Abstract
APOBEC3 proteins are cytidine deaminases which help defend cells against retroviral infections. One antiviral mechanism involves deaminating dC residues in minus-strand DNA during reverse transcription, resulting in G-to-A mutations in the coding strand. We investigated the effects of mouse APOBEC3 (mA3) and human APOBEC3G (hA3G) upon Moloney murine leukemia virus (MLV). We find that mA3 inactivates MLV but is significantly less effective against MLV than is hA3G. In contrast, mA3 is as potent against human immunodeficiency virus type 1 (HIV-1, lacking the protective Vif protein) as is hA3G. The two APOBEC3 proteins are packaged to similar extents in MLV particles. Dose-response profiles imply that a single APOBEC3 molecule (or oligomer) is sufficient to inactivate an MLV particle. The inactivation of MLV by mA3 and hA3G is accompanied by relatively small reductions in the amount of viral DNA in infected cells. Although hA3G induces significant levels of G-to-A mutations in both MLV and HIV DNAs, and mA3 induces these mutations in HIV DNA, no such mutations were detected in DNA synthesized by MLV inactivated by mA3. Thus, MLV has apparently evolved to partially resist the antiviral effects of mA3 and to totally resist the ability of mA3 to induce G-to-A mutation in viral DNA. Unlike the resistance of HIV-1 and human T-cell leukemia virus type 1 to hA3G, the resistance of MLV to mA3 is not mediated by the exclusion of APOBEC from the virus particle. The nature of its resistance and the mechanism of inactivation of MLV by mA3 are completely unknown.
Mammalian cells contain a number of mechanisms for protection against retroviral infection and retrotransposition. One of these mechanisms involves members of the APOBEC3 (APO) family of cytidine deaminases (19, 45). The existence of this mechanism was first discovered through experiments on the human immunodeficiency virus type 1 (HIV-1) accessory protein Vif; it has become clear in recent years that the intracellular binding of Vif to human APOBEC3G (hA3G) results in proteasomal degradation of the APO protein, thus preventing its incorporation into virions (35, 52). While the antiretroviral activity of hA3G was initially revealed in studies on HIV-1, it has recently become clear that hA3G is only one member of a family of cytidine deaminases (26) and that several APOBEC3 family members are able to inhibit infections by a remarkable variety of retroviruses. Indeed, it appears that endogenous retroviruses and retrotransposons, as well as exogenous retroviruses, are sensitive to the effects of these cellular proteins (6, 11, 12, 38). Interestingly, another retrovirus, human T-cell leukemia virus type 1 (HTLV-1), resembles HIV-1 in that it excludes hA3G from virions, but this exclusion involves an unusual acidic region in the nucleocapsid domain of its Gag protein (9) rather than a nonstructural protein like Vif.
Although this system has been studied intensively for several years, there are a number of outstanding questions that remain to be resolved. One of these is how APO proteins interfere with the viral replication cycle. One obvious factor that contributes to the inhibitory action of hA3G is the hypermutation of G residues to A residues; these mutations result from deamination by hA3G of C nucleotides in the single-stranded minus-strand DNA, the first product of reverse transcription during viral infection (18, 32, 53). However, quantitative studies suggest that this effect is often not sufficient to explain the potent inhibition of infection by hA3G (22, 40).
One viral system for which the effects of APOBEC3 are not yet clear is the murine leukemia viruses (MLVs). It is known that hA3G is capable of interfering with MLV infectivity (18, 32). However, as mice and mouse cells are permissive for MLV replication, one might expect MLV to be resistant to the effect of murine APOBEC3 (mA3), the sole member of the APOBEC3 family in mice. MLV is a “simple” retrovirus; that is, unlike HIV-1 and HTLV-1, MLV encodes only the three polyproteins that are assembled to form infectious progeny virions. Thus, if it is indeed resistant to mA3, this resistance cannot be attributed to an additional protein like Vif. Some reports indicate that mA3 is not incorporated into MLV particles (10, 29), perhaps because it is degraded by the viral protease (1), while others present evidence for efficient encapsidation of the protein without a significant antiviral effect (33).
In hopes of clarifying some of these questions, we have performed a quantitative comparison of the effects of mA3 and hA3G upon MLV and MLV-derived viral vectors. We now report that the murine protein is encapsidated to virtually the same extent as the human protein. It possesses significant inhibitory activity against MLV, but this activity is far lower than that of hA3G. The anti-MLV activity of mA3 does not seem to involve hypermutation.
MATERIALS AND METHODS
Cells and viruses.
Unless otherwise noted, all of the experiments described here were performed with a full-length infectious Moloney MLV proviral clone that was identical to previously described clone pRR88 (14), except for the removal of 5′ cellular flanking sequences. Virus particles were produced by transient transfection of 293T cells with Transit 293 (Mirus) in accord with the manufacturer's instructions. When viruses were to be analyzed for infectivity, pBABE-Luc, in which the firefly luciferase gene from pGL3 (Promega) has been inserted into the MLV-derived vector pBabe Puro (37, 44), was cotransfected with the MLV clone and with the appropriate APO plasmid. The total amount of plasmid DNA transfected into each culture was kept constant by the addition of plasmid pUC-CMV as required; a typical transfection would include, per 10-cm culture dish, 5 μg of MLV plasmid and 3 μg of pBABE-Luc DNA in addition to the indicated amounts of APO expression plasmid.
Infectivity was measured by infecting 293T cells expressing ecotropic MLV receptor mouse cationic amino acid transporter 1 (mCAT1; a kind gift of J. Cunningham, Harvard Medical School) (2) and assaying cell extracts for luciferase activity with the Luciferase Assay System (Promega) at 48 h after infection. (Similar results were also obtained in assays on NIH 3T3 cells.) Luciferase assays were always performed in triplicate. Virions were also assayed for the level of replication-competent MLV with the S+ L− focus assay (3) or for the titer of virus (derived from the pLXSH vector [36]) encoding hygromycin resistance. In the latter assays, NIH 3T3 cells were infected with serial dilutions of the sample. On the day after infection, the medium was replaced with fresh medium containing 200 μg/ml hygromycin B (Invitrogen). The fluid in the plates was changed periodically with hygromycin-containing medium, and the plates were finally fixed and stained 10 days after hygromycin treatment, when colonies of hygromycin-resistant cells were clearly visible. Except where specified otherwise, all of the techniques used were as previously described (42, 44).
The infectivity of HIV-1-derived viruses was assayed with a luciferase vector exactly as previously described (8). The plasmid backbone in the packaging plasmid was pCMVpA (i.e., pUC19 containing the cytomegalovirus immediate-early promoter and the simian virus 40 polyadenylation signal), and the vif gene was destroyed by site-directed mutagenesis, replacing the methionine codon at position 8 with a termination codon.
Plasmids expressing either hA3G or mA3 were a kind gift from Nathaniel Landau (New York University School of Medicine). The proteins were both expressed in plasmid pcDNA3.1 and tagged at their C termini with the hemagglutinin (HA) epitope (33). The mA3 protein encoded by the plasmid used here is the isoform lacking exon 5. For studies involving mutant APO proteins, the coding regions were transferred into pCMVpA and mutations were created with the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Wild-type, HA-tagged hA3G in pCMVpA was mutated to produce E67Q, E259Q, and the double mutant E67Q/E259Q; HA-tagged mA3 was mutated to E73Q, E253Q, and E73Q/E253Q. To express the mA3 isoform containing exon 5, we purchased a plasmid encoding mA3 cDNA from Open Biosystems Inc. The mA3 coding sequence in the plasmid (clone ID 3155422; accession no. BC003314) was amplified and subcloned into a cytomegalovirus-driven mammalian expression vector with a C-terminal HA epitope tag.
Immunoblotting.
Immunoblotting against MLV p30CA was performed with rabbit polyclonal anti-p30CA antiserum as previously described (39). Some immunoblotting experiments also used rabbit polyclonal anti-p15MA and rabbit anti-gp70SU antisera. Amounts of HA-tagged APO protein were similarly compared with mouse anti-HA monoclonal antibody 16B12 (Covance) as the primary antibody and horseradish peroxidase-conjugated horse anti-mouse immunoglobulin G (Cell Signaling) as the secondary antibody. For comparison of the amounts of HA-tagged APO protein in transfected cells, the cells were lysed in the culture dishes with loading buffer and the resulting extracts were sonicated. The total protein concentration in the lysates was determined by the bicinchoninic acid protein assay (Pierce). Aliquots of the lysates containing equal amounts of protein were then assayed by immunoblotting with anti-HA monoclonal antibody as described above. The lysates were also analyzed by immunoblotting with a monoclonal antibody against β-actin (Sigma) as the primary antibody and peroxidase-conjugated horse anti-mouse immunoglobulin G as the secondary antibody.
Subtilisin treatment.
Virus particles were prepared by centrifugation through 20% sucrose. They were then treated with subtilisin as previously described (43).
Virus fractionation.
Virus particles were first concentrated by centrifugation through 20% glycerol. Pellets were resuspended in 4 ml phosphate-buffered saline (PBS), and each sample was divided into two aliquots of 2 ml each. Each aliquot was layered on top of a step gradient consisting of 1 ml 10% sucrose-PBS over 1 ml 30% sucrose-PBS; in one of each pair of tubes, the 10% sucrose-PBS layer contained 0.1% Igepal CA-630 (Sigma). Samples were centrifuged in an SW50 rotor at 30,000 rpm for 90 min at 4°C. Pellets were then dissolved in loading buffer and analyzed by immunoblotting.
RT activity.
In some experiments, amounts of virus were compared with reverse transcriptase (RT) activity as a quantitative measurement of virus particle levels. Two methods were used for RT assays. In one, viruses were pelleted by ultracentrifugation and resuspended in an assay buffer containing 65 mM Tris (pH 8.0), 65 mM KCl, 0.65 mM MnCl2, 10 mM dithiothreitol, and 0.65% NP-40 together with 0.5 μCi [3H]TTP and primer/template-conjugated scintillation proximity assay beads from the Quan-T-RT [3H] Reverse Transcriptase Enzyme Assay Kit (GE Healthcare). After 3 h at 37°C, beads were processed according to the manufacturer's instructions. The signal was linearly proportional to the amount of sample assayed over at least a sevenfold range, and values were taken from the middle of this range. Alternatively, samples were analyzed for RT activity as previously described (15) following concentration with polyethylene glycol (16). This assay is linear over a wide range (13).
Analysis of reverse transcription complexes: real-time PCR and large-scale sequencing.
The ability of MLV-derived virus particles to perform DNA synthesis upon infecting new host cells was assayed as follows. Viruses were produced by transient transfection of 293T cells that had previously been stably transfected with pLXSH (36) and selected for hygromycin resistance. The virus particles obtained following the transient transfection were then used to infect 293T cells expressing mCAT1. Twenty-four hours later, the cells were lysed by the QIAamp DNA Mini Kit (Qiagen) and the cell extracts were assayed for hygromycin phosphotransferase (hph) DNA and two-long-terminal-repeat (2-LTR) circular DNA by real-time PCR as previously described (21, 44). The results were normalized for minor differences in the recovery of cellular DNA by comparing the copy numbers of the cellular gene CCR5 (44, 48).
Real-time PCR enumeration of luciferase DNA copies following infection with the HIV-1-derived luciferase vector was performed with Luc1006F (5′ ATCAGGCAAGGATATGGGCTCACT 3′) and Luc1130R (5′ TCCAGATCCACAACCTTCGCTTCA 3′) for primers and Luc1079P (5′ FAM-GCGCGGTCGGTAAAGTTGTTCCATTT-TAMRA 3′ [where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine]) for a probe. pBabe-Luc plasmid DNA (44) was used as a standard in these assays. Since these viruses were collected ∼48 h after transient transfection with plasmids including the luciferase vector, it was crucial to eliminate contaminating plasmid DNA from the virus before infecting the target cells and analyzing the reverse transcription products. Therefore, the virus-containing culture fluid was preincubated with 10 U/ml bovine pancreatic DNase I (Sigma) and 4 mM MgCl2 for 1 h at 37°C. An aliquot of the DNase-treated virus was inactivated by incubation at 68°C for 20 min. Cells were then infected with these virus preparations. The effectiveness of the DNase treatment was determined by measuring luciferase DNA sequences in the cells “infected” with the heat-inactivated virus; in all cases, the number of luciferase copies in these cells was ≤2% of that in the cells infected with active virus.
Bulk sequencing of reverse transcription products.
The hph DNA synthesized upon infection with MLV stocks that included the pLXSH vector was sequenced as follows. Cells were lysed as described above. The hph sequences from pLXSH were amplified from 0.5 to 2 μl of the lysate with primers hph 2050F (5′ AAAGCCTGAACTCACCGCGACGTC 3′) and hph 3030R (5′ CACGAGTGCTGGGGCGTCGGTTTC 3′) by using Pfu Turbo polymerase (Stratagene) for 20 to 30 cycles under the following PCR conditions: 95°C, 30 s; 67.5°C, 1 min; and 72°C, 1 min. Typically, the PCR products were fractionated by agarose gel electrophoresis in an ∼1.5% agarose gel. The ∼1-kb PCR product was then eluted and purified with a QIAquick gel extraction kit (Qiagen). Alternatively, it was purified away from small nonspecific PCR products with a mini Quick Spin column (Roche) and then ethanol precipitated. In either case, the purified DNA was then ligated into PCR Blunt II-TOPO (Invitrogen) and transformed into Top 10 cells (Invitrogen) and then colonies were selected by growth on kanamycin-containing medium. In order to minimize repeated amplification and cloning of the same DNA, transformed bacteria were only grown for 20 to 35 min before plating. Selected colonies were grown in 1 ml Terrific Broth in 96-well plates, and DNA was purified with the Qiagen Biorobot 3000. Fifteen microliters of the purified DNAs (representing ∼15% of the DNA) was sequenced with the SP6 primer.
DNA synthesized by viruses carrying the HIV-1-derived luciferase reporter vector was analyzed as follows. DNA was isolated from the infected cells as described above, and luciferase sequences were amplified with primers Luc 34F (5′ GCGCCATTCTATCCGCTGGAAGAT 3′) and Luc 1525R (5′ CGGTTGTTACTTGACTGGCGACGT 3′). The amplification included an initial treatment at 95°C for 1 min, followed by 30 cycles of 95°C, 30 s; 55°C, 30 s; and 72°C, 2 min; and finally 5 min at 72°C. The products were then cloned, sequenced, and analyzed as for the hph DNA.
Sequence data were analyzed for mutations by trimming all sequences to the same length and aligning them with Clustal W (EMBL-EBI at http://www.ebi.ac.uk/clustalw/#) (7) and Jalview. Typically, each clone yielded at least 800 nucleotides of sequence information, but only 750 nucleotides were used for analysis.
RESULTS
Effect of mA3 on MLV infectivity.
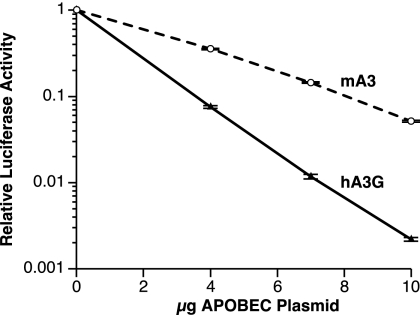
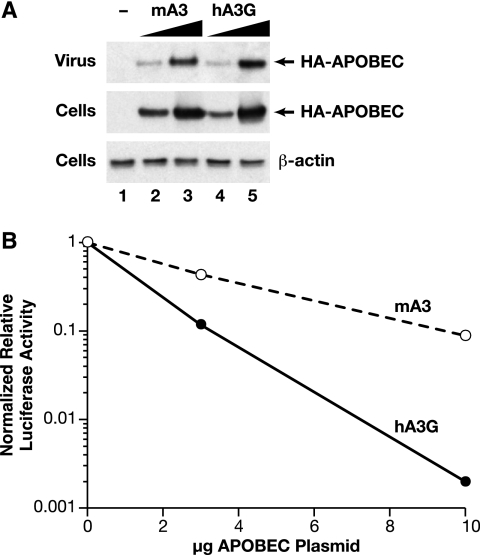
To assess the effects of mA3 and hA3G on MLV infectivity, we transiently transfected 293T cells (which do not express detectable APOBEC3G) with a mixture of an infectious wild-type clone of Moloney MLV, the reporter plasmid pBABE-Luc, and various amounts of the APO-expressing plasmids. We harvested the supernatants from the transfected cultures at ∼48 h after transfection and measured the levels of infectious virus capable of infecting 293T-mCAT1 cells and expressing the luciferase reporter. Thus, the particles analyzed here were composed of viral proteins encoded by the MLV helper plasmid, but some of the particles contained the pBABE-Luc vector genome. Typical results are shown in Fig. 1. It can be seen that increasing amounts of the mA3 plasmid in the transfection mixture caused a progressive decrease in the level of luciferase production in the infected cells. Figure 1 also shows that hA3G produced a significantly larger reduction in luciferase expression than mA3; for example, 10 μg of the hA3G plasmid reduced the luciferase level ∼300-fold, while the same level of the mA3 plasmid only resulted in an ∼15-fold decrease.
FIG. 1.
Effects of hA3G and mA3 on the infectivity of an MLV-derived luciferase vector. 293T cells were transiently transfected with an infectious MLV clone, the pBABE-Luc plasmid, and different doses of expression plasmids for hA3G or mA3. 293T-mCAT1 cells were then infected with culture fluids from the transfectants, and lysates of these cells were assayed for luciferase activity. The error bars show the standard deviations of the luciferase assay results.
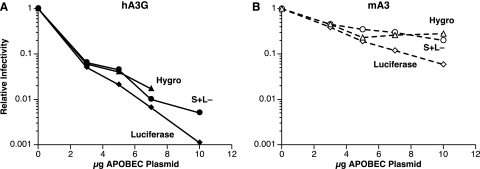
It was conceivable that this effect of mA3 and hA3G on virus infectivity was specific to the pBABE-Luc vector. To test the generality of the phenomenon, we cotransfected the MLV plasmid together with pBABE-Luc and the APO expression plasmids into 293T cells which had previously been stably transfected with another MLV-derived vector, i.e., pLXSH. Culture fluids harvested from the transfected cells were then assayed for luciferase-inducing activity as described in the legend to Fig. 1. In addition, they were assayed on NIH 3T3 cells for the ability to induce hygromycin resistance (a measurement of pLXSH infectivity) and on S+ L− cells (3) for infectious, replication-competent MLV produced from the wild-type MLV plasmid that rescued pBABE-Luc and pLXSH. The results of these assays are shown in Fig. 2. It is evident that the effects of hA3G (panel A) and mA3 (panel B) on pLXSH and MLV are quantitatively similar to their respective effects on pBABE-Luc. These results suggest that mA3 is capable of inactivating any MLV-derived virus, although its effects are considerably less severe than those of hA3G.
FIG. 2.
Comparison of the effects of hA3G and mA3 on the infectivity of a luciferase vector, an hph vector, and MLV itself. 293T cells that had been stably transfected with pLXSH were transiently transfected as described in the legend to Fig. 1. The culture fluids from the transfectants were then assayed for pBABE-Luc infectivity, pLXSH infectivity, and MLV infectivity as described in Materials and Methods. Hygro, hygromycin.
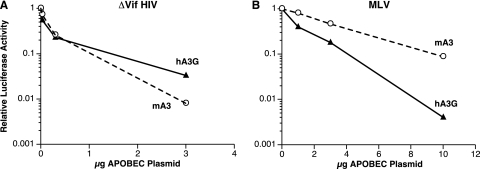
The stronger effect of hA3G than mA3 on the MLV-derived viruses (Fig. 1 and 2) might simply indicate that hA3G is a more potent general inhibitor of retroviral infections than mA3. Alternatively, MLV may exhibit specific partial resistance to mA3. To distinguish between these possibilities, we compared the two APOs with respect to their effects on an HIV-1 vector encoding luciferase. The HIV-1 packaging plasmid used here did not encode Vif, which normally protects wild-type HIV-1 against hA3G. As shown in Fig. 3A, the two APOs were roughly equivalent in the ability to inactivate an HIV-1-derived vector; in fact, the extent of inactivation by mA3 was consistently slightly greater than that of inactivation by hA3G. Thus, the mA3 encoded by the expression plasmid used here is not deficient with respect to overall antiretroviral activity.
FIG. 3.
Comparison of the effects of hA3G and mA3 on the infectivity of HIV-1-derived and MLV-derived luciferase vectors. A, HIV-1 vector; B, MLV vector.
It is important to note that the inactivation of MLV and MLV-derived vectors by both mA3 and hA3G showed a linear dose-response relationship on the semilogarithmic plots in Fig. 1, 2, and 3B, indicating an exponential inactivation pattern. In other words, all of the data could be fitted to the expression e−kx, where x is the micrograms of APO plasmid transfected into the culture. In Fig. 1, k = 0.622 for hA3G and 0.287 for mA3. This feature of these experiments was extremely reproducible; over nine titrations comparing the effects of hA3G and mA3 upon luciferase activity, the means of the R2 values for the fits of the hA3G and mA3 curves to the e−kx expression were 0.9878 and 0.9825, respectively. The average ratio of the hA3G exponent to the mA3 exponent was 2.41 ± 0.176 (mean ± standard deviation). The implications of the single-hit exponential character of these dose-response curves are considered in the Discussion.
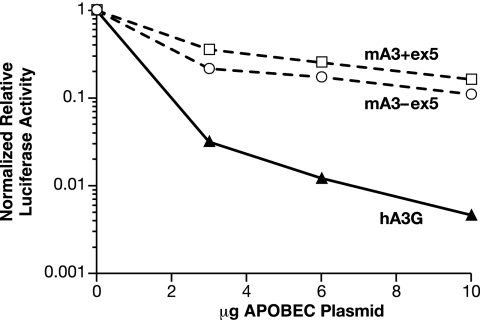
mA3 exists in two isoforms that differ by the presence or absence of exon 5 (29, 33). We compared the anti-MLV efficacies of these isoforms. As shown in Fig. 4, their effects on the infectivity of MLV appeared to be very similar.
FIG. 4.
Comparison of the effects of mA3 isoforms with and without exon 5 on the infectivity of an MLV-derived luciferase vector. Virions were produced as in the experiment shown in Fig. 1, with plasmids encoding mA3 lacking exon 5 (as in all of the other experiments described in this report), mA3 containing exon 5, or hA3G. The graph shows the luciferase-inducing activity divided by the virus particle concentration (as determined by RT activity following polyethylene glycol precipitation), so that the data represent the specific infectivity of the samples.
Packaging of APO proteins in MLV particles.
One possible explanation for the stronger anti-MLV effects of hA3G relative to mA3 is that the former protein is more efficiently incorporated into MLV particles during virus assembly. As both proteins were tagged with the HA epitope, we were able to compare their levels in MLV virions by immunoblotting with an anti-HA monoclonal antibody. We compared the packaging of hA3G and mA3 into MLV particles as follows. Cultures of 293T cells were transfected with 3 or 10 μg of either the hA3G or mA3 expression plasmid (as well as the MLV plasmid). Following harvest of the culture fluid, the cells were lysed for comparison of their APO levels. As shown in Fig. 5A (middle), the steady-state levels of hA3G and mA3 were similar in the cultures transfected with equivalent amounts of plasmid and were considerably higher in the cultures transfected with 10 μg than in those receiving only 3 μg of plasmid. As shown at the top, similar relationships were observed in the viral samples as well. These results show that the level of APO encapsidation is, under our experimental conditions, roughly proportional to the level of APO in the virus-producing cells and that the efficiencies of encapsidation of hA3G and mA3 are very similar to each other. In contrast, Fig. 5B confirms that encapsidated hA3G and mA3 are drastically different in their abilities to interfere with MLV infectivity, in agreement with the other results in this report.
FIG. 5.
Comparison of the encapsidation of mA3 and hA3G in MLV virions. Virus particles were produced by transient transfection in the presence of either 3 or 10 μg of either mA3 or hA3G expression plasmid or in the presence of an empty vector (lane −). Virions were assayed for RT activity following precipitation from the culture fluids with polyethylene glycol. (A) Equal amounts of virus were loaded into the five lanes and analyzed by immunoblotting with antiserum against the HA epitope tag on the APO proteins. The cells were also lysed, and equal amounts of lysate (as determined by protein concentration) were analyzed by immunoblotting with the anti-HA antiserum. As an additional loading control, equal amounts of the lysates were also analyzed by immunoblotting with anti-β-actin antiserum. The sample in lane 1 was produced without an APOBEC expression plasmid; those in lanes 2 and 3 were produced with 3 and 10 μg of mA3 plasmid, respectively; and those in lanes 4 and 5 were produced with 3 and 10 μg of hA3G plasmid. In addition to autoradiography, the chemiluminescence of the immunoblots of the viral samples was measured on an Alpha Innotech ChemiImager 5500. Two different exposures were analyzed in this way, and the percent integrated density values of the viral samples (means ± standard deviations) were as follows: 3 μg mA3, 13.6 ± 1.2; 10 μg mA3, 32.1 ± 1.0; 3 μg hA3G, 11.4 ± 2.8; 10 μg hA3G, 43.0 ± 3.0. (B) The infectivity of the viruses shown in panel A was analyzed by luciferase assays. The graph shows the luciferase-inducing activity divided by the virus particle concentration, and thus, the data represent the specific infectivity of the samples.
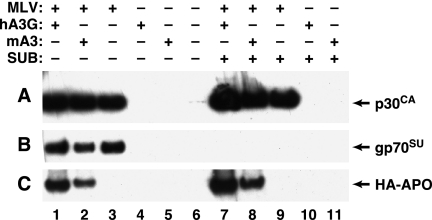
The viral pellets analyzed in Fig. 5 were purified only by centrifugation through a layer of 20% sucrose. It seemed possible that the APO proteins detected in these pellets were not really contained within virions; for example, they could have been released from the cells in association with large cellular debris. They might also be in the viral pellets simply because they adhere to virus particles. To critically test these possibilities, we compared the viral pellets with “mock” viral pellets, which were transfected with the APO expression plasmids but with an empty plasmid vector rather than MLV; in addition, we treated the pellets with subtilisin (43) to eliminate proteins that were not sequestered within particles.
The results of this experiment are shown in Fig. 6. There was no APO protein detectable in the mock viral pellets (lanes 4 and 5 and lanes 10 and 11, panel C); thus, cells expressing APO did not release it in pelletable form unless they were also producing virus particles. Aliquots of the subtilisin-digested and the mock-digested pellets were tested by immunoblotting with anti-p30CA and anti-gp70SU, as well as anti-HA, antisera in order to monitor the effects of subtilisin digestion. Comparison of lanes 7 to 9 with lanes 1 to 3 of panels A and B shows that the subtilisin treatment eliminated the gp70, but not the p30CA, from the pellets. This result demonstrates both the structural integrity of the particles and the efficacy of the digestion, as gp70 is exposed on the surface of intact virions while p30 is an internal viral protein. Finally, as shown in panel C (lanes 1 and 2 and lanes 7 and 8), the digestion also had no effect on the hA3G or mA3 band. Taken together, these results confirm that the APO proteins in the viral pellets are contained within virions.
FIG. 6.
Subtilisin resistance of mA3 in MLV virions. MLV particles produced in the presence of hA3G or mA3 plasmids were subjected to subtilisin (SUB) digestion as described in Materials and Methods and then analyzed by immunoblotting for intact p30CA (A), gp70SU (B), or HA-tagged APO (C) protein. Lanes: 1 and 7, virions produced by cells expressing MLV and hA3G; 2 and 8, virions produced by cells expressing MLV and mA3; 3 and 9, virions produced by cells expressing MLV; 4 and 10, “virions” produced by cells expressing hA3G; 5 and 11, “virions” produced by cells expressing mA3. The samples in lanes 7 to 11 were digested with subtilisin before immunoblotting.
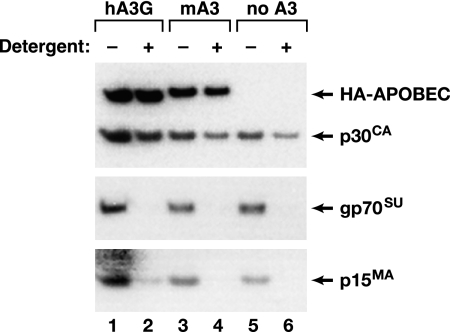
It was also possible that mA3 has lower efficacy against MLV than hA3G because, while it is within the virions (Fig. 6), it is excluded from the nucleoprotein core of the mature particle. To explore this possibility, we fractionated virions containing mA3 and hA3G by centrifuging them through a solution of 0.1% Igepal. This treatment will remove the lipid bilayer, matrix protein, and surface glycoprotein from the particles while preserving the viral core; the resulting pellet should then contain the CA and NC proteins but not the MA or Env protein. As shown in Fig. 7, the pellets in the detergent-treated samples (lanes 2, 4, and 6) lacked MA and gp70SU (second and third panels) but contained CA (top panel), as expected. Immunoblotting with the anti-HA antiserum (top panel) showed that when virions were produced in cells expressing mA3 or hA3G, these proteins were also present in the pellets. Thus, this experiment did not suggest any difference in the location of mA3 and hA3G within MLV particles.
FIG. 7.
Localization of APO proteins in the interior of MLV particles. MLV particles were prepared by transient transfection in the presence of 10 μg of hA3G (lanes 1 and 2), mA3 (lanes 3 and 4), or empty (lanes 5 and 6) expression plasmid. The virions in the culture fluids were fractionated as described in Materials and Methods, and the pellets were analyzed by immunoblotting against the HA epitope tag or p30CA (top), gp70SU (middle), or p15MA (bottom). In lanes 1, 3, and 5, the virions were sedimented with no detergent, while in lanes 2, 4, and 6, Igepal was present in the 10% sucrose layer during centrifugation.
MLV reverse transcription: comparison of mA3 and hA3G effects.
The antiviral effects of hA3G have been characterized in considerable detail. They include, but may not be limited to, the deamination of cytidine residues in minus-strand DNA, leading to G-to-A mutations in the provirus. It was obviously of interest to determine the mechanism of MLV inactivation by mA3.
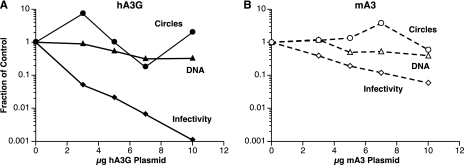
As a first step in the analysis of the block to MLV infection by the APO proteins, we measured the level of reverse transcription by the inactivated virions. 293T-mCAT1 cells were infected with virus from the experiment described in Fig. 2. Twenty-four hours after infection, the cells were lysed and assayed for the hph DNA that was synthesized during reverse transcription of the pLXSH particles (as described previously [44], this experimental design eliminates the possibility that plasmid molecules present in the inoculum will register in the DNA assays). DNA recovery was monitored by simultaneously measuring copies of the single-copy cellular gene CCR5. As shown in Fig. 8, synthesis of viral DNA was reduced only about 3-fold in the presence of maximal levels of mA3 and hA3G while infectivities were reduced ∼15-fold by mA3 and ∼1,000-fold by hA3G. Thus, interference with viral DNA synthesis may contribute to but does not seem to play a major role in MLV inactivation by the APO proteins.
FIG. 8.
DNA synthesis by APO-inactivated MLV. 293T-mCAT1 cells were infected with the culture fluids that were assayed for infectivity in Fig. 2. They were lysed 24 h later and assayed by real-time PCR for hph DNA and 2-LTR circles as described in Materials and Methods. Values are corrected for differences in CCR5 DNA copy numbers.
We also tested the effects of mA3 and hA3G upon the formation of 2-LTR closed circular viral DNA. These molecules, formed by ligation of the ends of the full-length viral DNA, are often taken as an indicator of the entry of the viral DNA into the nucleus of the newly infected cell. Figure 8 shows that the levels of 2-LTR circular DNA were not dramatically affected by either of the APO proteins tested here (the copy numbers of 2-LTR circle junctions measured here were <1,000; this low level reduces the accuracy of these measurements and is undoubtedly responsible for the “scatter” in these titrations).
Sequence analysis of reverse transcription products.
APO proteins are cytidine deaminases and are known to induce G-to-A hypermutation by deamination of C residues on the minus strand during reverse transcription by HIV-1 (18, 32, 53). To determine whether this property was responsible for inactivation of MLV, we also performed large-scale sequencing on a 750-nucleotide block of the hph DNA synthesized upon infection with MLVs made as described above. The results are summarized in Table 1. We found that MLV inactivated by hA3G (rows 4 and 5) had a G-to-A mutation frequency ∼50-fold higher than that seen in MLV produced in the absence of APO (row 2); the latter frequency was indistinguishable from that obtained by direct amplification and sequencing of pLXSH plasmid DNA (row 1). Interestingly, we observed virtually identical G-to-A mutation frequencies in the MLVs inactivated by the intermediate and maximal hA3G doses (rows 4 and 5), despite the fact that the maximal dose reduced virus-induced luciferase activity nearly 100-fold more than the intermediate dose. In striking contrast, however, MLV that had been inactivated by mA3 (row 3) did not show any elevation of G-to-A mutation frequencies. It is important to note that the relative infectivity of the mA3-inactivated MLV sample analyzed here (as assessed by its luciferase titer) was slightly lower than that of the sample inactivated with the intermediate hA3G dose; thus, if G-to-A mutation were responsible for MLV inactivation by mA3, the mutations would certainly have been detectable in the mA3-inactivated viral DNA. Taken together, the results show that inactivation of MLV by hA3G is accompanied by G-to-A mutation, but the mutation frequency is evidently not proportional to the level of viral inactivation. Inactivation of MLV by mA3 is not associated with detectable G-to-A mutations, at least within the “body” of the viral genome.
TABLE 1.
Induction of mutations by APO proteinsa
| Source of DNA | APOBEC treatment | Relative titer | No. of bases sequenced | No. of G-to-A mutations | No. of all other mutations | Frequency of G-to-A mutations |
|---|---|---|---|---|---|---|
| Plasmid pLXSH | None | NAb | 5,746 | 1 | 0 | 1.74 × 10−4 |
| MLV-infected cells | None | 1 | 56,481 | 8 | 22 | 1.42 × 10−4 |
| MLV-infected cells | 10 μg mA3 | ∼0.08 | 43,556 | 6 | 8 | 1.32 × 10−4 |
| MLV-infected cells | 3 μg hA3G | ∼0.17 | 22,176 | 165 | 5 | 7.44 × 10−3 |
| MLV-infected cells | 10 μg hA3G | ∼0.0035 | 31,017 | 227 | 32 | 7.32 × 10−3 |
| HIV-infected cells | None | 1 | 22,022 | 8 | 3.6 × 10−4 | |
| HIV-infected cells | 3 μg mA3 | 0.008 | 25,304 | 209 | 8.3 × 10−3 |
DNA was isolated from cells infected with MLV that had been produced by cotransfection with APO-expressing plasmids. hph DNA was amplified and sequenced as described in Materials and Methods. A similar analysis was also performed directly on pLXSH plasmid DNA. DNA was also isolated from cells infected with ΔVif HIV, and luciferase-encoding DNA was similarly amplified and sequenced. “Relative titers” are approximate for MLV because samples from two experiments with very similar results were analyzed and the results are pooled here. Mutations other than G-to-A transitions were not tabulated for the HIV samples.
NA, not applicable.
We also tested the ability of mA3 to induce G-to-A mutation in the ΔVif HIV-1-derived vector. As shown in Table 1, row 7, DNA synthesized by the HIV-derived particles carrying mA3 contained a very high G-to-A mutation level, in agreement with previous reports (5, 29, 33, 51). Thus, the inability of mA3 to induce detectable G-to-A mutations (Table 1, row 3) appears to be specific to MLV.
We also determined the immediate sequence context of the G-to-A mutations induced in MLV vectors by hA3G. At both hA3G doses, ∼95% of the mutated residues were followed by G while the remainder were followed by A, as expected for the effects of hA3G, in which the preferred target is CC (data not shown) (20, 51). In contrast, the great majority of the G-to-A mutations that appeared in ΔVif HIV-1 DNA synthesized in the presence of mA3 were followed by either G or A; the “hot spots” for G-to-A mutation nearly always contained an A residue 2 bases 3′ to the mutated G (data not shown), in excellent agreement with previous work on mA3 showing that the preferred target is T T/C C (5, 28, 51). The simplicity of the target sequences for both hA3G and mA3 means that the two DNA sequences analyzed here, i.e., hph (in MLV) and luciferase (in HIV-1), both contain many copies of each target sequence; thus, the lack of mA3-induced mutations in hph is not due to the absence of these targets.
Anti-MLV activities of mutant mA3 and hA3G molecules.
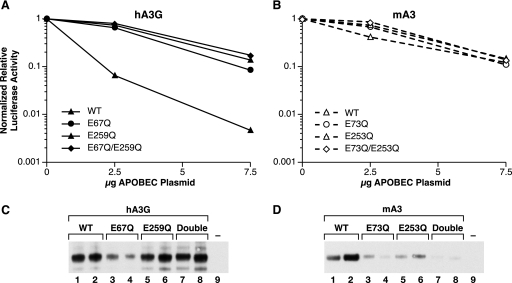
APO proteins have two zinc fingers, and in each finger a glutamic acid residue is the putative active site of the cytidine deaminase enzyme. In order to obtain some insight into the possible role of deamination in anti-MLV activity, we mutated these residues in hA3G and mA3 to glutamine and analyzed the effects of these mutant proteins as in Fig. 1. In these experiments, the level of virus in each sample was quantitated by RT assays and the luciferase activity was normalized to the level of virus. As shown in Fig. 9A, replacing either of the catalytic-site glutamic acids in hA3G with glutamine greatly reduced the ability of the protein to inhibit MLV infection; we saw no significant difference in activity between the two single mutants and the double mutant in which both glutamic acid residues are replaced.
FIG. 9.
Effects of mutants of hA3G and mA3 on the infectivity of an MLV-derived luciferase vector. Mutant and wild-type (WT) APO proteins were assayed as described in the legend to Fig. 1, except that the luciferase activities were corrected for variations in the amount of virus, so that the graphs represent the specific infectivities of the virus preparations. Virus preparations were also analyzed by immunoblotting against the HA tag in the APO proteins. In panels C and D, lanes 1, 3, 5, and 7 are the samples transfected with 2.5 μg of APO plasmid; lanes 2, 4, 6, and 8 are the samples produced with 7.5 μg; and lane 9 is the sample produced with no APO.
The anti-MLV activity of mA3 is, as described above, far lower than that of hA3G. As shown in Fig. 9B, there was little effect of the mutations upon the anti-MLV activity of mA3.
We also determined the efficiency with which the mutant APO proteins were encapsidated into MLV virions. Figure 9 shows the results of immunoblotting for the HA tag in hA3 (panel C) and mA3 (panel D). It can be seen that replacements of glutamic acid with glutamine had little effect on hA3G packaging (panel C), but encapsidation of mA3 was significantly reduced by the mutations.
DISCUSSION
Mammalian cells possess a number of defense mechanisms that protect them from infection by retroviruses. These defenses include the APOBEC3 family of proteins. The existence of this cellular system was only recently discovered (45), and many fundamental questions concerning its underlying mechanisms remain unanswered.
We have investigated the effects of hA3G and mA3 upon MLV and MLV-derived vectors. We found that mA3 blocks infection by these viruses, although hA3G induces a significantly more severe inhibition. In contrast, mA3 was as potent as hA3G as an inhibitor of infection by a ΔVif HIV-1-derived vector, in agreement with earlier work (18, 32). Thus, MLV appears to be partially resistant to mA3.
In our experiments, inactivation of MLV by either mA3 or hA3G always followed single-hit kinetics (Fig. 1, 2, 3B, and 5B). This dose-response pattern suggests that a single APOBEC3 molecule (or oligomer, assuming that intracellular APOBEC3 concentrations are high enough to ensure efficient oligomerization) is sufficient to inactivate an MLV particle. The ability of a single APOBEC3 molecule or oligomer to inactivate a virion is quite consistent with the results of Xu et al., who reported that HIV-1 particles produced in human peripheral blood mononuclear cells contain only about seven hA3G molecules (50). (While the titration data imply that a single molecule or oligomer can inactivate a virion, any number of APOBEC3 molecules might be present in each particle without contributing to the inactivation.) The respective slopes of the mA3 and hA3G titrations imply that, per microgram of plasmid transfected, hA3G inactivates MLV ∼2.5 times more efficiently than mA3. As the hA3G and mA3 protein levels in cells and virions are, for a given plasmid dose, similar to each other (Fig. 5), the results suggest that a molecule of hA3G blocks MLV infection ∼2.5 times as effectively as a molecule of mA3.
The ability of mA3 to inhibit infection by MLV-related genomes is obviously significantly lower than that of hA3G, and it might be suggested that this activity is only an artifact of the high level of expression of mA3 in our transfected cells. However, the data are not consistent with this suggestion; rather, the steady, progressive decline of infectivity with increasing dose of mA3 plasmid (Fig. 1 to 5) shows that the inhibition is not confined to cells with especially high mA3 concentrations, even though it is most obvious in these cells.
It might be imagined that MLV exhibits partial resistance to mA3 by partially excluding it from assembling virus particles. In fact, both HIV-1 and HTLV-1 have mechanisms for excluding hA3G from progeny virions, although these mechanisms are very different from each other; in HIV-1, resistance depends upon the accessory protein Vif, but in HTLV-1, the exclusion of both hA3G and mA3 involves the nucleocapsid domain of Gag (9, 35, 52). However, immunoblotting with anti-HA antibody (for detection of the HA-tagged APO proteins) showed that mA3 was incorporated into MLV particles as efficiently as hA3G (Fig. 5). Further analysis showed that mA3, like hA3G, is within the detergent-resistant ribonucleoprotein in the interior of MLV particles (Fig. 7).
Despite intensive investigations in a number of laboratories, the mechanism by which APOBEC3 molecules inactivate retroviruses is still not well understood. The cytidine deaminase activity of hA3G is well documented (17, 25, 47), and it is very clear that APOBEC3 proteins can induce G-to-A mutations in retroviral genomes (18, 30, 32, 33, 53). However, several situations have been described in which this activity does not seem sufficient to explain the inactivation (reviewed in reference 23). Evidence has been presented for additional inactivation mechanisms, including interference with reverse transcription or with the stability of the newly synthesized viral DNA (4, 23, 24, 31-34, 46, 49) and with integration of the viral DNA into cellular DNA (23, 34), possibly by impairing the normal removal of primer tRNA after the second-strand transfer step during reverse transcription (34) or by interfering with the function of a viral component such as IN or NC in the preintegration complex (23).
In the present work, we also addressed the mechanism by which mA3 and hA3G interfere with MLV infection. We found a slight reduction in the production of viral DNA following infection with APOBEC3-inactivated MLV (Fig. 8), but the magnitude of this effect was far too low to account for the loss of infectivity (23, 34). There was no significant effect on the formation of 2-LTR circular viral DNA (Fig. 8), implying that transport to the nucleus was unimpaired.
Most interestingly, we found no G-to-A mutations in the DNA synthesized upon infection with MLV inactivated by mA3, while these mutations were readily detected if the MLV contained hA3G, or in mA3-containing ΔVif HIV-1-derived vector particles (Table 1). The same frequency of G-to-A mutations was observed in DNA from MLV generated with two different doses of hA3G expression plasmid (3 and 10 μg, Table 1), although the specific infectivities of these two virus preparations differed by almost 100-fold. As 10 μg of mA3 plasmid inactivated the MLV as effectively as 3 μg of hA3G plasmid, G-to-A mutations should have been detectable in the former sample if virus inactivation proceeds via cytidine deamination. Both the absence of the mutations in mA3 and the discordance between mutation frequency and infectivity in hA3G strongly suggest that cytidine deamination cannot fully explain the inactivation of MLV by APOBEC3 proteins, as previously proposed by others for HIV-1 (see above). In addition, we observed that each of the APO proteins inactivates pLXSH and replication-competent MLV with similar efficiencies (Fig. 2); this is also difficult to reconcile with a simple deamination mechanism, since the minus-strand DNA of the latter virus would present a target for deamination roughly twice the size of that of the former.
We also tested the anti-MLV activity of mutant hA3G and mA3 molecules in which the glutamic acids at the deaminase active sites are replaced with glutamines. In hA3G, it was clear (Fig. 9A) that both glutamic acids are essential for the efficient inactivation of MLV. In contrast, the relatively inefficient inactivation of MLV induced by wild-type mA3 was only slightly reduced by elimination of either or both of the glutamic acid residues (Fig. 9B). We also noted that the mutations significantly reduced the amount of mA3 protein packaged into MLV (Fig. 9D). One explanation of these results might be that, under our conditions, most of the wild-type mA3 molecules packaged in virions are not participating in its antiviral activity. However, it is also possible that the mutations do significantly affect the antiviral activity of mA3 but that this effect is difficult to detect because the activity of wild-type mA3 is relatively low. In this case, the antiviral effects seen at the higher doses of the mA3 mutants may represent artifacts of high expression levels.
In summary, our data indicate that MLV is partially resistant to the antiviral effects of mA3 and completely resistant to the G-to-A hypermutation seen with many other retrovirus-APO combinations. Unlike the previously described resistance mechanisms (9, 35, 52), the mechanism by which MLV resists mA3 does not involve exclusion of the latter from the virion. We have no direct information on the nature of this mechanism; it seems possible that a constituent of MLV particles specifically inhibits the enzymatic activity of mA3 or the access of the latter to intracellular structures, such as the replication complex or the preintegration complex, that arise from the viral core during infection. Exploration of these possibilities is under way.
Other investigators have previously reported that MLV is unaffected by mA3 (10, 29, 33). Some reports have also indicated that mA3 is excluded from MLV particles (1, 10, 29). We do not know why our results are different from those of these other investigators. Nearly all of our experiments used the mA3 isoform lacking exon 5, and it has been reported that MLV is more resistant to mA3 containing this exon than to the more spliced isoform, evidently because the exon contains a site which is cleaved by MLV protease (1). However, MLV appeared to exhibit similar resistance to both isoforms in our experiments (Fig. 4). Further work is required to fully resolve the differences between the various experimental systems used to analyze mA3-MLV interactions.
Like MLV, mouse mammary tumor virus (MMTV) has been grown for many years in mouse cells. It is interesting that the recent report by Okeoma et al. (41) indicates that MMTV replication is impaired, although not completely prevented, by mA3 both in cultured cells and in mice. No G-to-A mutations were detected in the DNA synthesized by the MMTV inactivated by mA3. The similarity of these results and ours with MLV presumably reflects the similarity in the selective pressures to which the two viruses have been subjected. In contrast, G-to-A mutations are present in the genomes of some endogenous MLVs but not in others (27). The mutations bear all of the hallmarks of mA3 action. Thus, it appears that some, but not all, of the gammaretroviruses that have infected the mouse germ line were susceptible to deamination by mA3.
ADDENDUM
While the manuscript was in revision, a paper by E. P. Browne and D. R. Littman appeared, with results in general agreement with ours (6a).
Acknowledgments
This work was supported in part by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health, and has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400.
We thank Nathaniel Landau and Jim Cunningham for reagents; Judith Levin, David Ott, Vinay Pathak, and James Thomas for very helpful suggestions; Teresa L. Shatzer for technical assistance; and Richard Frederickson for artwork.
Footnotes
Published ahead of print on 30 April 2008.
REFERENCES
- 1.Abudu, A., A. Takaori-Kondo, T. Izumi, K. Shirakawa, M. Kobayashi, A. Sasada, K. Fukunaga, and T. Uchiyama. 2006. Murine retrovirus escapes from murine APOBEC3 via two distinct novel mechanisms. Curr. Biol. 161565-1570. [DOI] [PubMed] [Google Scholar]
- 2.Albritton, L. M., L. Tseng, D. Scadden, and J. M. Cunningham. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57659-666. [DOI] [PubMed] [Google Scholar]
- 3.Bassin, R. H., N. Tuttle, and P. J. Fischinger. 1971. Rapid cell culture assay for murine leukaemia virus. Nature 229564-566. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 808450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 141392-1396. [DOI] [PubMed] [Google Scholar]
- 6.Bogerd, H. P., H. L. Wiegand, A. E. Hulme, J. L. Garcia-Perez, K. S. O'Shea, J. V. Moran, and B. R. Cullen. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. USA 1038780-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Browne, E. P., and D. R. Littman. 2008. Species-specific restriction of Apobec3-mediated hypermutation. J. Virol. 821305-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 313497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derse, D., S. A. Hill, P. A. Lloyd, H. Chung, and B. A. Morse. 2001. Examining human T-lymphotropic virus type 1 infection and replication by cell-free infection with recombinant virus vectors. J. Virol. 758461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derse, D., S. A. Hill, G. Princler, P. Lloyd, and G. Heidecker. 2007. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl. Acad. Sci. USA 1042915-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doehle, B. P., A. Schafer, H. L. Wiegand, H. P. Bogerd, and B. R. Cullen. 2005. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 798201-8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433430-433. [DOI] [PubMed] [Google Scholar]
- 12.Esnault, C., J. Millet, O. Schwartz, and T. Heidmann. 2006. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 341522-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu, W., B. A. Ortiz-Conde, R. J. Gorelick, S. H. Hughes, and A. Rein. 1997. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J. Virol. 716940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 675443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorelick, R. J., D. J. Chabot, D. E. Ott, T. D. Gagliardi, A. Rein, L. E. Henderson, and L. O. Arthur. 1996. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J. Virol. 702593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorelick, R. J., S. M. Nigida, Jr., J. W. Bess, Jr., L. O. Arthur, L. E. Henderson, and A. Rein. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 643207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakata, Y., and N. R. Landau. 2006. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J. Biol. Chem. 28136624-36631. [DOI] [PubMed] [Google Scholar]
- 18.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113803-809. [DOI] [PubMed] [Google Scholar]
- 19.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4868-877. [DOI] [PubMed] [Google Scholar]
- 20.Harris, R. S., A. M. Sheehy, H. M. Craig, M. H. Malim, and M. S. Neuberger. 2003. DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses. Nat. Immunol. 4641-643. [DOI] [PubMed] [Google Scholar]
- 21.Hibbert, C. S., J. Mirro, and A. Rein. 2004. mRNA molecules containing murine leukemia virus packaging signals are encapsidated as dimers. J. Virol. 7810927-10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes, R. K., F. A. Koning, K. N. Bishop, and M. H. Malim. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation: comparisons with APOBEC3G. J. Biol. Chem. 2822587-2595. [DOI] [PubMed] [Google Scholar]
- 23.Holmes, R. K., M. H. Malim, and K. N. Bishop. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32118-128. [DOI] [PubMed] [Google Scholar]
- 24.Iwatani, Y., D. S. Chan, F. Wang, K. S. Maynard, W. Sugiura, A. M. Gronenborn, I. Rouzina, M. C. Williams, K. Musier-Forsyth, and J. G. Levin. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 357096-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwatani, Y., H. Takeuchi, K. Strebel, and J. G. Levin. 2006. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J. Virol. 805992-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79285-296. [DOI] [PubMed] [Google Scholar]
- 27.Jern, P., J. P. Stoye, and J. M. Coffin. 2007. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 32014-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jónsson, S. R., G. Hache, M. D. Stenglein, S. C. Fahrenkrug, V. Andresdottir, and R. S. Harris. 2006. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 345683-5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi, M., A. Takaori-Kondo, K. Shindo, A. Abudu, K. Fukunaga, and T. Uchiyama. 2004. APOBEC3G targets specific virus species. J. Virol. 788238-8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 3001112. [DOI] [PubMed] [Google Scholar]
- 31.Luo, K., T. Wang, B. Liu, C. Tian, Z. Xiao, J. Kappes, and X. F. Yu. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 817238-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 42499-103. [DOI] [PubMed] [Google Scholar]
- 33.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 11421-31. [DOI] [PubMed] [Google Scholar]
- 34.Mbisa, J. L., R. Barr, J. A. Thomas, N. Vandegraaff, I. J. Dorweiler, E. S. Svarovskaia, W. L. Brown, L. M. Mansky, R. J. Gorelick, R. S. Harris, A. Engelman, and V. K. Pathak. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 817099-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 2797792-7798. [DOI] [PubMed] [Google Scholar]
- 36.Miller, A. D., D. G. Miller, J. V. Garcia, and C. M. Lynch. 1993. Use of retroviral vectors for gene transfer and expression. Methods Enzymol. 217581-599. [DOI] [PubMed] [Google Scholar]
- 37.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 183587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muckenfuss, H., M. Hamdorf, U. Held, M. Perkovic, J. Lower, K. Cichutek, E. Flory, G. G. Schumann, and C. Munk. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 28122161-22172. [DOI] [PubMed] [Google Scholar]
- 39.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 985246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15166-170. [DOI] [PubMed] [Google Scholar]
- 41.Okeoma, C. M., N. Lovsin, B. M. Peterlin, and S. R. Ross. 2007. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature 445927-930. [DOI] [PubMed] [Google Scholar]
- 42.Oshima, M., D. Muriaux, J. Mirro, K. Nagashima, K. Dryden, M. Yeager, and A. Rein. 2004. Effects of blocking individual maturation cleavages in murine leukemia virus Gag. J. Virol. 781411-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ott, D. E., L. V. Coren, D. G. Johnson, B. P. Kane, R. C. Sowder II, Y. D. Kim, R. J. Fisher, X. Z. Zhou, K. P. Lu, and L. E. Henderson. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 26642-51. [DOI] [PubMed] [Google Scholar]
- 44.Rulli, S. J., Jr., D. Muriaux, K. Nagashima, J. Mirro, M. Oshima, J. G. Baumann, and A. Rein. 2006. Mutant murine leukemia virus Gag proteins lacking proline at the N-terminus of the capsid domain block infectivity in virions containing wild-type Gag. Virology 347364-371. [DOI] [PubMed] [Google Scholar]
- 45.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418646-650. [DOI] [PubMed] [Google Scholar]
- 46.Simon, J. H., and M. H. Malim. 1996. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 705297-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svarovskaia, E. S., H. Xu, J. L. Mbisa, R. Barr, R. J. Gorelick, A. Ono, E. O. Freed, W. S. Hu, and V. K. Pathak. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 27935822-35828. [DOI] [PubMed] [Google Scholar]
- 48.Thomas, J. A., T. D. Gagliardi, W. G. Alvord, M. Lubomirski, W. J. Bosche, and R. J. Gorelick. 2006. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology 35341-51. [DOI] [PubMed] [Google Scholar]
- 49.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 674945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu, H., E. Chertova, J. Chen, D. E. Ott, J. D. Roser, W. S. Hu, and V. K. Pathak. 2007. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology 360247-256. [DOI] [PubMed] [Google Scholar]
- 51.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11435-442. [DOI] [PubMed] [Google Scholar]
- 52.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 3021056-1060. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 42494-98. [DOI] [PMC free article] [PubMed] [Google Scholar]