Abstract
Toll-like receptor 3 (TLR3) can signal the production of a suite of cytokines and chemokines in response to double-stranded RNA (dsRNA) ligands or the dsRNA mimic poly(I-C). Using a human embryonic kidney 293T cell line to express human TLR3, we determined that poly(I-C)-induced signal could be significantly inhibited by single-stranded DNAs (ssDNAs), but not ssRNA or dsDNA. The ssDNA molecules that down-modulated TLR3 signaling did not affect TLR4 and do not require the hypomethylated CpG motif found in TLR9 ligands. The degree of modulation can be altered by the length, base sequence, and modification state of the ssDNAs. An inhibitory ssDNA was found to colocalize with TLR3 in transfected cells and in a cell line that naturally expresses TLR3. The inhibitory ssDNAs can compete efficiently with dsRNA for binding purified TLR3 ectodomains in vitro, while noninhibitory nucleic acids do not. The ssDNAs also decrease the levels of several cytokines produced by the human bronchial epithelial cell line BEAS-2B and by human peripheral blood mononuclear cells in response to poly(I-C) stimulation of native TLR3. These activities indicate that ssDNAs could be used to regulate the inflammatory response through TLR3.
The types and amounts of cytokines produced can dictate the outcome of pathogen infection and are responsible for inflammation-associated diseases, including colitis, asthma, psoriasis, and septic shock (13, 26, 39). The ability to modulate cytokine production is an important therapeutic approach for treating these conditions.
Innate immune receptors are promising targets to regulate the complex cascade of events that lead to cytokine production (1). These receptors recognize pathogenic and endogenous ligands through their molecular signatures and then use several signaling pathways to alter gene expression. The Toll-like receptors (TLRs) are a family of structurally related class I single-pass transmembrane proteins that serve as the sentries for pathogen infections (9, 14, 38). At least 11 TLRs have been identified in the mammalian genome and are classified according to the ligands that initially activate TLR-dependent signaling, including highly conserved pathogen proteins, cell wall components, and nucleic acids (8, 27).
Four TLRs can respond to nucleic acids. TLR7 and TLR8 recognize single-stranded RNAs (ssRNAs) (1, 15, 38); TLR9 recognizes ssDNA molecules that contain hypomethylated CpG motifs (4, 25); and TLR3, the focus of this study, recognizes double-stranded RNAs (dsRNAs) (2). The activation of TLR3 by ligands can lead to the nuclear translocation of the transcription factor NF-κB, conferring changes in gene expression (10, 21, 34). In laboratory studies, poly(I-C), a synthetic dsRNA analog, has served as a TLR3 ligand (30). Poly(I-C) stimulation of TLR3 can be assayed by using a luciferase reporter driven by NF-κB promoter elements (10, 34, 37). The effects of TLR3 could also be monitored by assessing the amounts of cytokines and chemokines produced by poly(I-C)-induced cells (20).
How TLR3 interacts with its ligand is not completely understood. In vitro, TLR3 can bind poly(I-C), a potent TLR3 agonist that is a dsRNA mimic. Recent models favor direct contact between TLR3 and poly(I-C) through an asparagine-rich surface in the TLR3 ectodomain (3ECD) (6, 34). Binding is also improved at lower pHs (6, 10, 34, 37), an observation that has led to the model that higher-affinity ligand binding by TLR3 may take place within the confines of acidic vesicles, sites where TLR3 is primarily localized (30). Ligand binding and the subsequent oligomerization of TLR3 will activate the TRIF adaptor, eventually leading to signaling by the NF-κB transcription factor (23).
Several TLRs can also regulate each other's signaling either through heterodimerization or through cross talk mediated by ligands and/or signaling molecules. ssDNA ligands containing an unmethylated CpG motif, such as oligodeoxynucleotide 2006 (ODN2006) (15, 16), are agonists for TLR9. Furthermore, ssDNAs have been reported to enhance the activation of murine chemokine and cytokine expression through the poly(I:C)- mediated pathway (3, 32). Unlike TLR3, TLR9 activation involves the MyD88 adaptor, also leading to NF-κB activation (4). We show that ssDNAs inhibited poly(I-C)-mediated TLR3 signaling in two human cell lines, unlike the enhancement observed in murine cells. Furthermore, the ssDNA also inhibited cytokine production in human peripheral blood mononuclear cells (PBMCs). This effect is unrelated to TLR9 since ssDNAs that do not activate TLR9 nonetheless have potent inhibitory activity against the TLR3 pathway.
MATERIALS AND METHODS
Reagents.
ODN2216, ODN2216c, ODN2006, and ODN2006c were purchased from InvivoGen (San Diego, CA) or synthesized by Invitrogen (Carlsbad, CA). The sequence of ODN2216 is 5′-ggG GGA CGA TCG TCg ggg gg-3′. The sequence of ODN2216c is 5′-ggG GGA GCA TGC TGg ggg gc-3′. The sequence of ODN2006 is 5′-tcg tcg ttt tgt cgt ttt gtc gtt-3′. The sequence of ODN2006c is 5′-tgc tgc ttt tgt gct ttt gtg ctt-3′. The bases in capital letters form phosphodiesters, and those in lowercase form phosphorothioates. Other chemically synthesized deoxyoligonucleotides were made by IDT (Coralville, IA). Poly(I-C) was purchased from GE Amersham (Piscataway, NJ) and reconstituted in phosphate-buffered saline (PBS) while being heated at 50°C. Lipofectamine 2000 was purchased from Invitrogen, Inc. Dual-Glo luciferase assay system reagents and plasmid phRL-TK were purchased from Promega Corp. (Madison, WI). Plasmids pNiFty-Luc and pUNO-hTLR3 were purchased from InvivoGen. The antibody specific to TLR3 (AF1487) was purchased from R&D Systems (Minneapolis, MN). The antibody specific to TLR3 used to stain cells in microscopy experiments was purchased from eBioscience. Unless stated otherwise, all medium components were purchased from Invitrogen or Sigma (St. Louis, MO). Ficoll-Paque Plus solution was purchased from GE Amersham or Invitrogen. A custom human 14-plex cytokine kit was purchased from Invitrogen, Inc.
Human TLR3 luciferase reporter assay.
Human embryonic kidney (HEK) 293T cells were harvested from an actively growing culture and plated in CoStar white 96-well plates at 4.4 × 104/ml for transfection. When the cells were ∼85 to 90% confluent, they were transfected with a mixture of Lipofectamine 2000 and plasmids pNiFty-Luc, pUNO-huTLR3, and phRL-TK that, respectively, code for the firefly luciferase reporter, full-length wild-type TLR3, and the Renilla luciferase transfection control. The cells were allowed to incubate for 24 h to allow expression from the plasmids. Poly(I-C) (2.5 μg/ml) was then added to appropriate sets of transfected cells to induce TLR3-dependent NF-κB activity. After another 24 h of incubation, the cells were harvested by using a Dual-Glo luciferase assay (Promega Corp.). Luminescence was measured by using a FLUOstar OPTIMA plate reader (BMG Labtech, Inc.). The data are usually presented as either a ratio, which was derived by dividing the results for NF-κB firefly luciferase in relative light units by the results for the control Renilla relative light units, or a determination of the level of induction, in which all treatment group luciferase ratios were divided by the luciferase ratio for unstimulated TLR3-transfected cells.
The TLR4 assay was performed similarly to the TLR3 assay except that pUNOTLR4, along with its cofactors pUNOCD14, pUNOMD2, and pUNOLBP, was used instead of the TLR3 plasmid.
Cytokine production by BEAS-2B cells.
BEAS-2B cells were grown in LHC-9 medium (Invitrogen). Cells were plated at 1 × 106/ml and 200 μl/well in collagen-coated 96-well plates (BD Biosciences). Cells were allowed to attach for 4 to 6 h before stimulation with 25 ng/ml poly(I-C) in the presence or absence of oligonucleotide. Supernatants were collected after 24 h and stored at −20°C until measurement of cytokine secretion by using a custom multiplex Luminex kit (Invitrogen). Sample acquisition and analysis were performed by using a Luminex 100 IS (Luminex Corp., Austin, TX) with StarStation software (Applied Cytometry Systems, Inc., Sacramento, CA).
Assays with human PBMCs.
Human PBMCs were isolated from whole blood collected from healthy donors. All necessary permissions, approvals, and licenses, including approval by a relevant third-party Institutional Review Board (IRB) of an informed consent form and protocol and any relevant study-related documentation required by the IRB, are collected and maintained for the sourcing, handling, storage, banking, transport, or use of biological samples from employee volunteers at Centocor, R&D, Inc. Blood was collected into CPT vacutainers containing heparin (BD Biosciences), which were centrifuged at 1,500 × g for 15 min at room temperature. The entire layer above the gel was collected into a 50-ml conical tube. The conical was filled with Hanks balanced salt solution and spun at 300 × g for 15 min. The cells were washed once more with Hanks balanced salt solution. After the final wash, the pellet was resuspended in complete medium: RPMI 1640 medium with Glutamax-1TM, 10% FBS, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 10 μg/ml gentamicin. An aliquot of the cells was removed and mixed with 50 μg/ml trypan blue to obtain a live-cell count. The cells were plated in 96-well plates at a concentration of 1 × 106 cells/well (0.2 ml/well).
Poly(I-C) or ssDNAs used in the analysis of cytokine/chemokine production were added to the cells simultaneously. The ssDNAs were reconstituted in water before use. Supernatants from cells were harvested after 24 h and frozen at −20°C until all of the samples were ready for quantification of cytokines. Cytokine levels were measured by using Luminex technology and a custom human 14-plex kit (Invitrogen).
Western blots.
293T cells expressing TLR3 and treated with various nucleic acids were lysed with passive lysis buffer (Promega, Inc.) and sonicated to degrade chromosomal DNA. Equal amounts of proteins from each sample, as determined by staining with Coomassie blue, were separated on NuPAGE 4 to 12% bis-Tris gels and blotted onto polyvinylidene difluoride membrane. The anti-TLR3 antibody AF1487 from R&D Systems was used as primary antibody for Western analysis. The blots were developed with peroxidase-conjugated secondary antibodies and an ECL-plus Western-blotting detection system. Samples from the 3ECD secretion assay were collected as follows. Amounts of 1 × 105 HEK 293T cells were transfected with plasmid pBETH-3ECD (33) by using Lipofectamine as described above. The medium was removed 24 h later and replaced with 500 μl Dulbecco's modified Eagle's medium or Dulbecco's modified Eagle's medium containing ssDNAs. Twenty-four hours later, the medium was harvested and the cells lysed with the passive lysis buffer. Both the medium and the cell lysate were subjected to Western analysis.
Biochemical analysis of TLR3 ECD binding to nucleic acid.
A recombinant 3ECD containing amino acid residues 27 to 704 was expressed and purified in 293T cells and used for the cross-linking assay (37). ODN2006 and IC18 were end labeled by using T4 polynucleotide kinase and [γ-32P]ATP. A standard cross-linking reaction mixture used a mixture of 3ECD (200 ng) with and without competitors in buffer X containing 50 mM sodium acetate (pH 5.6), 50 mM NaCl, and 4 mM MgCl2. Cross-linking was performed for 2 min at 3,600 μJ/cm2. The reaction mixture was separated in a 4 to 12% denaturing protein gel.
Biotinylated ODN2006 was bound to streptavidin resin overnight in buffer X. A 30-μl slurry of resin containing approximately 3 μg of biotinylated ODN2006 was mixed with 3 μg of 3ECD and 3 μg of bovine serum albumin (BSA) in the presence or absence of 3 μg of competitors (unless otherwise mentioned) and incubated on ice for 5 h or at room temperature, as specified. The unbound fraction was removed by centrifugation, and the pellet washed with the same buffer as described above. The final pellet was resuspended in 15 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer and separated on 4 to 12% denaturing protein gel along with the unbound fraction, followed by staining with Coomassie blue. This experiment yielded reproducible results in three independent assays.
Fluorescence microscopy.
Poly(I-C) was labeled with fluorophore by using a Cy5 labeling kit as recommended by the manufacturer (Mirus Bio Corp.). HEK 293T cells were transiently transfected with a pcDNA plasmid encoding wild-type TLR3 by using Lipofectamine 2000. Following a 24-h incubation, the medium was replaced and cells were seeded onto 12-mm coverslips coated with rat tail collagen I (BD Biosciences, San Diego, CA). After a 24-h incubation, the cells were either treated with 2 μM of ODN2006 modified at the 3′-end with fluorescein isothiocyanate (FITC) (ODN3′F) (InvivoGen) or left untreated for 24 h. All cells on coverslips were then gently washed with PBS, transferred to wells containing 4% paraformaldehyde diluted in PBS, and fixed for 30 min at room temperature. After two washes in PBS containing 0.05% Tween 20 (PBST), the cells were permeabilized for 15 min with 0.1% TX-100 diluted in PBS, washed once more, blocked for 30 min with Image-iTFX signal enhancer (Invitrogen), and further blocked for an additional 2 h at room temperature with 1× blocking buffer (Sigma). The permeabilized and fixed cells were incubated with a goat anti-human TLR3 antibody (3 μg/ml; eBioscience), diluted in blocking buffer overnight at 4°C, and then washed four times with PBST and incubated for 1 h at room temperature with Alexa Fluor 647-conjugated donkey anti-goat immunoglobulin G (2 μg/ml; Invitrogen) containing 1 μg/ml of 4′,6′-diamidino-2-phenylindole (DAPI; Sigma) diluted in 1× blocking buffer. The coverslips were next carefully washed an additional four times in PBST, followed by one wash in distilled water, inverted, placed on microscope slides containing Citifluor mounting medium (Ted Pella), and sealed with nail polish. Cells were imaged by using a 60× oil immersion objective (numerical aperture, 1.4), and optical slices of 0.2 μm were captured by using an UltraView ERS confocal microscope (Perkin Elmer).
FACS analysis.
Fluorescence-activated cell sorter (FACS) analyses were performed with 293T cells grown in six-well collagen-coated plates (BD Biosciences) at a concentration of 2 × 106 cells/well. The cells were transfected with 1 μg of the appropriate plasmids by using Lipofectamine 2000 (Invitrogen). The staining of cell surface-associated TLR3 used cells that were harvested 24 h after transfection and washed twice with ice-cold FACS buffer (1× PBS containing 10 mM phosphate, 150 mM NaCl, pH 7.4, 3% fetal bovine serum, 0.04% sodium azide) before suspension at ∼2 × 107 cells/ml in FACS buffer. The cells were stained for 30 min at 4°C with 1 μg of phycoerythrin-labeled anti-human TLR3 monoclonal antibody (MAb) (clone TLR3.7; eBioscience, San Diego, CA) or a negative-control mouse immunoglobulin G1 antibody. The antibodies were added to aliquots of the cells in 96-well plates and incubated for 30 min on ice in the dark. The cells were washed twice with FACS buffer to remove unbound antibody and then resuspended in FACS buffer. Viaprobe (BD Biosciences) was added to the cultures to exclude dead cells. The cells were transferred to the appropriate tubes and analyzed by using a FACSCalibur machine (BD Biosciences).
RESULTS AND DISCUSSION
ssDNAs can inhibit the poly(I-C) induction of TLR3 in an NF-κB reporter assay.
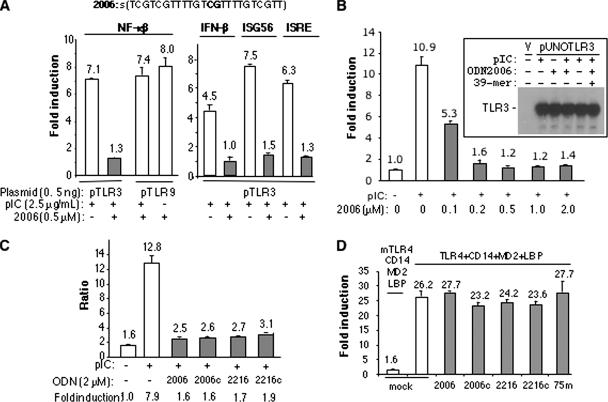
In examining whether ligands of TLR3 and TLR9 can influence their respective activities, we added a mixture of poly(I-C) and ODN2006 to reporter assays in transiently transfected HEK 293T cells, a cell line that has low levels of these receptors (2, 4). This ligand combination did not obviously affect TLR9 activity but significantly reduced NF-κB activity in cells expressing TLR3 (Fig. 1A). Furthermore, the inhibition of TLR3-dependent reporter activity was a function of the concentration of ODN2006, with 50% inhibition being observed at ∼0.1 μM of ODN2006 and the NF-κB signal equivalent to background at 2 μM (Fig. 1B). The inhibitory effects were also observed in cells transfected with different amounts of the TLR3 expression plasmids and with a number of TLR3 mutants that are partially defective due to amino acid substitutions in the ligand binding and/or dimerization domains (Table 1; data not shown) (34). Similar levels of inhibition were also observed with luciferase reporter constructs driven by the beta interferon (IFN-β), IFN-stimulated gene 56, and IFN-stimulated responsive element promoters, all of which can respond to TLR3 (Fig. 1A).
FIG. 1.
Single-stranded oligonucleotides can inhibit TLR3 activation of signaling. (A) ODN2006 inhibited the TLR3-dependent activation of the firefly luciferase reporter activity driven by the NF-κβ promoter, as well as that driven by the IFN-stimulated gene 56 (ISG56) and IFN-stimulated responsive element (ISRE) promoters. The sequence of the fully phosphorothioated ODN2006 is shown above the graph. Plasmids used to express TLR3 and TLR9 are in the pUNO vector that is driven by a human EF1/human T-cell leukemia virus promoter. (B) ODN2006 can inhibit TLR3 activation of the NF-κB luciferase activity in a concentration-dependent manner. Inset shows that the addition of ODN2006 or another ssDNA did not affect the level of TLR3 expression, as determined by Western blotting using a monoclonal antibody (AF1487) from R&D Systems directed to 3ECD. “V” indicates a sample transfected with the empty vector. (C) Other single-stranded ODNs can also inhibit TLR3 activation of the NF-κB reporter activity. (D) Signaling by TLR4 and its coadapters was not sensitive to the presence of several ssDNAs. “Mock” indicates the addition of naïve medium. TLR4 was induced by the addition of lipopolysaccharide at 1 μg/ml. mTLR4, murine TLR4; 75m, 75-mer. Each bar represents the mean of the results for activity in six independent luciferase assays, and 1 standard error is reported above the bars. The level of activity was calculated as follows. The ratio of the signals from firefly luciferase over Renilla luciferase (internal control driven from a constitutive promoter) in cells induced with poly(I-C) (final concentration, 2.5 μg/ml) was divided by the ratio of the signals from an uninduced sample. pIC, poly(I-C); 2006, ODN2006; +, present; −, absent.
TABLE 1.
A summary of the effects of mutations in TLR3 on inhibition by ODN2006
| Description of TLR3 protein | Amt (μM) of ODN2006 used | Activity (%) (SE) |
|---|---|---|
| Wild-type TLR3 | 0 | 100 (1) |
| Wild-type TLR3 | 0.2 | 13 (2) |
| K89A | 0 | 87 (2) |
| K89A | 0.2 | 12 (2) |
| L412F | 0 | 38 (1) |
| L412F | 0.2 | 12 (2) |
| E442A | 0 | 61 (2) |
| E442A | 0.2 | 10 (2) |
| N515A | 0 | 42 (1) |
| N515A | 0.2 | 9 (1) |
| R544A | 0 | 87 (4) |
| R544A | 0.2 | 23 (4) |
| R331A K418A | 0 | 84 (6) |
| R331A K418A | 0.2 | 13 (2) |
| R394A K418A | 0 | 52 (3) |
| R394A K418A | 0.2 | 13 (1) |
| R331A R394A K418A | 0 | 70 (5) |
| R331A R394A K418A | 0.2 | 14 (2) |
The ability of ODN2006 to inhibit TLR3 signaling was specific, as single-stranded poly(I), poly(C), and poly(U) all retained 70% or more of the level of TLR3 activity induced by poly(I-C) with wild-type TLR3 (Table 2). Plasmid DNAs and the dsRNA mimic poly(I-U) also did not significantly affect TLR3 activity. A 54-bp dsDNA at 0.2 μM (6.6 μg/ml) final concentration resulted in only a 15% reduction of TLR3 activity, in contrast to the sevenfold inhibition seen with 0.2 μM of ODN2006. These results demonstrate that ODN2006 can specifically inhibit the TLR3 pathway in comparison to the inhibition of the TLR3 pathway by other nucleic acid moieties.
TABLE 2.
dsDNA and ssRNAs are unable to inhibit TLR3 signaling
| Ligand or expression plasmid (μg/ml)a | Form | Amt (μg/ml) of poly(I-C) used | TLR3 activityb (%) |
|---|---|---|---|
| None | None | 15 | |
| None | 2.5 | 100 (7) | |
| Plasmid A (12.5) | dsDNA | 2.5 | 114 (9) |
| Plasmid A (25) | dsDNA | 2.5 | 100 (9) |
| Plasmid B (12.5) | dsDNA | 2.5 | 102 (3) |
| Plasmid B (25) | dsDNA | 2.5 | 114 (17) |
| 54-bp DNA (6.6) | dsDNA | 2.5 | 83 (9) |
| 54-bp DNA (17) | dsDNA | 2.5 | 63 (5) |
| Poly(I) (12.5) | ssRNA | 2.5 | 78 (3) |
| Poly(I) (25) | ssRNA | 2.5 | 75 (9) |
| Poly(C) (12.5) | ssRNA 2.5 | 2.5 | 91 (22) |
| Poly(C) (25) | ssRNA 2.5 | 2.5 | 78 (13) |
| Poly(U) (12.5) | ssRNA 2.5 | 2.5 | 110 (9) |
| Poly(U) (25) | ssRNA 2.5 | 2.5 | 109 (4) |
| Poly(I-U) (12.5) | ssRNA | 2.5 | 76 (3) |
| Poly(I-U) (25) | ssRNA | 2.5 | 73 (5) |
Plasmid A, 6.5-kb pcDNA-derived plasmid; plasmid B, 9.0-kb pcDNA-derived plasmid; none, no ligand or plasmid added.
Data are the means of the results of five sets of experiments. The numbers in parentheses denote 1 standard error.
To determine whether the inhibition of TLR3 activity was due to an effect on TLR3 expression, Western blot analysis was performed in the presence of various ligands (Fig. 1B, inset). None of the ligands significantly affected TLR3 protein levels, indicating that the inhibitory effect of ODN2006 on TLR3 is not at the level of TLR3 expression.
To determine if the TLR3 inhibitory effect is specific to ODN2006 and related to the activity of TLR9, we tested several other TLR9 stimulatory and nonstimulatory ssDNAs (Fig. 1C). ODN2006c has the internal CpG dinucleotide of ODN2006 replaced by a GpC, a change which is associated with a loss of TLR9 activation (4). ODN2216 is a type A human TLR9 ligand, while the variant ODN2216c contains a substitution in the CpG motif that renders ODN2216 a nonfunctional ligand of TLR9 (4). All four ssDNAs inhibited TLR3 to a similar extent (Fig. 1C), suggesting that the inhibition is not specific to the CpG sequence that characterizes TLR9 ligands and antagonists. These results demonstrate that the ability of ssDNAs to inhibit TLR3 activity is not directly related to signaling by TLR9.
To determine whether the ssDNAs can inhibit signaling by an unrelated TLR, we assessed their effects on TLR4. A 20-fold activation of the NF-κB promoter-driven luciferase reporter was observed in 293T cells transfected with plasmids expressing TLR4 and its coreceptors (CD14, MD2, and LBP) and induced with lipopolysaccharide 24 h after transfection (Fig. 1D). Several ssDNA inhibitors of TLR3 did not significantly affect TLR4-dependent luciferase production (Fig. 1D), and the lack of any observable effect on TLR4 signaling argues against TRIF being a target of inhibition by the ssDNAs, although it remains possible that TLR4 can signal through an MyD88-dependent pathway in our assay (22, 39).
Properties of inhibitory ssDNAs.
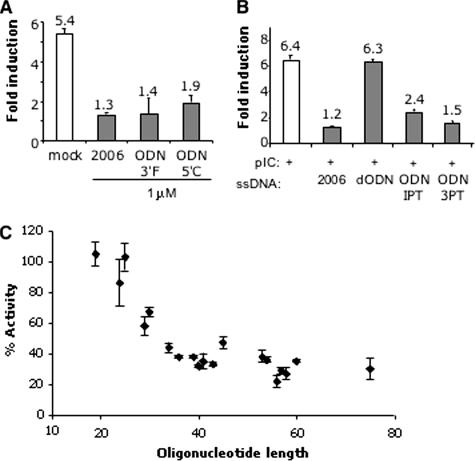
To address whether the inhibitory activities of the ssDNAs require unmodified 5′ and 3′ termini, ODN2006 was synthesized with either a 5′ Cy5 modification, resulting in a molecule named ODN5′C, or the modification forming ODN3′F. Both dyes are bulky in comparison to nucleotides, and an FITC attached to the 5′ terminus of a CpG-containing oligonucleotide is known to prevent its recognition by TLR9 (40). Both ODN5′C and ODN3′F inhibited TLR3 signaling, indicating that unmodified termini of ODN2006 are not required to inhibit TLR3 (Fig. 2A). This feature also differs from the recognition of ssDNA by TLR9, which requires an unblocked 5′ terminus in the ligands.
FIG. 2.
Properties of the ssDNAs needed to inhibit poly(I:C)-dependent TLR3 activation of reporter expression. (A) Fluorescent dyes attached to the 3′ or 5′ terminus of ODN2006 did not affect the inhibitory activity of ODN2006. These molecules were added to the cells at a final concentration of 1 μM, while poly(I-C) was added at 2.5 μg/ml. The numbers above the bars show the mean levels of induction for all of the independent trials, and the error bars show the range for 1 standard error. “Mock” indicates control media. (B) Comparison of the inhibitory properties of fully phosphorothioated ODN2006 and derivatives that vary only in the number of phosphorothioates. All ODNs were used at 2 μM. pIC, poly(I-C). (C) Correlation between the length of the unmodified ssDNAs and their inhibitory activity against TLR3-dependent activation of NF-κB reporter. All of the oligonucleotides used in this graph contained only phosphodiesters and were used at 2 μM. The sequences of the oligonucleotides are shown in Table 3. 2006, ODN2006.
ODN2216 and ODN2006 had, respectively, 6 or all 23 phosphodiester groups replaced with phosphorothioates. We examined whether phosphorothioates are necessary for the inhibitory activities of ssDNAs. A phosphodiester version of ODN2006 (named dODN) was unable to inhibit TLR3 (Fig. 2B), suggesting that phosphorothioates contribute to the ability to inhibit TLR3. ODN1PT, with phosphorothioates only at the two termini, and ODN3PT, with three phosphorothioates each at the two termini, had inhibitory activity against TLR3 signaling that was improved in comparison to the activity of dODN (Fig. 2B). These results indicate that while phosphorothioates in ODN2006 can contribute to TLR3 inhibition, not all of the phosphodiesters need to be modified (Fig. 2B).
Phosphorothioates can increase DNA stability in cells (36). If the potent inhibitory effect of ODN2006 is due to its stability, we reasoned that some unmodified ssDNAs should be inherently more resistant to degradation due to their structure in solution and/or their length. To test this prediction, a panel of unmodified ssDNAs that varied in sequence and length from 19 to 75 nucleotides (nt) were tested at 2 μM (Fig. 2C and Table 3). A range of TLR3 inhibitory activity was observed. Interestingly, the degree of inhibition correlated with the length of the ssDNA, with those of 25 nt or less having no obvious inhibitory activity (Fig. 2C). An examination of the predicted structures of the oligonucleotides did not reveal an obvious correlation to inhibitory activity (data not shown). These results demonstrate that ssDNAs lacking phosphorothioates can also inhibit TLR3 activity.
TABLE 3.
Correlation between oligonucleotide length and TLR3 inhibitory activity
| Length (nt) | Sequence | NF-κβ activitya (%) (SE) |
|---|---|---|
| 19 | CCTTACCCATAATCAACTC | 105 (8) |
| 24 | TTGGCCCTTACCCATAATCAACTC | 86 (15) |
| 25 | GTCGACAAGGGATTGAACCTCGTTC | 103 (8) |
| 29 | CAGTGTTGGCCCTTACCCATAATCAACTC | 58 (6) |
| 30 | TACGTACTTAGATATGTCTTCAAACCATAC | 67 (3) |
| 34 | CATAACAGTGTTGGCCCTTACCCATAATCAACTC | 44 (3) |
| 36 | GGCTTCACTTTCATAGACTGCAACGGTGCCTGAAAG | 38 (1) |
| 39 | ACAAACATAACAGTGTTGGCCCTTACCCATAATCAACTC | 38 (1) |
| 40 | GCTCTAGATTAAGATTTACCGATGTCGCTCACTAAGGACC | 32 (1) |
| 41 | AGTTCTAGACTACGTGACGACCTCCAGGTCAGCCGACATGC | 35 (5) |
| 43 | ACAACTTAAATGTGGTTGGTGCCGATTCCTTTGCTTGGCTTCC | 33 (1) |
| 45 | GAAGAACTGGATATCTTTGCCGCTTCATCTTTAAAAAAATTAGAG | 47 (4) |
| 53 | AGTGCTAGCAGTCATCCAACAGAATCATGTCGATGTCCACTACATGGACAGGC | 38 (4) |
| 54 | CAGTGCTAGCAGTCATCCAACAGAATCATGAATGGAATATTTGCTATTGAGAAG | 36 (2) |
| 56 | AGTGCTAGCAGTCATCCAACAGAATCATGGCCCCCATCACGTACTCCACCTATGGC | 22 (4) |
| 57 | AGTGCTAGCAGTCATCCAACAGAATCATGAGCACCTGGGTGCTGGTAGGCGGAGTCC | 29 (2) |
| 58 | AGTGCTAGCAGTCATCCAACAGAATCATGGCCTCACACCTCCCTTACATCGAAGACGG | 27 (4) |
| 60 | CTCGAGCAGAGGTCTCACACAGAGACAAGCGCATCAGTCAACAGAGATCCCTTGCGCTTC | 35 (1) |
| 75 | GTCGACCACGGTTCTGCTACTTGTTCTTTGTTTTTCACCAACAAAATGGTATGGTTTGAAGACATATCTAAGTAC | 30 (7) |
TLR3 activity obtained with poly(I-C) as the ligand was set at 100%.
To address further the features required for ssDNAs to be inhibitory against TLR3 signaling, we added six nucleotides to either terminus of dODN (lacking phosphorothioates) to allow for the formation of a hairpin clamp, resulting in an ODN named HP1 (Table 4). We also tested two additional oligonucleotides with clamps and lengths identical to those of HP1 but with an internal sequence of either homopolymeric thymidylates (HP2) or adenylates (HP3). While dODN was unable to inhibit TLR3, HP1 reduced TLR3-specific signaling to 35%. The effect of HP1 is not simply due to its length since HP2 and HP3, of identical length to HP1, had only minimal effects on TLR3 activity (Table 4). These results suggest that both the stability of the oligonucleotide and its sequence contribute to optimal inhibition of TLR3 signaling.
TABLE 4.
Unmodified oligonucleotides can inhibit TLR3 signaling
| Oligonucleotide | Sequence | Amt (μg/ml) of poly(I-C) used | TLR3 activitya (%) (SE) |
|---|---|---|---|
| None | None | 20 | |
| None | 2.5 | 100 (2) | |
| ODN2006 | TCGTCGTTTGTCGTTTTGTCGTT | 2.5 | 22 (2) |
| HP1 | CCGCCCTCGTCGTTTGTCGTTTTGTCGTTGGGCGG | 2.5 | 35 (1) |
| HP2 | CCGCCCTTTTTTTTTTTTTTTTTTTTTTTGGGCGG | 2.5 | 80 (1) |
| HP3 | CCGCCCAAAAAAAAAAAAAAAAAAAAAAAGGGCGG | 2.5 | 110 (6) |
The numbers in parentheses denote 1 standard error. TLR3 activity obtained with poly(I-C) as the ligand was set at 100%.
Functional relationship between ODN2006 and poly(I-C).
How do the ssDNAs affect TLR3 signaling? Several possibilities that are not mutually exclusive exist: (i) the ssDNAs could affect TLR3's distribution in the cell (30); (ii) the ssDNAs could affect poly(I-C) uptake; (iii) the ssDNAs could bind to TLR3 and prevent its binding to dsRNA; and (iv) the ssDNAs could act on adaptors downstream of TLR3 signaling.
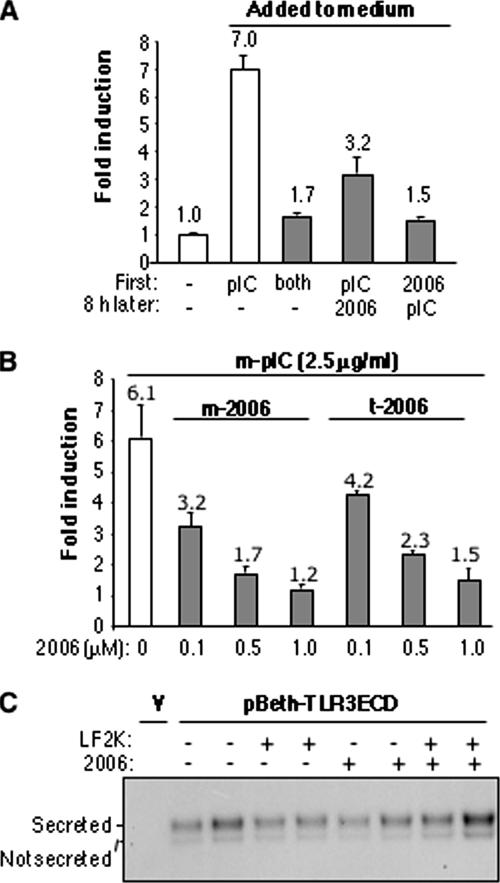
TLR3 can exist in two locations, the plasma membrane and cytoplasmic endosomes (31). To examine a possible effect of ODN2006 on TLR3 trafficking, the cell surface expression of TLR3 in the presence of ssDNA was detected by FACS analysis of intact cells. No major difference in the abundance on cells of surface-expressed TLR3, as detected by the fluorescence of an antibody recognizing TLR3, was found in the presence or absence of ssDNAs (Fig. 3A). To examine trafficking further, we used an assay for the secretion of 3ECD. 3ECD was documented as being secreted into the medium in a process that likely mimics trafficking from the endoplasmic reticulum to the plasma membrane (33). The presence of several ssDNAs did not affect 3ECD secretion into the medium, as detected by Western blots (Fig. 3B). In combination, we do not have evidence in support of ODNs affecting the process of TLR3 export to the plasma membrane.
FIG. 3.
Effects of ODNs on cell surface expression of TLR3 and secretion of 3ECD. (A) FACS analysis of the cell-surface expression of TLR3 in the presence of 1 μM ODN2006. The oligonucleotide of 18 cytidylates (rC18) that does not affect TLR3 signaling was used as a negative control. The times indicate the time the cells were fixed after the addition of the oligonucleotides. All results are for three independent samples, except for the 90-min time point at which we only had enough culture wells for two independent trials. The results from the two trials were within 10% of each other. 2006, ODN2006; MFI, mean fluorescence intensity; s.e., standard error. (B) Secretion of 3ECD is not affected by the addition of several different ssDNAs or poly(I-C) to the cell medium. The bands are from Western blots of the medium or the cell lysate probed with an MAb to TLR3 from R&D Systems. “+ Inh.” denotes a reaction where a known inhibitor of 3ECD secretion was added to the reaction mixture; pIC, poly(I-C); +, present; −, absent.
Next, whether ODNs affect poly(I-C) uptake was examined by manipulating the timing of the addition of ODN2006 and poly(I-C). 293T cells transfected to express TLR3 were treated by adding poly(I-C) to the medium at the same time as ODN2006, adding poly(I-C) 8 h prior to the addition of ODN2006, or adding them in the reverse order (Fig. 4A). The level of NF-κB activation was close to the background level when ODN2006 was added simultaneously with poly(I-C) or 8 h prior to poly(I-C). However, when ODN2006 was added 8 h after poly(I-C) treatment, the NF-κB-driven luciferase activity was reduced by more than half. These results suggest that ODN2006 can inhibit TLR3 activity even after poly(I-C) has had an opportunity to induce TLR3 activity.
FIG. 4.
Examination of the relationship between ODN2006 and poly(I-C) and their effects on TLR3 signaling. (A) Effects of the order of addition of poly(I-C) and ODN2006 on luciferase activity in transfected HEK 293T cells. The times of addition are shown at the bottom of the graph. (B) Comparison of the route of introduction and the inhibitory effects of ODNs. The ligands were either added to the cell culture medium (m-) or transfected into the cells by using Lipofectamine (t-), as indicated. The means of the results are shown above the bars, and the error bars show standard errors of the means. (C) The secretion of 3ECD is not affected by the transfection of ODN2006 or dODN into HEK cells. Where ODN2006 was used in the absence of Lipofectamine (LF2K), it was added to the cell culture medium. “V” indicates a sample transfected with the empty vector. The upper band is the glycosylated 3ECD that was secreted into the medium. The fainter lower band was found to be inside the cell (S. Hoose, data not shown). pIC, poly(I-C); 2006, ODN2006; +, present; −, absent.
To delve deeper into whether ODN inhibition is related to the poly(I-C) uptake, we examined whether the route of introduction of ODN2006 would affect its ability to inhibit TLR3 (Fig. 4B). ODNs were either added to the culture medium or transfected into cells by using Lipofectamine, and in both cases, TLR3 was induced by adding poly(I-C) to the medium. Both routes of introduction resulted in the inhibition of TLR3 signaling (Fig. 4B). In addition, neither route of introduction had a significant effect on the secretion of 3ECD into the culture medium (Fig. 4C). This result suggests that ODN2006 and poly(I-C) can both bypass any receptor at the plasma membrane to affect TLR3 signaling. Scavenger receptor class A has been identified as a receptor for the import of dsRNA, including poly(I-C), in endothelial cells (29). The transport function of scavenger receptor class A is thus not apparently affected by treatment with ODN2006.
Last, we visualized the localization of Cy5-labeled poly(I-C) in the presence and absence of ODN2006. In HEK 293T cells, Cy5-poly(I-C) was found in punctate forms in the absence and presence of TLR3 expression (see Fig. S1A in the supplemental material). However, this distribution was apparently not affected in the presence of ODN2006. As reported in the summary of the results in this section, we do not have evidence that ODN2006 affects poly(I-C) uptake or trafficking.
ODN2006 can localize in endosomes.
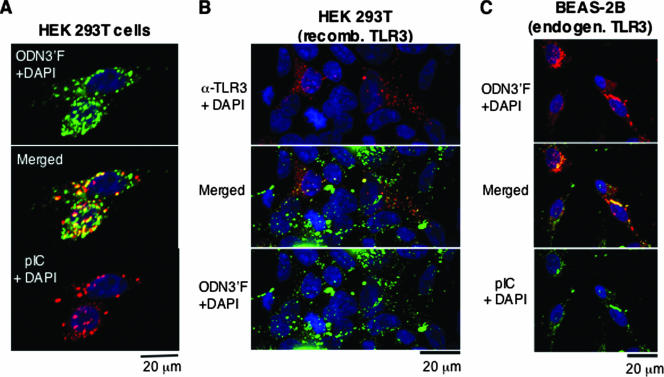
A third potential mechanism of action for ssDNAs is that they can bind TLR3 and prevent its activation. A prediction of this model is that the ssDNAs would localize at the sites of TLR3, endosomes that can be marked by the location of the TLR3 ligand poly(I-C). ODN3′F was used as the ssDNA since it retained inhibitory activity for TLR3. Poly(I-C) was labeled with Cy5 as described in Materials and Methods, and the labeled poly(I-C) was determined to be capable of activating NF-kB reporter expression in a TLR3-dependent manner (data not shown). The distribution of ODN3′F and poly(I-C) overlapped significantly (Fig. 5A; see Fig. S1B in the supplemental material). These results show that ODNs could traverse the membrane of HEK 293T cells to reach endosomes, consistent with previous reports (17, 23). In addition, this result further suggests that the ODN did not inhibit poly(I-C) uptake.
FIG. 5.
Colocalization of ODN2006 with TLR3. (A) Colocalization of the fluorescently labeled ODN2006 ODN3′F with fluorescently labeled poly(I-C). The three panels contain reconstructed confocal images of a typical field of transiently transfected HEK 293T cells. All three panels show the locations of cellular nuclei stained with DAPI (blue). Fluorescent poly(I-C) was added to the cells to a final concentration of 5 μg/ml, and ODN3′F was used at 2 μM. In this panel, poly(I-C) is in red, while ODN3′F is in bright green. Yellow denotes areas where the two signals merged. (B) Colocalization of ODN3′F with TLR3 in transfected HEK 293T cells. The upper panel shows cells stained for TLR3, as detected by a fluorescently labeled secondary antibody. The bottom panel shows ODN3′F localization in green. ODN3′F was used at 2 μM. The middle panel contains a merged image representing the locations of TLR3 in red, the locations of ODN3′F in bright green, and areas of colocalization in yellow. (C) Colocalization of ODN3′F with natively expressed TLR3 from a human lung epithelial cell line. Fluorescent poly(I-C) was added to the cells to a final concentration of 2 μg/ml, and ODN3′F was used at 2 μM. In this panel, ODN3′F is red, poly(I-C) is bright green, and areas of colocalization are yellow. pIC, poly(I-C); α, anti; recomb., recombinant; endogen., endogenous.
Next, we examined whether ODN3′F can colocalize with TLR3 in HEK 293T cells transfected to express TLR3 (Fig. 5B). ODN3′F was added for 24 h and then fixed, permeabilized, and stained with a TLR3-specific antibody in combination with a fluorescently conjugated secondary antibody. As to be expected in a transfection experiment, not all of the cells expressed TLR3. In those that did, a majority of TLR3 was localized to punctate spots indicative of intracellular vesicles (Fig. 5B). ODN3′F was also localized in punctate spots, even in untransfected cells that did not express TLR3. However, in the cells expressing TLR3, the fluorescence from ODN3′F was coincident in numerous cases with the locations of TLR3 (Fig. 5B).
Since 293T cells express TLR3 from transfected plasmids, we examined whether natively expressed TLR3 from a human bronchial epithelial cell line (BEAS-2B) can colocalize with ODN3′F added to the medium. Significant overlap in the fluorescence of TLR3 stained with fluorescently labeled MAb and ODN3′F was observed in punctate spots in the cytoplasm of the BEAS-2B cells (Fig. 5C). These results are consistent with a model in which the inhibitory effect of ODN2006 is due to its binding to TLR3 either directly or indirectly through an adaptor that is needed to transmit the signal from TLR3 or both.
ODN2006 binding to 3ECD.
The colocalization results are consistent with the possibility that ssDNAs can bind TLR3. ODN2006 could compete with poly(I-C) for the same binding site, allosterically affect poly(I-C) binding, and/or alter the consequence of TLR3 signaling in a manner independent of poly(I-C).
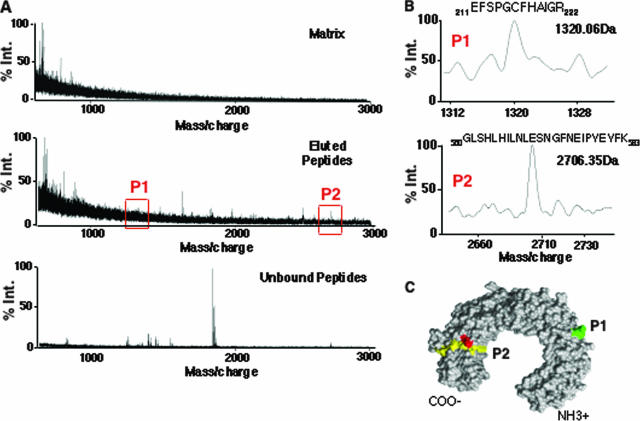
To obtain a preliminary indication of whether TLR3 could bind ssDNAs, we used a reversible cross-linking assay and purified, recombinant 3ECD made in 293T cells (Fig. 6) (37). This assay was previously used to map the RNA binding domain in the hepatitis C virus RNA polymerase and the severe-acute respiratory syndrome coronavirus endoribonuclease (7, 24). Briefly, ODN2006-resin was cross-linked to 3ECD by using formaldehyde and then subjected to extensive digestion with sequencing-grade trypsin. The unbound peptides were saved for analysis. The resin was then washed extensively prior to reversing the cross-linking with a 3-h treatment at 70°C. The peptides in the bound and unbound fractions were then identified by mass spectrometry.
FIG. 6.
Initial mapping of the interaction between ODN2006 and 3ECD. (A) Identification of peptides derived from 3ECD that can be reversibly cross-linked to ODN2006. The three mass spectra are of ions associated with the matrix, ions eluted from ODN2006-resin, and ions that were not cross-linked to the resin. Regions of the spectra in the eluted peptides match to theoretical peptides P1 and P2 that were generated from 3ECD (red boxes). % Int, percentage of peak intensity. (B) Expanded views of the P1 and P2 spectra, along with the residues of the two peptides assigned to these masses. (C) The locations of peptides P1 and P2 within the three-dimensional model of 3ECD (PDB accession number, 1ZIW). The peptide spanning residues 211 to 222 is in green, and the one spanning residues 560 to 583 is in yellow. Red denotes the location of residue H539, which has been identified as contacting dsRNA (6).
A number of peptides in the fraction eluted from ODN2006-resin were enriched in comparison to the amounts of the unbound peptides (Fig. 6A). The majority of the peptides did not match within 1 Da of the peptides expected from trypsin cleavage of the theoretical 3ECD, likely due to extensive modification of the 3ECD produced in human cells (34). However, two peptides were within 1 Da of the expected masses, a peptide of relatively low abundance that corresponds to residues 211 to 222 of the 3ECD (P1) and a more highly enriched peptide corresponding to residues 560 to 583 (P2) (Fig. 6B). The presence of multiple peptides indicates that ODN2006 could bind to more than one region of 3ECD. However, P2 maps to the asparagine-rich surface of 3ECD that has been demonstrated to be critical for TLR3 function (34) and adjacent to residues H539 and N541 that were previously demonstrated to contact poly(I-C) (Fig. 6C) (6). These results show that ssDNAs can bind to 3ECD, and the location of P2 suggests that ODN2006 can compete with poly(I-C) for binding TLR3.
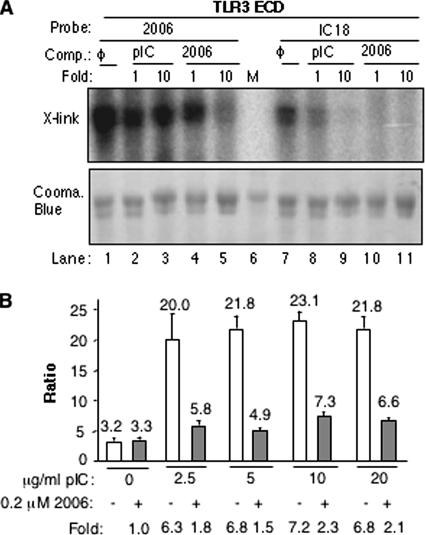
To examine further how ssDNA can compete with poly(I-C) for binding to 3ECD, we induced purified 3ECD binding with 5′-radiolabeled ODN2006 by using UV irradiation. A parallel reaction was performed with a radiolabeled 18-bp poly(I-C) named IC18. Both ODN2006 and IC18 could be cross-linked to 3ECD (Fig. 7A). While the same amounts of radiolabel and ligand were used, issues such as radiolabeling efficiency make it difficult to directly compare the cross-linking results. Also, it is possible that an 18-bp poly(I-C) may be too small to bind efficiently with TLR3 (28). Therefore, the relative cross-linking to radiolabeled ODN2006 or IC18 was examined in the presence of 1- or 10-fold excesses of unlabeled versions of ODN2006 and heterogeneous poly(I-C) that had lengths in excess of 200 bp (Fig. 7A). Cross-linking to radiolabeled ODN2006 was more resistant to challenge with heterogeneous poly(I-C) than the reverse combination (Fig. 7A, compare lanes 2 to 5 with 8 to 11), demonstrating that it is possible that ODN could not only bind directly to 3ECD but could do so with higher relative affinity than both poly(I-C) of the defined length of 18 bp and heterogeneous poly(I-C) of a length that is an efficient inducer of TLR3 signaling.
FIG. 7.
Examination of the interaction of 3ECD with poly(I-C) and ODN2006. (A) Recombinant 3ECD binding to ODN2006 and poly(I-C) in a UV-induced cross-linking (X-link) assay. ODN2006 and IC18 were radiolabeled by a kinase reaction, and the autoradiograph is shown in the upper panel. The lower panel contains a Coomassie (Cooma.) blue-stained gel that is intended to allow normalization for the amount of 3ECD used in each reaction. The competitors (Comp.) used were mock (φ), ODN2006, and heterogeneous poly(I-C). “M” corresponds to the size marker. (B) ODN2006 is a potent inhibitor of TLR3 signaling even at higher poly(I-C) concentrations. Since the experiment was performed with different concentrations of ligands, the basal level of luciferase activity is expected to change. Hence, the ratios of the levels of NF-κB-driven firefly luciferase to the levels of the putatively constitutive thymidine kinase promoter are graphed. The level of induction is listed below the graph. Note that there is a modest increase in TLR3 signaling at 10 and 20 μg/ml of poly(I-C) in comparison to the level at 2.5 μg/ml of poly(I-C). The means of the results are shown above the bars, and the error bars show standard errors of the means. pIC, poly(I-C); 2006, ODN2006; +, present; −, absent.
To examine the competition between ODN2006 and poly(I-C) for interaction with TLR3, we examined whether increasing concentrations of poly(I-C) (length, >200 bp) could overcome the effects of ODN2006 in 293T cells (Fig. 7B). Cells transfected to express TLR3 were treated with 2.5 to 20 μg/ml poly(I-C) with and without 0.2 μM ODN2006. A four- to eightfold-higher concentration of poly(I-C) resulted in only a slight reduction in the inhibitory activity of ODN2006 (Fig. 7B). We did not attempt higher poly(I-C) concentrations, as that could lead to responses that are not dependent on TLR3. These results are consistent with the model that ODN2006 is a more-potent antagonist of TLR3 signaling than poly(I-C) is an agonist. Furthermore, it will be useful to examine further how ssDNAs and poly(I-C) can compete for TLR3 binding.
Correlation between ssDNA binding to 3ECD and inhibition of signaling.
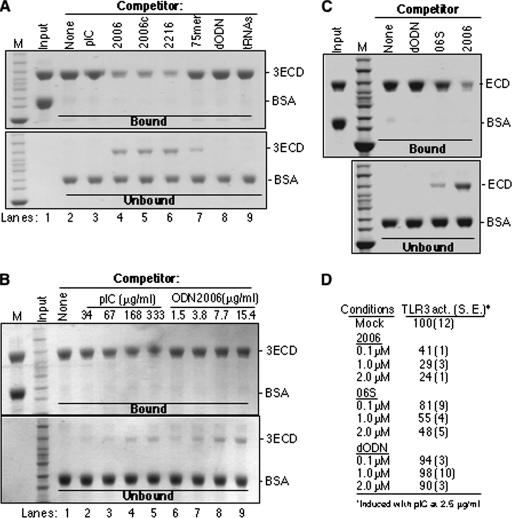
We characterized ligand binding by 3ECD by using a pull-down assay in the presence of ssDNA competitor. In this assay, biotinylated ODN2006 was bound to streptavidin resin and incubated with a mixture of 3ECD and BSA to provide an internal negative control. The assay is specific since BSA present in each reaction mixture is not bound (Fig. 8A). In addition, total yeast RNA was not able to prevent 3ECD from binding ODN2006 (Fig. 8A, lane 9). Poly(I-C) (length, >200 bp) was unable to prevent ECD binding to ODN2006, confirming that the ODN has higher affinity than poly(I-C) (Fig. 8A, lane 3). In contrast, ssDNAs ODN2006, ODN2006c, and ODN2216 that inhibited TLR3-specific reporter gene activation by poly(I-C) were effective inhibitors of 3ECD binding to ODN2006. The nonphosphorothioated 75-mer and dODN that had reduced inhibitory activity or lacked TLR3-inhibiting activity, respectively (Fig. 2B; Table 3), had reduced ability and inability to inhibit 3ECD binding to the ODN2006-resin (Fig. 8A, lanes 7 and 8).
FIG. 8.
ODN2006 is preferentially bound by TLR3. (A) An affinity pull-down assay was used to compare the binding of various ligands to 3ECD. The input used was a mixture of purified 3ECD and BSA. The mixture was then incubated with washed ODN2006-streptavidin resin in the presence of the competitor nucleic acid shown above the gel image. All competitors were used at 3 μg in the binding reaction mixtures. The gels were stained with Coomassie blue. Proteins bound and not bound to the resin are shown in the upper and lower gel images. The identities of the bands are shown to the right of the gel images. The binding reactions were performed at room temperature. (B) A comparison of the ability of heterogeneous poly(I-C) to compete with ODN2006 for binding of 3ECD. All concentrations are shown as μg/ml to facilitate comparisons. (C) ssDNA 06S that is partially defective for inhibiting TLR3 signaling is also defective for preventing 3ECD binding to ODN2006-resin. All competitors were added to the binding reaction mixture at 2 μM. (D) Results of reporter assay demonstrating that 06S is defective for inhibiting TLR3 signaling. The results are from the standard NF-κβ reporter assay of HEK 293T cells transfected to express TLR3. The cells were induced with poly(I-C) in the presence of the oligonucleotides at the concentrations shown. 2006, 2006c, and 2216 are ODNs. M, size marker; pIC, poly(I-C); act., activity; S.E., standard error.
Using the competition assay, we compared the ability of heterogeneous poly(I-C) to elute 3ECD bound to the ODN2006-resin to that of ODN2006. Poly(I-C) added to the reaction mixture at 168 to 333 μg/ml was able to elute approximately as much 3ECD as did 7.7 μg/ml of ODN2006 (Fig. 8B, compare lanes 4 and 5 to 8). These results are consistent with our previous observation that 3ECD preferentially binds ODN2006 over poly(I-C) (Fig. 7A). Furthermore, the experiment yielded the same results when performed either on ice or at room temperature.
To further extend the correlation between 3ECD ssDNA binding and the effects on TLR3 activation of signal transduction, we tested equimolar amounts of a phosphorothioated DNA that corresponds to the first 15 nucleotides of ODN2006, named 06S, which had reduced ability to compete for 3ECD binding to ODN2006 in vitro (Fig. 8C). Testing for its ability to inhibit TLR3 activation of the NF-kB reporter in cells showed that this activity was also reduced by a similar measure (Fig. 8D). All of these results demonstrate an excellent correlation between ssDNA binding by 3ECD and the ability to inhibit TLR3 activation of signaling. To summarize the results of all of the experiments probing the mechanism of action of the ssDNAs, we propose the following working model: ssDNAs can bind to TLR3 and either directly or indirectly prevent dsRNA binding. Furthermore, TLR3 in complex with ssDNA is unable to activate signaling, perhaps because ssDNA prevents the proper oligomerization and/or conformational changes that allow activation of the downstream adaptors.
ssDNA and cytokine production by BEAS-2B cells.
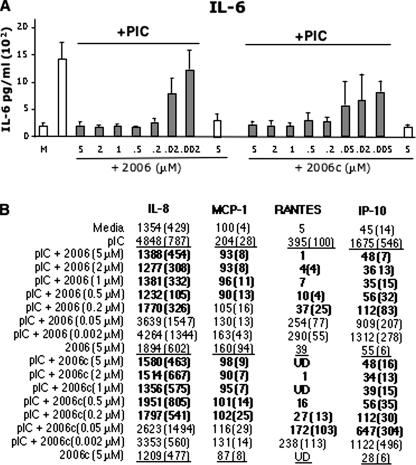
To better establish the physiological relevance of the ssDNA antagonists of TLR3 signaling, we examined cytokine production in the BEAS-2B cell line, which expresses native TLR3. BEAS-2B cells produced an average of 1,500 pg/ml of interleukin-6 (IL-6) upon induction with 25 ng/ml poly(I-C) (Fig. 9A), an amount ∼10-fold greater than that produced by the cells treated with medium alone. The presence of ODN2006 or ODN2006c alone did not induce cytokine production in the absence of poly(I-C) (Fig. 9A). In the presence of 0.2 μM ODN2006 and poly(I-C), IL-6 levels were reduced to less than 200 pg/ml. The inhibitory effect of ODN2006 was also dependent on its concentration (Fig. 9A). ODN2006c lacking the CpG motif required to activate TLR9 had the same inhibitory profile. Lastly, the same concentration-dependent inhibitory effect was observed for four additional cytokines and chemokines, IL-8, MCP-1, RANTES, and IP-10, that were previously reported to be produced by BEAS-2B cells (Fig. 9B) (12). Thus, the effects of ODN2006 in a bronchial epithelial cell line are consistent with those from reporter assays with transfected 293T cells.
FIG. 9.
ssDNA nucleotides can inhibit cytokine production in a human bronchial epithelial cell line, BEAS-2B. (A) Production of IL-6 with increasing concentrations of ODN2006 and ODN2006c. IL-6 and other ILs were measured by using Luminex beads and antibodies specific to the analyte. Poly(I-C) was added at 25 ng/ml in the samples whose results are shown with gray bars. “M” denotes the addition of unconditioned medium. Error bars show standard errors of the means. PIC, poly(I-C). (B) A summary of the production of several cytokines and chemokines in the presence of different concentrations of ODN2006 or ODN2006c in response to poly(I-C) treatment. The numbers in parentheses are the values for 1 standard error. The numbers in bold indicate the samples in which treatment with an ODN decreased the poly(I-C) induction of the cytokine or chemokine by 50%. The underlined numbers are values obtained with poly(I-C), 2006, and 2006c as the ligands. pIC, poly(I-C); UD, undetected.
Inhibition of cytokine production in human PBMCs.
To extend our observations of the effects of ssDNAs, we examined PBMCs from six independent donors. The results from all of the samples were consistent, and those from two are shown as representative results (Table 5) Cells from both donors showed little IFN-γ production when treated with the control medium or with several ssDNAs (Table 5; data not shown). However, a 24-h treatment with poly(I-C) at 5 μg/ml resulted in IFN-γ levels of approximately 1,200 to 6,600 pg/ml.
TABLE 5.
Effects of ssDNAs on IFN-γ production by PBMCs isolated from human donorsa
| ODN | Concn (μM) | IFN-γ production (pg/ml) of PBMCs treated with:
|
||||
|---|---|---|---|---|---|---|
| Mock (n = 6)
|
Poly(I-C) (n = 6)
|
|||||
| Range | Mean | Range | Mean | % | ||
| Mock | 1-27 | 14 | 1,160-6,674 | 4,214 | 100 | |
| 2006 | 1 | ND | 11-1,842 | 579 | 14 | |
| 5 | 1-42 | 25 | 6-56 | 31 | 1 | |
| 2006c | 1 | ND | 1-195 | 72 | 2 | |
| 5 | 1-56 | 25 | 13-95 | 44 | 1 | |
| 2216 | 1 | ND | 82-2,681 | 1,447 | 34 | |
| 5 | 4-64 | 38 | 55-631 | 287 | 7 | |
| 2216c | 1 | ND | 56-4,483 | 1,887 | 45 | |
| 5 | 16-135 | 53 | 25-3,266 | 1,278 | 30 | |
Effects of two concentrations of ssDNAs on IFN-γ production from six donors. Poly(I-C) was used at 5 μg/ml. ND, not done.
Next, the effects of ssDNAs on IFN-γ production were examined with PBMCs from both donors. Along with poly(I-C) at 5 μg/ml, ODN2006 and ODN2006c at 1 μM reduced IFN-γ levels significantly and at 5 μM reduced IFN-γ levels to background for both donors (Table 5). The effects of ODN2216 and ODN2216c were more complex. IFN-γ production by PBMCs from donor A was inhibited to background by all concentrations of ODN2216 tested. The IFN-γ level was less sensitive to ODN2216c, especially in donor B, although higher concentrations of ODN2216 and ODN2216c did reduce IFN-γ production. These results demonstrate that ssDNAs can inhibit IFN-γ production to various degrees in PBMCs isolated from different individuals.
To further extend the examination of the effects of ssDNAs, the production of several cytokines and chemokines by human PBMCs was quantified (Table 6). Of the 48 different samples treated with ODNs at 5 μM, only four did not reduce the levels of IL-12, IL-1β, and MIG by at least 50% (Table 6). In the absence of poly(I-C), only slight induction was seen for IL-12, IL-1β, and MIG. However, the levels of IL-6 in response to the ODNs were quite complex (see Fig. S2 in the supplemental material). In the absence of poly(I-C), treatment with three of the ODNs (2006, 2006c, and 2216) resulted in an increase in IL-6 levels, while treatment with ODN2216c had no effect on two of the PBMC samples and resulted in very high levels in samples from two donors (see Fig. S2 in the supplemental material). Treatment with poly(I-C) and ODN2216c did not seem to further affect IL-6 levels except in two samples. Furthermore, given that the ssDNAs lacking the CpG motif also had effects, there is no correlation with activation of TLR9 signaling. These results demonstrate that while ssDNAs can inhibit the production of cytokines and chemokines in human cells in response to poly(I-C), there are differences associated with the ssDNA sequence and, possibly, with the genotypic differences of the donors. We further note that IL-6 production in BEAS-2B cells was repressed in a concentration-dependent manner (Fig. 9A). While we do not understand these differences, we speculate that there are several receptors that can respond to dsRNA (1). The distribution of these receptors may also differ between cell types, and it is possible that cross talk between them could result in the more-complex cytokine pattern observed in human PBMCs. These downstream events are likely quite complex and should be the objective of a future study.
TABLE 6.
Summary of the effects of four ssDNAs on cytokine production
| Treatment | Amt (pg/ml) of cytokine produced by PBMC of each donora
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL-12
|
IL-1β
|
MIG
|
||||||||||
| A | B | C | D | A | B | C | D | A | B | C | D | |
| Poly(I-C) | 4,858 | 8,756 | 4,743 | 4,220 | 549 | 421 | 303 | 316 | 903 | 1,035 | 580 | 317 |
| Poly(I-C) plus ODN2006 | 85 | 128 | 95 | 74 | 28 | 447 | 34 | 39 | 47 | 49 | 21 | 35 |
| Poly(I-C) plus ODN2006c | 78 | 224 | 104 | 94 | 118 | 191 | 104 | 83 | 49 | 47 | 23 | 34 |
| Poly(I-C) plus ODN2216 | 1,150 | 1,991 | 701 | 105 | 145 | 237 | 112 | 55 | 603 | 376 | 68 | 39 |
| Poly(I-C) plus ODN2216c | 151 | 231 | 234 | 97 | 126 | 165 | 242 | 81 | 74 | 61 | 33 | 36 |
| Medium | 23 | 60 | 37 | 34 | 21 | 25 | 24 | 32 | 34 | 38 | 7 | 29 |
| Medium plus ODN2006 | 57 | 125 | 82 | 67 | 27 | 77 | 32 | 39 | 38 | 45 | 16 | 34 |
| Medium plus ODN2006c | 76 | 119 | 79 | 91 | 112 | 46 | 74 | 82 | 49 | 43 | 22 | 35 |
| Medium plus ODN2216 | 86 | 123 | 114 | 69 | 31 | 80 | 42 | 45 | 82 | 87 | 28 | 36 |
| Medium plus ODN2216c | 105 | 117 | 135 | 96 | 123 | 39 | 105 | 65 | 68 | 46 | 30 | 37 |
The numbers in bold show samples for which treatment with 5 μM ssDNA reduced cytokine production by more than 50%.
Summary and final comments.
We have demonstrated that several ssDNAs can inhibit TLR3-specific activation of the NF-κB reporter gene in 293T cells but do not affect TLR9 or TLR4. The inhibitory effects are also observed in cultured human bronchial epithelial cells and in human PBMCs that natively express TLR3 and can respond to poly(I-C). In terms of the mechanism of action, the ssDNAs can bind to purified 3ECD in vitro with higher apparent affinity than with poly(I-C) and colocalize with TLR3 in cells. There is also a strong correlation between ssDNA binding and inhibition of TLR3 activity. Whether the ssDNAs bind to the same location as poly(I-C) within 3ECD remains to be determined.
It is possible that ssDNAs with the ability to inhibit TLR3 could also act on other innate immune receptors and/or nucleic acid binding proteins to affect other signaling pathways. In fact, there is precedent for ssDNAs to affect the responses of TLRs. CpG-containing DNAs could activate STAT1 and IFN-β in mouse macrophage and dendritic cells (35). In addition, oligonucleotides consisting of homothymidines inhibited TLR7-specific signaling that was induced with Resiquimod (18, 19). Dorn et al. (11) had reported that oligonucleotides that lack CpG motifs could suppress IL-8 production in human keratinocytes. The oligonucleotides in their assay required phosphorothioates and accumulated in perinuclear regions of the cells. We have also observed a suppression of IL-8 production in poly(I-C)-stimulated BEAS-2B cells (Fig. 9), but phosphorothioates are not absolutely required for oligonucleotides to have inhibitory activity in 293T cells and the ssDNAs accumulated primarily in vesicles.
It is important to note that the TLR3-modulating ssDNAs include ones that are unable to function as potentiators or antagonists to TLR9. First, TLR9 expression is not detectable in HEK 293T cells. Second, a CpG motif is not required for the ssDNAs to have inhibitory activity. Third, blocking groups at the 5′ terminus of ODN2006 that abolish TLR9 activation did not affect the inhibitory activity against TLR3. It remains to be determined whether other innate receptors respond to these ssDNAs and perhaps contribute to the complex response of cytokine levels in human PBMCs. We do note that TLR4 signaling was not apparently affected by ODNs in 293T cells (Fig. 1D), suggesting that there is some specificity in the effects of ODNs on TLR3. All of the results from localization studies, binding studies using purified human 3ECD, and competition studies with reporter constructs consistently support a model where the ssDNAs bind to one or more sites in the 3ECD and prevent activation by poly(I-C). While TLR3 activation requires poly(I-C)s of lengths in excess of 40 bp, ODN2006 is only 24 nucleotides long. In addition, a derivative of ODN2006 of 15 nucleotides, 06S, can reduce TLR3 signaling by half when added to the medium of 293T cells at approximately 2 μM (Fig. 8D). A modified ssDNA of 15 to 24 nucleotides may not be able to span oligomers of TLR3, suggesting that the ssDNAs bind to TLR3 monomers. One possible outcome of this binding is the inhibition of TLR3 oligomerization. Modulation of signaling by an innate receptor could be important in disease outcomes, an example being the frequent bacterial infections of influenza patients that often exacerbate disease symptoms (5).
An intriguing property of the ssDNAs is that their inhibitory activity can differ according to their number of phosphorothioates, length, and also, base sequence. While further work is necessary to produce ssDNAs with improved specificity, this class of molecules holds promise for studies of signaling from the innate immune receptors and for modulating cytokine responses, perhaps in combination with the use of monoclonal antibodies specific to TLR3 (12).
Supplementary Material
Acknowledgments
We are grateful for the helpful discussions with L. San Mateo, R. Goldschmidt, and members of the Kao Lab at Texas A&M University. We thank K. Bhardwaj for help with molecular modeling. The manuscript is dedicated to Loren Snyder of Michigan State University upon his retirement from teaching.
Footnotes
Published ahead of print on 19 May 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124783-801. [DOI] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413732-738. [DOI] [PubMed] [Google Scholar]
- 3.Bagchi, A., E. A. Herrup, H. S. Warren, J. Trigilio, H. S. Shin, C. Valentine, and J. Hellman. 2006. MyD88-dependent and MyD88-independent pathways in synergy, priming, and tolerance between TLR agonists. J. Immunol. 1781164-1171. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, S., C. J. Kirschnin, H. Hacker, V. Redecke, S. Hausmann, S. Akira, H. Wagner, and G. B. Lipford. 2001. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. USA 989237-9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beadling, C., and M. K. Slifka. 2004. How do viral infections predispose patients to bacterial infections? Curr. Opin. Infect. Dis. 17185-191. [DOI] [PubMed] [Google Scholar]
- 6.Bell, J. K., J. Askins, P. R. Hall, and D. Davies. 2006. The dsRNA binding site of human Toll-like receptor 3. Proc. Natl. Acad. Sci. USA 1038792-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhardwaj, K., S. Palaninathan, J. M. Alcantara, L. Li Yi, L. Guarino, J. C. Sacchettini, and C. C. Kao. 2008. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease nsp15. J. Biol. Chem. 2833655-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter, S., and L. A. O'Neill. 2007. How important are Toll-like receptors for antimicrobial responses? Cell Microbiol. 91891-1901. [DOI] [PubMed] [Google Scholar]
- 9.Cook, D. N., D. S. Pisetsky, and D. A. Schwartz. 2004. Toll-like receptors in the pathogenesis of human disease. Nat. Immunol. 5975-979. [DOI] [PubMed] [Google Scholar]
- 10.de Bouteiller, O., E. Merck, U. A. Hasan, S. Hubac, B. Benguigui, G. Trinchieri, E. E. M. Bates, and C. Caux. 2005. Recognition of double-stranded RNA by human Toll-like receptor 3 and downstream receptor signaling requires multimerization and an acidic pH. J. Biol. Chem. 28038133-38145. [DOI] [PubMed] [Google Scholar]
- 11.Dorn, A., R. J. Ludwig, A. Bock, D. Thaci, K. Hardt, J. Bereiter-Hahn, R. Kaufmann, A. Bernd, and S. Kippenberger. 2007. Oligonucleotides suppress IL-8 skin keratinocytes in vitro and offer anti-inflammatory properties in vivo. J. Investig. Dermatol. 127846-854. [DOI] [PubMed] [Google Scholar]
- 12.Duffy, K. E., R. J. Lamb, L. R. San Mateo, J. L. Jordan, G. Canziani, M. Brigham-Burke, J. Korteweg, M. Cunningham, H. S. Beck, J. Carton, J. Giles-Kumar, C. Duchala, R. T. Sarisky, and M. L. Mbow. 2007. Down modulation of human TLR3 function by a monoclonal antibody. Cell. Immunol. 248103-114. [DOI] [PubMed] [Google Scholar]
- 13.Gaspari, A. A. 2006. Innate and adaptive immunity in the pathophysiology of psoriasis. J. Am. Acad. Dermatol. 54S67-S80. [DOI] [PubMed] [Google Scholar]
- 14.Gay, N. J., and M. Gangloff. 2007. Structure and function of Toll receptors and their ligands. Annu. Rev. Biochem. 76141-165. [DOI] [PubMed] [Google Scholar]
- 15.Hartmann, G., and A. M. Krieg. 2000. Mechanism and function of a newly identified CpG DNA motif in human primary cells. J. Immunol. 164944-952. [DOI] [PubMed] [Google Scholar]
- 16.Hartmann, G., R. D. Weeratna, Z. K. Ballas, P. Payette, S. Blackwell, I. Suparto, W. L. Rasmussen, M. Waldschmidt, D. Sajuthi, R. H. Purcell, H. L. Davis, and A. M. Krieg. 2000. Delineation of a CpG phosphorothioate oligodeoxynucleotide for activating primate immune responses in vitro and in vivo. J. Immunol. 1641617-1624. [DOI] [PubMed] [Google Scholar]
- 17.Heeg, K., A. Dalpke, M. Peter, and S. Zimmermann. 2007. Structural requirements for uptake and recognition of CpG oligonucleotides. Int. J. Med. Microbiol. 29833-38. [DOI] [PubMed] [Google Scholar]
- 18.Jurk, M., A. Kritzler, B. Schulte, S. Tluk, C. Schetter, A. M. Krieg, and J. Vollmer. 2006. Modulating responsiveness of human TLR7 and 8 to small molecule ligands with T-rich phosphorothiate oligodeoxynucleotides. Eur. J. Immunol. 361815-1826. [DOI] [PubMed] [Google Scholar]
- 19.Jurk, M., F. Heil, J. Vollmer, C. Schetter, A. M. Krieg, H. Wagner, G. Lipford, and S. Bauer. 2002. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat. Immunol. 3499. [DOI] [PubMed] [Google Scholar]
- 20.Kabelitz, D. 2007. Expression and function of Toll-like receptors in T lymphocytes. Curr. Opin. Immunol. 1939-45. [DOI] [PubMed] [Google Scholar]
- 21.Kariki, K., H. Ni, J. Capodici, M. Lamphier, and D. Weissman. 2004. mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem. 27912542-12550. [DOI] [PubMed] [Google Scholar]
- 22.Kawai, T., and S. Akira. 2005. Pathogen recognition with Toll-like receptors. Curr. Opin. Immunol. 17338-344. [DOI] [PubMed] [Google Scholar]
- 23.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7131-137. [DOI] [PubMed] [Google Scholar]
- 24.Kim, Y. C., W. K. Russell, C. T. Ranjith-Kumar, M. Thomson, D. H. Russell, and C. C. Kao. 2005. Functional analysis of RNA binding by the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 28038011-38019. [DOI] [PubMed] [Google Scholar]
- 25.Kindrachuk, J., J. E. Potter, R. Brownlie, A. D. Ficzyc, P. J. Griebel, N. Mookherjee, G. K K. Mutwiri, L. A. Babiuk, and S. Napper. 2007. Nucleic acids exert a sequence-independent cooperative effect on sequence-dependent activation of Toll-Like receptor 9. J. Biol. Chem. 28213944-13953. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi, M., Y. Tsuda, T. Yoshida, D. Takeuchi, T. Utsunomiya, H. Takahashi, and F. Suzuki. 2006. Bacterial sepsis and chemokines. Curr. Drug Targets 7119-134. [DOI] [PubMed] [Google Scholar]
- 27.Lancaster, G. I., Q. Khan, P. Drysdale, F. Wallace, A. E. Jeukendrup, M. T. Drayson, and M. Gleeson. 2005. The physiological regulation of Toll-like receptor expression and function in humans. J. Physiol. 563945-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leonard, J. N., R. Ghiriando, J. Askins, J. K. Bell, D. H. Margulies, D. R. Davies, and D. M. Segal. 2008. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc. Natl. Acad. Sci. USA 105258-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Limmon, G. V., M. Arredouani, K. L. McCann, R. A. C. Minor, L. Kobzik, and F. Imani. 2008. Scavenger receptor class-A is a novel cell surface receptor for double-stranded RNA. FASEB J. 22159-167. [DOI] [PubMed] [Google Scholar]
- 30.Matsukura, S., F. Kokubu, M. Kurokawa, M. Kawaguchi, K. Ieki, H. Kuga, M. Odaka, S. Suzuki, S. Watanabe, H. Takeuchi, T. Kasama, and M. Adachi. 2006. Synthetic double-stranded RNA induces multiple genes related to inflammation through Toll-like receptor 3 depending on NF-kappaB and/or IRF-3 in airway epithelial cells. Clin. Exp. Allergy 361049-1062. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto, M., K. Funami, M. Tanabe, H. Oshiumi, M. Shingai, Y. Seto, A. Yamamoto, and T. Seya. 2006. Subcellular localization of Toll-like receptor 3 in human dendritic cells. J. Immunol. 1713154-3162. [DOI] [PubMed] [Google Scholar]
- 32.Napolitani, G., A. Rinaldi, F. Bertoni, F. Sallusto, and A. Lanzavecchia. 2006. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat. Immunol. 6769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ranjith-Kumar, C. T., W. Miller, J. Sun, J. Xiong, J. Santos, I. Yarbrough, R. J. Lamb, J. Mills, K. E. Duffy, S. Hoose, M. Cunningham, A. Holzenburg, M. L. Mbow, R. T. Sarisky, and C. C. Kao. 2007. Effects of single nucleotide polymorphisms on Toll-like receptor 3 activity and expression in cultured cells. J. Biol. Chem. 28217696-176705. [DOI] [PubMed] [Google Scholar]
- 34.Ranjith-Kumar, C. T., W. Miller, J. Xiong, W. K. Russell, R. Lamb, J. Santos, K. E. Duffy, L. Cleveland, M. Park, K. Bhardwaj, Z. Wu, D. H. Russell, R. T. Sarisky, M. L. Mbow, and C. C. Kao. 2007. Biochemical and functional analyses of the human Toll-like receptor 3 ectodomain. J. Biol. Chem. 2827668-7678. [DOI] [PubMed] [Google Scholar]
- 35.Schroder, K., M. Spille, A. Pilz, J. Lattin, K. A. Bode, K. M. Irvine, A. D. Burrows, T. Ravasi, H. Weighardt, K. J. Stacey, D. A. Hume, A. H. Dalpke, and M. J. Sweet. 2007. Differential effects of CpG DNA on IFN-induction and STAT1 activation in murine macrophages versus dendritic cells: alternatively activated STAT1 negatively regulates TLR signaling in macrophages. J. Immunology 1793495-3503. [DOI] [PubMed] [Google Scholar]
- 36.Shinozuka, K., T. Morita, and H. Sawai. 1991. Synthesis and nuclease susceptibility of alpha-oligodeoxyribonucleotide phosphorothioate. Nucleic Acids Symp. Ser. 25101-102. [PubMed] [Google Scholar]
- 37.Sun, J., K. E. Duffy, C. T. Ranjith-Kumar, J. Xiong, R. J. Lamb, J. Santos, H. Masarapu, M. Cunningham, A. Holzenburg, R. T. Sarisky, M. L. Mbow, and C. Kao. 2006. Structural and functional analyses of the human Toll-like receptor 3. Role of glycosylation. J. Biol. Chem. 28111144-11151. [DOI] [PubMed] [Google Scholar]
- 38.Uematsu, S., and S. Akira. 2006. Toll-like receptors and innate immunity. J. Mol. Med. 84712-725. [DOI] [PubMed] [Google Scholar]
- 39.Underhill, D. M. 2004. Toll-like receptors and microbes take aim at each other. Curr. Opin. Immunol. 16483-487. [DOI] [PubMed] [Google Scholar]
- 40.Yu, D., E. R. Kandimalla, Y. Cong, J. Tang, J. Y. Tang, Q. Zhao, and S. Agrawal. 2002. Design, synthesis, and immunostimulatory properties of CpG DNAs containing alkyl-linker substitutions: role of nucleosides in the flanking sequences. J. Med. Chem. 454540-4548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.