Abstract
Cells of Mus minutoides, an African pygmy mouse of the subgenus Nannomys, are susceptible to ecotropic Moloney and Friend mouse leukemia viruses (MLVs) but not to AKV-type MLVs. Transfected MA139 ferret cells expressing the mCAT-1 cell surface receptor, with the minCAT-1 substitutions K222Q and V233L, did not restrict AKV MLV. The resistance of M. minutoides cells to AKV MLV was not relieved by inhibitors of glycosylation or by the introduction of NIH 3T3 mCAT-1. Resistance is thus not mediated by receptor sequence variation, expression level, or glycosylation. M. minutoides cells are also infectible with LacZ pseudotypes having AKV Env and Moloney MLV (MoMLV) Gag proteins, further indicating that AKV Env sequence variations do not contribute to the observed block. The pattern of virus resistance in M. minutoides differs from that of the known variants of the Fv1 postentry resistance gene; M. minutoides is equally resistant to N-, B-, and NR-tropic AKV viruses and is equally susceptible to NR- and NB-tropic Friend MLVs. This novel resistance blocks replication before reverse transcription, whereas Fv1 generally restricts replication after reverse transcription; M. minutoides cells produce 2-long-terminal-repeat viral DNA circles and linear viral DNA after infection with MoMLV but not with AKV MLV. Analysis of MoMLV-AKV MLV chimeras determined that the target of resistance is in the virus capsid gene. Mutagenesis demonstrated that restriction is mediated by two amino acid substitutions, H117L and A110R; substitutions at these sites can also be targeted by the resistance genes Fv1 and TRIM5α. M. minutoides cells thus have a novel postentry resistance to AKV MLVs.
Numerous cellular genes that inhibit retrovirus replication have been identified. For the mouse gammaretroviruses, specific restriction genes that interfere with replication at different stages of the viral life cycle are known; these include host genes that affect virus binding and entry; genes that inhibit the early stages of replication, involving DNA synthesis, trafficking, and integration; and genes that block the late stages, involving virus protein synthesis and assembly. The postentry, preintegration stage of virus replication is the most poorly understood part of the viral life cycle, but efforts to describe these processes are aided by the identification of host genes that restrict postentry replication. In particular, genetic studies done more than 30 years ago described the prototype of this intrinsic type of cellular immunity (6), Fv1. This gene acts shortly after virus entry to reduce the formation of proviral insertions (32, 41).
Alleles at the Fv1 locus control the relative sensitivities of mouse cells to different subgroups of mouse leukemia viruses (MLVs), and this resistance was initially described as a 100- to 1,000-fold reduction in virus titer (16). The restricted viruses can be classed as N tropic if they replicate best in cells with the Fv1n allele, B tropic if they replicate best in Fv1b cells, or NB tropic if they grow equally well in both cell types. A third restriction allele, termed Fv1nr, affects susceptibility to B-tropic viruses as well as to some, but not all, viruses restricted by Fv1b cells (20, 24). A null allele, Fv1o, has been identified in wild mouse species and is not associated with virus restriction (15, 24, 26). There is also evidence for additional Fv1 variants with less dramatic effects on virus replication, for example, in DBA/2 mice or some wild mouse species (24, 25). Among the laboratory mouse strains, Fv1n and Fv1b alleles are most common, but most wild mouse species tested either lack Fv1 restriction or have an Fv1nr phenotype (24).
Early studies on Fv1 demonstrated that the restriction is characterized by “two-hit” titration curves (11, 33) and by the abrogation of Fv1 restriction by preexposure of cells to the restricted virus (2); this suggests that Fv1 encodes a saturable inhibitor of MLV replication. The Fv1 gene was cloned in 1996 and is related to the gag gene of the human endogenous retrovirus L family (5) and a related family of mouse endogenous retroviruses, MuERV-L (3). Sequence comparisons of the various restriction alleles and mutagenesis studies showed that restriction is associated with a segment in the middle of the Fv1 coding region and with several amino acid residues in the C-terminal one-third (7).
It has been demonstrated that the Fv1 gene targets the virus capsid (CA). A single amino acid at CA position 110 distinguishes the N- and B-tropic viruses restricted by Fv1n and Fv1b (25). Additional mutagenesis studies identified targets for Fv1 NR and NB tropism at additional sites in the N-terminal domain of CA (12, 20, 27, 39, 44). Despite the identification of the Fv1 gene and its target, the mechanism of virus restriction has not been determined. Efforts to determine how the Fv1 gene product interferes with virus replication have been impeded by the low level of Fv1 expression (5) and the failure to demonstrate Fv1 binding to capsid (6).
While only Mus species have Fv1 resistance, primates and other mammals carry another, unrelated retroviral resistance factor that restricts human immunodeficiency virus (HIV) as well as N-tropic MLVs (17, 22, 31, 43, 49). This factor, TRIM5α, targets the same MLV capsid residue that is targeted by Fv1b, R110 (43). Comparisons of the Fv1 and TRIM5α blocks to replication indicate that the TRIM5α block generally occurs before reverse transcription whereas the Fv1 block generally occurs after reverse transcription (19, 43, 48). The TRIM gene family is large, and there is no clearly identifiable orthologue of human TRIM5 in mouse; the mouse TRIM family member most closely related to human TRIM5, based on sequence similarity and chromosome location, has not been shown to restrict virus (40).
Although studies on blocks to the replication of mouse gammaretroviruses in their natural host have been largely limited to analysis of the various inbred mouse strains, these mice are not representative of the genus Mus, as shown by our earlier analysis of Fv1-type resistance in species closely related to the laboratory mice (24). To extend our knowledge of the nature and evolution of genes affecting virus susceptibility and disease induction in natural populations, we tested a more divergent set of wild mouse species for their sensitivities to ecotropic MLVs (E-MLVs). We report here the identification of a novel postentry resistance to E-MLVs, termed Minr, in the African pygmy mouse, Mus minutoides.
MATERIALS AND METHODS
Cells and viruses.
E-MLV isolates were obtained from J. W. Hartley (NIAID, Bethesda, MD) and included AKV MLV, WN1802B, AKR-L1, Gross Passage A MLV, Rauscher MLV (RaMLV), Moloney MLV (MoMLV), and two Friend MLV (FrMLV) isolates, F-S MLV and FBLV. F-S MLV is an FrMLV isolate that, like AKR-L1, is not restricted in cells with the Fv1nr allele; these viruses are designated NR tropic. FBLV (NB-tropic FrMLV) is a biologically cloned virus originally provided by R. Risser (University of Wisconsin, Madison). AKR-L1 was originally isolated from the spleen, thymus, and lymph nodes of an AKR mouse with leukemia (33). WN1802B N′B was isolated after forced passage of WN1802B in NIH Swiss embryo fibroblasts; it grows to equal titers in NIH 3T3 and BALB/c cells. An additional E-MLV, pLU7, was generated by introducing the CAgag mutation R110L into AKV MLV clone pAKV34 (25). Virus stocks were made by collecting culture fluids from virus-infected cells or, in the case of pLU7, from cells transfected with viral DNA.
The XC overlay test (37) was used to test susceptibility to virus infection in various cell lines, including a Mus dunni cell line (26), NIH 3T3, SC-1 (15), and a cell line derived from tail fibroblasts of the wild mouse species M. minutoides that was obtained from J. Rodgers (Baylor College of Medicine, Houston, TX). Embryo fibroblasts were also prepared from mice of strains 129/J and BALB/cJ, which were obtained from The Jackson Laboratory (Bar Harbor, ME). Subconfluent cell cultures were infected with virus dilutions in the presence of Polybrene (4 μg/ml; Aldrich, Milwaukee, WI). After 4 to 5 days, cultures were UV irradiated with UV light from germicidal bulbs to kill the cells and were then overlaid with 106 rat XC cells/60-mm dish (37). Plates were fixed and stained 3 days later and were then examined for plaques of XC cell syncytia to identify foci of infected cells.
Cells were treated prior to virus infection by two inhibitors of N-linked glycosylation, tunicamycin (0.2 μg/ml) and 2-deoxy-d-glucose (10 to 50 mM) (Sigma-Aldrich, St. Louis, MO). Inhibitors were added to cultures that had been seeded the previous day and were not removed when virus was added, 18 to 24 h later.
Virus was also introduced into mouse cells by transfection using molecular clones of MoMLV (pNCA-HA [30]) and AKV MLV (pGEM-AKV, containing the 8.2-kb PstI insert from pAKV34 [28]; obtained from J. Lenz, Albert Einstein College of Medicine, Bronx, NY). NIH 3T3, SC-1, and M. minutoides cells were transfected using TransIT-LT1 transfection reagent (Mirus, Madison, WI). Cells were passed 2 days later and at regular intervals after that into duplicate plates, one of which was tested for virus by the XC overlay test.
minCAT-1 sequencing and mutagenesis of mCAT-1.
The full-length CAT-1 receptor gene of M. minutoides cells was amplified by reverse transcription-PCR using primers minCAT-F and minCAT-R (Table 1). cDNA was synthesized using the SuperScript II reverse transcriptase kit (Invitrogen, CA), and PCR was performed with 2 μl of cDNA as a template using 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 1 min, and extension at 72°C for 4 min. The 1.87-kb PCR product was sequenced directly.
TABLE 1.
Primers used for mutagenesis and PCR
| Primer | Sequencea | Position (GenBank accession no.) |
|---|---|---|
| minCAT-F | 5′-ATGGGCTGCAAAAACCTGCTCGGTCTGG | 199-226 |
| minCAT-R | 5′-TCATTTGCACTGGTCCAAGTTGCTGTCAG | 2039-2067 (M26687) |
| E1-F | 5′-CTGGCAGCTCACGGAGcAAAATTTCTCCTGTAAC | 846-879 |
| E2-R | 5′-GTTACAGGAGAAATTTTgCTCCGTGAGCTGCCAG | 846-879 (M26687) |
| E3-F | 5′-AACAACGACACAAACcTGAAATACGGTGAGG | 880-910 |
| E4-R | 5′-CCTCACCGTATTTCAgGTTTGTGTCGTTGTT | 880-910 (M26687) |
| p30F1 | 5′-TATTGCCTATGAACCCCCTCCGTG | 902-925 |
| p30R1 | 5′-CTTTTCCTCTGTCTCCCTTCTAACG | 1964-1988 (J01998) |
| AK-F | 5′-GAACTCTTTGTCGACGAGAAGCAGG | 3711-3735 |
| AK-R | 5′-GGTTGAGATgCAtGGAGTAAGTCCAGTAC | 7047-7075 (J01998) |
| Pro-F | 5′-GACTTCCGGATCCAGCAC | 3237-3254 |
| Pro-R | 5′-AAACCCAGGGATCCAGAG | 3546-3563 (J01998) |
| LP5-F | 5′-AGTCCTCCGATAGACTGAG | 6-24 |
| LP6-R | 5′-CCCTTTTTATAGAGCTGGG | 8269-8287 (J01998) |
| Ak-ca1F | 5′-CCCTCGAGCGTCCCGATTGGGATTACAC | 1581-1600 |
| Ak-ca2R | 5′-GGTCGCGATCAAGTTGGGGCCTCCTTCGCT | 2113-2134 (J01998) |
| OMo-F | 5′-CCCTCGAGCGCCCAGACTGGGATTACAC | 1558-1585 |
| OMo-R | 5′-CtTTTATTAAAGATCcTTTCTGCCTCTCTAACCAAATC | 1884-1921 (J02255) |
| OAK-F | 5′-GAAAGGATCTTTAATAAAAGAGAAACTCCAG | 1917-1947 |
| OAK-R | 5′-AAATCGCGATCAAGTTGGGGCCTCCTTCGC | 2114-2134 (J01998) |
| Ak-ca1F | 5′-CCCTCGAGCGTCCCGATTGGGATTACAC | 1581-1600 |
| AK129R | 5′-GGTGGGGCTTCtGCCCGCGTTTTGGAGACCTGCTA | 1648-1682 (J01998) |
| MO129F | 5′-CCAAAACGCGGGCAGAAGCCCCACCAATTTG | 1643-1673 |
| MONCR | 5′-TCGCGATCGAGTTGGGACCTCC | 2104-2125 (J02255) |
| A110R-F | 5′-GGGATTACACCACCCAaagAGGTAGGAACCACCTAG | 1576-1611 |
| A110R-R | 5′-CTAGGTGGTTCCTACCTcttTGGGTGGTGTAATCCC | 1576-1611 (J02255) |
| H117L-F | 5′-AGGAACCACCTAGTCCtCTATCGCCAGTTGCTCCTAGC | 1599-1636 |
| H117L-R | 5′-GCTAGGAGCAACTGGCGATAGaGGACTAGGTGGTTCCT | 1599-1636 (J02255) |
| R110A-F | 5′-GGGATTACACCACCCAggcAGGTAGGAACCACCTAG | 1591-1626 |
| R110A-R | 5′-CTAGGTGGTTCCTACCTgccTGGGTGGTGTAATCCC | 1591-1626 (J01998) |
| L117H-F | 5′-AGGAACCACCTAGTTCaCTATCGCCAGTTGCTTTTAGC | 1614-1651 |
| L117H-R | 5′-GCTAAAAGCAACTGGCGATAGtGAACTAGGTGGTTCCT | 1614-1651 (J01998) |
| K214R-F | 5′-TGGTTAGAGAGGCAGAAAgGATCTTTAATAAACGAGAAACC | 1888-1928 |
| K214R-R | 5′-GGTTTCTCGTTTATTAAAGATCcTTTCTGCCTCTCTAACCA | 1888-1928 (J02255) |
| I229V-F | 5′-GAAGAAAGAGAGGAACGTgTtAGGAGAGAAACAGAGGAA | 1932-1970 |
| I229V-R | 5′-TTCCTCTGTTTCTCTCCTaAcACGTTCCTCTCTTTCTTC | 1932-1970 (J02255) |
| D244E-F | 5′-GAACGCCGTAGGACAGAGGAgGAGCAGAAAGAGAAAGAA | 1977-2015 |
| D244E-R | 5′-TTCTTTCTCTTTCTGCTCcTCCTCTGTCCTACGGCGTTC | 1977-2015 (J02255) |
Lowercase letters represent introduced base substitutions. Underlined bases represent restriction sites: SalI (AK-F), NsiI (AK-R), XhoI (Ak-ca1F, OMo-F), and NruI (Ak-ca2R, OAK-R, MONCR).
Two mutations (K222Q and V233L) identified in the third extracellular loop of the minCAT-1 gene were introduced into an expression plasmid containing the hemagglutinin (HA)-tagged NIH 3T3 mCAT-1 (pcDNA3-MCAT; a gift of J. Cunningham, Harvard Medical School, Boston, MA) using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). V233L was introduced using primers E3-F and E4-R (Table 1). The K222Q mutation was then introduced into the K222Q clone using primers E1-F and E2-R (Table 1). Mutations were confirmed by sequencing.
The CAT-1 constructs were introduced into M. minutoides cells or MA139 ferret cells obtained from J. Hartley (NIAID, Bethesda, MD) using the FuGene 6 transfection reagent (Roche Applied Science, Indianapolis, IN). Stable transfectants expressing CAT-1 were isolated in 0.8 mg/ml Geneticin (Invitrogen, Grand Island, NY), and expression was confirmed by Western blotting. Protein was extracted using M-PER mammalian protein extraction reagent (Pierce, Rockford, IL), and CAT-1 expression was identified using a mouse anti-HA monoclonal antibody (clone 12CA5) and anti-mouse immunoglobulin G2b conjugated with horseradish peroxidase as a secondary antibody (both from Roche, Indianapolis, IN).
Sequencing of viral CA genes.
RNA was extracted from cultures of M. dunni cells infected with WN1802B N′B, F-S MLV, or FBLV. A segment of the viral gag gene was amplified by reverse transcription-PCR using primers p30F1 and p30R1. A PCR product of 1.1 kb was isolated. The FBLV and F-S MLV PCR products were sequenced directly; the WN1802B N′B product was cloned into pCR2.1-TOPO and sequenced.
Pseudotype assay.
LacZ pseudotype virus was generated by cotransfection of human 293 cells with pCLMFG-LacZ (Imgenex Co., San Diego, CA) and expression vectors containing three different E-MLV env genes. The pCL-eco retrovirus packaging vector (Imgenex Co., San Diego, CA) was used to generate pseudotypes with MoMLV ecotropic Env. The SU gene of FrMLV57 was cloned into pCL-eco as described previously (21). A third ecotropic env expression vector (pCL-AK) was generated by replacing the 3.35-kb SalI-NsiI fragment of pCL-eco with the corresponding region of AKV MLV. This fragment includes part of pol and most of SU. Primers AK-F and AK-R (Table 1) were used to amplify the fragment from pGEM-AKV and to introduce a novel NsiI site. The reaction was performed for 30 cycles with denaturation at 94°C for 30 s, annealing at 53°C for 1 min, and extension at 72°C for 6 min. The PCR product was digested with SalI and NsiI and then cloned into the pCL-eco vector from which the corresponding SalI-NsiI fragment had been deleted.
Supernatants containing pseudotype virus were collected from transfected 293 cells, filtered, and used to infect cells that had been plated in 12-well culture dishes. The cells were infected with appropriate dilutions of pseudotype virus in the presence of 4 to 8 μg/ml Polybrene. The cells were fixed 32 to 48 h after infection with 0.4% glutaraldehyde and were assayed for β-galactosidase activity by using as a substrate 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 2 mg/ml; ICN Biomedicals, Aurora, OH). Infectious titers were expressed as the number of blue cells per 100 μl of virus supernatant.
Extraction and analysis of unintegrated viral DNA.
Unintegrated viral DNA was extracted from virus-infected cultures using the Hirt fractionation method (18). The DNAs were digested with SacII, electrophoresed on 0.4% agarose gels, transferred to nylon membranes (Hybond N+; Amersham, Piscataway, NJ), and hybridized with a radiolabeled probe. A 327-bp segment of the AKV pol gene (positions 3237 to 3563 of GenBank accession number J01998) was used as a hybridization probe.
Hirt DNA from virus-infected cultures was used as a template to amplify long-terminal-repeat (LTR) junction fragments by PCR from circular viral DNA using primers LP5-F and LP6-R. After denaturation at 94°C for 5 min, 35 cycles with denaturation at 94°C for 30 s, annealing at 55°C for 1 min, and extension at 72°C for 1 min were performed.
Construction of capsid chimeras and mutants.
All chimeras were constructed using the MoMLV clone pNCA-HA (30) as a backbone, and all primers are given in Table 1. To generate chimera CA1, a 0.56-kb region of AKV CA was amplified from pGEM-AKV using primers Ak-ca1F and Ak-ca2R. XhoI and NruI sites were introduced at the 5′ ends of the two primers as indicated. Thirty cycles of PCR were performed with 30 s of denaturation at 94°C, 30 s of annealing at 55°C, and 1 min of extension at 72°C. After digestion with XhoI and NruI, the PCR product was ligated into pNCA-HA from which the corresponding fragment had been removed. The resulting clone, CA1, contains 165 amino acids of the C terminus (positions 99 to 263) of AKV CA and the first 22 amino acids of the AKV nucleocapsid (NC).
CA2 was constructed using overlapping PCR. The first two primers (OMo-F and OMo-R) were used to amplify 0.36 kb of MoMLV capsid, and the second pair of primers (OAK-F and OAK-R) was used to amplify 0.22 kb of AKV capsid. Then primers OMo-F and OAK-R were used to amplify a 0.56-kb product with a mixture of the two smaller products as the template. The final PCR product was digested with XhoI and NruI and then ligated into pNCA-HA. CA2 contains 50 amino acids of AKV CA (positions 214 to 263) and the first 22 amino acids of AKV NC.
CA3 was constructed using two primers to amplify 90 bp of AKV capsid (AK-ca1F and AK129R); a second pair of primers was used to amplify 0.47 kb of MoMLV capsid (MO129F and MONCR). Then primers AK-ca1F and MONCR were used to amplify 0.56 kb with a mixture of two smaller products as the template. The final PCR product was digested with XhoI and NruI and then ligated into pNCA-HA. CA3 includes 31 amino acids from positions 99 through 129 of the AKV MLV capsid.
We introduced six mutations into pNCA-HA with the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). A single-site mutation at position 110 (A110R) was made with forward primer A110R-F and reverse primer A110R-R. The H117L mutant was made with primers H117L-F and H117L-R. A third mutant (A110R H117L) was generated with both substitutions. Primers K214R-F and K214R-R were used to make the K214R mutant, primers I229V-F and I229V-R were used to make the I229V mutant, and primers D244E-F and D244E-R were used to make the D244E mutant. PCR was done for 16 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 1 min, and extension at 68°C for 12 min.
We made three reciprocal mutations in AKV MLV using pGEM-AKV. To generate R110A in the AKV capsid, we used primers R110A-F and R110A-R. The second mutation, L117H, was made with primers L117H-F and L117H-R. The third mutated AKV clone contained both R110A and L117H.
All mutations and chimeras were confirmed by sequencing. Pools of infectious virus were prepared by collecting culture fluids of transfected NIH 3T3 cells or, for the R110A mutations in pGEM-AKV, by collecting culture fluid from transfected SC-1 cells.
RESULTS
Novel resistance to AKV ecotropic gammaretrovirus.
Mouse cells were infected with three different isolates of E-MLVs: AKV MLV, FBLV, and MoMLV. Susceptibility to productive infection was quantitated using the XC plaque overlay test (Table 2, Expt 1). In this assay, clusters of infected cells expressing ecotropic Env glycoprotein are identified by plaques of syncytia formed by overlaid rat XC cells (37). NIH 3T3 cells show the prototypical susceptibility to all three E-MLVs; this pattern is characteristic of cells from the common strains of laboratory mice.
TABLE 2.
Virus titers of ecotropic gammaretroviruses on untreated mouse cells and cells transfected with the mCAT-1 receptor or treated with the glycosylation inhibitor tunicamycin
| Expt and cellsa | Log10 virus titerb
|
||
|---|---|---|---|
| AKV MLV | MoMLV | FBLV | |
| Expt 1 | |||
| NIH 3T3 | 4.7 | 5.1 | 5.8 |
| M. dunni | 4.3 | 2.3 | 5.0 |
| M. minutoides | <0 | 3.5 | 3.2 |
| Expt 2 | |||
| NIH 3T3 | 4.1 | 5.2 | ND |
| M. dunni | 4.4 | 1.5 | ND |
| M. dunni (mCAT-1) | 5.4 | 4.7 | ND |
| M. minutoides | <0 | 3.8 | ND |
| M. minutoides (mCAT-1) | <0 | 4.9 | ND |
| Expt 3 | |||
| NIH 3T3 | 5.1 | 5.8 | ND |
| NIH 3T3 + Tu | 5.2 | 5.2 | ND |
| M. dunni | 5.0 | 2.7 | ND |
| M. dunni + Tu | 4.9 | 4.0 | ND |
| M. minutoides | <0 | 3.2 | ND |
| M. minutoides + Tu | <0 | 3.8 | ND |
Cells stably transfected with mCAT-1 are indicated. Tunicamycin (Tu) was used at 0.2 μg/ml.
Virus titers were determined by the XC overlay test, in which the indicated cells were infected with virus dilutions, irradiated 4 days later, and overlaid with XC cells to identify clusters of virus-infected cells (37). Titers represent the number of XC PFU in 0.2 ml in representative experiments. Infections were done four times for the first experiment, three times for the second, and three times for the third. ND, not done.
We observed two different patterns of susceptibility to these three viruses among cells of wild mouse species (Table 2, Expt 1). The Asian wild mouse (M. dunni) cell line shows the previously described resistance to MoMLV (13), although it is susceptible to AKV and FBLV. Cells derived from the African wild mouse species, M. minutoides, showed a second novel pattern of virus resistance. Susceptibility to infection by MoMLV and FBLV as measured by the number of XC plaques was reduced nearly 100-fold relative to the XC titers observed in NIH 3T3 cells. No XC plaques at all were detected in M. minutoides cells infected with AKV MLV.
AKV MLV and MoMLV were also introduced into NIH 3T3 and M. minutoides cells by transfection of molecular clones of both viruses. In two experiments, the transfected cells were passed two and three times and were tested for infectious virus by the XC overlay test at each passage. By the second passage in both experiments, the cultures all showed high proportions of infected cells (too many XC plaques to count) except for M. minutoides cells transfected with AKV MLV; these cells had no detectable virus by the XC plaque test at any time after transfection (data not shown).
CAT-1 receptor of M. minutoides.
AKV MLV, MoMLV, and FBLV are all E-MLVs, but they show considerable sequence heterogeneity in the receptor binding domains (RBDs) of their env genes. The AKV MLV RBD shows about 77% amino acid identity to the RBDs of MoMLV and FBLV, whereas MoMLV and FBLV are more closely related (88% identity). To determine if the unusual absolute resistance to AKV MLV infection is an entry-related resistance mediated by a wild mouse variant of the E-MLV receptor, we sequenced the M. minutoides CAT-1 receptor, minCAT-1. Limited studies on CAT-1 receptor genes in wild mouse species have previously identified one functional variant of CAT-1 (dCAT-1) in M. dunni (13), a species that is more closely related to the laboratory mouse than is M. minutoides (Fig. 1). The unusual resistance of M. dunni to MoMLV has been attributed to the polymorphism of the dCAT-1 receptor and to its altered glycosylation (13, 14). Sequence analysis of minCAT-1 identified 11 substitutions relative to the sequence of the NIH 3T3 gene, mCAT-1 (Fig. 2A). These differences included five substitutions in putative transmembrane domains, two in intracellular loops, and four in extracellular loops, of which two substitutions are in the third extracellular loop, which is known to carry the critical amino acids needed for virus binding and entry (1, 50). One of these substitutions, V233L, is within the cluster of amino acids identified as critical for binding (36, 50). The second substitution, K222Q, is adjacent to N223, one of the two glycosylation sites in this loop.
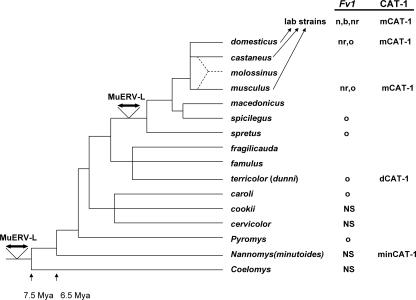
FIG. 1.
Schematic representation of the evolution of Mus. This synthetic evolutionary tree is based on morphological characteristics, DNA sequence data, and analysis of karyotypic rearrangements (8, 45). All species from Mus domesticus through Mus cervicolor are subgenus Mus. M. dunni is part of the Mus terricolor group. The most recent node of the tree represents the house mouse Mus musculus complex; Mus molossinus is a natural hybrid of Mus castaneus and M. musculus. The common strains of the laboratory mouse are derived from three species of house mouse, with the major contribution from M. domesticus (47). CAT-1 variants and Fv1 restriction phenotypes are indicated to the right; NS indicates species that have been tested for susceptibility to Fv1-restricted viruses but that have generally reduced virus susceptibility or nonstandard patterns of virus susceptibility. The horizontal double arrows indicate the two copy-number expansions of endogenous MuERV-L genomes (4).
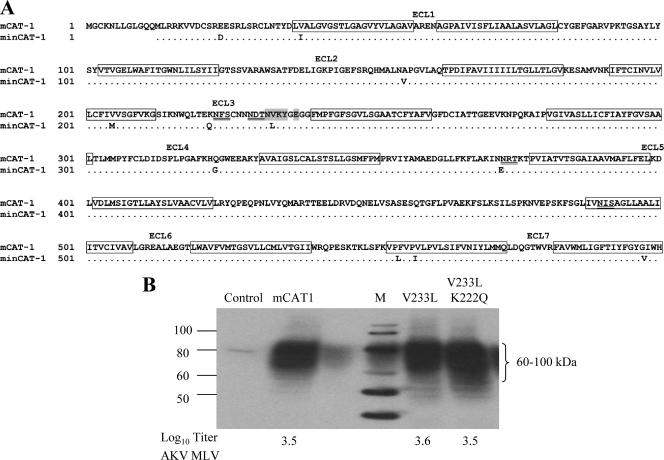
FIG. 2.
Sequence comparison of the CAT-1 receptor genes of NIH 3T3 and M. minutoides. (A) Amino acid sequence comparison. Putative transmembrane domains are boxed; the seven extracellular loops (ECL) are indicated. Glycosylation sites are underlined, and the critical region for virus binding is shaded. (B) Substitutions identified in ECL3 of minCAT-1 were introduced into mCAT-1. The clones were transfected into MA139 ferret cells, and expression was determined by immunoblot analysis. Cells were infected with AKV MLV, and virus susceptibility was quantitated by the XC overlay test. The numbers at the bottom represent log10 virus titers. Control, untransfected ferret cells; M, marker.
Site-specific mutagenesis was used to introduce one or both of these substitutions into a molecular clone of mCAT-1, and the constructs were expressed in MA139 ferret cells. As shown in Fig. 2B, the CAT-1 receptors were all expressed as heterogeneously glycosylated proteins, as expected (23). The transfected cells were all infected by AKV MLV; equivalent XC titers were produced in cells expressing mCAT1, mCAT1 with V233L, and mCAT-1 with both V233L and K222Q. Thus, these minCAT-1 receptor sequence variations do not account for resistance to AKV MLV.
We next considered the possibility that resistance may be related to suboptimal receptor expression levels or to posttranslational glycosylation specific to M. minutoides cells. We introduced mCAT-1 by transfection into the two wild mouse cell lines naturally resistant to E-MLVs (Table 2, Expt 2). Transfected M. dunni cells showed increased susceptibility to MoMLV, as previously demonstrated (13). Introduction of mCAT-1 into M. minutoides cells resulted in a 10-fold increase in susceptibility to MoMLV, but no replication of AKV MLV was detected in these cells.
Previous studies on CAT-1-mediated virus entry showed that glycosylation inhibitors can relieve the resistance to E-MLVs in rat and hamster cells (42, 46) as well as the resistance to MoMLV in M. dunni cells (14). Therefore, we treated M. minutoides cells with the inhibitor tunicamycin (Table 2, Expt 3). This reduced the resistance of M. dunni cells to MoMLV, as expected. Tunicamycin increased the XC plaque titer of MoMLV on M. minutoides, but this treatment did not result in detectable infection of M. minutoides cells with AKV MLV. Comparable results were obtained with the glycosylation inhibitor 2-deoxy-d-glucose (data not shown).
To further investigate the observed differences in virus infectivity for cells expressing different CAT-1 genes, we assessed infectivity using viral pseudotypes in a single-round infectivity assay (Table 3). We infected NIH 3T3 cells and M. minutoides cells with pCLMFG-LacZ pseudotypes constructed with MoMLV Gag-Pol and with the Env of FrMLV57, MoMLV, or AKV MLV. NIH 3T3 cells were readily infectible by all three pseudotypes, as was M. minutoides, although the wild mouse cells showed reduced susceptibility to all three pseudotypes. The greatest reduction in the pseudotype titer in M. minutoides cells was that for the AKV pseudotype, but this reduction does not account for the absolute resistance to infectious AKV MLV. This reduction, however, does suggest that Env may modulate the resistance to AKV MLV.
TABLE 3.
Titers of E-MLV LacZ pseudotypes in mouse cells
| Cell type | Log10 LacZ pseudotype titera
|
||
|---|---|---|---|
| AKV MLV | MoMLV | FrMLV57 | |
| NIH 3T3 | 4.5 | 4.9 | 4.7 |
| M. minutoides | 2.3 | 3.7 | 3.2 |
Measured as the number of blue cells in 0.1 ml of virus. Cells were tested three times.
These data, taken together, suggest that this novel resistance is not mediated by the minCAT-1 receptor variant, by cell-specific glycosylation, or by receptor level. The ability of the AKV LacZ pseudotypes to enter these cells further indicated that the AKV envelope does not mediate the resistance and suggested that the block to AKV MLV replication occurs postentry.
Restriction of AKV MLV occurs before reverse transcription.
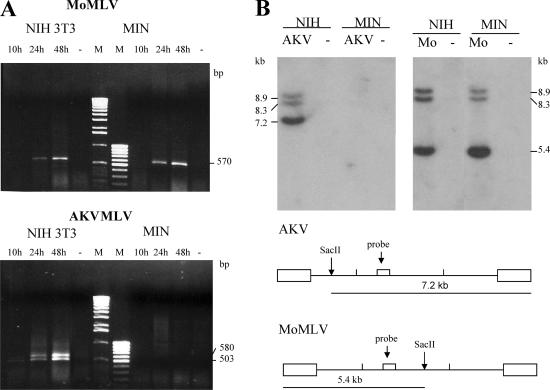
To determine if AKV MLV infection is blocked during early stages of replication, we did two experiments. Low-molecular-weight Hirt DNA was extracted at various time points from NIH 3T3 and M. minutoides cells infected with virus at a multiplicity of infection of 1 to 2. The DNAs were used for PCR with primers designed to identify 2-LTR viral circles (Fig. 3A). The expected PCR product of 570 bp was identified in MoMLV-infected NIH 3T3 and M. minutoides cells. A PCR product doublet was also identified in AKV-infected NIH 3T3 cells and corresponded to LTR fragments with one or two enhancers, as confirmed by sequencing (data not shown). No PCR products were identified in M. minutoides cells infected with AKV MLV. Two-LTR circles are hallmarks of nuclear entry, and their absence in AKV MLV-infected M. minutoides cells establishes that there is an early block to replication.
FIG. 3.
Restriction of AKV MLV in M. minutoides cells occurs before reverse transcription. (A) Hirt DNA extracts prepared at several time points after virus infection were used to detect 2-LTR viral DNA circles in infected NIH 3T3 and M. minutoides (MIN) cells. PCR identified a doublet in the NIH 3T3 cells that sequencing confirmed to represent LTRs with one or two enhancer copies. Marker lanes are labeled M. Dashes represent uninfected cells. (B) Southern blot analysis of SacII-cut Hirt DNA extracted 3 days after virus infection using a pol-specific hybridization probe. NIH 3T3 and MIN cells were infected with AKV MLV or MoMLV. Dashes represent uninfected cells. Diagrams at the bottom show the positions of the SacII sites and the hybridization probe in the two virus genomes.
Hirt DNAs from virus-infected NIH 3T3 and M. minutoides cells were cleaved with the restriction enzyme SacII; this enzyme cleaves AKV MLV and MoMLV DNAs once at different sites to generate diagnostic fragments (Fig. 3B). Southern blotting of DNAs from MoMLV-infected cells identified three fragments using a pol-specific hybridization probe; fragments of 8.3 and 8.9 kb represent linearized 1- and 2-LTR circles, and a 5.4-kb fragment represents the 5′ end of SacII-cleaved linear viral DNA. All three fragments were identified in both types of cells infected with MoMLV. AKV MLV-infected NIH 3T3 DNA produced the expected three fragments representing linearized circles and the 7.2-kb fragment of the 3′ end of cleaved linear DNA, but no viral DNA was detected in M. minutoides infected with AKV MLV. These results indicate that the block to AKV MLV replication in M. minutoides cells occurs before reverse transcription.
Resistance does not resemble that of known Fv1 alleles.
The mouse Fv1 resistance gene restricts virus replication after entry and before proviral integration. While it is typically described as a post-reverse transcription block, some combinations of virus and Fv1 allele result in reduced levels of viral DNA (9, 48), indicating that the Fv1 block can reduce replication before or during reverse transcription. To compare the M. minutoides restriction with that of Fv1, we infected cells with a panel of viruses restricted by one or more of the known Fv1 alleles. These viruses included AKV-type E-MLVs of various tropisms, as well as NR-tropic and NB-tropic FrMLVs. As shown in Table 4, AKV MLVs with three different tropisms all failed to replicate in M. minutoides cells, while NB- and NR-tropic FrMLVs replicated in these cells. Four additional virus isolates were tested on NIH 3T3 and M. minutoides cells; M. minutoides cells supported the replication of NB-tropic RaMLV but not that of three additional AKV MLV-derived viruses. This set of restricted AKV-related viruses included NR-tropic Gross Passage A MLV, WN1802B N′B (NB-tropic), and pLU7, a virus of indeterminate Fv1 tropism carrying the capsid mutation R110L (25) (data not shown). Viruses to which M. minutoides was susceptible all showed reduced XC titers in these cells relative to those in fully susceptible NIH 3T3 or M. dunni cells; this reduction, however, was not associated with the type of two-hit titration curves typically observed for Fv1-restricted virus.
TABLE 4.
Virus titers of E-MLVs of known Fv1 tropism on mouse cells carrying different Fv1 alleles
| Cells | Fv1 | Log10 virus titera (tropism)
|
||||
|---|---|---|---|---|---|---|
| AKV (N) | AKR-L1 (NR) | WN1802B (B) | FBLV (NB) | F-S MLV (NR) | ||
| M. minutoides | <0 | <0 | <0 | 3.3 | 3.9 | |
| NIH 3T3 | n | 4.3 | 4.3 | 1.0 | 4.0 | 5.8 |
| BALB/c-EF | b | 1.5 | 2.3 | 4.0 | 5.1 | 1.6 |
| 129/J-EF | nr | 2.0 | 4.2 | <0 | 4.0 | 5.8 |
| SC-1 | o | 4.5 | 4.4 | 4.8 | 4.5 | 5.9 |
Titers were determined as the number of XC plaques in 0.2 ml. In cells with known Fv1 alleles, underlined titers are reduced more than 95% (1.3 log10) relative to those in susceptible SC-1 cells.
Thus, the resistance of M. minutoides cells, here termed Minr, does not resemble that of any known Fv1 allele in several ways: the Minr pattern of resistance to this panel of viruses is unique; Minr resistance is absolute, whereas Fv1 inhibits infection by 10- to 1,000-fold; the Minr block occurs before reverse transcription rather than, as is typically observed for Fv1, after reverse transcription.
Identification of the target of resistance.
The three resistance alleles of Fv1 target specific residues in the virus capsid. Capsid amino acid 110 distinguishes N- and B-tropic viruses (25), but various substitutions in amino acid positions 82 to 117 of the CA N-terminal domain can generate NR- or NB-tropic viruses (12, 20, 27, 39, 44).
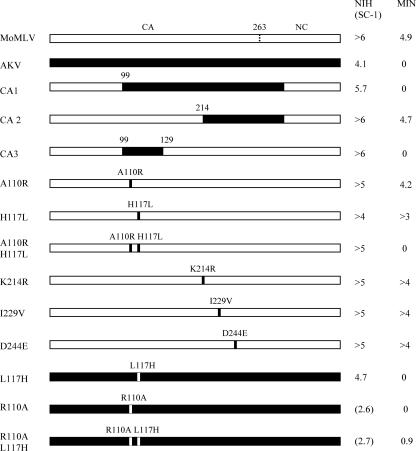
In order to identify the target of Minr resistance, we focused on two viruses, AKV and MoMLV, and made three chimeras using full-length molecular clones. Segments of CAgag and NCgag of AKV MLV were introduced into a MoMLV clone and transfected into NIH 3T3 cells. Virus collected from these cultures was tested for relative infectivity on NIH 3T3 and M. minutoides cells (Fig. 4). The failure of CA3 to replicate in MIN cells suggests that the target of this resistance is within a 30-amino-acid segment of the AKV CA that overlaps the Fv1 target region. This region contains only two substitutions that distinguish MoMLV and AKV MLV: H117L and A110R. These substitutions were introduced into MoMLV separately or together. The individual substitutions did not generate a virus restricted in M. minutoides cells, but the combination of H117L with A110R produced a virus that had a high XC titer (>105) in NIH 3T3 cells but did not replicate in M. minutoides cells (Fig. 4).
FIG. 4.
Susceptibilities of NIH 3T3, SC-1, and M. minutoides (MIN) cells to MoMLV and AKV MLV or to viruses with mutated or chimeric CA and NC genes. The numbers at the right represent virus titers in 0.2 ml as determined by the XC overlay assay.
We then introduced the reciprocal changes L117H and R110A, separately and in combination, into pGEM-AKV. Clones carrying the mutations were transfected into NIH 3T3 (L117H) or SC-1 (R110A, R110A L117H) cells. Both viruses with R110A were grown and tested in SC-1 instead of NIH 3T3 cells, because previous work indicated that this mutation is at least partly restricted in NIH 3T3 cells (39). The L117H mutation alone did not alter the AKV MLV tropism (Fig. 4). Both viruses carrying R110A had reduced titers on SC-1 cells. The R110A mutation alone had no effect on the inability of AKV MLV to replicate in M. minutoides cells, but the combination of R110A and L117H generated a virus that was able to replicate in these cells; in four experiments, the XC titer of virus carrying this double mutation was reduced 10- to 100-fold in M. minutoides cells compared to SC-1 cells, a reduction that is comparable to the reduced susceptibility of the wild mouse cells to MoMLV. These results are consistent with the results obtained using MoMLV carrying reciprocal mutations, and they indicate that the target of the Minr restriction includes these two amino acid residues in the virus capsid.
Sequences of the Fv1 target regions of 10 viruses used in these studies are shown in Fig. 5. Among the six Minr-restricted viruses, only three share the R110 L117 combination identified as the target of Minr restriction by the mutagenesis results; among the four nonrestricted viruses, only MoMLV has the A110 H117 combination associated with a lack of restriction. This suggests that, as has been shown for NB- and NR-tropic viruses, other substitutions can generate Minr-restricted MLVs. Sequence comparison of the Fv1-sensitive regions of all 10 CA genes failed to identify any specific residue or obvious combination of residues that distinguishes the restricted from the unrestricted viruses. Outside the Fv1-sensitive region, we identified common substitutions at only three sites, positions 214, 229, and 244. We generated three additional MoMLV mutants carrying the AKV residue at these sites (Fig. 4), but viruses with all three of these changes replicated efficiently in M. minutoides cells. These results indicate that the Minr resistance factor targets two amino acids in the Fv1-sensitive region of capsid and that viruses with the restricted tropism may be generated by additional substitutions or combinations of substitutions.
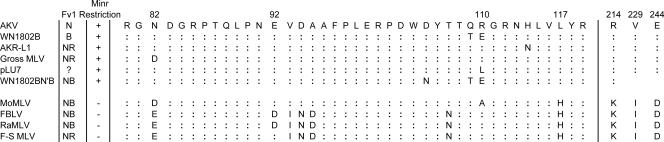
FIG. 5.
Amino acid sequence comparison of the Fv1 target regions of CA of six viruses restricted in M. minutoides cells and four viruses not restricted in M. minutoides cells. Sequences for seven of the viruses have been determined previously: AKV (GenBank accession number J01998), WN1802B and pLU7 (25), AKR-L1 (20), Gross (39), MoMLV (GenBank accession number J02255), and RaMLV (GenBank accession number U94692).
DISCUSSION
Cells of the African pygmy mouse show a novel resistance to the naturally occurring AKV-type E-MLVs but are susceptible to infection with laboratory-derived Friend, Moloney, and Rauscher E-MLVs. The block to replication affects an early stage in virus replication and targets the virus CA. While there is some resemblance to Fv1-type resistance, this unusual wild-mouse type of intrinsic immunity, here termed Minr, differs from Fv1 in several important ways. (i) The Minr pattern of E-MLV resistance does not resemble that of any known Fv1 allele. (ii) Fv1 reduces the titer of the restricted virus 10- to 1,000-fold, whereas the Minr block is absolute. (iii) Fv1 restriction occurs post-reverse transcription, whereas the Minr block is generally before reverse transcription.
While resistance to AKV MLVs in M. minutoides cells is clearly due to a postentry block, we also noted that the infectibility of these cells by Friend, Moloney, and Rauscher viruses was reduced relative to that of NIH 3T3 cells. This reduction was seen by the XC plaque test and by infectibility with LacZ pseudotypes; it may be due in part to receptor density, since transfection with mCAT-1 increased susceptibility to infection with MoMLV. M. minutoides has an altered E-MLV CAT-1 receptor, minCAT-1, which has two substitutions in the third extracellular loop, containing the critical residues for virus entry. The positions of these two substitutions, V233L and K222Q, suggested a potential role in the reduced infectibility. K222Q is adjacent to one of two extracellular glycosylation sites in this protein at N223. The second site, V233L, is within the receptor critical region (10). Previous studies have suggested that mutations at V233 can have a major effect on infectibility (1, 36, 50). Our results, however, indicate that V233L alone or in combination with K222Q does not produce an inactive receptor or have more than a small effect on the efficiency of infection with AKV MLV.
The postentry Minr resistance targets the virus CA, and mutagenesis identified two critical amino acid residues in restricted AKV MLV: L117 and R110. Residues at both of these CA positions also govern sensitivity to two known resistance factors, Fv1 and TRIM5α. CA110 is the target for Fv1n (E110) and Fv1b (R110) restriction (25), and MLVs bearing R110, A110, and H117 are restricted by human TRIM5α (49). Studies on naturally occurring or mutated MLVs with altered tropism have identified at least eight additional positions within the CA82-214 segment that can alter sensitivity to Fv1 or TRIM5α, and these tropism-determining residues can be masked or modulated when combined (12, 20, 27, 39, 44). A similar ability to target multiple capsid residues or combinations of residues may also explain why the 2-amino-acid Minr target identified in AKV MLV (R110 L117) was not present in three of the six restricted viruses. Further mutagenesis may uncover additional residues or combinations of residues that generate Minr-restricted viruses.
The Minr restriction bears some similarities to Fv1 restriction, and characterization of the M. minutoides Fv1 gene is in progress. M. minutoides is not closely related to the wild mouse progenitors of the laboratory strains that carry the known Fv1 allelic variants (Fig. 1). The genus Mus originated in the Indian subcontinent about 7.5 million years ago, and the African pygmy mouse subgenus (Nannomys) diverged about 6.5 million years ago. Fv1-related sequences have been identified in all Mus species, including M. minutoides (35); the Fv1 genetic sequence is related to the gag gene of the MuERV-L family of retroviruses (3). Multiple copies of this ERV family are present in all mice, but there have been two major copy number expansions of MuERV-L during Mus evolution: one expansion occurred at the base of the Mus phylogenetic tree, and the other at the later palearctic radiation, which predates the appearance of Fv1 restriction (4).
Studies on the species and tissue tropisms of retroviruses and lentiviruses have uncovered several host factors other than Fv1 that are responsible for Minr-like examples of intrinsic immunity. Two of the features that define Minr, the targeted capsid residues and the post-reverse transcription block in replication, are also hallmarks of TRIM5α restriction (49). Although there have been a few studies of TRIM5α in nonprimate species (43), mice have no clearly recognizable orthologue of the human TRIM5 gene, and the most closely related mouse gene (9230105E10Rik) does not restrict HIV type 1 (HIV-1) (40).
In addition to TRIM5α and Fv1, other host retrovirus resistance factors have been identified that could be considered candidates for Minr (38). Lv2 and Lv3 restrict late entry stages in infection with HIV-2 in human cells and HIV-1 in macaque cells, respectively (29, 34). The Lv2 restriction, like Fv1 and TRIM5α restriction, targets the viral capsid gene, but both Lv2 and Lv3 differ from Fv1 and TRIM5α in that restriction is also dependent on Env and/or entry pathways. Although we found no evidence that Minr restriction is at the level of entry, our observation that LacZ pseudotypes with AKV Env are reduced 10-fold relative to pseudotypes with MoMLV or FrMLV57 Env suggests that either Env modulates the Minr restriction or there is a secondary block at an Env-dependent step preceding reverse transcription.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, NIAID.
We thank Esther Shaffer and Qingping Liu for expert technical assistance, Alicia Buckler-White for sequencing, and Caroline Ball for editorial assistance in the preparation of the manuscript. We also thank Jonathan Silver for helpful discussions.
Footnotes
Published ahead of print on 16 April 2008.
REFERENCES
- 1.Albritton, L. M., J. W. Kim, L. Tseng, and J. M. Cunningham. 1993. Envelope-binding domain in the cationic amino acid transporter determined the host range of ecotropic murine retroviruses. J. Virol. 672091-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassin, R. H., G. Duran-Troise, B. I. Gerwin, and A. Rein. 1978. Abrogation of Fv-1b restriction with murine leukemia viruses inactivated by heat or by gamma irradiation. J. Virol. 26306-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bénit, L., N. De Parseval, J.-F. Casella, I. Callebaut, A. Cordonnier, and T. Heidmann. 1997. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol. 715652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bénit, L., J.-B. Lallemand, J.-F. Casella, H. Philippe, and T. Heidmann. 1999. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J. Virol. 733301-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best, S., P. LeTossier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382826-829. [DOI] [PubMed] [Google Scholar]
- 6.Bieniasz, P. D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 51109-1115. [DOI] [PubMed] [Google Scholar]
- 7.Bishop, K. N., M. Bock, G. Towers, and J. P. Stoye. 2001. Identification of the regions of Fv1 necessary for murine leukemia virus restriction. J. Virol. 755182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boursot, P., J.-C. Auffray, J. Britton-Davidian, and F. Bonhomme. 1993. The evolution of house mice. Annu. Rev. Ecol. Syst. 24119-152. [Google Scholar]
- 9.Chinsky, J., and R. Soeiro. 1981. Fv-1 host restriction of Friend leukemia virus: analysis of unintegrated proviral DNA. J. Virol. 4045-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davey, R. A., Y. Zuo, and J. M. Cunningham. 1999. Identification of a receptor-binding pocket on the envelope protein of Friend murine leukemia virus. J. Virol. 733758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Declève, A., O. Niwa, E. Gelmann, and H. S. Kaplan. 1975. Replication kinetics of N- and B-tropic murine leukemia viruses on permissive and nonpermissive cells in vitro. Virology 65320-332. [DOI] [PubMed] [Google Scholar]
- 12.Doi, K., A. Kawana, A. Iwamoto, H. Yoshikura, and T. Odawara. 1997. One base change is sufficient for host range conversion of murine leukemia virus from B to NB tropism. Arch. Virol. 1421889-1894. [DOI] [PubMed] [Google Scholar]
- 13.Eiden, M. V., K. Farrell, J. Warsowe, L. C. Mahan, and C. A. Wilson. 1993. Characterization of a naturally occurring ecotropic receptor that does not facilitate entry of all ecotropic murine retroviruses. J. Virol. 674056-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eiden, M. V., K. Farrell, and C. A. Wilson. 1994. Glycosylation-dependent inactivation of the ecotropic murine leukemia virus receptor. J. Virol. 68626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartley, J. W., and W. P. Rowe. 1975. Clonal cell lines from a feral mouse embryo which lack host-range restrictions for murine leukemia viruses. Virology 65128-134. [DOI] [PubMed] [Google Scholar]
- 16.Hartley, J. W., W. P. Rowe, and R. J. Huebner. 1970. Host-range restrictions of murine leukemia viruses in mouse embryo cell cultures. J. Virol. 5221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 10110774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26365-369. [DOI] [PubMed] [Google Scholar]
- 19.Jolicoeur, P., and E. Rassart. 1980. Effect of Fv-1 gene product on synthesis of linear and supercoiled viral DNA in cells infected with murine leukemia virus. J. Virol. 33183-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung, Y. T., and C. A. Kozak. 2000. A single amino acid change in the murine leukemia virus capsid gene responsible for the Fv1nr phenotype. J. Virol. 745385-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung, Y. T., T. Wu, and C. A. Kozak. 2004. Novel host range and cytopathic variant of ecotropic Friend murine leukemia virus. J. Virol. 7812189-12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keckesova, Z., L. M. J. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 10110780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J. W., and J. M. Cunningham. 1993. N-linked glycosylation of the receptor for murine ecotropic retroviruses is altered in virus-infected cells. J. Biol. Chem. 26816316-16320. [PubMed] [Google Scholar]
- 24.Kozak, C. A. 1985. Analysis of wild-derived mice for the Fv-1 and Fv-2 murine leukemia virus restriction loci: a novel wild mouse Fv-1 allele responsible for lack of host range restriction. J. Virol. 55281-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak, C. A., and A. Chakraborti. 1996. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology 225300-305. [DOI] [PubMed] [Google Scholar]
- 26.Lander, M. R., and S. K. Chattopadhyay. 1984. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J. Virol. 52695-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lassaux, A., M. Sitbon, and J.-L. Battini. 2005. Residues in the murine leukemia virus capsid that differentially govern resistance to mouse Fv1 and human Ref1 restrictions. J. Virol. 796560-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenz, J., R. Crowther, A. Staceski, and W. Haseltine. 1982. Nucleotide sequence of Akv env gene. J. Virol. 42519-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchant, D., S. J. D. Neil, K. Aubin, C. Schmitz, and A. McKnight. 2005. An envelope-determined, pH-independent endocytic route of viral entry determines the susceptibility of human immunodeficiency virus type 1 (HIV-1) and HIV-2 to Lv2 restriction. J. Virol. 799410-9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ou, W., N. Lu, S. S. Yu, and J. Silver. 2006. Effect of epitope position on neutralization by anti-human immunodeficiency virus monoclonal antibody 2F5. J. Virol. 802539-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 10111827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pincus, T., J. W. Hartley, and W. P. Rowe. 1971. A major genetic locus affecting resistance to infection with murine leukemia viruses. I. Tissue culture studies of naturally occurring viruses. J. Exp. Med. 1331219-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pincus, T., J. W. Hartley, and W. P. Rowe. 1975. A major genetic locus affecting resistance to infection with murine leukemia viruses. IV. Dose-response relationships in Fv-1 sensitive and resistant cell cultures. Virology 65333-342. [DOI] [PubMed] [Google Scholar]
- 34.Pineda, M. J., B. R. Orton, and J. Overbaugh. 2007. A TRIM5α-independent post-entry restriction to HIV-1 infection of macaque cells that is dependent on the path of entry. Virology 363310-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi, C.-F., F. Bonhomme, A. Buckler-White, C. Buckler, A. Orth, M. R. Lander, S. K. Chattopadhyay, and H. C. Morse III. 1998. Molecular phylogeny of Fv1. Mamm. Genome 91049-1055. [DOI] [PubMed] [Google Scholar]
- 36.Qian, Z., R. Donald, H. Wang, Q. Chen, and L. M. Albritton. 2003. Identification of a critical basic residue on the ecotropic murine leukemia virus receptor. J. Virol. 778596-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowe, W. P., W. E. Pugh, and J. W. Hartley. 1970. Plaque assay techniques for murine leukemia viruses. Virology 421136-1139. [DOI] [PubMed] [Google Scholar]
- 38.Schmitz, C., D. Marchant, S. J. D. Neil, K. Aubin, S. Reuter, M. T. Dittmar, and A. McKnight. 2004. Lv2, a novel postentry restriction, is mediated by both capsid and envelope. J. Virol. 782006-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevens, A., M. Bock, S. Ellis, P. LeTissier, K. N. Bishop, M. W. Yap, W. Taylor, and J. P. Stoye. 2004. Retroviral capsid determinants of Fv1 NB and NR tropism. J. Virol. 789592-9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. USA 1035514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sveda, M. M., and R. Soeiro. 1976. Host restriction of Friend leukemia virus: synthesis and integration of the provirus. Proc. Natl. Acad. Sci. USA 732356-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tavoloni, N., and A. Rudenholz. 1997. Variable transduction efficiency of murine leukemia retroviral vector on mammalian cells: role of cellular glycosylation. Virology 22949-56. [DOI] [PubMed] [Google Scholar]
- 43.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 9712295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ulm, J. W., M. Perron, J. Sodroski, and R. C. Mulligan. 2007. Complex determinants within the Moloney murine leukemia virus capsid modulate susceptibility of the virus to Fv1 and Ref1-mediated restriction. Virology 363245-255. [DOI] [PubMed] [Google Scholar]
- 45.Veyrunes, F., G. Dobigny, F. Yang, P. C. M. O'Brien, J. Catalan, T. J. Robinson, and J. Britton-Davidian. 2006. Phylogenomics of the genus Mus (Rodentia: Muridae): extensive genome repatterning is not restricted to the house mouse. Proc. R. Soc. B 2732925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, C. A., and M. V. Eiden. 1991. Viral and cellular factors governing hamster cell infection by murine and gibbon ape leukemia viruses. J. Virol. 655975-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, H., T. A. Bell, G. A. Churchill, and F. Pardo-Manuel de Villena. 2007. On the subspecific origin of the laboratory mouse. Nat. Genet. 391100-1107. [DOI] [PubMed] [Google Scholar]
- 48.Yang, W. K., J. O. Kiggans, D.-M. Yang, C.-Y. Ou, R. W. Tennant, A. Brown, and R. H. Bassin. 1980. Synthesis and circularization of N- and B-tropic retroviral DNA in Fv-1 permissive and restrictive mouse cells. Proc. Natl. Acad. Sci. USA 772994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 10110786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshimoto, T., E. Yoshimoto, and D. Meruelo. 1993. Identification of amino acid residues critical for infection with ecotropic murine leukemia retrovirus. J. Virol. 671310-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]