we have shown that an insect DNA virus, the baculovirus BmNPV, relocates host cell chromatin to the nuclear periphery. Unlike infection with a mammalian DNA virus (herpes simplex virus type 1), baculovirus infection induced cellular chromatin exclusion not only from the DNA replication compartment, the VS, but also from the non-DNA replication compartment, the PR. Moreover, we have found that the subnuclear structures induced by transfection with ie1, lef3, p143, and hr exclude cellular chromatin in uninfected cells. The IE1/LEF3/P143-associated subnuclear structures possessed less DNA (Fig. 5B and D). These results suggest that DNA accumulations are dispensable for compartment formation or chromatin exclusion. When viral DNA synthesis was inhibited in baculovirus-infected cells, VS expansion was also inhibited, suggesting that the increase of DNA content in the compartment generates the mechanical force necessary for chromatin displacement. On the other hand, chromatin exclusion by DNA-lacking compartments, which we have shown here, suggests that non-DNA compartments are also capable of generating the mechanical force that displaces chromatin, albeit by an as-yet-unknown mechanism.
Based on studies using mammalian viruses in which the DNA replication compartment excludes host chromatin, we would expect that the VS would exclude the host cell chromatin during baculovirus infection. We were somewhat surprised, however, by the fact that the PR also exhibited chromatin exclusion. Since the beginning of electron microscopic studies on baculoviruses, the VS has been known as a virus-induced subnuclear compartment or structure (22). In contrast, until recently, the PR has not been considered a subnuclear compartment bounded by a distinct border to discriminate between itself and others. Rather, in electron microscopic observations, it has been regarded as an unlimited external area of the VS where intranuclear envelopment of ODV occurs. Hence, we were surprised by the PR-mediated chromatin exclusion. However, the nature of the PR-based chromatin exclusion that we have shown here obviously indicates that the PR has a clear border for maintaining its shape and is one of the discrete virus-induced subnuclear compartments. Therefore, we can now define the PR as a baculovirus-induced subnuclear structure distinct from the VS that produces ODVs and that has a chromatin exclusion property, although we cannot eliminate the possibility that a third chromatin-exclusive structure is present in baculovirus-infected cells. In our studies, we used P91, ODV-E25, and P74 as PR markers, which are ODV associated and localize outside the VS (4, 18, 19, 21). Thus, we have confirmed the suitability of these proteins for use as PR markers, because the distribution of the three proteins was relevant to the region in which chromatin, as well as the VS, was excluded.
It is well known that baculoviruses produce two types of virions, ODV and BV, but the process leading to “differentiation” between the two virions has not yet been defined. Whereas the mechanisms for acquiring viral membranes are completely different (i.e., intranuclear envelopment versus budding from the cytoplasmic membrane), the nucleocapsids of the two virions are likely to assemble in similar ways within the VS, since ODV and BV contain an identical DNA genome and an identical major capsid component, VP39. Although we do not yet know whether or not they have already differentiated within the VS, if their nucleocapsids are identical, then egress of the nucleocapsids from the VS might be the most important step for their differentiation. As shown in Fig. 1 and 2, from 8 to 16 hpi, the VS gradually expanded and consequently occupied almost the entire nucleus, whereas the PR was absent or showed no more than a limited distribution. During this period, nucleocapsids released from the VS are able to easily reach the nuclear membrane in order to become BV. In contrast, from 16 to 24 hpi, because the VS is enclosed by the PR, nucleocapsids must enter the PR as soon as they exit from the VS. Once entrapped by the PR, nucleocapsids might be unable to leave the PR and thus become enveloped within the PR, where they finally undergo occlusion in polyhedra to become ODV. Although this model is based on the assumption that nucleocapsids of the two virion types are identical, it is the simplest model to explain how they are differentiated and therefore it may be useful for future studies on their differentiation. Also, if this model is correct, it suggests that the timing and position of PR formation are key factors in ODV formation.
Normal cell nuclei, as well as baculovirus-infected cell nuclei, are highly compartmentalized into functional units or nuclear bodies. In contrast to cytoplasmic organelles, nuclear bodies are nonmembrane structures; consequently, continuous barriers for holding the bodies, such as lipid bilayers, are unlikely to account for maintenance of their organization. Whereas the maintenance mechanisms of each structure are not yet determined, it is possible that various nuclear structures share common mechanisms to establish nuclear compartments. In this study we have shown that there are two types of virus-induced or viral protein-induced subnuclear structures; one possesses chromatin-exclusive properties, but the other does not. As described above, it is possible that chromatin exclusion facilitates nuclear compartmentalization; however, other mechanisms must be functional, because nuclear structures lacking chromatin-exclusive properties exist. Before viral DNA synthesis (∼4 hpi), IE1 exhibits a focal distribution in the nuclei of infected cells. A similar distribution of IE1 can be represented by cotransfection of uninfected cells with ie1 and hr. These IE1-associated structures (IE1 foci), which are relatively small structures, are likely to have no chromatin-exclusive properties, suggesting that chromatin exclusion is not essential for formation of virus-induced subnuclear structures. Some mammalian cells have a similar small nuclear structure, the PML nuclear body, which has also no chromatin-exclusive property (1). While a number of proteins localize to this structure, PML is the primary essential component of the structure (11). Recently, a model for formation of the PML nuclear body has been proposed (20). According to this model, creation of a PML-mediated protein network constitutes the nucleation event for subsequent recruitment of other proteins to the PML nuclear bodies. It is likely, therefore, that network formation of primary components is an important mechanism to organize nuclear structures. Baculovirus IE1 might also be able to generate a network to form the IE1-accociated focus structures. In contrast to the IE1 foci, the IE1/LEF3/P143-associated structures exhibited a chromatin-exclusive property. Nevertheless, it is highly possible that their formation depends on a protein network, since it appears that the IE1/LEF3/P143-associated structures comprise many focus structures similar to the IE1 foci (Fig. 5). One possibility is that chromatin exclusion serves to assemble network-based focus structures in order to form larger nuclear structures. Relatively larger nuclear structures, such as the IE1/LEF3/P143-associated structures, and more complex structures that contain various subcompartments such as the VS, the PR, and possibly the nucleoli may apply chromatin exclusion for their own stability. Compared to protein networks, chromatin exclusion may be able to provide a less rigid and more dynamic organization, and this may be one of the reasons why more complex structures require chromatin exclusion.
In addition to maintenance of compartments, we can also expect other functions of chromatin exclusion from the VS and PR. For instance, the lack of cellular DNA within the VS could prevent nonspecific binding of viral DNA replication factors and transcription factors to chromatin. Moreover, although the nucleosomal structure of host chromatin is considered to be maintained during infection (23), chromatin marginalization by the compartments might affect various chromatin activities such as cellular gene expression or cell cycle progression. Thus, because chromatin exclusion seems to be a key concept in baculovirus infection and possibly in cell biology, future studies will focus on analyzing the molecular mechanisms of chromatin exclusion to better understand the nuclear organization of not only infected cells but normal cells.
Acknowledgments
We thank Masaaki Kurihara for help with the culture of BmN cells and J. Joe Hull for critical reading of the manuscript.
This research was supported by the Bioarchitect Research Program and the Chemical Biology Research Program from RIKEN.
Footnotes
Published ahead of print on 23 April 2008.
REFERENCES
- 1.Ascoli, C. A., and G. G. Maul. 1991. Identification of a novel nuclear domain. J. Cell Biol. 112785-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braunagel, S. C., J. K. Burks, G. Rosas-Acosta, R. L. Harrison, H. Ma, and M. D. Summers. 1999. Mutations within the Autographa californica nucleopolyhedrovirus FP25K gene decrease the accumulation of ODV-E66 and alter its intranuclear transport. J. Virol. 738559-8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braunagel, S. C., R. Parr, M. Belyavskyi, and M. D. Summers. 1998. Autographa californica nucleopolyhedrovirus infection results in Sf9 cell cycle arrest at G2/M phase. Virology 244195-211. [DOI] [PubMed] [Google Scholar]
- 4.Faulkner, P., J. Kuzio, G. V. Williams, and J. A. Wilson. 1997. Analysis of p74, a PDV envelope protein of Autographa californica nucleopolyhedrovirus required for occlusion body infectivity in vivo. J. Gen. Virol. 783091-3100. [DOI] [PubMed] [Google Scholar]
- 5.Friesen, P. D. 1997. Regulation of baculovirus early gene expression, p. 141-166. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, NY.
- 6.Funk, C. J., S. C. Braunagel, and G. F. Rohrmann. 1997. Baculovirus structure, p. 7-32. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, NY.
- 7.Greber, U. F., M. Suomalainen, R. P. Stidwill, K. Boucke, M. W. Ebersold, and A. Helenius. 1997. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 165998-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hang, X., W. Dong, and L. A. Guarino. 1995. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein. J. Virol. 693924-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda, M., and M. Kobayashi. 1999. Cell-cycle perturbation in Sf9 cells infected with Autographa californica nucleopolyhedrovirus. Virology 258176-188. [DOI] [PubMed] [Google Scholar]
- 10.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawasaki, Y., S. Matsumoto, and T. Nagamine. 2004. Analysis of baculovirus IE1 in living cells: dynamics and spatial relationships to viral structural proteins. J. Gen. Virol. 853575-3583. [DOI] [PubMed] [Google Scholar]
- 13.McDougal, V. V., and L. A. Guarino. 2000. The Autographa californica nuclear polyhedrosis virus p143 gene encodes a DNA helicase. J. Virol. 745273-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monier, K., J. C. G. Armas, S. Etteldorf, P. Ghazal, and K. F. Sullivan. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2661-665. [DOI] [PubMed] [Google Scholar]
- 15.Nagamine, T., Y. Kawasaki, T. Iizuka, and S. Matsumoto. 2005. Focal distribution of baculovirus IE1 triggered by its binding to the hr DNA elements. J. Virol. 7939-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagamine, T., Y. Kawasaki, and S. Matsumoto. 2006. Induction of a subnuclear structure by the simultaneous expression of baculovirus proteins, IE1, LEF3, and P143 in the presence of hr. Virology 352400-407. [DOI] [PubMed] [Google Scholar]
- 17.Okano, K., V. S. Mikhailov, and S. Maeda. 1999. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J. Virol. 73110-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosas-Acosta, G., S. C. Braunagel, and M. D. Summers. 2001. Effects of deletion and overexpression of the Autographa californica nuclear polyhedrosis virus FP25K gene on synthesis of two occlusion-derived virus envelope proteins and their transport into virus-induced intranuclear membranes. J. Virol. 7510829-10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell, L. Q., and G. F. Rohrmann. 1997. Characterization of P91, a protein associated with virions of an Orgyia pseudotsugata baculovirus. Virology 233210-223. [DOI] [PubMed] [Google Scholar]
- 20.Shen, T. H., H.-K. Lin, P. P. Scaglioni, T. M. Yung, and P. P. Pandolfi. 2006. The mechanisms of PML-nuclear body formation. Mol. Cell 24331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slack, J. M., E. M. Dougherty, and S. D. Lawrence. 2001. A study of the Autographa californica multiple nucleopolyhedrovirus ODV envelope protein p74 using a GFP tag. J. Gen. Virol. 822279-2287. [DOI] [PubMed] [Google Scholar]
- 22.Williams, G. V., and P. Faulkner. 1997. Cytological changes and viral morphogenesis during baculovirus infection, p. 61-107. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, NY.
- 23.Wilson, M. J., and L. K. Miller. 1986. Changes in the nucleoprotein complexes of a baculovirus DNA during infection. Virology 151315-328. [DOI] [PubMed] [Google Scholar]