Abstract
A phase I clinical vaccine study of a human immunodeficiency virus type 1 (HIV-1) vaccine regimen comprising a DNA prime formulation (5-valent env and monovalent gag) followed by a 5-valent Env protein boost for seronegative adults was previously shown to induce HIV-1-specific T cells and anti-Env antibodies capable of neutralizing cross-clade viral isolates. In light of these initial findings, we sought to more fully characterize the HIV-1-specific T cells by using polychromatic flow cytometry. Three groups of participants were vaccinated three times with 1.2 mg of DNA administered intradermally (i.d.; group A), 1.2 mg of DNA administered intramuscularly (i.m.; group B), or 7.2 mg of DNA administered i.m. (high-dose group C) each time. Each group subsequently received one or two doses of 0.375 mg each of the gp120 protein boost vaccine (i.m.). Env-specific CD4 T-cell responses were seen in the majority of participants; however, the kinetics of responses differed depending on the route of DNA administration. The high i.m. dose induced the responses of the greatest magnitude after the DNA vaccinations, while the i.d. group exhibited the responses of the least magnitude. Nevertheless, after the second protein boost, the magnitude of CD4 T-cell responses in the i.d. group was indistinguishable from those in the other two groups. After the DNA vaccinations and the first protein boost, a greater number of polyfunctional Env-specific CD4 T cells (those with ≥2 functions) were seen in the high-dose group than in the other groups. Gag-specific CD4 T cells and Env-specific CD8 T cells were seen only in the high-dose group. These findings demonstrate that the route and dose of DNA vaccines significantly impact the quality of immune responses, yielding important information for future vaccine design.
A preventative human immunodeficiency virus (HIV) vaccine is urgently needed to curtail the global AIDS epidemic. Clinical trials of HIV vaccines have shown that the induction of type-specific binding-antibody responses alone is not sufficient to elicit protective immunity in humans (28, 34). This finding is true despite the fact that antibody responses correlate with efficacy for the majority of currently licensed vaccines (5, 8, 48). Furthermore, nonhuman primate studies suggest that antibody responses can prevent infection following mucosal challenge, an outcome that may not be possible with T-cell-based vaccines (2, 7, 72). The recent failure of a vaccine, designed primarily to induce T-cell responses without Env-specific antibody responses (16, 17), highlights the importance of trying to develop immunogens that will elicit broad Env-specific antibodies. Prior attempts using an Env glycoprotein-based vaccine design have been hampered either by poor immunogenicity (1) or by the lack of an antibody response that can effectively neutralize several circulating strains of HIV (56). This result may be due partly to the use of recombinant Env glycoproteins based on a single viral isolate (38, 40, 47, 52, 59). Attempts at vaccinating with recombinant viral vectors encoding Env glycoprotein and boosting with gp120 recombinant proteins have not been successful in generating broadly reactive antibodies capable of neutralizing primary viral isolates (35).
DNA vaccines represent an alternative approach which has been shown to induce cell-mediated immune (CMI) responses (13, 41, 75). Although DNA vaccines prime CMI and humoral responses, these responses have typically been low in magnitude in humans. Other strategies have combined a DNA prime vaccine regimen with a viral vector boost (13, 37, 58) in order to enhance immunogenicity. However, these approaches have been only marginally successful in the rhesus macaque animal model system, possibly because they did not induce protective antibody responses (46). Another approach has been to use a viral vector prime followed by a recombinant protein boost, and this strategy induced low-level CMI responses and/or neutralizing antibodies to laboratory-adapted viral strains (10, 26, 39, 66, 68). Similar to the latter vaccine strategy, a DNA prime/protein boost approach has the potential for eliciting both cellular and humoral immunity. Additionally, priming with a DNA vaccine potentially allows for the immune response to focus on HIV epitopes by ignoring competing epitopes expressed by the recombinant viral vector (27, 29). Recent preclinical studies with small animals and nonhuman primates demonstrated that a DNA prime and protein boost vaccine combination is effective in eliciting CMI and humoral responses (19, 60-62). In addition, this strategy is more effective at inducing neutralizing antibodies against primary HIV isolates than the recombinant-protein approach alone (81). In prior preclinical testing (84), the DNA prime and protein boost strategy, using polyvalent primary Env antigens from clades A, B, C, D, and E as immunogens, was found to be more effective than the monovalent Env antigen in eliciting broad neutralizing antibodies in rabbits.
In a phase I clinical study, a polyvalent gp120 DNA prime/protein boost approach induced antibody responses capable of neutralizing cross-clade viral isolates, in addition to T cells capable of secreting gamma interferon (IFN-γ) (83). The robustness of these responses led us to further characterize the T cells generated by this polyvalent gp120 DNA prime/protein boost vaccine regimen. DNA vaccinations were able to generate dose-dependent polyfunctional Env-specific CD4 T-cell responses. Additionally, vaccine-induced CD4 T-cell responses were lower in magnitude when the DNA vaccine was administered intradermally (i.d.). The high-dose DNA vaccination induced T cells with the greatest breadth, magnitude, and number of functions. These data highlight the importance of the dose and route of administration of DNA vaccines in determining the character of CD4 T cells, further enhancing our understanding of the human immune response to immunization.
MATERIALS AND METHODS
Polyvalent DNA prime and protein boost DP6-001 formulation.
The DP6-001 vaccine used in this study contained equal amounts of six individual DNA plasmids: five codon-optimized gp120 plasmids from primary HIV type 1 (HIV-1) isolates, subtypes A (92UG037.8), B (92US715.6 [B] and Bal), C (96ZM651), and E (93TH976.17), and a codon-optimized gag gene from subtype C (96ZM651) (82). The protein boost contained equal amounts of the five homologous gp120 proteins corresponding to the plasmids used for DNA priming (62).
Study design.
Complete details of the study design are described elsewhere (J. S. Kennedy, M. Co, S. Green, K. Longtine, J. Longtine, M. A. O'Neill, J. P. Adams, A. L. Rothman, Q. Yu, R. J. Leva, R. Pal, S. Wang, S. Lu, and P. Markham, submitted for publication). Briefly, 28 healthy HIV-1-seronegative subjects who participated in the trial were vaccinated at three time points (days 0, 28, and 84) with one of two different doses of DNA (1.2 and 7.2 mg) administered i.d. or intramuscularly (i.m.) (Table 1). DNA vaccinations were followed by one or two protein boosts (0.375 mg i.m. on days 140 and 196). Participants in groups A and B received two protein boosts; however, group C participants received only a single protein boost. This study was approved by the Institutional Review Board at the University of Massachusetts Medical School (UMMS) and the Protocol Evaluation and Review Committee at the Division of AIDS, National Institutes of Health (NIH). This study was conducted at the Center for Infectious Diseases and Vaccine Research at the UMMS. Informed consent was obtained from all the participants in this study.
TABLE 1.
Study design showing the doses of DNA used for priming and the routes of immunization
| Groupa | No. of volunteers | DNAb dose (mg) | Route of DNA immunization |
|---|---|---|---|
| A | 11 | 1.2 | i.d. at 4 sites |
| B | 11 | 1.2 | i.m. at 2 sites |
| C | 6 | 7.2 | i.m. at 2 sites |
Each group received 5-valent gp120 in either a single boost of 0.375 mg (group C) or two boosts of 0.375 mg each (groups A and B).
The DNA vaccine contained DNA for 5-valent gp120 (from subtypes A, B, C, E, and Bal) and a single Gag protein (from subtype C).
Peptides.
Envelope (gp120) antigens from clade A isolate 92UG037.8 (Env-A) and clade B isolate 92US715.6 (Env-B) used in this study were synthesized at the UMMS peptide core facility. These peptides were 20-mers overlapping by 10 amino acids. Pools of peptides for Env-A (49 peptides) and Env-B (47 peptides) were made, and each peptide in the pool was used at a final concentration of 2 μg/ml. Three peptide pools comprising selected peptides from (i) cytomegalovirus, Epstein-Bar virus, and influenza virus (CEF; catalog no. 9808); (ii) HIV-1 HXB2 Pol (catalog no. 4358); and (iii) HIV-1 subtype C Gag (catalog no. 3993) were obtained from the NIH AIDS Research and Reference Reagent Program. Both the Gag and Pol peptide sets were also 20-mers overlapping by 10 amino acids. Optimized 10-mer CD8 epitopes were synthesized by the Mimitopes, Inc., peptide core facility (Raleigh, NC).
PFC.
Cryopreserved peripheral blood mononuclear cells (PBMC) were analyzed at five time points: the baseline, 2 weeks after the final DNA prime vaccination (day 98), 2 weeks after the first protein boost (day 154, or Prot-1), 2 weeks after the second protein boost (day 210, or Prot-2), and 24 weeks after the second protein boost (day 364, or closeout). The samples were stimulated with homologous gp120 (subtype A and B) and HIV-1 Czm Gag (GagCzm) peptide pools and analyzed for the production of cytokines (IFN-γ, interleukin-2 [IL-2], and tumor necrosis factor alpha [TNF-α]), the upregulation of the CD40 ligand (CD154), and degranulation (as indicated by CD107 expression) by polychromatic flow cytometry (PFC).
PBMC were washed twice in RPMI medium containing 10% human AB sera (R-10 medium) by centrifugation at 250 × g for 10 min each time at room temperature. Costimulatory monoclonal antibodies (anti-CD28 and anti-CD49d; Becton Dickinson, San Jose, CA) at 1 μg/ml each and 50 U of Benzonase (Novagen, Madison, WI)/ml were added to each tube containing 106 PBMC in 500 μl of R-10 medium. For coculture, CD107a/CD107b-fluorescein isothiocyanate (FITC) (a/b) and CD154-phycoerythrin (PE; Becton Dickinson, San Jose, CA) were added. Cells were then pulsed with the appropriate peptide pool (each peptide, 2 μg/ml; UMMS peptide core facility) for 1 h. Monensin (Becton Dickinson, San Jose, CA) was then added at 10 μg/ml, and the addition was followed by a 5-h incubation at 37°C. The Env-A, Env-B, and Gag peptides used in the pools were 20-mers overlapping by 10 amino acids. Staphylococcal enterotoxin B (SEB; 1 μg/ml) and CEF (NIH AIDS Research and Reference Reagent Program) were used as positive controls. An irrelevant peptide pool (HIV-1 Pol peptides from the NIH AIDS Research and Reference Reagent Program) was used as a negative control. The cells were washed twice with fluorescence-activated cell sorter (FACS) wash (phosphate-buffered saline-1% fetal bovine serum) before Cytofix/Cytoperm reagent (Becton Dickinson, San Jose, CA) was added. After 20 min of incubation in the dark at room temperature, the cells were washed twice with perm/wash buffer. The cells were then labeled in a single step with the surface and intracellular antibodies.
For the analysis of the surface phenotype, the samples were stained with anti-CD3-Pacific blue and anti-CD8-peridinin chlorophyll protein-Cy5.5 (Becton Dickinson, San Jose, CA) and anti-CD4-Alexa 610 (Caltag, Burlingame, CA). For the intracellular cytokine staining, anti-IL-2-allophycocyanin (APC), anti-TNF-α-PE-Cy7, and anti-IFN-γ-Alexa Fluor 700 conjugated antibodies (Becton Dickinson, San Jose, CA) were used. The cells were washed once with perm/wash buffer and fixed in 2% paraformaldehyde. CD3+ events (≥100,000) were acquired on an LSR II flow cytometer (Becton Dickinson, San Jose, CA), and the data were analyzed using FlowJo version 8.1.1 software (TreeStar, San Carlos, CA). Lymphocytes were analyzed based on forward- and side-scatter profiles, and the gates were set based on the data for the medium control (irrelevant peptides). These gates were applied to all samples from the same individual for each time point. Cytokines that were produced were measured based upon the CD3+ CD4+ or the CD3+ CD8+ gates relative to the medium control values. The total event count for CD4 or CD8 T cells in each assay was ≥60,000. A response was considered positive for a cytokine or CD154 if the value was greater than or equal to the mean +3 standard deviations for all vaccine samples in each group, as determined relative to the medium control values. For the response to be considered positive, the value also had to be more than two times the medium control value for each individual with a response magnitude of ≥0.01%. Only participant samples found to have a positive response (any cytokine or CD154 upregulation) were further analyzed for polyfunctional responses. The polyfunctionality of the T-cell responses was determined using PESTLE and SPICE software, provided courtesy of Mario Roederer, Vaccine Research Center, Bethesda, MD.
For memory and homing markers, PBMC samples for each antigen were stained in two tubes: tube 1 (CD3-Pacific blue, CD4-Alexa 750, CD127-PE, CD45RA-APC, CCR7-PE-Cy7, IFN-γ-Alexa 700, and IL-2-FITC) and tube 2 (CD3-Pacific blue, CD4-Alexa 750, CD27-PE, CD43-FITC, CD45RO-PE-Cy7, IFN-γ-Alexa 700, and IL-2-APC). For these experiments, a two-step intracellular cytokine staining assay was used in which the surface markers were stained first, followed by the intracellular cytokines. This strategy was used for all markers except CCR7, which was stained prior to the two-step assay. All fluorochrome-conjugated antibodies were obtained from Becton Dickinson, San Jose, CA, except CD4-Alexa 750 (eBioscience, Inc., San Diego, CA).
Statistics.
Comparisons of continuous variables within each group were made using the nonparametric Wilcoxon test. Analyses of variables between each group were made using the nonparametric Mann-Whitney test. Differences in the responder frequencies were compared using Fisher's exact test.
RESULTS
Induction of HIV-specific CD4 and CD8 T-cell responses.
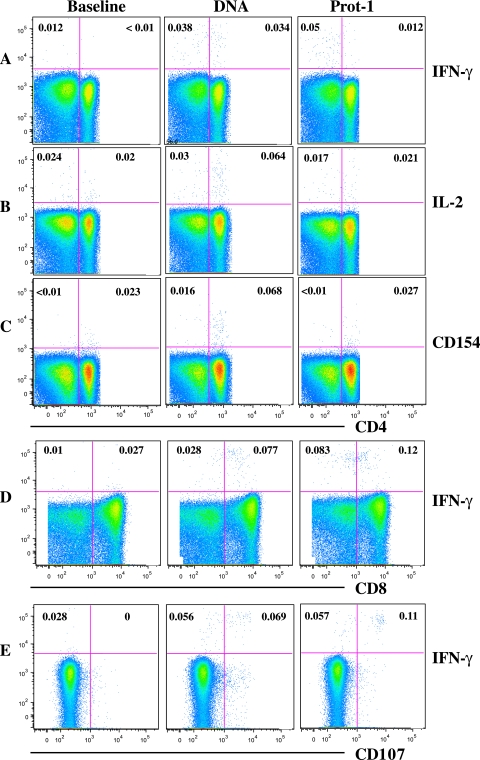
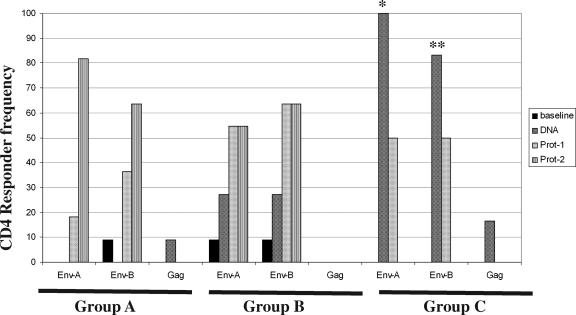
Using PFC, we evaluated the T-cell phenotypes (i.e., CD4 versus CD8) and functional capacities of vaccine-generated T-cell responses by measuring degranulation (as indicated by CD107 expression), the upregulation of the CD40 ligand (CD154), and the production of cytokines (IFN-γ, IL-2, and TNF-α) in response to Env-A, Env-B, and GagCzm antigens (Fig. 1). To determine the frequency of participants responding to the vaccine, we evaluated the presence of IFN-γ-producing CD4 T cells induced after DNA prime or protein boost vaccinations (Fig. 2). Env-specific CD4 T-cell responses were detected in all three vaccine groups; however, the kinetics of the responses in the three groups were different. In group A (given DNA i.d.), vaccine-induced CD4 T cells were not detected following the DNA vaccinations and the peak frequency of responses to Env was observed after the second protein boost. In contrast, those subjects receiving the DNA i.m. (i.e., those in groups B and C) had CD4 responses after DNA priming alone. A significantly greater percentage of participants (83 to 100%) receiving the high-dose DNA vaccine (group C) than of participants in groups A and B had vaccine-induced CD4 T-cell responses at this post-DNA priming time point (P, ≤0.001 and 0.05, respectively). Low-frequency CD4 Gag-specific responses were seen in group C after the DNA priming. Similarly, Env-B-specific CD8 T-cell responses were observed only in this high-dose group after the DNA prime regimen (67% of the subjects were responders) (data not shown). The CD8 T cells in these responses tended to be monofunctional, producing IFN-γ (in two vaccinees) or IL-2 or TNF-α (in one vaccinee each). The magnitude of the CD8 T-cell responses in the four responders after the DNA priming ranged from 0.02 to 0.24%. Env-specific CD8 T-cell populations declined following the first protein boost and were not detectable at the time of the study closeout (data not shown).
FIG. 1.
Induction of Env-specific CD4 and CD8 T-cell responses. Representative intracellular cytokine staining data show the production of cytokines IFN-γ (A) and IL-2 (B), the upregulation of CD154 by CD4 T cells (C), and the production of IFN-γ by CD8 (D) and CD8+ CD107+ (E) T cells. Data represent Env-specific responses in a group C vaccinee at baseline, day 98 (DNA), and day 154 (Prot-1).
FIG. 2.
Frequency of CD4 T-cell responses in vaccinees. The percentages of subjects in each of the three groups, group A (given a low dose i.d.), group B (given a low dose i.m.), and group C (given a high dose i.m.), that were positive for IFN-γ at various time points postvaccination are shown. Env-A- and Env-B-specific responses were significantly more frequent in group C than in the other groups (*, P ≤ 0.001, and **, P ≤ 0.05, respectively; Fisher's exact test). DNA, day 98.
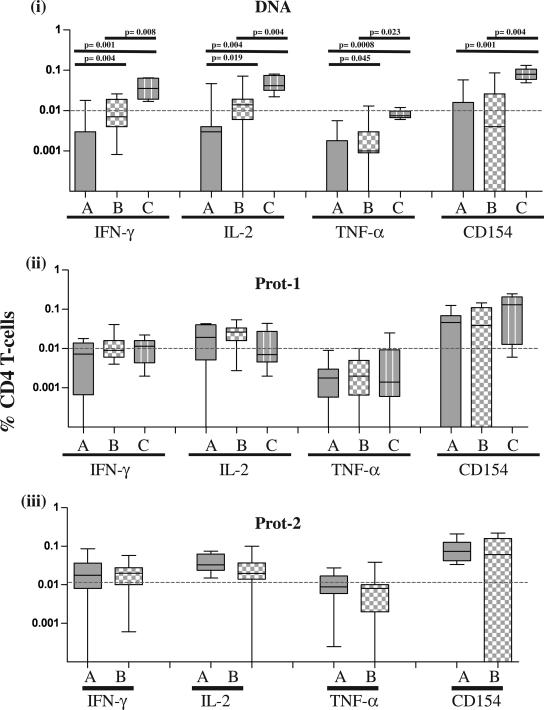
Following DNA vaccinations, group C subjects also had higher magnitudes of Env-specific CD4 T cells than group A or B subjects for each of the four functions analyzed (P ≤ 0.023) (Fig. 3, panel i). The magnitude of CD4 T cells in group B (given DNA i.m.) was also higher than that in group A (given DNA i.d.) for each of the three cytokines (P ≤ 0.045). Following the first protein boost (for groups A, B, and C) and the second protein boost (for groups A and B only), the magnitudes of vaccine-induced CD4 T-cell responses in the groups were indistinguishable (Fig. 3, panels ii and iii). The differences in magnitude among the three groups were independent of whether Env-A or Env-B was used for PBMC stimulation.
FIG. 3.
Comparison of the magnitudes of the Env-specific CD4+ T-cell responses. The production of IFN-γ, IL-2, and TNF-α and the upregulation of CD154 in each of the three groups, i.e., groups A, B, and C, at 2 weeks after DNA administration and 2 weeks after each of the two protein booster doses (Prot-1 and Prot-2) were analyzed. Statistical comparisons were made using the Mann-Whitney U test.
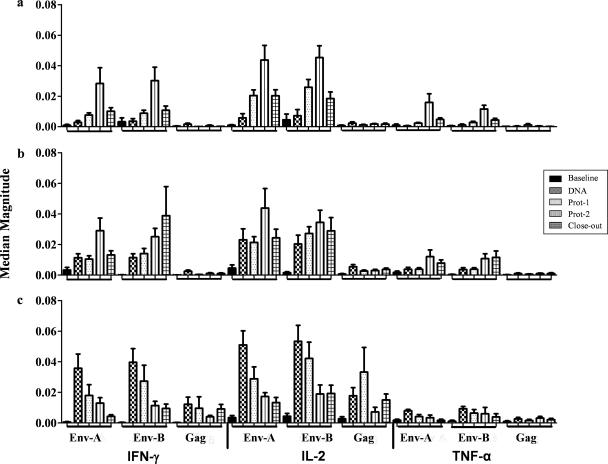
CD154 (CD40 ligand) upregulation after a short-term in vitro stimulation with antigens was recently shown to be a good marker for the assessment of recently activated antigen-specific TH cells (15, 32). Additionally, the expression of the CD40 ligand by CD4 T cells is critical for the initiation of a T-cell-dependent antibody response (6, 12, 77). Env-specific CD154+ CD4 T cells producing IFN-γ, IL-2, or TNF-α were detected in all three vaccine groups postvaccination (Fig. 4). The dominant cytokines produced by CD154+ CD4+ T cells in response to any of the three antigens tested (i.e., Env-A, Env-B, and GagCzm) were IFN-γ and IL-2. In group A, the peak response to Env-A or Env-B was observed after the second protein boost (Prot-2) (Fig. 4a). In group B, the magnitude of the response increased significantly after the DNA vaccine (day 98); however, similar to that in group A, this response peaked after the second protein boost (Fig. 4b). In group C, the magnitude of the response peaked after the DNA prime vaccinations, but a single protein boost did not increase the magnitude of the CD4 T-cell response (Fig. 4c). Gag-specific CD4 T-cell responses were lower in magnitude than Env-specific CD4 T-cell responses and were seen only in group C (Fig. 4c).
FIG. 4.
Magnitudes of CD154+ CD4+ cytokine-producing T cells. The production of IFN-γ, IL-2, or TNF-α by CD154+ CD4+ T cells in groups A, B, and C after stimulation with Env-A, Env-B, and Gag antigens was measured. Data represent the median responses with interquartile ranges. For each antigen, data are shown for the baseline, 2 weeks after DNA administration, 2 weeks after each of the protein booster doses (Prot-1 and Prot-2), and closeout as five vertical bars on the x axis. The horizontal bars on the x axis represent all five time points for each antigen. Responses were significant (P < 0.05; Wilcoxon text) in all instances in which the interquartile range at baseline did not overlap with the median for the DNA, Prot-1, Prot-2, or study closeout time point.
The vaccine induces polyfunctional Env-specific CD4 T cells.
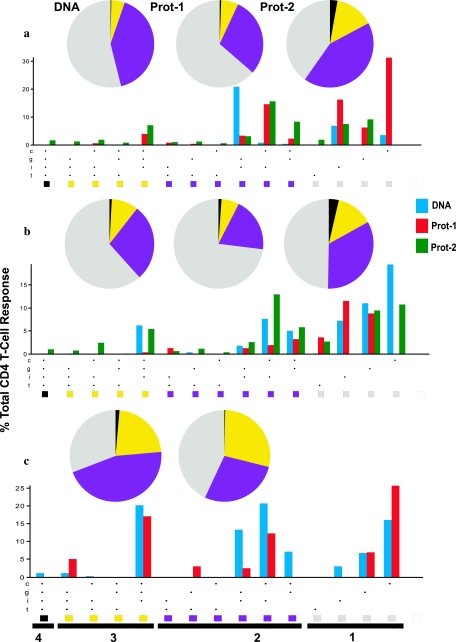
Next, we measured the contributions of one to four functions (the upregulation of CD154 and the production of IFN-γ, IL-2, and TNF-α) of antigen-specific CD4 T cells in the three vaccine groups (Fig. 5). After volunteers received the three DNA immunizations i.d. (group A; day 98), there were few T-cell responses and the T cells possessed mainly one to two functions (Fig. 5a); however, the functional response improved significantly (i.e., cells demonstrated ≥3 functions) following the second protein boost (P = 0.005). Participants in group B had Env-specific polyfunctional CD4 T-cell responses whose pattern was slightly different from that of group A responses (Fig. 5b). The DNA given i.m. (to groups B and C) tended to induce a greater frequency of CD4 T cells with ≥3 functions after the DNA priming than the DNA vaccination given to group A. Also, polyfunctional responses were not increased after the first protein boost, while the second protein boost induced a higher frequency of CD4 T cells with four functions in the group B vaccinees. Interestingly, participants receiving the high-dose DNA had a significantly greater percentage of vaccine-induced CD4 T cells that were able to fulfill ≥3 functions after the DNA vaccination regimen alone than those in groups A and B (P = 0.0007 and 0.0002, respectively) (Fig. 5c). The level of polyfunctionality of these responses (the frequency of cells with ≥3 functions) in group C remained higher than that in group B (P = 0.023) but not that in group A after the first protein boost.
FIG. 5.
Induction of polyfunctional CD4 T cells. Pie charts demonstrate the functions of the CD4 T cells (IFN-γ, IL-2, and/or TNF-α production and/or CD154 upregulation) 2 weeks after DNA administration (DNA) and 2 weeks after each of the two protein vaccinations (Prot-1 and Prot-2) for group A (a), group B (b), and group C (c). Underneath each set of pie charts, bar graphs show all possible combinations of the one to four functions on the x axis, with the bold line along the x axis covering all the possible subcombinations for a given function. All subcombinations for a given function are identified by a single color that is also represented as a slice in the pie chart. For each of the 15 functions shown, a bar represents the median response expressed as a percentage of the total CD4 T-cell response (y axis) at post-DNA administration (DNA; blue), Prot-1 (red), and Prot-2 (green) time points. For group C, data for time points following the administration of the DNA vaccine and the first protein booster dose only are shown, as this group received a single protein booster dose. c, CD154; g, IFN-γ; i, IL-2; t, TNF-α.
The CD4 T-cell responses were characterized by an effector memory T-cell phenotype.
T cells can be classified based on costimulatory (CD27), homing (CCR7), and activation (CD45RA and CD45RO) markers into naïve, effector memory, central memory, and terminally differentiated effector cells (3, 33, 43, 44, 50, 51, 70, 71, 87). A recent study (45) also showed that activation markers such as CD27 and CD43 define three memory populations of CD8 T cells (those with high-level CD27 expression [CD27hi] and low-level CD43 expression [CD43lo], CD27hi CD43hi cells, and CD27lo CD43lo cells) and that the quality of the recall response is predicted by phenotypes in the following order, starting with the phenotype corresponding to the best recall response: CD27hi CD43lo > CD27hi CD43hi > CD27lo CD43lo. CD43 was also found to play a key role in the differentiation of human CD4+ T cells into a TH1 pattern (65). CD127 (IL-7 receptor) is another marker that has also been used by numerous groups to distinguish CD8+ and CD4+ memory T cells in HIV-1 patients (18, 54, 67, 85).
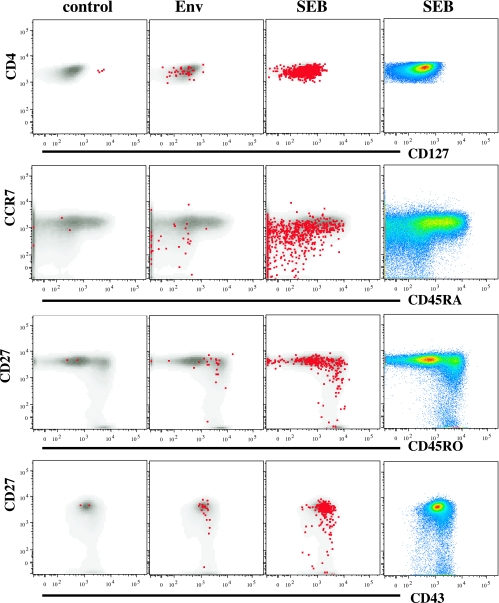
We characterized the memory cell phenotype markers on responder CD4 T cells from a subset of individuals from each group (a total of eight: three in group A, three in group B, and two in group C) at time points after DNA (day 98; n = 6) and glycoprotein boost (day 210; n = 8) vaccinations. For all groups, the CD4 T cells making IL-2 in response to Env-A or Env-B antigen on day 98 were predominately those with CD127hi CD45RA− CCR7− (effector memory), CD27+ CD45RO+ (memory), and intermediate-level-CD43-expression CD27hi phenotypes (Fig. 6). The patterns did not differ among groups, except that CD127lo IFN-γ+ CD4+ T cells were detectable only in group C following the DNA vaccinations (data not shown). The memory phenotype pattern did not change after the two glycoprotein boosts, despite an increase in the response magnitude (data not shown). Hence, the overall memory phenotype in the three groups (CD45RA− CD45RO+ CCR7− CD127+ CD27+) suggests that these CD4 T cells largely represent an effector memory phenotype, making both IFN-γ and IL-2 (42).
FIG. 6.
Surrogate markers of memory phenotypes on Env-specific CD4+ T cells. Data shown are representative of the phenotypic characterization of antigen-specific CD4 T cells after the second protein boost in a volunteer from group B (given a low dose i.m.). Surface markers for memory on Env-specific CD4+ T cells in select combinations (CD127, CCR7 versus CD45RA, CD27 versus CD45RO, and CD27 versus CD43) are shown. The production of IL-2 (red dots) over that of the base population (gray) in response to Env antigen and staphylococcal enterotoxin B (SEB), along with data for the unstimulated sample (control), is shown. Pseudocolor plots of the select combinations are shown in the far-right column.
DISCUSSION
To date, only two studies have reported the use of a DNA prime and protein boost strategy in humans. In the first study, a DNA vaccine encoding hepatitis B virus surface antigen given i.d., followed by a recombinant protein boost given i.m., elicited an antibody response only after the protein boost (74). In the second study, which investigated a malaria vaccine using a DNA and protein boost vaccine approach (80), antibody and T-cell responses were induced. A single function (IFN-γ production) measured by an enzyme-linked immunospot assay was used to assess the T-cell responses in that study. The present study simultaneously characterized several functions of the vaccine-induced T cells seen with a polyvalent HIV-1 DNA prime/glycoprotein boost vaccine strategy that was previously shown to induce cross-clade neutralizing antibody responses in humans (83). We demonstrated that this vaccine regimen induced predominantly Env-specific CD4 T-cell responses, irrespective of the dose and route of administration. However, the magnitudes and functional kinetics of the responses differed significantly among groups. CD4+ T-cell responses were very weak in the participants receiving the DNA vaccine i.d. but were detected in the majority of participants receiving the same DNA vaccine dose i.m. The high-dose DNA given i.m. was unique, inducing both CD4 T cells with greater function than those induced by the lower dose and Env-specific CD8 T cells. Similarly, Gag-specific CD4 T cells were reliably detected only in the high-dose DNA group.
The high proportion of IL-2-producing CD4 T cells following vaccination is in accordance with published data that protein-based vaccines induce IL-2-secreting cells whereas infections induce more IFN-γ-secreting cells (21, 22). The effector memory CD4 T cells generated in this study (CD127hi CD45RA− CCR7−) were multifunctional, with CD154-expressing CD4 T cells having the capacity to secrete IFN-γ and IL-2. This phenotype has been reported previously for long-term nonprogressors and aviremic patients on highly active antiretroviral therapy (42, 86). Cells with such a memory phenotype have proliferative potential and can be the source of the rapid generation and maturation of CD4 T cells with an effector function (42). Stubbe and colleagues (73) also demonstrated that hepatitis B vaccination results in the generation of a polyfunctional effector memory T-cell phenotype. Although polyfunctional CD4 T cells are better at effector functions than monofunctional cells and are needed for optimal and sustained protection, repeated vaccine administration may be necessary to maintain such responses when immunogens other than live and replicating vectors are used (20).
Antigen-specific CD8 T-cell responses were lower in frequency, seen only in the high-dose DNA group, and the T cells in these cases expressed IFN-γ or CD107, suggesting the possibility that CD8 T-cell responses were underestimated by using pools of 20-mers. We therefore tested an A2- and A11-restricted Env-B peptide (TK-10 peptide), TVYYGVPVWK (amino acids 4 to 13), in four subjects. The frequency of CD8 T cells producing IFN-γ in response to this 10-mer was similar to the frequency of such cells in response to the 20-mer peptide encompassing it (data not shown). This skewing toward a CD4 T-cell response is consistent with the findings of two recent studies in which DNA vaccines induced antigen-specific CD4 and CD8 T cells in about 90 and 40% of participants, respectively (13, 14).
We previously demonstrated that following the DNA vaccine priming, antibody responses were reliably induced with only the high-dose preparation (83). While numerous preclinical data have demonstrated a dose effect with DNA vaccines, there are limited clinical trial data on this issue. Part of the explanation for this lack of information is the historically poor immunogenicity of DNA vaccines when given alone to humans (23, 25, 57, 63). Nevertheless, in trials of DNA vaccines in which immune responses have been induced, a dose-dependent increase of T-cell responses has been shown in one study (79) but not in others (41, 55). Our finding that CD4 T cells generated by the higher DNA vaccination dose were greater in magnitude and function (i.e., the ability to secrete three cytokines and upregulate CD154) demonstrates that the dose is an important factor in determining the quality of immune responses. Despite the improved CD4 T-cell responses induced by the high-dose DNA, the subsequent administration of a recombinant gp120 protein did not boost this response, indicating either the ineffectiveness of the protein along with the QS-21 adjuvant to boost CD4 without subsequent priming or a null or even detrimental effect of repeated vaccinations with recombinant Env on T-cell memory. The former possibility may be explained by the observation that CD4 T cells have an inherently lower proliferation potential than CD8 T cells (30).
It is interesting that the i.m. route of DNA vaccination was more effective than the i.d. route in eliciting CD4 T cells. This was true even though the doses of the vaccine administered in the i.d. and i.m. groups (1.2 mg) were identical. The protein boost effect, however, tended to be greater in the i.d. than the i.m. groups, possibly due to the longer persistence of DNA given i.d. (62). Prior studies have compared the i.d. and i.m. vaccination routes in clinical trials, with disparate effects on the magnitudes of immune responses (4, 9, 24, 31, 36, 49, 53, 64, 69, 78). These trials have generally used particle-based vaccines, but one trial demonstrated that antibody responses in volunteers receiving a hepatitis B vaccine i.d. were diminished compared to those in subjects receiving the vaccine i.m. (76). However, in that trial, the i.d. dose was 1/10 of the i.m. dose. We are not aware of any previous study that has compared the i.d. and i.m. routes of delivery of DNA vaccines in humans. Our findings suggest that DNA vaccines and particle-based vaccines may behave differently depending on the route of administration and demonstrate that such information cannot simply be extrapolated from prior studies based on other forms of vaccines.
The identification of positive neutralizing antibody responses (titers of >1:20) against laboratory-adapted and pseudotyped viruses expressing Env from primary isolates from clades A, B, C, D, and E, as well as the gp120-specific immunoglobulin G titers (83), matched the onset and maturation of T-cell responses as measured in this study (data not shown). However, the magnitude of Env-specific CD4 T-cell responses as measured by any of the four functions did not significantly correlate with the magnitude of binding or neutralizing antibodies (data not shown). The induction of antibody responses is likely the result of a complex series of interactions, and our data indicate that they are impacted by factors in addition to CD4 T cells.
In summary, this polyvalent HIV-1 DNA prime/protein boost vaccine regimen induced different qualities of immune response following the DNA priming depending upon the dose and route of administration. This information may be useful for the future application of DNA vaccines to be used alone or in combination with other vaccines. The protein boosts resulted in CD4 T cells that were similar among the low-dose groups, consistent with the antibody responses observed. The responses induced by the DNA, particularly with the high dose, are encouraging and suggest that this vaccine formulation could be used as an effective prime vaccine for a recombinant vectored vaccine boost. This finding is particularly important information when one considers that the vaccine-induced T-cell responses observed in clinical trials to date have been significantly lower than those observed in natural infection or primate models of vaccine protection.
Acknowledgments
S.L. and S.W. declare a financial conflict of interest in the subject area of this study. The other authors (A.B., B.J., K.W., J.S.K., and P.A.G.) have no conflict of interest with the research described in this report.
This work was supported by HIV Vaccine Design and Development Teams contracts AI05394; AI049126, AI073103-01, and A1064060 (P.A.G.); and AI065250 (S.L.).
We appreciate the help provided by Marion Spell for flow cytometric acquisition and technical assistance from Betty Squyres. Thanks to S. Sabbaj and S. L. Heath for critiquing the manuscript.
Footnotes
Published ahead of print on 30 April 2008.
REFERENCES
- 1.Ackers, M. L., B. Parekh, T. G. Evans, P. Berman, S. Phillips, M. Allen, and J. S. McDougal. 2003. Human immunodeficiency virus (HIV) seropositivity among uninfected HIV vaccine recipients. J. Infect. Dis. 187879-886. [DOI] [PubMed] [Google Scholar]
- 2.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 29269-74. [DOI] [PubMed] [Google Scholar]
- 3.Appay, V., P. R. Dunbar, M. Callan, P. Klenerman, G. M. Gillespie, L. Papagno, G. S. Ogg, A. King, F. Lechner, C. A. Spina, S. Little, D. V. Havlir, D. D. Richman, N. Gruener, G. Pape, A. Waters, P. Easterbrook, M. Salio, V. Cerundolo, A. J. McMichael, and S. L. Rowland-Jones. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 8379-385. [DOI] [PubMed] [Google Scholar]
- 4.Arbizu, E. A., R. B. Marugan, J. Y. Grijalba, P. L. Serrano, L. G. Grande, and S. Del Campo Terron. 2003. Intramuscular versus intradermal administration of anti-hepatitis B vaccine in non-cirrhotic hepatitis C patients. Vaccine 212747-2750. [DOI] [PubMed] [Google Scholar]
- 5.Banatvala, J. E., and P. Van Damme. 2003. Hepatitis B vaccine—do we need boosters? J. Viral Hepat. 101-6. [DOI] [PubMed] [Google Scholar]
- 6.Banchereau, J., F. Bazan, D. Blanchard, F. Briere, J. P. Galizzi, C. van Kooten, Y. J. Liu, F. Rousset, and S. Saeland. 1994. The CD40 antigen and its ligand. Annu. Rev. Immunol. 12881-922. [DOI] [PubMed] [Google Scholar]
- 7.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415335-339. [DOI] [PubMed] [Google Scholar]
- 8.Barr, E., and G. Tamms. 2007. Quadrivalent human papillomavirus vaccine. Clin. Infect. Dis. 45609-617. [DOI] [PubMed] [Google Scholar]
- 9.Belshe, R. B., F. K. Newman, K. Wilkins, I. L. Graham, E. Babusis, M. Ewell, and S. E. Frey. 2007. Comparative immunogenicity of trivalent influenza vaccine administered by intradermal or intramuscular route in healthy adults. Vaccine 256755-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belshe, R. B., C. Stevens, G. J. Gorse, S. Buchbinder, K. Weinhold, H. Sheppard, D. Stablein, S. Self, J. McNamara, S. Frey, J. Flores, J. L. Excler, M. Klein, R. E. Habib, A. M. Duliege, C. Harro, L. Corey, M. Keefer, M. Mulligan, P. Wright, C. Celum, F. Judson, K. Mayer, D. McKirnan, M. Marmor, and G. Woody. 2001. Safety and immunogenicity of a canarypox-vectored human immunodeficiency virus type 1 vaccine with or without gp120: a phase 2 study in higher- and lower-risk volunteers. J. Infect. Dis. 1831343-1352. [DOI] [PubMed] [Google Scholar]
- 11.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 1074781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanchard, D., C. Gaillard, P. Hermann, and J. Banchereau. 1994. Role of CD40 antigen and interleukin-2 in T cell-dependent human B lymphocyte growth. Eur. J. Immunol. 24330-335. [DOI] [PubMed] [Google Scholar]
- 13.Catanzaro, A. T., R. A. Koup, M. Roederer, R. T. Bailer, M. E. Enama, Z. Moodie, L. Gu, J. E. Martin, L. Novik, B. K. Chakrabarti, B. T. Butman, J. G. Gall, C. R. King, C. A. Andrews, R. Sheets, P. L. Gomez, J. R. Mascola, G. J. Nabel, and B. S. Graham. 2006. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 candidate vaccine delivered by a replication-defective recombinant adenovirus vector. J. Infect. Dis. 1941638-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catanzaro, A. T., M. Roederer, R. A. Koup, R. T. Bailer, M. E. Enama, M. C. Nason, J. E. Martin, S. Rucker, C. A. Andrews, P. L. Gomez, J. R. Mascola, G. J. Nabel, and B. S. Graham. 2007. Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine 254085-4092. [DOI] [PubMed] [Google Scholar]
- 15.Chattopadhyay, P. K., J. Yu, and M. Roederer. 2005. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat. Med. 111113-1117. [DOI] [PubMed] [Google Scholar]
- 16.Cohen, J. 2007. AIDS research. Did Merck's failed HIV vaccine cause harm? Science 3181048-1049. [DOI] [PubMed] [Google Scholar]
- 17.Cohen, J. 2007. AIDS research. Promising AIDS vaccine's failure leaves field reeling. Science 31828-29. [DOI] [PubMed] [Google Scholar]
- 18.Colle, J. H., J. L. Moreau, A. Fontanet, O. Lambotte, M. Joussemet, J. F. Delfraissy, and J. Theze. 2006. CD127 expression and regulation are altered in the memory CD8 T cells of HIV-infected patients: reversal by highly active anti-retroviral therapy (HAART). Clin. Exp. Immunol. 143398-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cristillo, A. D., S. Wang, M. S. Caskey, T. Unangst, L. Hocker, L. He, L. Hudacik, S. Whitney, T. Keen, T. H. Chou, S. Shen, S. Joshi, V. S. Kalyanaraman, B. Nair, P. Markham, S. Lu, and R. Pal. 2006. Preclinical evaluation of cellular immune responses elicited by a polyvalent DNA prime/protein boost HIV-1 vaccine. Virology 346151-168. [DOI] [PubMed] [Google Scholar]
- 20.Darrah, P. A., D. T. Patel, P. M. De Luca, R. W. Lindsay, D. F. Davey, B. J. Flynn, S. T. Hoff, P. Andersen, S. G. Reed, S. L. Morris, M. Roederer, and R. A. Seder. 2007. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 13843-850. [DOI] [PubMed] [Google Scholar]
- 21.De Rosa, S. C., F. X. Lu, J. Yu, S. P. Perfetto, J. Falloon, S. Moser, T. G. Evans, R. Koup, C. J. Miller, and M. Roederer. 2004. Vaccination in humans generates broad T cell cytokine responses. J. Immunol. 1735372-5380. [DOI] [PubMed] [Google Scholar]
- 22.Divekar, A. A., D. M. Zaiss, F. E. Lee, D. Liu, D. J. Topham, A. J. Sijts, and T. R. Mosmann. 2006. Protein vaccines induce uncommitted IL-2-secreting human and mouse CD4 T cells, whereas infections induce more IFN- gamma-secreting cells. J. Immunol. 1761465-1473. [DOI] [PubMed] [Google Scholar]
- 23.Dunachie, S. J., M. Walther, J. E. Epstein, S. Keating, T. Berthoud, L. Andrews, R. F. Andersen, P. Bejon, N. Goonetilleke, I. Poulton, D. P. Webster, G. Butcher, K. Watkins, R. E. Sinden, G. L. Levine, T. L. Richie, J. Schneider, D. Kaslow, S. C. Gilbert, D. J. Carucci, and A. V. Hill. 2006. A DNA prime-modified vaccinia virus Ankara boost vaccine encoding thrombospondin-related adhesion protein but not circumsporozoite protein partially protects healthy malaria-naive adults against Plasmodium falciparum sporozoite challenge. Infect. Immun. 745933-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egemen, A., S. Aksit, Z. Kurugol, S. Erensoy, A. Bilgic, and M. Akilli. 1998. Low-dose intradermal versus intramuscular administration of recombinant hepatitis B vaccine: a comparison of immunogenicity in infants and preschool children. Vaccine 161511-1515. [DOI] [PubMed] [Google Scholar]
- 25.Epstein, J. E., E. J. Gorak, Y. Charoenvit, R. Wang, N. Freydberg, O. Osinowo, T. L. Richie, E. L. Stoltz, F. Trespalacios, J. Nerges, J. Ng, V. Fallarme-Majam, E. Abot, L. Goh, S. Parker, S. Kumar, R. C. Hedstrom, J. Norman, R. Stout, and S. L. Hoffman. 2002. Safety, tolerability, and lack of antibody responses after administration of a PfCSP DNA malaria vaccine via needle or needle-free jet injection, and comparison of intramuscular and combination intramuscular/intradermal routes. Hum. Gene. Ther. 131551-1560. [DOI] [PubMed] [Google Scholar]
- 26.Evans, T. G., M. C. Keefer, K. J. Weinhold, M. Wolff, D. Montefiori, G. J. Gorse, B. S. Graham, M. J. McElrath, M. L. Clements-Mann, M. J. Mulligan, P. Fast, M. C. Walker, J. L. Excler, A. M. Duliege, and J. Tartaglia. 1999. A canarypox vaccine expressing multiple human immunodeficiency virus type 1 genes given alone or with rgp120 elicits broad and durable CD8+ cytotoxic T lymphocyte responses in seronegative volunteers. J. Infect. Dis. 180290-298. [DOI] [PubMed] [Google Scholar]
- 27.Fischer, M. A., D. C. Tscharke, K. B. Donohue, M. E. Truckenmiller, and C. C. Norbury. 2007. Reduction of vector gene expression increases foreign antigen-specific CD8+ T-cell priming. J. Gen. Virol. 882378-2386. [DOI] [PubMed] [Google Scholar]
- 28.Flynn, N. M., D. N. Forthal, C. D. Harro, F. N. Judson, K. H. Mayer, and M. F. Para. 2005. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 191654-665. [DOI] [PubMed] [Google Scholar]
- 29.Follenzi, A., L. Santambrogio, and A. Annoni. 2007. Immune responses to lentiviral vectors. Curr. Gene Ther. 7306-315. [DOI] [PubMed] [Google Scholar]
- 30.Foulds, K. E., L. A. Zenewicz, D. J. Shedlock, J. Jiang, A. E. Troy, and H. Shen. 2002. Cutting edge: CD4 and CD8 T cells are intrinsically different in their proliferative responses. J. Immunol. 1681528-1532. [DOI] [PubMed] [Google Scholar]
- 31.Frazer, I. H., B. Jones, M. Dimitrakakis, and I. R. Mackay. 1987. Intramuscular versus low-dose intradermal hepatitis B vaccine. Assessment by humoral and cellular immune response to hepatitis B surface antigen. Med. J. Aust. 146242-245. [PubMed] [Google Scholar]
- 32.Frentsch, M., O. Arbach, D. Kirchhoff, B. Moewes, M. Worm, M. Rothe, A. Scheffold, and A. Thiel. 2005. Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nat. Med. 111118-1124. [DOI] [PubMed] [Google Scholar]
- 33.Fritsch, R. D., X. Shen, G. P. Sims, K. S. Hathcock, R. J. Hodes, and P. E. Lipsky. 2005. Stepwise differentiation of CD4 memory T cells defined by expression of CCR7 and CD27. J. Immunol. 1756489-6497. [DOI] [PubMed] [Google Scholar]
- 34.Gilbert, P. B., M. L. Peterson, D. Follmann, M. G. Hudgens, D. P. Francis, M. Gurwith, W. L. Heyward, D. V. Jobes, V. Popovic, S. G. Self, F. Sinangil, D. Burke, and P. W. Berman. 2005. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J. Infect. Dis. 191666-677. [DOI] [PubMed] [Google Scholar]
- 35.Goepfert, P. A., G. D. Tomaras, H. Horton, D. Montefiori, G. Ferrari, M. Deers, G. Voss, M. Koutsoukos, L. Pedneault, P. Vandepapeliere, M. J. McElrath, P. Spearman, J. D. Fuchs, B. A. Koblin, W. A. Blattner, S. Frey, L. R. Baden, C. Harro, and T. Evans. 2007. Durable HIV-1 antibody and T-cell responses elicited by an adjuvanted multi-protein recombinant vaccine in uninfected human volunteers. Vaccine 25510-518. [DOI] [PubMed] [Google Scholar]
- 36.Gomber, S., R. Sharma, and V. G. Ramachandran. 2004. Immunogenicity of low dose intradermal hepatitis B vaccine and its comparison with standard dose intramuscular vaccination. Indian Pediatr. 41922-926. [PubMed] [Google Scholar]
- 37.Goonetilleke, N., S. Moore, L. Dally, N. Winstone, I. Cebere, A. Mahmoud, S. Pinheiro, G. Gillespie, D. Brown, V. Loach, J. Roberts, A. Guimaraes-Walker, P. Hayes, K. Loughran, C. Smith, J. De Bont, C. Verlinde, D. Vooijs, C. Schmidt, M. Boaz, J. Gilmour, P. Fast, L. Dorrell, T. Hanke, and A. J. McMichael. 2006. Induction of multifunctional human immunodeficiency virus type 1 (HIV-1)-specific T cells capable of proliferation in healthy subjects by using a prime-boost regimen of DNA- and modified vaccinia virus Ankara-vectored vaccines expressing HIV-1 Gag coupled to CD8+ T-cell epitopes. J. Virol. 804717-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorse, G. J., L. Corey, G. B. Patel, M. Mandava, R. H. Hsieh, T. J. Matthews, M. C. Walker, M. J. McElrath, P. W. Berman, M. M. Eibl, and R. B. Belshe for the National Institute of Allergy and Infectious Disease Aids Vaccine Evaluation Group. 1999. HIV-1MN recombinant glycoprotein 160 vaccine-induced cellular and humoral immunity boosted by HIV-1MN recombinant glycoprotein 120 vaccine. AIDS Res. Hum. Retrovir. 15115-132. [DOI] [PubMed] [Google Scholar]
- 39.Gorse, G. J., G. B. Patel, and R. B. Belshe. 2001. HIV type 1 vaccine-induced T cell memory and cytotoxic T lymphocyte responses in HIV type 1-uninfected volunteers. AIDS Res. Hum. Retrovir. 171175-1189. [DOI] [PubMed] [Google Scholar]
- 40.Gorse, G. J., G. B. Patel, M. Mandava, P. W. Berman, and R. B. Belshe for the National Institute of Allergy and Infectious Disease Aids Vaccine Evaluation Group. 1999. MN and IIIB recombinant glycoprotein 120 vaccine-induced binding antibodies to native envelope glycoprotein of human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retrovir. 15921-930. [DOI] [PubMed] [Google Scholar]
- 41.Graham, B. S., R. A. Koup, M. Roederer, R. T. Bailer, M. E. Enama, Z. Moodie, J. E. Martin, M. M. McCluskey, B. K. Chakrabarti, L. Lamoreaux, C. A. Andrews, P. L. Gomez, J. R. Mascola, and G. J. Nabel. 2006. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 DNA candidate vaccine. J. Infect. Dis. 1941650-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harari, A., V. Dutoit, C. Cellerai, P. A. Bart, R. A. Du Pasquier, and G. Pantaleo. 2006. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunol. Rev. 211236-254. [DOI] [PubMed] [Google Scholar]
- 43.Harari, A., F. Vallelian, and G. Pantaleo. 2004. Phenotypic heterogeneity of antigen-specific CD4 T cells under different conditions of antigen persistence and antigen load. Eur. J. Immunol. 343525-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hendriks, J., L. A. Gravestein, K. Tesselaar, R. A. van Lier, T. N. Schumacher, and J. Borst. 2000. CD27 is required for generation and long-term maintenance of T cell immunity. Nat. Immunol. 1433-440. [DOI] [PubMed] [Google Scholar]
- 45.Hikono, H., J. E. Kohlmeier, S. Takamura, S. T. Wittmer, A. D. Roberts, and D. L. Woodland. 2007. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J. Exp. Med. 2041625-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horton, H., T. U. Vogel, D. K. Carter, K. Vielhuber, D. H. Fuller, T. Shipley, J. T. Fuller, K. J. Kunstman, G. Sutter, D. C. Montefiori, V. Erfle, R. C. Desrosiers, N. Wilson, L. J. Picker, S. M. Wolinsky, C. Wang, D. B. Allison, and D. I. Watkins. 2002. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J. Virol. 767187-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim, J. H., P. Pitisuttithum, C. Kamboonruang, T. Chuenchitra, J. Mascola, S. S. Frankel, M. S. DeSouza, V. Polonis, R. McLinden, A. Sambor, A. E. Brown, B. Phonrat, K. Rungruengthanakit, A. M. Duliege, M. L. Robb, J. McNeil, and D. L. Birx. 2003. Specific antibody responses to vaccination with bivalent CM235/SF2 gp120: detection of homologous and heterologous neutralizing antibody to subtype E (CRF01.AE) HIV type 1. AIDS Res. Hum. Retrovir. 19807-816. [DOI] [PubMed] [Google Scholar]
- 48.Klinman, D. M. 2006. Adjuvant activity of CpG oligodeoxynucleotides. Int. Rev. Immunol. 25135-154. [DOI] [PubMed] [Google Scholar]
- 49.Lankarani, K. B., A. R. Taghavi, S. Agah, and A. Karimi. 2001. Comparison of intradermal and intramuscular administration of hepatitis B vaccine in neonates. Indian J. Gastroenterol. 2094-96. [PubMed] [Google Scholar]
- 50.Lanzavecchia, A., and F. Sallusto. 2002. Progressive differentiation and selection of the fittest in the immune response. Nat. Rev. Immunol. 2982-987. [DOI] [PubMed] [Google Scholar]
- 51.Lanzavecchia, A., and F. Sallusto. 2005. Understanding the generation and function of memory T cell subsets. Curr. Opin. Immunol. 17326-332. [DOI] [PubMed] [Google Scholar]
- 52.Lee, S. A., R. Orque, P. A. Escarpe, M. L. Peterson, J. W. Good, E. M. Zaharias, P. W. Berman, H. W. Sheppard, and R. Shibata. 2001. Vaccine-induced antibodies to the native, oligomeric envelope glycoproteins of primary HIV-1 isolates. Vaccine 20563-576. [DOI] [PubMed] [Google Scholar]
- 53.Levitz, R. E., B. W. Cooper, and H. C. Regan. 1995. Immunization with high-dose intradermal recombinant hepatitis B vaccine in healthcare workers who failed to respond to intramuscular vaccination. Infect. Control Hosp. Epidemiol. 1688-91. [DOI] [PubMed] [Google Scholar]
- 54.MacPherson, P. A., C. Fex, J. Sanchez-Dardon, N. Hawley-Foss, and J. B. Angel. 2001. Interleukin-7 receptor expression on CD8+ T cells is reduced in HIV infection and partially restored with effective antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 28454-457. [DOI] [PubMed] [Google Scholar]
- 55.Martin, J. E., N. J. Sullivan, M. E. Enama, I. J. Gordon, M. Roederer, R. A. Koup, R. T. Bailer, B. K. Chakrabarti, M. A. Bailey, P. L. Gomez, C. A. Andrews, Z. Moodie, L. Gu, J. A. Stein, G. J. Nabel, and B. S. Graham. 2006. A DNA vaccine for Ebola virus is safe and immunogenic in a phase I clinical trial. Clin. Vaccine Immunol. 131267-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montefiori, D. C., B. Metch, M. J. McElrath, S. Self, K. J. Weinhold, and L. Corey. 2004. Demographic factors that influence the neutralizing antibody response in recipients of recombinant HIV-1 gp120 vaccines. J. Infect. Dis. 1901962-1969. [DOI] [PubMed] [Google Scholar]
- 57.Mulligan, M. J., N. D. Russell, C. Celum, J. Kahn, E. Noonan, D. C. Montefiori, G. Ferrari, K. J. Weinhold, J. M. Smith, R. R. Amara, and H. L. Robinson. 2006. Excellent safety and tolerability of the human immunodeficiency virus type 1 pGA2/JS2 plasmid DNA priming vector vaccine in HIV type 1 uninfected adults. AIDS Res. Hum. Retrovir. 22678-683. [DOI] [PubMed] [Google Scholar]
- 58.Mwau, M., I. Cebere, J. Sutton, P. Chikoti, N. Winstone, E. G. Wee, T. Beattie, Y. H. Chen, L. Dorrell, H. McShane, C. Schmidt, M. Brooks, S. Patel, J. Roberts, C. Conlon, S. L. Rowland-Jones, J. J. Bwayo, A. J. McMichael, and T. Hanke. 2004. A human immunodeficiency virus 1 (HIV-1) clade A vaccine in clinical trials: stimulation of HIV-specific T-cell responses by DNA and recombinant modified vaccinia virus Ankara (MVA) vaccines in humans. J. Gen. Virol. 85911-919. [DOI] [PubMed] [Google Scholar]
- 59.Nitayaphan, S., C. Khamboonruang, N. Sirisophana, P. Morgan, J. Chiu, A. M. Duliege, C. Chuenchitra, T. Supapongse, K. Rungruengthanakit, M. deSouza, J. R. Mascola, K. Boggio, S. Ratto-Kim, L. E. Markowitz, D. Birx, V. Suriyanon, J. G. McNeil, A. E. Brown, and R. A. Michael for the AFRIMS-RIHES Vaccine Evaluation Group. 2000. A phase I/II trial of HIV SF2 gp120/MF59 vaccine in seronegative Thais. Vaccine 181448-1455. [DOI] [PubMed] [Google Scholar]
- 60.Pal, R., V. S. Kalyanaraman, B. C. Nair, S. Whitney, T. Keen, L. Hocker, L. Hudacik, N. Rose, I. Mboudjeka, S. Shen, T. H. Wu-Chou, D. Montefiori, J. Mascola, P. Markham, and S. Lu. 2006. Immunization of rhesus macaques with a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 vaccine elicits protective antibody response against simian human immunodeficiency virus of R5 phenotype. Virology 348341-353. [DOI] [PubMed] [Google Scholar]
- 61.Pal, R., S. Wang, V. S. Kalyanaraman, B. C. Nair, S. Whitney, T. Keen, L. Hocker, L. Hudacik, N. Rose, A. Cristillo, I. Mboudjeka, S. Shen, T. H. Wu-Chou, D. Montefiori, J. Mascola, S. Lu, and P. Markham. 2005. Polyvalent DNA prime and envelope protein boost HIV-1 vaccine elicits humoral and cellular responses and controls plasma viremia in rhesus macaques following rectal challenge with an R5 SHIV isolate. J. Med. Primatol. 34226-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pal, R., Q. Yu, S. Wang, V. S. Kalyanaraman, B. C. Nair, L. Hudacik, S. Whitney, T. Keen, C. L. Hung, L. Hocker, J. S. Kennedy, P. Markham, and S. Lu. 2006. Definitive toxicology and biodistribution study of a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 (HIV-1) vaccine in rabbits. Vaccine 241225-1234. [DOI] [PubMed] [Google Scholar]
- 63.Peters, B. S., W. Jaoko, E. Vardas, G. Panayotakopoulos, P. Fast, C. Schmidt, J. Gilmour, M. Bogoshi, G. Omosa-Manyonyi, L. Dally, L. Klavinskis, B. Farah, T. Tarragona, P. A. Bart, A. Robinson, C. Pieterse, W. Stevens, R. Thomas, B. Barin, A. J. McMichael, J. A. McIntyre, G. Pantaleo, T. Hanke, and J. Bwayo. 2007. Studies of a prophylactic HIV-1 vaccine candidate based on modified vaccinia virus Ankara (MVA) with and without DNA priming: effects of dosage and route on safety and immunogenicity. Vaccine 252120-2127. [DOI] [PubMed] [Google Scholar]
- 64.Playford, E. G., P. G. Hogan, A. S. Bansal, K. Harrison, D. Drummond, D. F. Looke, and M. Whitby. 2002. Intradermal recombinant hepatitis B vaccine for healthcare workers who fail to respond to intramuscular vaccine. Infect. Control Hosp. Epidemiol. 2387-90. [DOI] [PubMed] [Google Scholar]
- 65.Ramirez-Pliego, O., D. L. Escobar-Zarate, G. M. Rivera-Martinez, M. G. Cervantes-Badillo, F. R. Esquivel-Guadarrama, G. Rosas-Salgado, Y. Rosenstein, and M. A. Santana. 2007. CD43 signals induce type one lineage commitment of human CD4+ T cells. BMC Immunol. 830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russell, N. D., B. S. Graham, M. C. Keefer, M. J. McElrath, S. G. Self, K. J. Weinhold, D. C. Montefiori, G. Ferrari, H. Horton, G. D. Tomaras, S. Gurunathan, L. Baglyos, S. E. Frey, M. J. Mulligan, C. D. Harro, S. P. Buchbinder, L. R. Baden, W. A. Blattner, B. A. Koblin, and L. Corey. 2007. Phase 2 study of an HIV-1 canarypox vaccine (vCP1452) alone and in combination with rgp120: negative results fail to trigger a phase 3 correlates trial. J. Acquir. Immune Defic. Syndr. 44203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sabbaj, S., S. L. Heath, A. Bansal, S. Vohra, J. M. Kilby, A. J. Zajac, and P. A. Goepfert. 2007. Functionally competent antigen-specific CD127hi memory CD8+ T cells are preserved only in HIV-infected individuals receiving early treatment. J. Infect. Dis. 195108-117. [DOI] [PubMed] [Google Scholar]
- 68.Sabbaj, S., M. J. Mulligan, R. H. Hsieh, R. B. Belshe, J. R. McGhee, et al. 2000. Cytokine profiles in seronegative volunteers immunized with a recombinant canarypox and gp120 prime-boost HIV-1 vaccine. AIDS 141365-1374. [DOI] [PubMed] [Google Scholar]
- 69.Sabchareon, A., P. Chantavanich, S. Pasuralertsakul, C. Pojjaroen-Anant, V. Prarinyanupharb, P. Attanath, V. Singhasivanon, W. Buppodom, and J. Lang. 1998. Persistence of antibodies in children after intradermal or intramuscular administration of preexposure primary and booster immunizations with purified Vero cell rabies vaccine. Pediatr. Infect. Dis. J. 171001-1007. [DOI] [PubMed] [Google Scholar]
- 70.Sallusto, F., J. Geginat, and A. Lanzavecchia. 2004. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 22745-763. [DOI] [PubMed] [Google Scholar]
- 71.Sallusto, F., D. Lenig, R. Forster, M. Lipp, and A. Lanzavecchia. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401708-712. [DOI] [PubMed] [Google Scholar]
- 72.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415331-335. [DOI] [PubMed] [Google Scholar]
- 73.Stubbe, M., N. Vanderheyde, M. Goldman, and A. Marchant. 2006. Antigen-specific central memory CD4+ T lymphocytes produce multiple cytokines and proliferate in vivo in humans. J. Immunol. 1778185-8190. [DOI] [PubMed] [Google Scholar]
- 74.Tacket, C. O., M. J. Roy, G. Widera, W. F. Swain, S. Broome, and R. Edelman. 1999. Phase 1 safety and immune response studies of a DNA vaccine encoding hepatitis B surface antigen delivered by a gene delivery device. Vaccine 172826-2829. [DOI] [PubMed] [Google Scholar]
- 75.Tavel, J. A., J. E. Martin, G. G. Kelly, M. E. Enama, J. M. Shen, P. L. Gomez, C. A. Andrews, R. A. Koup, R. T. Bailer, J. A. Stein, M. Roederer, G. J. Nabel, and B. S. Graham. 2007. Safety and immunogenicity of a Gag-Pol candidate HIV-1 DNA vaccine administered by a needle-free device in HIV-1-seronegative subjects. J. Acquir. Immune Defic. Syndr. 44601-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turchi, M. D., C. M. Martelli, M. L. Ferraz, A. E. Silva, D. das Dores de Paula Cardoso, P. Martelli, and L. J. Oliveira. 1997. Immunogenicity of low-dose intramuscular and intradermal vaccination with recombinant hepatitis B vaccine. Rev. Inst. Med. Trop. Sao Paulo 3915-19. [DOI] [PubMed] [Google Scholar]
- 77.van Kooten, C., and J. Banchereau. 2000. CD40-CD40 ligand. J. Leukoc. Biol. 672-17. [DOI] [PubMed] [Google Scholar]
- 78.Vlassopoulos, D. A., D. K. Arvanitis, D. S. Lilis, K. I. Louizou, and V. E. Hadjiconstantinou. 1999. Lower long-term efficiency of intradermal hepatitis B vaccine compared to the intramuscular route in hemodialysis patients. Int. J. Artif. Organs 22739-743. [PubMed] [Google Scholar]
- 79.Wang, R., D. L. Doolan, T. P. Le, R. C. Hedstrom, K. M. Coonan, Y. Charoenvit, T. R. Jones, P. Hobart, M. Margalith, J. Ng, W. R. Weiss, M. Sedegah, C. de Taisne, J. A. Norman, and S. L. Hoffman. 1998. Induction of antigen-specific cytotoxic T lymphocytes in humans by a malaria DNA vaccine. Science 282476-480. [DOI] [PubMed] [Google Scholar]
- 80.Wang, R., J. Epstein, Y. Charoenvit, F. M. Baraceros, N. Rahardjo, T. Gay, J. G. Banania, R. Chattopadhyay, P. de la Vega, T. L. Richie, N. Tornieporth, D. L. Doolan, K. E. Kester, D. G. Heppner, J. Norman, D. J. Carucci, J. D. Cohen, and S. L. Hoffman. 2004. Induction in humans of CD8+ and CD4+ T cell and antibody responses by sequential immunization with malaria DNA and recombinant protein. J. Immunol. 1725561-5569. [DOI] [PubMed] [Google Scholar]
- 81.Wang, S., J. Arthos, J. M. Lawrence, D. Van Ryk, I. Mboudjeka, S. Shen, T. H. Chou, D. C. Montefiori, and S. Lu. 2005. Enhanced immunogenicity of gp120 protein when combined with recombinant DNA priming to generate antibodies that neutralize the JR-FL primary isolate of human immunodeficiency virus type 1. J. Virol. 797933-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang, S., D. J. Farfan-Arribas, S. Shen, T. H. Chou, A. Hirsch, F. He, and S. Lu. 2006. Relative contributions of codon usage, promoter efficiency and leader sequence to the antigen expression and immunogenicity of HIV-1 Env DNA vaccine. Vaccine 244531-4540. [DOI] [PubMed] [Google Scholar]
- 83.Wang, S., J. S. Kennedy, K. West, D. C. Montefiori, S. Coley, J. Lawrence, S. Shen, S. Green, A. L. Rothman, F. A. Ennis, J. Arthos, R. Pal, P. Markham, and S. Lu. 2008. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine 261098-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang, S., R. Pal, J. R. Mascola, T. H. Chou, I. Mboudjeka, S. Shen, Q. Liu, S. Whitney, T. Keen, B. C. Nair, V. S. Kalyanaraman, P. Markham, and S. Lu. 2006. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology 35034-47. [DOI] [PubMed] [Google Scholar]
- 85.Wherry, E. J., C. L. Day, R. Draenert, J. D. Miller, P. Kiepiela, T. Woodberry, C. Brander, M. Addo, P. Klenerman, R. Ahmed, and B. D. Walker. 2006. HIV-specific CD8 T cells express low levels of IL-7Rα: implications for HIV-specific T cell memory. Virology 353366-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Younes, S. A., B. Yassine-Diab, A. R. Dumont, M. R. Boulassel, Z. Grossman, J. P. Routy, and R. P. Sekaly. 2003. HIV-1 viremia prevents the establishment of interleukin 2-producing HIV-specific memory CD4+ T cells endowed with proliferative capacity. J. Exp. Med. 1981909-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yue, F. Y., C. M. Kovacs, R. C. Dimayuga, P. Parks, and M. A. Ostrowski. 2004. HIV-1-specific memory CD4+ T cells are phenotypically less mature than cytomegalovirus-specific memory CD4+ T cells. J. Immunol. 1722476-2486. [DOI] [PubMed] [Google Scholar]