Abstract
Infant acute lymphoblastic leukemia (ALL) with MLL gene rearrangements is characterized by early pre-B phenotype (CD10−/CD19+) and poor treatment outcome. The t(4;11), creating MLL-AF4 chimeric transcripts, is the predominant 11q23 chromosome translocation in infant ALL and is associated with extremely poor prognosis as compared with other 11q23 translocations. We analyzed an infant early preB ALL with ins(5;11)(q31;q13q23) and identified the AF5q31 gene on chromosome 5q31 as a fusion partner of the MLL gene. The AF5q31 gene, which encoded a protein of 1,163 aa, was located in the vicinity of the cytokine cluster region of chromosome 5q31 and contained at least 16 exons. The AF5q31 gene was expressed in fetal heart, lung, and brain at relatively high levels and fetal liver at a low level, but the expression in these tissues decreased in adults. The AF5q31 protein was homologous to AF4-related proteins, including AF4, LAF4, and FMR2. The AF5q31 and AF4 proteins had three homologous regions, including the transactivation domain of AF4, and the breakpoint of AF5q31 was located within the region homologous to the transactivation domain of AF4. Furthermore, the clinical features of this patient with the MLL-AF5q31 fusion transcript, characterized by the early pre-B phenotype (CD10−/CD19+) and poor outcome, were similar to those of patients having MLL-AF4 chimeric transcripts. These findings suggest that AF5q31 and AF4 might define a new family particularly involved in the pathogenesis of 11q23-associated-ALL.
The MLL gene (also called ALL-1, HRX, and HTRX-1) has been identified in 11q23 translocations (1–4) and is considered to be a transcriptional maintenance factor (5). At least 30 chromosomal regions for partners of 11q23 have been observed, such as t(4;11), t(9;11), and t(11;19) (6). MLL gene rearrangement, which is found in the majority of infant (7, 8) and therapy-related leukemias (9, 10) is strongly associated with poor outcome in infants with acute lymphoblastic leukemia (ALL) compared with older children and adults with ALL, or with acute myeloid leukemia (AML). Of these, t(4;11)(q21;q23) creating a MLL-AF4 fusion transcript was reported to be the most important prognostic factor in infant ALL (11). Currently almost 20 partner genes for MLL have been cloned from leukemia cells with various types of reciprocal 11q23 translocations (12). The functions of some genes have been revealed, including those for a Ras-binding protein (AF6) (13), an RNA polymerase II elongation factor (ELL/MEN) (14), transcriptional coactivator/histone acetyltransferase (CBP and p300) (15–17), and ABL and eps8-binding protein (ABI-1) (18). It has been shown both by “knock-in” mice and by retroviral transformation of murine bone marrow that MLL and the partner gene both are required for leukemogenesis (19, 20).
Hitherto, only a few genes have been identified as fusion partners of the MLL gene from ALL with 11q23 translocations (12). In the present study, we analyzed an infant ALL with ins(5;11)(q31;q13q23) and identified a novel AF4-related gene as a fusion partner of the MLL gene.
Materials and Methods
Patient.
A 4-month-old girl was diagnosed as having ALL. Leukemic cells expressed CD15, CD19, and HLA-DR but not CD10, and were cytogenetically characterized as ins(5;11)(q31;q13q23), i(17q). She achieved a complete remission by intensive chemotherapy, but relapsed three times and died 20 months after diagnosis.
Southern Blot Analysis.
High molecular weight DNA was extracted from bone marrow cells from the patient by proteinase K digestion and phenol/chloroform extraction. Ten micrograms of DNA was digested with appropriate restriction enzymes, subjected to electrophoresis on 0.8% agarose gels, transferred to charged nylon filters (Amersham Pharmacia), and hybridized to DNA probes labeled by the random hexamer method (15). A 0.9-kb BamHI fragment (designated probe x) derived from MLL cDNA was used as a probe (21).
Preparation of mRNA and cDNA Libraries.
Poly(A)+ RNA from frozen cells was extracted with a Fast Track mRNA Isolation Kit (Invitrogen). A cDNA library was constructed with poly(A)+ mRNA from patient cells and the BALM14 cell line following established procedures (18). Briefly, random hexanucleotide-primed synthesized cDNAs were ligated with EcoRI adaptors and cloned into the EcoRI-digested λgt10 cloning vector (Promega). After packaging with commercial packaging kits (Epicentre Technologies, Madison, WI), phage plaques were screened with probes labeled by using a random primer synthesis kit (Stratagene). The probe x was used for screening the patient cDNA library, and the 114-bp AF5q31 cDNA probe derived from the MLL-AF5q31 chimeric clone was used for screening the BALM14 and human placenta cDNA libraries.
Reverse Transcription–PCR and Genomic PCR.
Total cellular RNA was extracted from bone marrow cells of the patient by the acid guanidine isothiocyanate-phenol-chloroform method (22). Four micrograms of total RNA was reverse transcribed to cDNA in a total volume of 20 μl with random hexamers and 20 units of reverse transcriptase (avian myeloblastosis virus) (Boehringer Mannheim). One-twentieth of the cDNA was amplified by PCR in a total volume of 100 μl with 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris⋅HCl (pH 9.0 at room temperature), 25 pmol of each primer, 75 μM of each dNTP, and 1 unit of Taq polymerase (Applied Biosystems). After 35 rounds of PCR (30 sec at 94°C, 30 sec at 55°C, and 1 min at 72°C), 5 μl of PCR product was electrophoresed in a 3% agarose gel. The primers used were as follows: MLL-7S, 5′-TCCTCAGCACTCTCTCCAAT-3′; MLL-11A, 5′-TTTGCCTGGAGTTGTGGATC-3′; 5–1A, 5′-CCATCACTGTCTTCACTGCT-3′; and 5–13S, 5′-ACACCATGCAAAACAGAACCT-3′.
One hundred nanograms of genomic DNA derived from the patient’s leukemic cells was amplified under the same condition as reverse transcription–PCR. The primers used were MLL-7S and 5–21A, 5′-TGCCAAATCTAAATGACCTGG-3′.
Nucleotide Sequencing.
PCR products were cloned into the TA cloning vector (Invitrogen). Nucleotide sequences of phage clones and PCR products were determined by the fluorometric method (18) (Dye Terminator Cycle Sequencing Kit, Applied Biosystems).
Northern Blot Analysis.
Multiple human tissue Northern blots (CLONTECH) were hybridized with 32P-labeled 0.6-kb AF5q31 cDNA probe, which covered nucleotides 1,135 to 1,735, and 0.6-kb AF4 cDNA probe, which covered nucleotides 476 to 1,117 (GenBank accession number L13773), respectively.
Fluorescence in Situ Hybridization (FISH) Analysis.
Chromosomal mapping of the genomic clone λH17–6 was performed by the FISH method (23). The phage clone was labeled by the standard nick-translation method using biotin-16-dUTP (Boehringer Mannheim). Chromosomal in situ suppression hybridization, washing, and signal detection procedures were performed as described (23). Preparations were analyzed under a conventional fluorescence microscope (BX40-RF; Olympus, Tokyo), and images were captured with a charge-coupled device camera (SenSys0400-G1, Photometrics, Tucson, AZ). Each chromosomal band was identified based on 4′,6-diaminido-2-phenylindole dihydrochloride (DAPI) staining properties.
Results
Isolation of the MLL Fusion cDNAs in ins(5;11)(q31;q13q23).
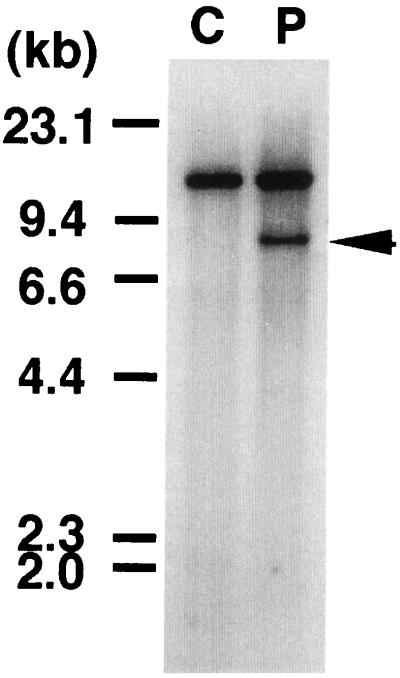
Southern blot analysis of DNA prepared from the leukemic cells of the patient using probe x revealed a chromosomal breakpoint within the breakpoint cluster region of the MLL gene at 11q23 (Fig. 1), which spanned exons 8–14 in the MLL gene (nomenclature according to ref. 24). To isolate fusion transcripts of MLL, we prepared a cDNA library from mRNA of the patient’s leukemic cells. Four cDNA clones were isolated by screening with probe x, and one (clone 17–6) of them was found to represent a fusion transcript of MLL. Clone 17–6, 534 bp in size, contained a 420-bp sequence corresponding to exons 8–10 in the MLL gene at the 5′ region, and the remaining 114-bp sequence did not match the MLL gene or the partner genes of MLL previously cloned (Fig. 2a). We could not isolate any 3′-MLL fusion clones.
Figure 1.
Southern blot of DNA digested with HindIII and probed with the 0.9-kb fragment of the MLL gene. C, normal peripheral lymphocytes. P, leukemic cells from the patient. The patient exhibited a rearranged band (arrow) with this probe.
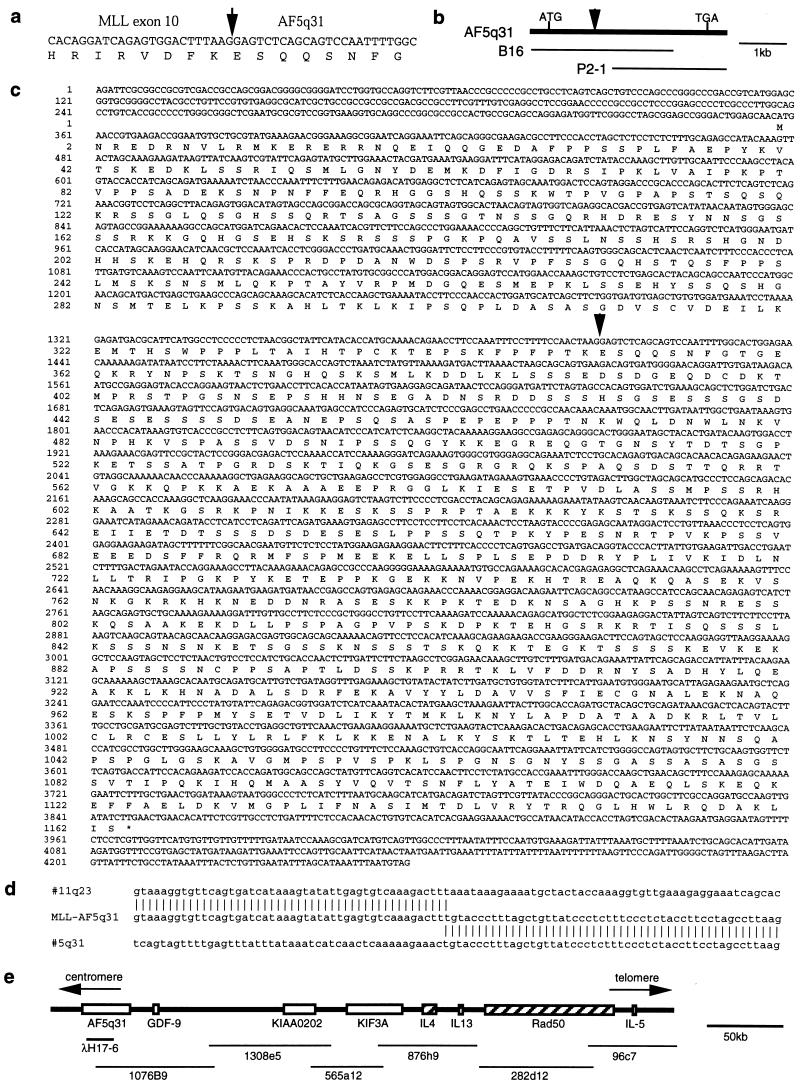
Figure 2.
(a) Partial sequence of the junction of the MLL-AF5q31 chimeric transcript. The arrow indicates the fusion point. (b) Cloning of AF5q31 cDNA clones. (c) Sequencing of the AF5q31 cDNA. * indicates a termination codon. The arrow indicates the breakpoint in the patient. (d) Sequence of the ins(5;11)(q31;q13q23) breakpoint region in the patient. (e) Physical map of the chromosome 5q31-cytokine cluster region and P1 clones.
Isolation of the AF5q31 Gene Encoding a Protein That Is Similar to AF4 Protein.
The 114-bp sequence identified in the chimeric clone was used as a probe to screen cDNA libraries from the BALM14 cell line and human placenta. We isolated two overlapping clones that spanned the complete coding region and encoded a protein of 1,163 aa with a predicted molecular mass of 127,457 Da (Fig. 2 b and c). Sequence comparisons of the predicted AF5q31 protein using the blast file showed partial similarity to the AF4 protein, which is a fusion partner of the MLL gene in t(4;11)(q21;q23) (Fig. 3a).
Figure 3.
(a) Comparison of predicted coding sequence of AF5q31 with other related sequences. (b) Schematic representation of homology between AF5q31 and AF4 proteins. The percentage of amino acid homology between corresponding regions of AF5q31 and AF4 is indicated. Arrowheads indicate the fusion points with MLL.
Detection of the MLL-AF5q31 Fusion Transcripts and the Genomic Junction of the Breakpoint.
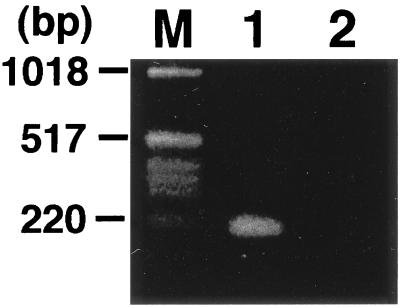
Using a sense primer from MLL exon 10 (MLL-7S) and an antisense primer from AF5q31 (5–1A), we obtained a PCR product of 214 bp from the patient (Fig. 4). However, reciprocal PCR products of AF5q31-MLL fusion transcripts were not generated by reverse transcription–PCR (Fig. 4). Furthermore, we cloned a genomic junction of the breakpoint by genomic PCR followed by sequencing (Fig. 2d).
Figure 4.
Detection of the MLL-AF5q31 chimeric transcripts by reverse transcription–PCR. Primers used were MLL-7S and 5–1A (lane 1), and 5–13S and MLL-11A (lane 2), respectively. M, size marker.
Expression of the AF5q31 Gene Compared with AF4 Gene in Normal Tissues.
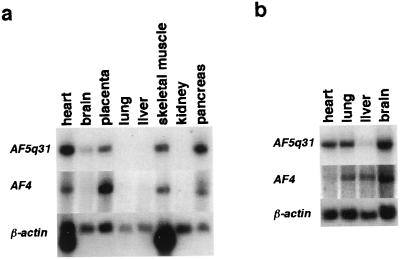
To examine for expression of the AF5q31 gene, we performed Northern blot analysis on poly(A)+ RNA from various human tissues. Expression of the 9.5-kb transcript was detected in adult heart, placenta, skeletal muscle, and pancreas, and fetal heart, lung, and brain at relatively high levels, and in adult brain and fetal liver at low levels (Fig. 5). We also performed Northern blot analysis on the same blots using AF4 cDNA probe. Expression of the 9.5-kb transcript was detected in adult heart, placenta, skeletal muscle, and pancreas, and fetal lung, liver, and brain at relatively high levels, and in fetal heart at a low level (Fig. 5).
Figure 5.
Northern blot analysis of RNAs from adult (a) and fetal (b) human tissues. UAF5q31 and AF4 cDNA fragments were used as probes for the Northern blots in the upper and middle figures, respectively. Membranes were rehybridized to the β-actin probe for the lower figures. The organs from which tissues were analyzed are indicated on top of each lane.
Chromosomal Assignment of the AF5q31 Gene.
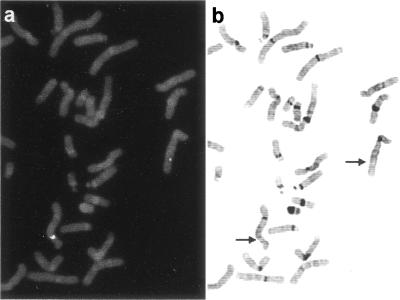
To assign the chromosomal location for the AF5q31 gene, we obtained a phage clone (λH17–6) after screening of a genomic library from human placental DNA using an AF5q31 cDNA probe. The phage clone showed specific signals at band 5q31.1 in all 25 metaphase cells tested (Fig. 6).
Figure 6.
Chromosomal localization of the AF5q31 gene. (a) A fluorescence in situ hybridization pattern obtained with the phage clone λH17–6 as a probe. (b) The same metaphase spread stained with 4′,6-diaminido-2-phenylindole dihydrochloride (DAPI). A DAPI image was inverted and enhanced in terms of band image contrast. The AF5q31 gene was assigned to band 5q31.1 as indicated by arrows.
AF5q31 Gene Is Located in the Vicinity of the Cytokine Cluster Region of Chromosome 5q31.
By blast search of GenBank, we found that the sequence of a P1 clone from chromosome 5q31 partially matched the sequence of AF5q31 cDNA. Sequence comparison between AF5q31 and this P1 clone revealed that the P1 clone contained the 3′ region of the breakpoint in the AF5q31 gene and that the AF5q31 gene contained at least 16 exons. Furthermore, six overlapping P1 clones covered a genomic region of about 380 kb, containing the GDF-9, KIF3A, IL-4, IL-13, RAD50, and IL-5 genes on chromosome 5q31 (Fig. 2e).
Discussion
In the present study, we isolated a novel fusion partner of the MLL gene, AF5q31, in an infant ALL with ins(5;11)(q31;q13q23). In the patient’s leukemic cells, only an MLL-AF5q31 fusion transcript was detected, but not an AF5q31-MLL transcript. Although we could not exclude the possibility of the presence of any other 3′-MLL fusion transcripts, the 5′-MLL-AF5q31-3′ transcript is thought to be critical in leukemogenesis, as described (19, 20).
The AF5q31 protein is homologous to the AF4 protein (Fig. 3 a and b). The AF5q31 and AF4 proteins have three homologous regions, including the transactivation domain of AF4 (25, 26) (Fig. 3b). Although the functions of the other two regions remain unknown, these homologous regions may include potential common functional domains of AF5q31 and AF4. The AF4 gene was cloned from ALL with t(4;11)(q21;q23) (3). Patients with the t(4;11) are characterized by very young age, hyperleukocytosis, early pre-B phenotype (CD10−/CD19+), and poor treatment outcome for infants and patients aged >10 years (27). The t(4;11) is the predominant 11q23 chromosome translocation in infant ALL (7, 8). As compared with other MLL chimeric transcripts in ALL and AML, MLL-AF4 chimeric transcripts in infant ALL have been reported to be associated with extremely poor prognosis, regardless of advances in therapy for childhood leukemia (11). The clinical features of this patient with MLL-AF5q31 fusion transcript, characterized by early pre-B phenotype (CD10−/CD19+) and poor outcome, were similar to those of the patients having MLL-AF4 chimeric transcripts. t(5;11)(q31;q23) has been described in only a few patients with de novo ALL (28, 29) and therapy-related AML (30). Although it remains unknown whether MLL-AF5q31 is associated with only ALL or not, these findings suggest that AF5q31 and AF4 might define a new family particularly involved in the pathogenesis of 11q23-associated-ALL. Further accumulation of these patients with this MLL-AF5q31 transcript may clarify the association between t(5;11)-ALL and t(4;11)-ALL.
Hitherto, two AF4-related genes had been identified, LAF4 (25) and FMR2 (31, 32), and sequencing analysis revealed that both LAF4 and FMR2 are also homologous to AF5q31 (Fig. 3a). LAF4, isolated as a lymphoid-restricted nuclear protein from a subtracted cDNA library, was expressed at high levels in lymphoid tissues and at lower levels in brain and lung (25). In human and mouse lymphoid cell lines, LAF4 expression was highest in pre-B cells, intermediate in mature B cells, and absent in plasma cells, thus suggesting that LAF4 plays a potential regulatory role in early lymphoid development. FMR2 originally was identified from chromosome Xq28 as a gene associated with FRAXE mental retardation (31, 32). FMR2 encodes a nuclear protein of 1,311 aa with putative nuclear transcription transactivation potential. Expression of the FMR2 gene was found in adult brain and placenta, and fetal brain, lung, and kidney (33). The expression pattern of the FMR2 gene in fetal tissues was similar to that of the AF5q31 gene, suggesting that both AF5q31 and FMR2 may play common roles in fetal development. Particularly, expression of the AF5q31 gene was decreased in adult lung, liver, and brain compared with that in fetal tissues, suggesting that AF5q31 may play critical roles in the fetal development of these tissues.
The 5q31 chromosomal region is known to be a very critical region in which many cytokine genes are clustered (34, 35). This region also has been suggested to be associated with interstitial deletion of 5q observed in AML and myelodysplastic syndrome (MDS), suggesting that a tumor suppressor gene for AML and MDS may be present in this region (34, 36). Therefore, this region has been under investigation for a long time by many researchers, and large amounts of sequence data from the region have been stored in databases. Interestingly, it was predicted that another AF4-related gene was present in the chromosome 5q31 region based on a homology search of databases by Frestedt et al. (37). Gecz et al. (33) showed that FMR2 is homologous to two expressed sequence tags (W26686 and AA025630) mapped to chromosome 5q31. AF5q31 is likely to be the gene predicted by those authors. However, the location of the AF5q31 gene is outside of the commonly deleted region previously reported in AML and MDS (36).
A few papers about the function of MLL fusion proteins have been published (19, 20, 38–41). However, the function of the MLL-AF4 fusion protein has not been analyzed yet. Functional analyses of the AF5q31 and MLL-AF5q31 fusion proteins may provide new insights into the function of the MLL-AF4 fusion protein and also the leukemogenesis of 11q23-associated-ALL.
Acknowledgments
We thank M. Seto, Aichi Cancer Center Research Institute, for providing the MLL cDNA probe (probe x). We express appreciation to S. Sohma and H. Soga for technical assistance. This work was supported by a Grant-in-Aid for Cancer Research from the Ministry of Health and Welfare of Japan, a Grant-in-Aid for Scientific Research on Priority Areas, and Grant-in-Aid for Scientific Research (B) and (C) from the Ministry of Education, Science, Sports, and Culture of Japan.
Abbreviations
- ALL
acute lymphoblastic leukemia
- AML
acute myeloid leukemia
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF197927).
References
- 1.McCabe N R, Burnett R C, Gill H J, Thirman M J, Mbangkollo D, Kipiniak M, van Melle E, Ziemin-van der Poel S, Rowley J D, Diaz M O. Proc Natl Acad Sci USA. 1992;89:11794–11798. doi: 10.1073/pnas.89.24.11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tkachuk D C, Kohler S, Cleary M L. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 3.Gu Y, Nakamura H, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 4.Djabali M, Selleri L, Parry P, Bower M, Young B D, Evans G A. Nat Genet. 1992;2:113–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 5.Yu B D, Hanson R D, Hess J L, Horning S E, Korsmeyer S J. Proc Natl Acad Sci USA. 1998;95:10632–10636. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowley J D. Annu Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 7.Rubnitz J E, Link M P, Shuster J J, Carroll A J, Hakami N, Frankel L S, Pullen D J, Cleary M L. Blood. 1994;84:570–573. [PubMed] [Google Scholar]
- 8.Taki T, Ida K, Bessho F, Hanada R, Kikuchi A, Yamamoto K, Sako M, Tsuchida M, Seto M, Ueda R, Hayashi Y. Leukemia. 1996;10:1303–1307. [PubMed] [Google Scholar]
- 9.Super H J, McCabe N R, Thirman M J, Le Beau M M, Pedersen-Bjergaard J, Philip P, Diaz M O, Rowley J D. Blood. 1993;82:3705–3711. [PubMed] [Google Scholar]
- 10.Hunger S P, Tkachuk D C, Amylon M D, Link M P, Carroll A J, Welborn J L, Willman C L, Cleary M L. Blood. 1993;81:3197–3203. [PubMed] [Google Scholar]
- 11.Heerema N A, Sather H N, Ge J, Arthur D C, Hilden J M, Trigg M E, Reaman G H. Leukemia. 1999;13:679–686. doi: 10.1038/sj.leu.2401413. [DOI] [PubMed] [Google Scholar]
- 12.Rubnitz J E, Behm F G, Downing J R. Leukemia. 1996;10:74–82. [PubMed] [Google Scholar]
- 13.Yamamoto T, Harada N, Kawano Y, Taya S, Kaibuchi K. Biochem Biophys Res Commun. 1999;259:103–107. doi: 10.1006/bbrc.1999.0731. [DOI] [PubMed] [Google Scholar]
- 14.Shilatifard A, Lane W S, Jackson K W, Conaway R C, Conaway J W. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 15.Taki T, Sako M, Tsuchida M, Hayashi Y. Blood. 1997;89:3945–3950. [PubMed] [Google Scholar]
- 16.Sobulo O M, Borrow J, Tomek R, Reshmi S, Harden A, Schlegelberger B, Housman D, Doggett N A, Rowley J D, Zeleznik-Le N J. Proc Natl Acad Sci USA. 1997;94:8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ida K, Kitabayashi I, Taki T, Taniwaki M, Noro K, Yamamoto M, Ohki M, Hayashi Y. Blood. 1997;90:4699–4704. [PubMed] [Google Scholar]
- 18.Taki T, Shibuya N, Taniwaki M, Hanada R, Morishita K, Bessho F, Yanagisawa M, Hayashi Y. Blood. 1998;92:1125–1130. [PubMed] [Google Scholar]
- 19.Corral J, Lavenir I, Impey H, Warren A J, Foster A, Larson T A, Bell S, McKerzie A N J, King G, Rabbitts T H. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 20.Lavau C, Szilvassy S J, Slany R, Cleary M L. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto K, Seto M, Komatsu H, Iida S, Akao Y, Kojima S, Kodera Y, Nakazawa S, Ariyoshi Y, Takahashi T, Ueda R. Oncogene. 1993;8:2617–2625. [PubMed] [Google Scholar]
- 22.Ida K, Taki T, Bessho F, Kobayashi M, Taira F, Hanada R, Yamamoto K, Okimoto Y, Seto M, Ueda R, Hayashi Y. Med Pediatr Oncol. 1997;28:325–332. doi: 10.1002/(sici)1096-911x(199705)28:5<325::aid-mpo1>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 23.Taniwaki M, Nishida K, Ueda Y, Misawa S, Nagai M, Tagawa S, Yamagami T, Sugiyama H, Abe M, Fukuhara S, Kashima K. Blood. 1995;85:3223–3228. [PubMed] [Google Scholar]
- 24.Nilson I, Lochner K, Siegler G, Greil J, Beck J D, Fey G H, Marschalek R. Br J Haematol. 1996;93:966–972. doi: 10.1046/j.1365-2141.1996.d01-1748.x. [DOI] [PubMed] [Google Scholar]
- 25.Ma C, Staudt L M. Blood. 1996;87:734–745. [PubMed] [Google Scholar]
- 26.Li Q, Frestedt J L, Kersey J H. Blood. 1998;92:3841–3847. [PubMed] [Google Scholar]
- 27.Pui C H, Frankel L S, Carroll A J, Raimondi S C, Shuster J J, Head D R, Crist W M, Land V J, Pullen D J, Steuber C P, et al. Blood. 1991;77:440–447. [PubMed] [Google Scholar]
- 28.Kearney L, Bower M, Gibbons B, Das S, Chaplin T, Nacheva E, Chessells J M, Reeves B, Riley J H, Lister T A, Young B D. Blood. 1992;80:1659–1665. [PubMed] [Google Scholar]
- 29.Rubnitz J E, Link M P, Shuster J J, Carroll A J, Hakami N, Frankel L S, Pullen D J, Cleary M L. Blood. 1994;84:570–573. [PubMed] [Google Scholar]
- 30.Hoyle C F, de Bastos M, Wheatley K, Sherrington P D, Fischer P J, Rees J K, Gray R, Hayhoe F G. Br J Haematol. 1989;72:45–53. doi: 10.1111/j.1365-2141.1989.tb07650.x. [DOI] [PubMed] [Google Scholar]
- 31.Gecz J, Gedeon A K, Sutherland G R, Mulley J C. Nat Genet. 1996;13:105–108. doi: 10.1038/ng0596-105. [DOI] [PubMed] [Google Scholar]
- 32.Gu Y, Shen Y, Gibbs R A, Nelson D L. Nat Genet. 1996;13:109–113. doi: 10.1038/ng0596-109. [DOI] [PubMed] [Google Scholar]
- 33.Gecz J, Bielby S, Sutherland G R, Mulley J C. Genomics. 1997;44:201–213. doi: 10.1006/geno.1997.4867. [DOI] [PubMed] [Google Scholar]
- 34.Le Beau M M, Lemons R S, Espinosa R, 3rd, Larson R A, Arai N, Rowley J D. Blood. 1989;73:647–650. [PubMed] [Google Scholar]
- 35.Marsh D G, Neely J D, Breazeale D R, Ghosh B, Freidhoff L R, Ehrlich-Kautzky E, Schou C, Krishnaswamy G, Beaty T H. Science. 1994;264:1152–1156. doi: 10.1126/science.8178175. [DOI] [PubMed] [Google Scholar]
- 36.Fairman J, Chumakov I, Chinault A C, Nowell P C, Nagarajan L. Proc Natl Acad Sci USA. 1995;92:7406–7410. doi: 10.1073/pnas.92.16.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frestedt J L, Hilden J M, Kersey J H. DNA Cell Biol. 1996;15:669–678. doi: 10.1089/dna.1996.15.669. [DOI] [PubMed] [Google Scholar]
- 38.Slany R K, Lavau C, Cleary M L. Mol Cell Biol. 1998;18:122–129. doi: 10.1128/mcb.18.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joh T, Hosokawa Y, Suzuki R, Takahashi T, Seto M. Oncogene. 1999;18:1125–1130. doi: 10.1038/sj.onc.1202400. [DOI] [PubMed] [Google Scholar]
- 40.Maki K, Mitani K, Yamagata T, Kurokawa M, Kanda Y, Yazaki Y, Hirai H. Blood. 1999;93:3216–3224. [PubMed] [Google Scholar]
- 41.Dobson C L, Warren A J, Pannell R, Forster A, Lavenir I, Corral J, Smith A J, Rabbitts T H. EMBO J. 1999;18:3564–3574. doi: 10.1093/emboj/18.13.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]