Abstract
The Drosophila melanogaster circadian oscillator comprises interlocked per/tim and Clk transcriptional feedback loops. In the per/tim loop, CLK-CYC-dependent transcriptional activation is rhythmically repressed by PER or PER-TIM to control circadian gene expression that peaks around dusk. Here we show that rhythmic transcription of per and tim involves time-of-day-specific binding of CLK-CYC and associated cycles in chromatin modifications. Activation of per and tim transcription occurs in concert with CLK-CYC binding to upstream and/or intronic E-boxes, acetylation of histone H3-K9, and trimethylation of histone H3-K4. These events are associated with RNA polymerase II (Pol II) binding to the tim promoter and transcriptional elongation by Pol II that is constitutively bound to the per promoter. Repression of per and tim transcription is associated with PER-dependent reversal of these events. Rhythms in H3-K9 acetylation and H3-K4 trimethylation are also associated with CLOCK-BMAL1-dependent transcription in mammals, indicating that the mechanism that controls rhythmic transcription is a conserved feature of the circadian clock even though feedback repression is mediated by different proteins.
In Drosophila melanogaster, circadian oscillator function is initiated by the basic helix-loop-helix PERIOD (PER)-ARNT-SIM transcription factors CLOCK (CLK) and CYCLE (CYC). These two proteins form a heterodimer that binds CACGTG E-box elements to activate transcription of target genes including per, tim, vri, and Pdp1ɛ during midday (zeitgeber time ∼6 h [ZT 6]) (2, 7, 9, 19, 34, 48). At this time, CLK-CYC is expected to promote histone modifications associated with transcriptional activation such as acetylation and H3-K4 trimethylation. As per mRNA accumulates to peak levels around dusk, PER accumulates in the cytoplasm, where it associates with TIM (11, 16, 21, 23, 35, 37, 43, 50, 51) and then enters the nucleus and inhibits CLK-CYC activity (5, 9, 35, 44). This inhibition is expected to involve histone modifications such as deacetylation and H3-K9 dimethylation, which are typically involved in transcriptional repression. PER is degraded early in the light phase to relieve transcriptional inhibition, thereby allowing the next cycle of CLK-CYC transcription to begin (11, 17, 26, 50, 51).
The mouse homologs of Drosophila CLK-CYC, CLOCK-BMAL1, have been shown to bind rhythmically to E-box elements in Dbp upstream and intronic sequences (41) but are constantly bound to E-boxes upstream of Per1 (28). Apparently, time-specific interactions between CRYPTOCHROME (CRY) or PER-CRY (PER-CRY) and CLOCK-BMAL1 inhibit transcription either by removing CLOCK-BMAL1 from E-boxes or by blocking CLOCK-BMAL1 transactivation. In Drosophila, CLK-CYC binds rhythmically to E-boxes in the per circadian regulatory sequence (CRS) and the tim upstream sequence (49), indicating that PER (or PER-TIM) inhibits transcription by removing CLK-CYC from E-boxes. Additional E-boxes in the per upstream, per intron 1, and tim intron 1 regions may also contribute to the proper phase and/or amplitude of per and tim mRNA cycling (19, 34, 48), though CLK-CYC binding to these E-boxes has not been determined. In mice, CLOCK-BMAL1 binding is associated with histone modifications that promote transcriptional activity including acetylation of histones H3 and H4 (6, 10, 12, 38, 41) and trimethylation of H3-K4 (41). Transcriptional repression by CRY is associated with methylation of H3-K27 (13), histone deacetylation (38), and dimethylation of H3-K9 (41). Given that PER and CRY are the key feedback repressors in flies and mammals, respectively, it is possible that some of the rhythmic transitions in histone modifications may be different between flies and mammals.
Histone modifications regulate transcription by controlling RNA polymerase II (Pol II) binding and activity. In mice, CLOCK-BMAL1 activity and associated rhythms in chromatin modifications occur in concert with Pol II binding to the Per1, Per2, and Cry1 promoters (6, 12). The C-terminal domain (CTD) of Pol II consists of multiple heptad repeats that mediate phosphorylation-dependent changes in Pol II activity. The Pol II CTD is phosphorylated at serine 5 (S5) by TFIIH as transcription is initiated at the 5′ end of the transcribed region and is phosphorylated at S2 by Ctk kinase as Pol II moves toward the 3′ end of the transcription unit (3). The phosphorylated Pol II CTD recruits factors important for productive Pol II elongation, RNA processing, and mRNA export (18). Constitutive binding of Pol II to rhythmically expressed genes might have suggested that Pol II activity is regulated, but this was not seen for Per1, Per2, and Cry1 in mice.
Circadian transcriptional regulation corresponds well with CLK-CYC binding to the per CRS and tim upstream E-boxes (49). Here we show that CLK-CYC rhythmically binds E-boxes in per intron 1 but doesn't bind other E-boxes in the per upstream region or in tim intron 1. These rhythms in CLK-CYC binding are associated with H3-K9 acetylation and H3-K4 trimethylation, thus demonstrating that these histone modifications are a conserved feature of the clock mechanism. PER-dependent transcriptional inhibition coincides with the reversal of these histone modifications. Rhythms in H3-K9 acetylation and H3-K4 trimethylation coincide with Pol II binding to the tim promoter. However, constitutive binding of Pol II to the per promoter suggests that rhythms in per transcription are controlled at the level of Pol II elongation.
MATERIALS AND METHODS
Fly stocks.
The Canton Special strain was used as the wild type for all experiments. The per01 and cyc01 fly strains were used to assess chromatin modifications when CLK-CYC is constitutively bound or not bound to E-boxes, respectively (49).
Antibodies.
Antibodies against histone modifications were acetyl-histone H3-K9 (Abcam; ab4441), acetyl-histone H3-K14 (Millipore; 07-353), acetyl-histone H4 (Millipore; 06-866), dimethyl-histone H3-K9 (Millipore; 07-441), trimethyl-histone H3-K4 (Abcam; ab8580), and phosphoryl-histone H3-S10 (Millipore; 05-817). Either rabbit immunoglobulin G (IgG; Sigma; I-5006) or rabbit serum (Sigma; R-9133) was used as a negative control for chromatin immunoprecipitation (ChIP) experiments for these antibodies depending of whether the selective antibody was purified IgG or was in serum. For purified antibodies, 3 μg antibody or control IgG was used for ChIP experiments. For antibodies in serum, 5 μl antibody or control serum was used for ChIP experiments.
Pol II, clone H14 (Covance; MMS-134R), which recognizes the heptapeptide repeat phosphorylated at serine 5 in the CTD, and Pol II, clone 8WG16 (Upstate, 05-952), which recognizes the unphosphorylated heptapeptide repeat in the CTD, were used for ChIP analysis. The H14 antibody was also used for Western blot analysis along with actin (Abcam; ab8224) as a loading control. For clone H14, rabbit anti-mouse IgM (Zymed; 61-6800) was used as a linker. Mouse IgG (Sigma; I-5318) was used as a negative control for 8WG16, and no IgG was used as a negative control for H14 for these ChIP experiments.
ChIP protocol.
ChIP experiments were done as described by Yu et al. (49) with some modifications. For ChIP with Pol II H14, a rabbit anti-mouse IgM linker was needed. After the cross-linking was reversed, the DNA was purified using a QIAquick PCR purification kit (Qiagen; 28104). For the positions and sequences of primers used for semiquantitative PCR and quantitative real-time PCR (qPCR) of ChIP samples see Table S1 in the supplemental material.
Semiquantitative PCR.
Semiquantitative PCR was carried out as described by Yu et al. (49) except that the PCR was performed for 18 cycles using a concentration of 0.5 μM of hot primers throughout the reaction.
qPCR.
ChIP samples and controls were diluted 1:10 and inputs were diluted 1:100 in Qiagen elution buffer. The qPCR was prepared as described previously (1), with the following modifications. For a 1× reaction the following were combined: 5 μl Sybr green PCR Master Mix (Applied Biosystems; 4309155), 0.66 μl forward primer (10 μM), 0.66 μl reverse primer (10 μM), 1.68 μl water, 2 μl of DNA. The ABI 7500 real-time PCR system was run in fast 7500 mode and set up as follows: stage 1, 1 cycle for 10 min at 95°C; stage 2, 40 cycles of 30 s at 95°C, 30 s at 53 to 60°C (depending upon the primer set), and 30 s at 60°C. The data collection step was set to stage 2, step 3, at 30 s. For each plate, the average of the cycle threshold values (CTs) was calculated for each input, sample, and control. The input CT was subtracted from the corresponding sample and control CTs. The following formula was then applied: power·1.9·negative ln (subtracted value). This value was used for further calculations. Each sample and control was normalized by dividing both numbers by the highest value so that each ChIP experiment was scaled from 0 to 1. For sample minus control values, a negative number was replaced with a zero.
Western blot analysis.
Whole-head protein samples were prepared from wild-type flies collected at ZT 2, 6, 10, 14, 18, and 22 by extraction in RBS buffer (49). Sonicated cross-linked nuclear extract (SXN) samples used in ChIP experiments were also used for Western analysis. For sodium dodecyl sulfate-polyacrylamide gel electrophoresis, 100 μg of whole-head sample or 10 μl of SXN sample was run on Criterion precast gels (Bio-Rad) and transferred to Hybond-P membranes (Amersham). To determine Pol II S5 levels, immunoblots were probed with Pol II H14 antibody at a dilution of 1:5,000 and a peroxidase-conjugated goat anti-mouse IgM (Sigma; A8786) secondary antibody at a dilution of 1:1,000 or probed with actin antibody at a dilution of 1:50,000 and a peroxidase-conjugated goat anti-mouse IgG (Sigma; A5278) secondary antibody at a dilution of 1:1,000 as a loading control. Phosphoryl-histone H3-S10 antibody was also used at a dilution of 1:2,000 and a peroxidase-conjugated goat anti-rabbit IgG (Sigma; A6154) secondary antibody at a dilution of 1:1,000. All probing was done for 1 h at room temperature. Immunoblots were visualized with ECL Plus (Amersham). Protein levels were calculated by densitometry analysis using Quantity One software (Bio-Rad). Background was removed by subtracting an area of the lane with no bands from an equal-size area containing the specific band. Levels were normalized to the actin loading control value. Relative levels were calculated by dividing each sample value by the peak sample value for each time course.
Statistical analysis.
For statistical analysis STATISTICA 7 software was used. The sample values minus control values were used in one-way analysis of variance, with post hoc analysis using the Fisher least significant difference to determine significant differences.
RESULTS
CLK-CYC binding to per and tim E-boxes.
Previous work has demonstrated that CLK-CYC binds an E-box upstream of per in the CRS and to an E-box upstream of tim (49). Several other canonical CACGTG E-boxes are present in per upstream and intron 1 sequences (Fig. 1), and a second canonical E-box is present in tim intron 1 (Fig. 2). To determine which per and tim E-boxes are bound by CLK-CYC in vivo, ChIP was done using CLK antiserum. ChIP analysis was performed on flies collected during light-dark (LD) cycles since per and tim mRNA cycling amplitude is high under these conditions yet reflects regulatory processes occurring in constant darkness (21, 43, 49).
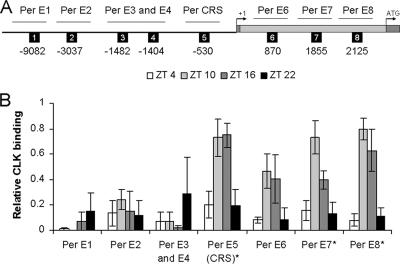
FIG. 1.
CLK rhythmic binding to per E-boxes. (A) Schematic representation of per indicating regions analyzed for CLK binding E-boxes. Upstream sequence, thick horizontal line; transcription start, arrow with +1; exons, dark gray bars; intron 1, light gray bar; translation start, arrow with ATG; E-boxes, black squares numbered 1 through 8 and marked by the position of the first nucleotide relative to the transcription start site; regions analyzed by ChIP, thin horizontal lines marked by the E-boxes they contain. (B) ChIP was performed in wild-type flies collected at ZT 4, ZT 10, ZT 16, and ZT 22 during a 12:12 LD cycle. The relative level of CLK binding for each E-box-containing region was determined by qPCR analysis of samples immunoprecipitated with anti-CLK or guinea pig serum (GPS). The relative levels of CLK binding have been corrected for nonspecific binding by subtracting GPS values from CLK values. These data were derived from three independent experiments. *, statistically significant (P < 0.05) rhythm.
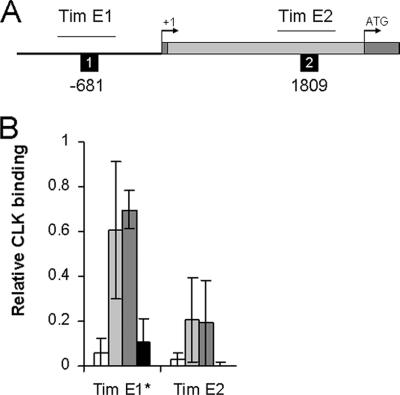
FIG. 2.
CLK rhythmic binding to tim E-boxes. (A) Schematic representation of tim indicating regions analyzed for CLK binding E-boxes. Symbols are as described in the Fig. 1A legend. (B) ChIP was performed as described for Fig. 1B. The relative level of CLK binding for each E-box-containing region was determined and quantified as described for Fig. 1B. These data were derived from three independent experiments. *, statistically significant (P < 0.05) rhythm.
As shown previously, strong binding to the per CRS E-box was detected and displayed significant (P < 0.02) diurnal cycling (Fig. 1) (49). However, CLK showed little if any binding to the E-boxes upstream of the CRS but exhibited moderate rhythmic binding to Per E-box 6 and strong rhythmic binding to Per E-box 7 (P < 0.001) and Per E-box 8 (P < 0.01), all of which are located in intron 1 (Fig. 1). The resolution of the ChIP assay is not sufficient to distinguish between CLK binding to Per E-box 7 only, to Per E-box 8 only, or to both Per E-box 7 and 8. CLK also strongly binds the E-box upstream of tim in a rhythmic manner (P < 0.03) (Fig. 2), as was shown previously (49). In contrast, only weak rhythmic binding of CLK to the E-box in tim intron 1 was detected (Fig. 2). These two E-boxes were far enough away from each other (>1 kb) to be distinguishable by the resolution of the ChIP assay. The strong binding of CLK to the E-box upstream of tim may not be due solely to the canonical CACGTG E-box since noncanonical E-boxes in tim upstream sequences also contribute to tim mRNA cycling (34). These results demonstrate that CLK, and by extension CLK-CYC, bind rhythmically to multiple E-boxes located upstream of, and within, the per gene and to an E-box upstream of tim.
Histone modifications in wild-type flies.
CLK-CYC strongly binds to per and tim E-boxes at ZT 10 and/or ZT 16 but weakly at ZT 4 and ZT 22 (Fig. 1 and 2) (49). Such binding coincides with high levels of target gene transcription; thus, chromatin associated with target gene DNA is expected to contain modifications that promote transcriptional activity. The acetylation and methylation of histones are known to alter transcriptional activity and occur in parallel to transcriptional rhythms in Per1, Per2, Cry1, and Dbp in mammals (6, 10, 12, 38, 41). To determine if histone modifications are also associated with rhythmic transcription in Drosophila, ChIP was performed in wild-type flies at ZT 4, 10, 16, and 22 to assess rhythmic histone modifications. Primers were designed to analyze regions near or including E-boxes and sites of transcriptional and translational initiation for per and tim (Fig. 3 and 4). Acetylation, trimethylation of H3-K4, and phosphorylation were expected to promote transcriptional activation, and dimethylation of H3-K9 was expected to be favorable for transcriptional repression (14). Antibodies were used to probe acetylation of H3-K9, H3-K14, and H4; dimethylation of H3-K9; trimethylation of H3-K4; and phosphorylation of H3-S10.
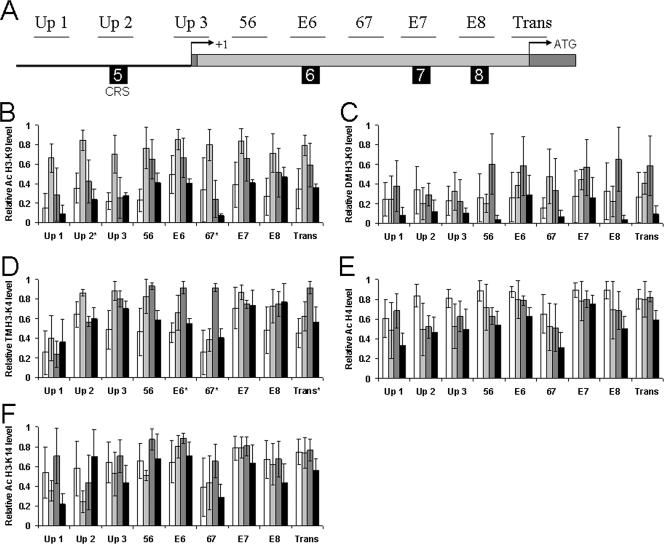
FIG. 3.
Histone modifications in the per upstream and intron 1 regions. (A) Schematic representation of sites analyzed for histone modifications. The upstream sequence, transcription start, exons, intron 1, translation start, and E-boxes are depicted as described for Fig. 1A. Only E-boxes that show rhythmic CLK binding are shown. The thin horizontal lines labeled Up 1, Up 2, Up 3, 56, E6, 67, E7, E8, and Trans represent per genomic regions analyzed by ChIP. (B to F) ChIP was performed as described for Fig. 1B. Relative levels of (B) acetyl (Ac)-H3-K9, (C) dimethyl (DM)-H3-K9, (D) trimethyl (TM)-H3-K4, (E) acetyl-H3-K14, and (F) acetyl-H4 were determined by qPCR analysis. The levels of each histone modification have been corrected for nonspecific binding by subtracting the rabbit IgG or rabbit serum ChIP values at each time point. These data were derived from three independent experiments. *, statistically significant (P < 0.05) rhythm.
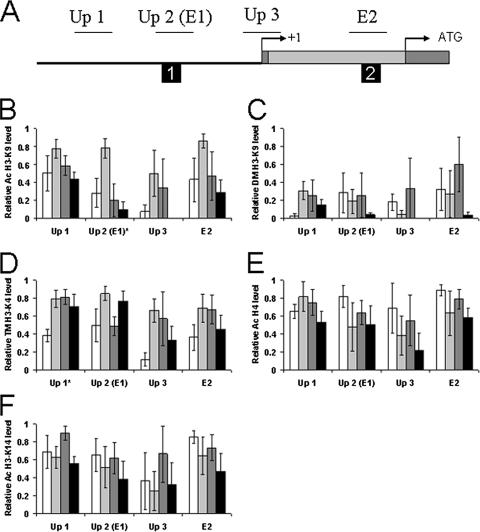
FIG. 4.
Histone modifications in the tim upstream and intron 1 regions. (A) Schematic representation of sites analyzed for histone modifications. Symbols are depicted as described for Fig. 3A, except that the regions analyzed by ChIP are labeled Up 1, Up 2, Up 3, and E2. (B to F) ChIP was performed as described for Fig. 1B. Histone modifications are abbreviated as in Fig. 3B. Relative levels of (B) acetyl-H3-K9, (C) dimethyl-H3-K9, (D) trimethyl-H3-K4, (E) acetyl-H3-K14, and (F) acetyl-H4 were determined by qPCR analysis. The levels of each histone modification have been corrected for nonspecific binding as described for Fig. 3B. These data were derived from three independent experiments. *, statistically significant (P < 0.05) rhythm.
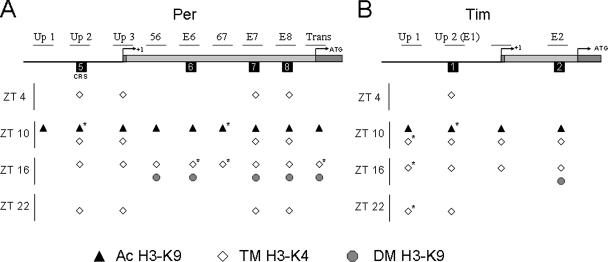
Phosphorylation of H3-S10 was not detected for any per or tim region (data not shown), whereas acetylation of H3-K14 and H4 was relatively constant for every per and tim region tested (Fig. 3 and 4). Acetylation of H3-K9 was rhythmic, with a peak at ZT 10 for most per and tim sites analyzed, though rhythmicity was significant for only the Per Up 2 (P < 0.03), Per 67 (P < 0.04), and Tim Up 2 (P < 0.02) regions (Fig. 3 and 4). Given that acetylation is associated with transcriptional activation, this result is consistent with high levels of CLK-CYC binding and per and tim transcription at ZT 10 (45, 49). Trimethylation of H3-K4 was rhythmic, constant, or very low, depending upon the region analyzed (Fig. 3 and 4). Rhythmic trimethylation of per and tim peaked at ZT 10 or ZT 16 and was significant for Per E-box 6 (P < 0.02), Per 67 (P < 0.01), Per Translation (P < 0.04), and Tim Up 1 (P < 0.02). The peak phase of trimethylation coincides with CLK binding and transcriptional activation of per and tim (45, 49), consistent with the known role of H3-K4 trimethylation (14). Dimethylation of H3-K9 was found to be rhythmic only in the first introns of per and tim (Fig. 3 and 4). Although the rhythm amplitude was relatively low and not statistically significant, a consistent peak was observed at ZT 16. Since this time corresponds to the end of CLK-CYC activation of per and tim, it is not clear whether this modification is involved in activation or repression. Dimethylation of H3-K9 within the coding region has been shown in some cases to be associated with Pol II elongation (47), but experiments with mammals have shown that dimethylation is associated with the repressed state of the circadian gene Dbp (41). Thus, in wild-type flies, H3-K14 and H4 acetylation is constantly high and S10 phosphorylation is not detected in per and tim upstream or intron 1 sequences, whereas H3-K9 acetylation, H3-K9 dimethylation, and H3-K4 trimethylation are rhythmic in some per and tim upstream or intron 1 sequences and not others (Fig. 5). Importantly, rhythms in H3-K9 acetylation, H3-K9 dimethylation, and H3-K4 trimethylation parallel rhythms in per and tim transcriptional activity.
FIG. 5.
Summary of per and tim histone modifications in wild-type flies. Histone modifications present at ZT 4, 10, 16, and 22 are shown for the upstream and intron 1 regions of per (A) and tim (B). Ac H3-K9, acetylation of H3-K9; TM H3-K4, trimethylation of H3-K4; DM H3-K9, dimethylation of H3-K9. *, modification with statistically significant (P < 0.05) rhythm. Schematic representations of the per and tim gene regions that were analyzed by ChIP are labeled as in Fig. 3A and 4A, respectively.
Histone modifications in per01 and cyc01 flies.
As an independent means of identifying sites of H3-K9 acetylation, H3-K9 dimethylation, and H3-K4 trimethylation associated with per and tim transcription, ChIP experiments were carried out with per01 and cyc01 flies. In per01 flies, PER is not present and can't interact with and inhibit CLK-CYC activation of target genes. Consequently, histone modifications associated with CLK-CYC-dependent transcriptional activation would be expected in per01 mutants. In contrast, since cyc01 flies lack CYC and thus CLK-CYC activity, histone modifications associated with transcriptional inactivity would be expected. Measurements of H3-K9 acetylation, H3-K9 dimethylation, and H3-K4 trimethylation were made at a time point during the light (ZT 10) and a time point during the dark (ZT 22). Light-induced differences in per and tim transcription and CLK-CYC-dependent E-box binding have not been detected in per01 flies (45, 49); thus, no differences in histone modifications are expected in per01 and cyc01 flies at ZT 10 and ZT 22.
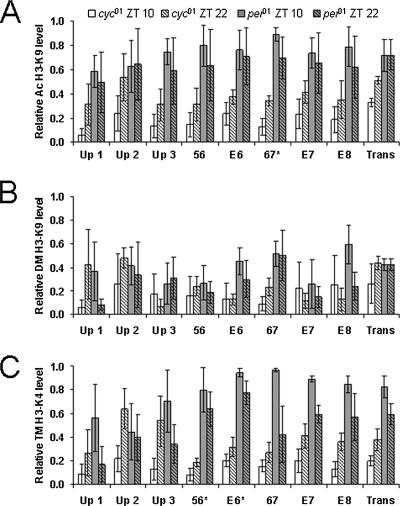
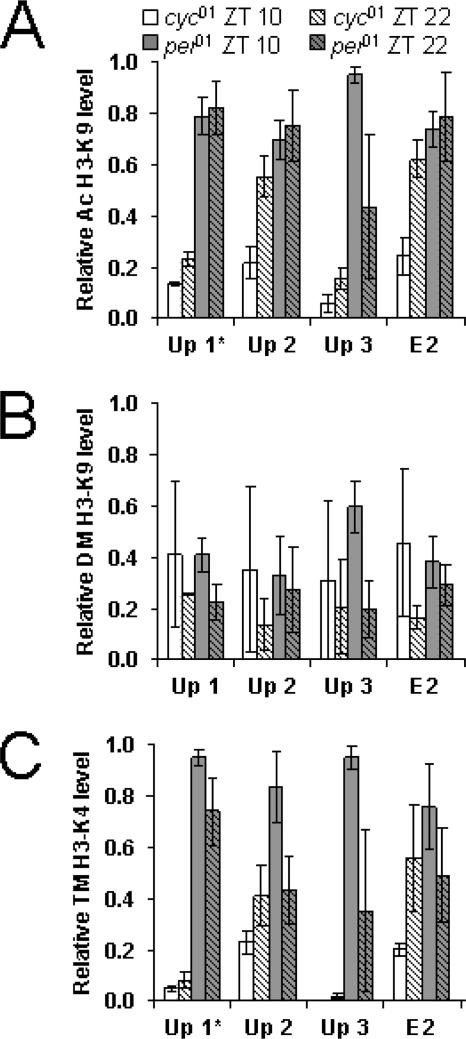
For most per and tim upstream/intron 1 regions, H3-K9 acetylation was higher in per01 flies than in cyc01 flies (Fig. 6 and 7). However, the only regions in which H3-K9 acetylation was significantly higher in per01 flies than in cyc01 flies at both times were Per 67 (P < 0.04) and Tim Up 1 (P < 0.001). Likewise, H3-K4 trimethylation in most per and tim upstream/intron 1 regions was higher in per01 flies than in cyc01 flies but was only significantly higher in the Per 56 (P < 0.04), Per E-box 6 (P < 0.002), and Tim Up 1 (P < 0.001) regions at both times (Fig. 6 and 7). The levels of H3-K9 dimethylation in per01 and cyc01 flies were similar; none of the per or tim regions tested showed significant differences (Fig. 6 and 7). Although H3-K9 acetylation, H3-K9 dimethylation, and H3-K4 trimethylation levels were similar within a given region of per and tim at both time points in cyc01 and per01 flies, the levels of H3-K9 acetylation and H3-K4 trimethylation tended to be somewhat higher at ZT 22 in cyc01 flies and somewhat lower at ZT 22 in per01 flies.
FIG. 6.
Histone modifications in per upstream and intron 1 regions from per01 and cyc01 flies. ChIP was performed in per01 and cyc01 flies collected at ZT 10 and ZT 22 during a 12:12 LD cycle. Relative levels of (A) acetyl (Ac)-H3-K9, (B) dimethyl (DM)-H3-K9, and (C) trimethyl (TM)-H3-K4 binding were determined for the regions depicted in Fig. 3A by qPCR analysis. The levels of each histone modification have been corrected for nonspecific binding by subtracting the rabbit IgG ChIP values at each time point. These data were derived from three independent experiments. *, statistically significant (P < 0.05) difference.
FIG. 7.
Histone modifications in tim upstream and intron 1 regions from per01 and cyc01 flies. ChIP was performed as described for Fig. 6. Relative levels of (A) acetyl (Ac)-H3-K9, (B) dimethyl (DM)-H3-K9, and (C) trimethyl (TM)-H3-K4 binding were determined for the regions depicted in Fig. 4A by qPCR analysis. The levels of each histone modification have been corrected for nonspecific binding as described for Fig. 6. These data were derived from three independent experiments. *, statistically significant (P < 0.05) difference.
H3-K9 acetylation and/or H3-K4 trimethylation was significantly higher in per01 than cyc01 mutants for sites within the per transcribed region (i.e., Per 56, Per E-box 6, and Per 67). Two of these sites (Per E-box 6 and Per 67) coincide with those that showed high levels of H3-K9 acetylation and/or H3-K4 trimethylation when per is highly transcribed in wild-type flies, further implicating these modifications in promoting per transcription. Moreover, the location of these chromatin modifications within the per transcribed region suggests that they may affect Pol II elongation. In contrast, H3-K9 acetylation and H3-K4 trimethylation were significantly higher in per01 than cyc01 mutants for a site (Tim Up 1) in the tim upstream region. This corresponds well with high levels of H3-K9 acetylation and/or H3-K4 trimethylation at Tim Up 1 and Tim Up 2 during tim transcription in wild-type flies, indicating that these sites are involved in activating tim transcription.
Pol II binding.
CLK-CYC binding to E-boxes coincides with H3-K9 acetylation, H3-K4 trimethylation, and activation of per and tim transcription. Consequently, these histone modifications are expected to promote Pol II recruitment and/or activity. Pol II binding and activity at the per and tim loci were assessed with antibodies that recognize either the non-actively transcribing form of Pol II having an unphosphorylated CTD (Pol II CTD) or the actively transcribing form of Pol II having an S5 phosphorylated CTD (Pol II S5 CTD). The Pol II S5 CTD antibody (H14) has been reported to bind specifically to either S5 or to S5 plus S2 and not to S2 alone, and the Pol II CTD antibody (8WG16) has been reported to bind with highest affinity to the unphosphorylated CTD and with weaker affinity to the phosphorylated forms (24). Both antibodies are able to recognize Pol II at the promoter region, indicating that 8WG16 can recognize either the unphosphorylated or S5-phosphorylated CTD (36). Neither of these antibodies has been reported to recognize Pol II in the transcribed region, where S2 phosphorylation predominates (36). These Pol II antibodies were used for ChIP analysis of various regions of per and tim in wild-type flies collected at ZT 4, 10, 16, and 22. For these experiments two gene-specific primer sets were used for the per and tim promoters (Up 3 and Up 3-2) and single primer sets were used for the upstream (Up 1) and coding (MT or LE) regions.
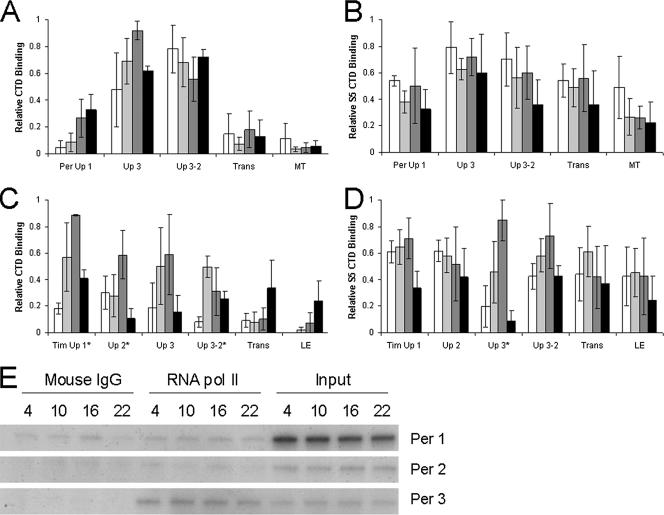
Relatively high levels of Pol II CTD are bound to the per promoter region at all times tested, where Per Up 3 shows a low-amplitude rhythm (albeit not statistically significant) with a peak at ZT 16 and Per Up 3-2 shows no rhythm in binding (Fig. 8A). Compared to those for the Per Up 3/Per Up 3-2 region, low binding levels were detected in the other per regions tested (Fig. 8A). Similarly, Pol II S5 CTD is constantly bound to the Per Up 3 region, though no rhythm in binding is apparent (Fig. 8B). The background of the Pol II S5 antibody makes comparisons of levels of binding to various regions difficult; however, comparisons can be made when using the negative control as a reference. For the Per Up 3/Per Up 3-2 region, Pol II S5 binding is >10-fold higher than that for the negative control, whereas it is <4-fold higher than that for the negative control for all other regions tested (see Fig. S2 in the supplemental material). Neither of these antibodies has been reported to recognize Pol II beyond the promoter region (36); thus, negative results are expected for the transcribed region. In contrast, Pol II CTD binds the Tim Up 3/Tim Up 3-2 region rhythmically; high levels of binding are seen at ZT 10 and 16, and significantly (P < 0.03) lower binding levels (for the Tim Up 3-2 primer set) are seen at ZT 4 and ZT 22 (Fig. 8C). Likewise, Pol II S5 CTD binding to Tim Up 3 shows a significant daily cycle (P < 0.03), though both Tim Up 3 and Tim Up 3-2 primer sets show peak levels of binding at ZT 16 and low levels of binding at both ZT 4 and ZT 22 (Fig. 8D), consistent with nuclear run-on experiments (45). Regions within the tim transcribed region show low Pol II CTD binding, while regions upstream show rhythmic Pol II CTD binding with statistically significant daily cycles for Tim Up 1 (P < 0.04) and Tim Up 2 (P < 0.05), suggesting that Pol II interacts with proteins associated with these regions (Fig. 8C). In comparison with Pol II S5 binding, negative control levels are much lower for the Tim Up 3 region and the Tim Up 1 region (see Fig. S3 in the supplemental material). Pol II S5 binding does not seem to correlate with protein levels, as Pol II S5 levels do not appear to be rhythmic (see Fig. S4 in the supplemental material), indicating that binding is regulated. Since Pol II was not expected to constitutively bind the per promoter region, additional ChIP assays were carried out to compare levels of Pol II CTD binding via semiquantitative PCR in the Per Up 1, Per Up 2, and Per Up 3 regions. Although little or no Pol II CTD binding to Per Up 1 or Per Up 2 was detected, strong binding to Per Up 3 was detected at all time points (Fig. 8E). These ChIP analyses demonstrate strong constitutive Pol II binding that is specific to the per promoter. These results also suggest that Pol II does not interact with proteins in the Per Up 1 and Per Up 2 regions; otherwise, Pol II would pull down those regions, as was seen with Tim Up 1 and 2 (Fig. 8).
FIG. 8.
Pol II binding to the per and tim genes. ChIP was performed in wild-type flies collected at ZT 4, 10, 16, and 22 during a 12:12 LD cycle. Relative levels were determined by qPCR analysis and are shown for samples immunoprecipitated with antibodies against Pol II CTD (A and C), with mouse IgG as a negative control, or Pol II S5 CTD (B and D), with no IgG as a negative control. The per (A and B) and tim (C and D) gene regions that were analyzed by ChIP are labeled as in Fig. 3A and 4A, respectively, with an additional primer set for the per and tim promoters, Per Up 3-2 and Tim Up 3-2, a primer set for the tim translation start (Trans), a primer set for the per translated region flanking intron 5 (MT), and a primer set for the last exon of tim (LE). The levels of each histone modification have been corrected for nonspecific binding by subtracting the mouse IgG or no IgG ChIP values at each time point. These data were derived from three independent experiments. *, statistically significant (P < 0.05) rhythm. (E) Semiquantitative PCR results for unphosphorylated CTD of Pol II binding to per. Shown are regions from Per Up 1, Per Up 2, and Per Up 3.
DISCUSSION
CLK-CYC binding to E-boxes is essential for rhythmic transcription of the core circadian oscillator genes per and tim in Drosophila. Previous analysis demonstrated that CLK-CYC binds rhythmically to the per CRS E-box in wild-type flies and that this binding is PER dependent (49). Although the per CRS E-box is sufficient to drive circadian transcription (8, 20, 32), other canonical CACGTG E-boxes present in per upstream and intron 1 sequences may also contribute to per transcriptional rhythms. ChIP analysis shows that CLK (and by extension CLK-CYC) binds rhythmically to E-boxes in per intron 1 (Fig. 1), indicating that multiple E-boxes are involved in rhythmic per transcription. This observation may explain rescue of behavioral rhythmicity by constructs that lack the per promoter but still contain E-boxes in intron 1 (15, 31). CLK-CYC binding to a region containing the tim upstream E-box is strongly rhythmic (Fig. 2) (49), in contrast to binding to the E-box-containing region in tim intron 1, which is weak (Fig. 2). These results are consistent with studies showing that the tim upstream region is sufficient to drive circadian transcription and that tim intron 1 is not able to activate CLK-CYC-dependent transcription in S2 cells (25, 34, 48). Although the canonical CACGTG E-box upstream of tim is important for rhythmic transcription, noncanonical E-boxes close to the canonical E-box contribute substantially to high-amplitude rhythmicity (34). Thus, rhythmic CLK binding to tim upstream sequences is likely due to canonical and noncanonical E-box elements. Taken together, these results suggest that cooperative binding of CLK-CYC to canonical and noncanonical E-boxes promotes high-amplitude cycling of tim transcription. We speculate that cooperative binding of CLK-CYC to multiple canonical and possibly noncanonical E-boxes also mediates high-amplitude cycling of per transcription.
Binding of CLK-CYC to per and tim regulatory sequences coincides with histone modifications that are known to alter transcriptional activity. Acetylation of H3-K9 was rhythmic, with a peak at ZT 10 for each per and tim region tested (Fig. 3 to 5). Such acetylation is consistent with transcriptional activation since CLK-CYC binding, per and tim transcription, and per and tim RNA accumulation are all high at ZT 10. Indeed, relatively low levels of H3-K9 acetylation are detected within/upstream of per and tim in cyc01 flies (Fig. 6 and 7), which exhibit constant low levels of CLK-CYC binding and express little or no per and tim mRNA (42, 49). Similar results are seen for Dbp in mice, where circadian expression is associated with rhythmic CLOCK-BMAL1 binding to multiple extra- and intragenic E-boxes and acetylation of H3-K9 (41). Likewise, Per1 activation is coincident with H3 acetylation (6, 10, 12, 38), and CRY-mediated repression likely occurs through histone deacetylation since histone deacetylase (HDAC) is recruited into the complex by CRY and CBP overexpression relieves repression by CRY (6, 38). A similar situation is likely to exist in Drosophila since H3-K9 acetylation decreases as PER accumulates, though an interaction between PER and an HDAC has not been identified. The levels of H3-K9 acetylation in per01 flies are consistent with PER-dependent recruitment of HDAC. In per01 flies H3-K9 acetylation is constantly high, indicating that deacetylation is PER dependent (Fig. 6).
Our results suggest that CLK-CYC binding leads to histone acetylation. Recent work has provided information as to the histone acetyltransferases (HATs) that may be involved. One possibility is that CLK itself is a HAT, as has been shown to be the case with the mammalian CLOCK (10). Another possibility is that CLK interacts with other HATs. One HAT shown to interact with CLK is the CBP ortholog NEJIRE (22, 29). In one case NEJIRE/CBP was found to act as a transcriptional coactivator (22), but other evidence shows that overexpression of NEJIRE has a negative effect on CLK-CYC-mediated transcription (29). It is possible that the reduced levels of CLK-CYC-dependent gene expression observed after nejire overexpression are due to circadian feedback compensation and that nejire/CBP is indeed a coactivator (22).
As with H3-K9 acetylation, a strong rhythm in H3-K4 trimethylation was observed just after the first exon of per (Per 56, Per E6, Per 67) and near the tim transcription initiation site (Tim Up 1, Tim Up 3). This rhythm in H3-K4 trimethylation consistently peaked at ZT 16, which was 6 h after the peak in H3-K9 acetylation and just after the peak in per and tim mRNAs. Rhythmic H3-K4 trimethylation in these regions suggests that this modification may be involved in Pol II elongation for per and Pol II recruitment for tim. Although trimethylation of H3-K4 is expected to be involved in transcriptional activation, analysis of cross talk among histone modifications indicates that this modification can also be present during the repressed state (14). This modification may allow for HATs, HDACs, histone methyltransferases, or other modifiers to selectively target histones in a key regulatory position. Rhythmic trimethylation of H3-K4 in wild-type flies was observed in several upstream/intron 1 regions of per and tim but was either constantly high or constantly low in others. However, regions of per and tim where H3-K4 trimethylation is rhythmic in wild-type flies showed either constant high or low levels of H3-K4 trimethylation in per01 and cyc01 flies, respectively (Fig. 6 and 7), consistent with a role in activating transcription.
Another modification analyzed was dimethylation of H3-K9. Although no dimethylation was detected in upstream regions of per and tim, low-amplitude rhythmic dimethylation was observed in introns 1 of per and tim, peaking at ZT 16. Cycling of H3-K9 dimethylation was antiphase to H3-K9 acetylation and was associated with repression of Dbp in mice (41), but this was not the case for per and tim in flies. Since dimethylation of H3-K9 in the transcribed region is associated with transcriptional elongation in mice (47), somewhat higher levels of H3-K9 dimethylation in the first introns of per and tim are expected, as their transcription peaks might suggest that dimethylation of H3-K9 promotes elongation. The levels of H3-K9 dimethylation in per01 and cyc01 mutants were lower, higher, or unchanged compared to that for wild-type flies depending on the time point assayed (Fig. 3, 4, 6, and 7), thus making it difficult to correlate this modification with transcriptional activation or repression. Low-amplitude H3-K9 dimethylation rhythms in wild-type flies prevented meaningful interpretations regarding the function of this chromatin modification. Similarly, both H3-K9 acetylation and dimethylation at the Dbp locus are also reduced in arrhythmic Per1−/−; Per2Brdm/Brdm mutant mice, though a high-amplitude antiphase cycling of H3-K9 acetylation and dimethylation suggests that H3-K9 dimethylation represses transcription (41).
In contrast to the rhythmic chromatin modifications in per and tim upstream/intron 1 sequences, H3-K14 and H4 acetylation was generally at constant high levels. Acetylation of these sites may be involved in creating a chromatin structure that allows access to proteins that regulate circadian transcription, perhaps as a developmental modification that occurred during clock cell determination. Recent work has shown that histone modifications such as acetylation and methylation have a crucial role in development (30, 33). For instance, sequential H3-K4 methylation, H3 acetylation, and H4 acetylation are associated with the expression of HoxD4 genes during mouse neurogenesis (40) and sequential H3-K9 deacetylation, H3-K4 demethylation, and H3-K9 methylation are associated with downregulation of the mouse terminal transferase gene during thymocyte maturation (46). Although the phospho-histone H3-S10 antibody has been validated for use in ChIP experiments and phosphorylation of H3 often accompanies H3 acetylation (4, 39), phosphorylation of H3-S10 was also negative for all regions of per and tim tested. These negative results could mean that either H3-S10 is not phosphorylated or that the antibody could not detect phosphorylation at levels present in fly head nuclear extracts. This antibody recognizes a band corresponding to H3 (17 kDa) on an immunoblot containing the sonicated SXN samples used in ChIP experiments (see Fig. S1 in the supplemental material). This result suggests that the phospho-histone H3-S10 antibody recognizes phosphorylated H3-S10 in fly extracts, and thus the negative ChIP result is more likely a result of no phosphorylation of H3-S10 in the transcriptional regulation of per or tim.
The levels of H3-K9 acetylation and H3-K4 trimethylation in per and tim upstream/intron 1 sequences were constitutively high in per01 flies and constitutively low in cyc01 flies. However, the levels of these modifications were consistently, though not significantly, different at ZT 10 and ZT 22 in each mutant. In cyc01 flies, H3-K9 acetylation and H3-K4 trimethylation were higher at ZT 22 than at ZT 10, but in per01 flies, H3-K9 acetylation was lower at ZT 22 than at ZT 10 for several regions (the transcribed region of per and Tim Up 3) and H3-K4 trimethylation was also lower at ZT 22 than at ZT 10 (Fig. 6 and 7). ChIP was carried out on mutants collected in LD cycles, suggesting that light may alter the levels of H3-K9 acetylation and H3-K4 trimethylation. Since time-dependent differences in H3-K9 acetylation and H3-K4 trimethylation are not unidirectional in per01 and cyc01 flies, they appear to be regulated rather than acute responses to light. These time-dependent differences in H3-K9 acetylation and H3-K4 trimethylation do not lead to changes in transcription since per and tim mRNAs are expressed at constant levels in per01 and tim01 flies collected during LD cycles (21, 43).
High levels of Pol II are always present on the per promoter, which suggests that rhythms in per transcription are regulated by events associated with Pol II elongation. One such event is H3-K4 trimethylation, which exhibits high-amplitude rhythms in per intron 1. Deleting a specific regulator of H3-K4 trimethylation in Saccharomyces cerevisiae, Bur2 kinase, significantly impairs the recruitment of the PAF elongation factor, thus linking H3-K4 trimethylation with Pol II elongation (27). Since cross talk among chromatin modifications can occur at the level of a single histone tail or at the level of nucleosomes (14), H3-K4 trimethylation along with H3-K9 acetylation may promote per transcription by promoting Pol II elongation, whereas H3-K4 trimethylation along with H3-K9 deacetylation may inhibit per transcription by promoting Pol II pausing. The coincidence between CLK-CYC binding and high levels of H3-K9 acetylation and H3-K4 trimethylation suggests that there may be cross talk between these modifications. Although previous results from cultured mammalian cells suggested that the antibodies used in these experiments would not detect high levels of nonphosphorylated and S5-phosphorylated Pol II in per and tim transcribed regions, it is possible that the Pol II CTD modification associated with per and tim transcription elongation is S2 phosphorylation of Pol II (36).
In contrast to the situation with per, Pol II binding is rhythmic in the tim promoter region. Since H3-K4 trimethylation and H3-K9 acetylation are strongly rhythmic in tim upstream and promoter sequences and show more-modest rhythms in tim intron 1, it is possible that both Pol II recruitment and elongation are involved in regulating tim transcriptional rhythms. The rhythmic recruitment of Pol II, possibly in combination with Pol II elongation rhythms, may account for the high rate and amplitude of tim transcription compared to those of per transcription. In mammals, rhythmic histone acetylation at the Per1, Per2, and Cry1 promoters correlates with Pol II binding and transcriptional cycling (6, 12), indicating that rhythmic recruitment of Pol II is a conserved mechanism for regulating transcriptional rhythms in mammals. Pol II was also shown to interact with tim upstream sequences (Tim Up 1 and Tim Up 2), presumably through Pol II interactions with proteins associated with these regions (Fig. 8). In addition to rhythmic H3-K9 acetylation and/or H3-K4 trimethylation in wild-type flies, there is strong H3-K9 acetylation and H3-K4 trimethylation of Tim Up 1 and Tim Up 2 in per01 flies and weak H3-K9 acetylation and H3-K4 trimethylation in cyc01 flies. The resolution of ChIP is about 500 bp, and Tim Up 2 is less than 500 bp from Tim Up 1 and Tim Up 3; therefore, results showing Pol II interaction with the Tim Up 2 region may be due to crossover. This is consistent with lower Pol II CTD levels for Tim Up 2 than for Tim Up 1 (Fig. 8) and levels of Pol II S5 CTD that are closer to negative control levels for Tim Up 2 than for Tim Up 1 or 3 (see Fig. S3A to C in the supplemental material).
Results from these experiments demonstrate rhythmic binding of CLK-CYC to per intronic E-boxes; rhythmic and constitutive histone modifications in per and tim upstream, promoter, and transcribed regions; and rhythmic or constitutive Pol II binding to the tim and per promoters. The activation and repression of these genes may be directly or indirectly dependent upon CLK-CYC binding or other circadian proteins that interact with CLK-CYC, such as PER. Many of the histone modifications associated with rhythmic transcription in flies (e.g., H3-K9 acetylation, H3-K4 trimethylation, and H3-K9 dimethylation) are conserved in mammals, which suggests that PER and CRY repress transcription through similar mechanisms. Our results imply that histone-modifying enzymes such as HATs, HDACs, histone methyltransferases, ubiquitinases, and sumoylases and Pol II recruitment and elongation factors rhythmically interact with the CLK-CYC complex to regulate transcriptional cycling. Identifying proteins that interact with CLK-CYC during times of transcriptional activity or repression will advance our understanding of the mechanisms that regulate circadian transcription.
Supplementary Material
Acknowledgments
We thank X. Li from M. Biggin's lab for advice on which Pol II antibodies to use for ChIP assays and the linker antibody to use with anti-S5 CTD. We are grateful for advice from F. Ng on qPCR assays used to quantify ChIP assays.
This work was supported by Public Health Service grant NS052854 from the National Institute of Neurological Disorders and Stroke.
Footnotes
Published ahead of print on 12 May 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aparicio, O., J. V. Geisberg, and K. Struhl. 2004. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo, p. 17.7.1-17.7.23. In J. S. Bonifacio, M. Dasso, J. B. Harford, J. Lippincott-Schwartz, and K. M. Yamada (ed.), Current protocols in cell biology. John Wiley & Sons, Hoboken, NJ. [DOI] [PubMed]
- 2.Blau, J., and M. W. Young. 1999. Cycling vrille expression is required for a functional Drosophila clock. Cell 99661-671. [DOI] [PubMed] [Google Scholar]
- 3.Buratowski, S. 2003. The CTD code. Nat. Struct. Biol. 10679-680. [DOI] [PubMed] [Google Scholar]
- 4.Cheung, P., K. G. Tanner, W. L. Cheung, P. Sassone-Corsi, J. M. Denu, and C. D. Allis. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5905-915. [DOI] [PubMed] [Google Scholar]
- 5.Curtin, K. D., Z. J. Huang, and M. Rosbash. 1995. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron 14365-372. [DOI] [PubMed] [Google Scholar]
- 6.Curtis, A. M., S. B. Seo, E. J. Westgate, R. D. Rudic, E. M. Smyth, D. Chakravarti, G. A. FitzGerald, and P. McNamara. 2004. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J. Biol. Chem. 2797091-7097. [DOI] [PubMed] [Google Scholar]
- 7.Cyran, S. A., A. M. Buchsbaum, K. L. Reddy, M. C. Lin, N. R. Glossop, P. E. Hardin, M. W. Young, R. V. Storti, and J. Blau. 2003. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112329-341. [DOI] [PubMed] [Google Scholar]
- 8.Darlington, T. K., L. C. Lyons, P. E. Hardin, and S. A. Kay. 2000. The period E-box is sufficient to drive circadian oscillation of transcription in vivo. J. Biol. Rhythms 15462-471. [DOI] [PubMed] [Google Scholar]
- 9.Darlington, T. K., K. Wager-Smith, M. F. Ceriani, D. Staknis, N. Gekakis, T. D. Steeves, C. J. Weitz, J. S. Takahashi, and S. A. Kay. 1998. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 2801599-1603. [DOI] [PubMed] [Google Scholar]
- 10.Doi, M., J. Hirayama, and P. Sassone-Corsi. 2006. Circadian regulator CLOCK is a histone acetyltransferase. Cell 125497-508. [DOI] [PubMed] [Google Scholar]
- 11.Edery, I., L. J. Zwiebel, M. E. Dembinska, and M. Rosbash. 1994. Temporal phosphorylation of the Drosophila period protein. Proc. Natl. Acad. Sci. USA 912260-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Etchegaray, J. P., C. Lee, P. A. Wade, and S. M. Reppert. 2003. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421177-182. [DOI] [PubMed] [Google Scholar]
- 13.Etchegaray, J. P., X. Yang, J. P. DeBruyne, A. H. Peters, D. R. Weaver, T. Jenuwein, and S. M. Reppert. 2006. The polycomb group protein EZH2 is required for mammalian circadian clock function. J. Biol. Chem. 28121209-21215. [DOI] [PubMed] [Google Scholar]
- 14.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15172-183. [DOI] [PubMed] [Google Scholar]
- 15.Frisch, B., P. E. Hardin, M. J. Hamblen-Coyle, M. Rosbash, and J. C. Hall. 1994. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron 12555-570. [DOI] [PubMed] [Google Scholar]
- 16.Gekakis, N., L. Saez, A. M. Delahaye-Brown, M. P. Myers, A. Sehgal, M. W. Young, and C. J. Weitz. 1995. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science 270811-815. [DOI] [PubMed] [Google Scholar]
- 17.Grima, B., A. Lamouroux, E. Chelot, C. Papin, B. Limbourg-Bouchon, and F. Rouyer. 2002. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature 420178-182. [DOI] [PubMed] [Google Scholar]
- 18.Hahn, S. 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11394-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao, H., D. L. Allen, and P. E. Hardin. 1997. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol. Cell. Biol. 173687-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao, H., N. R. Glossop, L. Lyons, J. Qiu, B. Morrish, Y. Cheng, C. Helfrich-Forster, and P. Hardin. 1999. The 69 bp circadian regulatory sequence (CRS) mediates per-like developmental, spatial, and circadian expression and behavioral rescue in Drosophila. J. Neurosci. 19987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardin, P. E., J. C. Hall, and M. Rosbash. 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343536-540. [DOI] [PubMed] [Google Scholar]
- 22.Hung, H. C., C. Maurer, S. A. Kay, and F. Weber. 2007. Circadian transcription depends on limiting amounts of the transcription co-activator nejire/CBP. J. Biol. Chem. 28231349-31357. [DOI] [PubMed] [Google Scholar]
- 23.Hunter-Ensor, M., A. Ousley, and A. Sehgal. 1996. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell 84677-685. [DOI] [PubMed] [Google Scholar]
- 24.Jones, J. C., H. P. Phatnani, T. A. Haystead, J. A. MacDonald, S. M. Alam, and A. L. Greenleaf. 2004. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J. Biol. Chem. 27924957-24964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, E. Y., K. Bae, F. S. Ng, N. R. Glossop, P. E. Hardin, and I. Edery. 2002. Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron 3469-81. [DOI] [PubMed] [Google Scholar]
- 26.Ko, H. W., J. Jiang, and I. Edery. 2002. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420673-678. [DOI] [PubMed] [Google Scholar]
- 27.Laribee, R. N., N. J. Krogan, T. Xiao, Y. Shibata, T. R. Hughes, J. F. Greenblatt, and B. D. Strahl. 2005. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr. Biol. 151487-1493. [DOI] [PubMed] [Google Scholar]
- 28.Lee, C., J. P. Etchegaray, F. R. Cagampang, A. S. Loudon, and S. M. Reppert. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107855-867. [DOI] [PubMed] [Google Scholar]
- 29.Lim, C., J. Lee, C. Choi, J. Kim, E. Doh, and J. Choe. 2007. Functional role of CREB-binding protein in the circadian clock system of Drosophila melanogaster. Mol. Cell. Biol. 274876-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, W., and S. Y. Dent. 2006. Functions of histone-modifying enzymes in development. Curr. Opin. Genet. Dev. 16137-142. [DOI] [PubMed] [Google Scholar]
- 31.Liu, X., Q. A. Yu, Z. S. Huang, L. J. Zwiebel, J. C. Hall, and M. Rosbash. 1991. The strength and periodicity of D. melanogaster circadian rhythms are differentially affected by alterations in period gene expression. Neuron 6753-766. [DOI] [PubMed] [Google Scholar]
- 32.Lyons, L. C., T. K. Darlington, H. Hao, J. Houl, S. A. Kay, and P. E. Hardin. 2000. Specific sequences outside the E-box are required for proper per expression and behavioral rescue. J. Biol. Rhythms 15472-482. [DOI] [PubMed] [Google Scholar]
- 33.Margueron, R., P. Trojer, and D. Reinberg. 2005. The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 15163-176. [DOI] [PubMed] [Google Scholar]
- 34.McDonald, M. J., M. Rosbash, and P. Emery. 2001. Wild-type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol. Cell. Biol. 211207-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer, P., L. Saez, and M. W. Young. 2006. PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science 311226-229. [DOI] [PubMed] [Google Scholar]
- 36.Morris, D. P., G. A. Michelotti, and D. A. Schwinn. 2005. Evidence that phosphorylation of the RNA polymerase II carboxyl-terminal repeats is similar in yeast and humans. J. Biol. Chem. 28031368-31377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers, M. P., K. Wager-Smith, A. Rothenfluh-Hilfiker, and M. W. Young. 1996. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science 2711736-1740. [DOI] [PubMed] [Google Scholar]
- 38.Naruse, Y., K. Oh-hashi, N. Iijima, M. Naruse, H. Yoshioka, and M. Tanaka. 2004. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 246278-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowak, S. J., and V. G. Corces. 2004. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 20214-220. [DOI] [PubMed] [Google Scholar]
- 40.Rastegar, M., L. Kobrossy, E. N. Kovacs, I. Rambaldi, and M. Featherstone. 2004. Sequential histone modifications at Hoxd4 regulatory regions distinguish anterior from posterior embryonic compartments. Mol. Cell. Biol. 248090-8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ripperger, J. A., and U. Schibler. 2006. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38369-374. [DOI] [PubMed] [Google Scholar]
- 42.Rutila, J. E., V. Suri, M. Le, W. V. So, M. Rosbash, and J. C. Hall. 1998. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93805-814. [DOI] [PubMed] [Google Scholar]
- 43.Sehgal, A., A. Rothenfluh-Hilfiker, M. Hunter-Ensor, Y. Chen, M. P. Myers, and M. W. Young. 1995. Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science 270808-810. [DOI] [PubMed] [Google Scholar]
- 44.Shafer, O. T., M. Rosbash, and J. W. Truman. 2002. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J. Neurosci. 225946-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.So, W. V., and M. Rosbash. 1997. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 167146-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su, R. C., K. E. Brown, S. Saaber, A. G. Fisher, M. Merkenschlager, and S. T. Smale. 2004. Dynamic assembly of silent chromatin during thymocyte maturation. Nat. Genet. 36502-506. [DOI] [PubMed] [Google Scholar]
- 47.Vakoc, C. R., S. A. Mandat, B. A. Olenchock, and G. A. Blobel. 2005. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell 19381-391. [DOI] [PubMed] [Google Scholar]
- 48.Wang, G. K., A. Ousley, T. K. Darlington, D. Chen, Y. Chen, W. Fu, L. J. Hickman, S. A. Kay, and A. Sehgal. 2001. Regulation of the cycling of timeless (tim) RNA. J. Neurobiol. 47161-175. [DOI] [PubMed] [Google Scholar]
- 49.Yu, W., H. Zheng, J. H. Houl, B. Dauwalder, and P. E. Hardin. 2006. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 20723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng, H., P. E. Hardin, and M. Rosbash. 1994. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J. 133590-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zerr, D. M., J. C. Hall, M. Rosbash, and K. K. Siwicki. 1990. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J. Neurosci. 102749-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.