Abstract
5S rRNA genes from Saccharomyces cerevisiae were examined by Miller chromatin spreading, representing the first quantitative analysis of RNA polymerase III genes in situ by electron microscopy. These very short genes, ∼132 nucleotides (nt), were engaged by one to three RNA polymerases. Analysis in different growth conditions and in strains with a fourfold range in gene copy number revealed regulation at two levels: number of active genes and polymerase loading per gene. Repressive growth conditions (presence of rapamycin or postexponential growth) led first to fewer active genes, followed by lower polymerase loading per active gene. The polymerase III elongation rate was estimated to be in the range of 60 to 75 nt/s, with a reinitiation interval of ∼1.2 s. The yeast La protein, Lhp1, was associated with 5S genes. Its absence had no discernible effect on the amount or size of 5S RNA produced yet resulted in more polymerases per gene on average, consistent with a non-rate-limiting role for Lhp1 in a process such as polymerase release/recycling upon transcription termination.
In eukaryotes, nuclear RNA transcription is carried out by one of three specialized RNA polymerases, Pol I, II, and III, with Pol III dedicated to making short, untranslated RNAs, including tRNAs, 5S rRNA, and many others (12). Consistent with the central role of these RNAs in protein synthesis, Pol III is an essential enzyme whose activity is regulated by various growth control pathways and is deregulated in human cancer cells and during cardiac hypertrophy (23, 67). Many years of study have led to detailed knowledge of the different types of promoters and of the trans-acting factors involved in Pol III gene expression (20, 31, 56). For the Saccharomyces cerevisiae 5S RNA genes studied herein, three factors are required: TFIIIA, which specifically binds the internal promoter of 5S genes, and TFIIIB and C, which are required for all yeast Pol III genes. Preinitiation complex formation begins with the deposition of TFIIIA on the internal promoter, followed by recruitment of the six-subunit TFIIIC complex, which contacts TFIIIA as well as sequences within the gene (Fig. 1A). TFIIIC recruits the three-subunit TFIIIB complex, which binds a conserved upstream regulatory sequence and is necessary and sufficient for Pol III transcription (30). TFIIIB, which contains TATA binding protein, recruits Pol III to the promoter and plays an essential role in the formation of the open initiation complex. The stability of TFIIIB, which restructures the DNA and recruits Pol III through multiple rounds of transcription (7, 30), renders these genes very highly transcribed and the most prominent binding sites for TATA binding protein in the cell (51). The internally bound factors do not form a barrier to Pol III elongation.
FIG. 1.
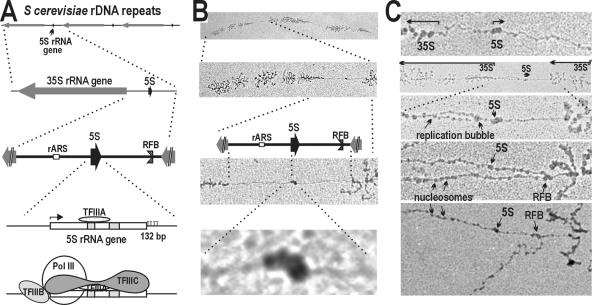
Identification of active S. cerevisiae 5S rRNA genes in rDNA intergenic spacers. (A) Schematic of three tandem rDNA repeats, shown at increasing magnification with a focus on the 5S gene in the intergene spacer between 35S genes. The middle panel shows elements identified in the spacer: origin of replication, the 5S gene, and replication fork barrier. The two bottom panels show the location of internal control elements in the gene (shaded areas) and the general size and position of TFIIIA, -B, and -C. TFIIIC has a flexible dumbbell shape that varies depending on the distance between its two binding sites. (B) EM image of three tandem rDNA repeats, shown at increasing magnification to parallel the schematics in panel A. The EM image of an intergene spacer in the third panel is shown aligned with a schematic of the spacer drawn to scale, showing the structure identified as an active 5S gene at the expected position, with larger view in bottom panel. (C) Additional EM images of rDNA spacers, showing that 5S genes can be distinguished from small replication bubbles opening at the origin of replication (at two magnifications in the second and third panels), from the replication fork barrier, where replication forks are paused (two bottom panels), and from nucleosomes (two bottom panels). rARS, replication fork barrier; RFB, replication fork barrier.
We used Miller chromatin spreading (39) to directly visualize the 5S gene population in actively growing yeast cells. This electron microscopy (EM) method has frequently been applied to the study of Pol I and Pol II gene expression. Pol III genes, however, proved difficult to unambiguously identify due to their small size, though Scheer (54) visualized short repeated genes that he speculated (almost certainly correctly) were Pol III genes. In S. cerevisiae, the 5S genes are located in the spacers between the tandemly repeated Pol I-transcribed 35S rRNA genes. Since the active 35S genes are easy to identify in chromatin spreads (19), it is likewise easy to identify the structures in the spacers that correspond to active 5S genes, allowing us to quantitatively analyze 5S genes at the level of both the individual gene and the gene population. Our results provide the first definitive information on Pol III loading per gene, allowing us to estimate in vivo elongation and reinitiation rates. In addition, visual analysis was informative regarding the phenomenon of facilitated recycling in which a polymerase repeatedly transcribes the same gene without dissociating from it (14, 15) and regarding the question of whether or not Pol I and III transcription are coregulated in the ribosomal DNA (rDNA) array (9). Our results also complement the results of genome-wide occupancy studies for Pol III and its factors (26, 40, 51), which have found that TFIIIB occupancy generally correlates with Pol III occupancy and that all Pol III genes can be detected in association with these proteins, consistent with their high rate of expression. Genome-wide occupancy studies are less informative in the case of multigene families, though the lower relative occupancy of 5S genes by these factors (40) is consistent with only a subset of the ∼150 gene copies being active, in agreement with the results of footprinting studies (8) and with results herein.
La is an abundant, conserved nuclear protein that associates with newly synthesized Pol III transcripts via their characteristic UUU-OH 3′ termini and has clearly defined functions as a chaperone in the maturation and stability of these transcripts (37, 71, 73). As discussed previously (37, 71), an additional role for La during Pol III transcription was proposed years ago (25) but remains controversial. That is, while the results of most in vitro studies find no transcriptional role for La (27, 34, 44, 66, 73), those of others indicate a role for La in transcript release and Pol III termination/reinitiation (25, 36, 38), and human La has been found in the Pol III holoenzyme (64) and at Pol III genes in vivo (17). Although La is essential in flies and mammals (2, 45), the La homolog in yeast is not essential (63, 72), and importantly, transcription continues normally in the absence of the La homolog, Lhp1, for those Pol III genes tested (44, 73). We found similarly that 5S RNA synthesis is not affected in lhp1Δ cells. However, we found by using chromatin immunoprecipitation (ChIP) analysis that Lhp1 is present on 5S genes, and in its absence, 5S genes have a higher-than-expected average number of polymerases per gene (an extra polymerase on 30 to 40% of the active genes). These results are consistent with a cotranscriptional but non-rate-limiting role of La in a process such as transcript release or polymerase recycling.
MATERIALS AND METHODS
For EM analysis, control and mutant strains (Table 1) were grown in yeast extract-peptone-dextrose plus 1 M sorbitol at 30°C. Cells were harvested and prepared for EM as described previously (19, 43). For analysis of 5S gene populations, all examples of active 35S/5S gene regions seen on EM grids were photographed, mapped, and included in the compilation. In counting polymerases/gene, those genes with an apparent polymerase mass between that of 1 and 2 polymerases (1+) or that of 2 and 3 polymerases (2+) (Fig. 2A) were counted as having 1 or 2 polymerases, respectively. Log-phase and postlog cultures were harvested at A600s of 0.35 to 0.55 and 2.5, respectively. When used, rapamycin (200 ng/ml) was added to cultures 30 min before cell lysis. When used, heparin (200 to 300 μg/ml) was added to dispersed cell contents at the time of cell lysis.
TABLE 1.
Yeast strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| NOY886 | MATα rpa135Δ::LEU2 ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 fob1Δ::HIS3 pNOY117 (CEN RPA135 TRP1); rDNA repeats, ≅42 | 19 |
| NOY1051 | MATaade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 fob1Δ::HIS3; rDNA repeats, ≅143 | 19 |
| NOY388 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100; rDNA repeats, ≅177 | 55 |
| NOY2167 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 spt4Δ::HIS3MX6; rDNA repeats, ≅64 | 55 |
| CY1 | MATα ura3 lys2 ade2 trp1 his3 leu2 LHP1; rDNA repeats, ≅190 | 73; this study |
| CY4 | MATaura3 lys2 ade2 trp1 his3 leu2 lhp1::LEU2; rDNA repeats, ≅140 | 73; this study |
| LC382 | MATα rex1::TRP1 ura3 lys2 ade2 trp1 his3 leu2 | This study |
| LC421 | MATarex1::TRP1 ura3 lys2 ade2 trp1 his3 leu2 lhp1::LEU2 | This study |
| LS44.4 | MATaura3 lys2 ade2 trp1 his3 leu2 rrp6::kanr; constructed using PCR to replace nt 118 to 1500 of the Rrp6 coding sequence with kanr | 35; this study |
| LS43.2 | MATα ura3 lys2 ade2 trp1 his3 leu2 lhp1::LEU2 rrp6::kanr; constructed as described for LS44.4 | This study |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 70 |
| BY4741 Δlhp1 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 lhp1::KAN | 70 |
FIG. 2.
Examples of active and inactive 5S genes. (A) Gallery of EM images showing the various configurations that occur at the position of the 5S gene, interpreted as genes with 0 to 3 polymerases (indicated by labeling on the left). The first column is a view of an entire spacer; the second column is a higher-magnification view of the gene in the first column. The third and fourth columns are additional examples of the type, with rare examples in the fourth column chosen because they clearly show individual polymerase-sized particles. The row labeled TF? shows possible transcription factors. 1+, size between that of 1 and 2 polymerases; 2+, size between that of 2 and 3 polymerases. (B) Particles identified as engaged polymerases are stable in the presence of 200 and 300 μg/ml heparin, confirming their identification as polymerases. One view of a spacer (top) and four additional 5S gene examples are shown (bottom). Heparin was added at the time of cell lysis, when initiation and elongation events are frozen, and incubation was for 20 min or longer. +, present. (C) Comparison of appearance of Pol I and Pol III on neighboring 35S and 5S genes. Two examples are shown in views of full spacers (first and third rows) and higher magnifications of polymerases (second and fourth rows). Note that in Miller chromatin spreads, nascent RNAs are typically too short to appear protruding from polymerases that have transcribed a length equivalent to the 5S gene. (D) Examples of active 5S genes in which there is an apparent physical interaction between the 5′ and 3′ ends of the gene (5′ end is to the left in all panels). One full view of a spacer is shown (top) with a higher magnification of that gene plus nine additional examples. The 5S promoter is to the left in all EM images.
To estimate rDNA copy numbers, Southern hybridization was performed as described previously (55).
ChIP analysis was performed as described previously (55), with the following modifications. Cells were treated with formaldehyde for 10 min to cross-link proteins to nucleic acids. Before immunoprecipitation (IP), cell extracts were split in half. One half was left untreated (no RNase), and the other was treated with 3 mg/ml RNase A. The extracts were incubated for 30 min at 20°C prior to IP (or freezing of input samples). Input and IP DNA samples were quantified by real-time PCR (7900HT; Applied Biosystems), using Sybr green as a detector. Each DNA sample was diluted 1/10, 1/100, and 1/1,000. Linear data over this range were plotted. Error bars equal plus or minus 1 standard deviation of these data.
For metabolic labeling of RNA, strains CY1 and CY4 were transformed with the URA3-containing vector YCplac33 (22) and grown in 40 ml of synthetic complete medium (SC)-uracil at 24°C to an A600 of 0.4. After being washed with SC-uracil, cells were resuspended in 400 μl of SC-uracil and pulsed with 100 μCi [3H]uracil (47 Ci/mmol; GE Healthcare) for 2 min, diluted 10-fold with SC containing 35 μg/ml uracil, and incubated for 0, 3, 12, and 30 min. RNA was extracted with acid phenol and sodium dodecyl sulfate at 65°C (1), and 60,000 cpm per sample was fractionated in a 5% polyacrylamide, 8.3 M urea gel. After being fixed for 1 h in 10% acetic acid, 30% methanol, the gel was incubated with En3hance (PerkinElmer) for 1 h, washed with water for 20 min, dried, and subjected to autoradiography.
For Northern analyses, strains were grown in yeast extract-peptone-dextrose to an A600 of 0.5. After total RNA was extracted as described above, 5 μg of RNA from each strain was fractionated in 5% polyacrylamide-8 M urea gels, transferred to Hybond N membranes (G.E. Healthcare), and hybridized with [γ32P]ATP-labeled oligonucleotides as described previously (59). The oligonucleotides used were pre-5S RNA, 5′ AAAAAGAAATAAAGATTGCAGCACC 3′; mature 5S RNA, 5′ CCAGCTTAACTACAGTTGATCGGACGGGAA 3′; and signal recognition protein RNA, 5′ TCAACGTATCCCATCCCAC 3′.
RESULTS
5S rRNA genes were identified in the spacers between active 35S rRNA genes.
Figure 1A shows schematics of yeast rDNA at increasingly higher levels of resolution, with a focus on the 5S RNA gene. The small 5S genes occur in the spacers between the 35S rRNA genes, resulting in divergently transcribed Pol I and Pol III promoters. The two bottom panels in Fig. 1A show features of the 132-nucleotide (nt) 5S gene, including internal promoter regions necessary for binding of TFIIIA and C, an upstream element bound by TFIIIB, and a simplified schematic of the transcription factor complex that recruits Pol III to the promoter (4). Figure 1B (top) shows three rDNA repeat units from exponentially growing yeast cells, followed by a higher magnification of one repeat unit. The spacers between actively transcribed Pol I genes exhibited no obvious RNA transcripts. Increasing magnification of the spacer region (Fig. 1B, third EM panel, shown aligned with a spacer schematic drawn to scale) reveals a characteristic structure precisely at the position of the 5S gene sequence. The bottom panel is a closer view of the structure at the position of the 5S gene, and is one of several typical configurations for an active 5S gene. The genes varied somewhat in appearance but did not display obvious nascent RNAs protruding from polymerases, due to the very short length of the transcripts. (Note that the rest of the figures showing EM images will show either the spacer region between two 35S genes or a higher magnification of the 5S gene region, similar to the views in the two bottom panels of Fig. 1B. The 5S promoter is always to the left.)
Views of additional rDNA spacers are shown in Fig. 1C. In the bottom four panels, DNA was in the process of replication. These images show that the 5S gene could be distinguished from the nearby origin of replication (as shown by the position of a newly initiated replication bubble; at low and high magnification in the second and third panels), from the replication fork barrier (as shown in the fourth and fifth panels by paused replication forks at the expected position downstream of active 35S genes) (41), and from nucleosomes, which are retained in some of our preparations (fourth and fifth panels).
The number of polymerases on individual 5S genes ranged from none to three.
To our knowledge, this visualization of active yeast 5S genes represents the first unambiguous identification by EM of any Pol III gene in situ, enabling us to directly address questions such as polymerase occupancy. Figure 2 shows a gallery of the types of structures seen at the position of the 5S gene. The majority of 5S genes in normal yeast strains, ∼70 to 80%, showed nothing bound (beyond the normal nucleoprotein strand seen in these spreads) (Fig. 2A, row 0) and thus appeared to be inactive. (The distinction between active and inactive genes becomes uncertain when dealing with such short genes, in that an inactive gene could either be epigenetically inactive or epigenetically active but unengaged by a polymerase at the moment of cell lysis. As will be discussed, combined evidence supports the conclusion that most 5S genes that contribute to 5S RNA production appear to be bound by polymerases in this EM approach. This includes the results of previous footprinting studies [8] that agree with our estimates of active gene number.)
In rare cases, particles smaller than polymerases were seen at the 5S gene position, possibly corresponding to transcription factors (Fig. 2A, row TF?) Their scarcity agrees with studies showing that genes with transcription factors bound are also likely to have Pol III bound (26). About 20 to 30% of 5S genes in control yeast strains appeared to be active genes, as variations on a structure lying along the DNA with the approximate width of one polymerase (15 to 20 nm) and more variable in length, from ∼15 to ∼50 nm (Fig. 2A, rows 1 to 3). In rare cases, one could discern from one to three distinct polymerase-sized particles at the position of the 5S gene (Fig. 2A, rightmost column), supporting our conclusion that the elongated structures represent genes bound by multiple polymerases. In the most-typical case, in which all the components appeared to meld into one elongated curved complex, it was not possible to distinguish its constituents. However, we presume that these structures are comprised of one to three polymerases, the nascent 5S RNA, and probably, associated transcription factors and RNP proteins. Many of the active genes, such as those in rows 1+ and 2+ in Fig. 2A, appeared to contain extra mass in addition to one or two polymerases, respectively.
Because TFIIIC is a large complex (520 kDa) and thus might be confused with Pol III (700 kDa), we treated dispersed chromatin samples with concentrations of heparin that dissociate TFIIIC from DNA, but not TFIIIB or engaged Pol III (30). As shown for five examples in Fig. 2B, the structures identified as probable polymerases on 5S genes were retained after heparin treatment (in both structure and frequency), including genes with one to three polymerases, supporting the conclusion that these are indeed polymerases actively engaged in transcription. Additional support comes from the appearance of these putative Pol IIIs as compared to the Pol I molecules at the extreme 5′ end of the neighboring 35S gene (Fig. 2C). The two types of polymerases were similar in appearance at this level of resolution, and after ∼150 nt or less were transcribed from the 5′ end, neither displayed nascent transcripts that were long enough to be detectable by this EM method.
High rates of Pol III transcription are achieved through facilitated recycling of Pol III, which involves the reinitiation of a terminating polymerase preferentially at the 5′ end of the same gene (14, 15), mediated either directly or indirectly through TFIIIB-Pol III interactions (18). This phenomenon has been studied on several different Pol III genes with intragenic promoters (e.g., tRNA genes), and although it has not been studied for 5S genes, it has been argued that it is likely to exist on 5S genes, which contain all the participating components and the same high in vivo transcription rate (15). Facilitated recycling would predict a physical interaction (perhaps transient) between the 5′ end of the gene where TFIIIB is located and the terminating polymerase. Some EMs showed an apparent interaction between the 5′ and 3′ ends of individual 5S genes (Fig. 2D), and thus may be instances of this phenomenon. The interaction may be facilitated by the natural curvature of the active genes, which presumably has contributions from the bending properties of TFIIIA, TFIIIB, and Pol III, and from the A/T-rich nature of the termination region (5, 50). However, we cannot rule out the possibility that these apparently looped genes are instead distorted or entangled genes.
These results showed that yeast 5S genes can be in an apparently inactive state or can be engaged by 1, 2, or 3 polymerases. Three polymerases per 5S gene corresponds to one polymerase/44 nt on this 132-nt gene. This agrees well with the ∼37 base pairs (bp) of DNA that are cross-linked to arrested Pol III in a ternary complex (3) and with the results of studies of Pol I genes where the maximum density observed was one polymerase/41 bp (19).
The activity of neighboring 5S genes and 35S genes was largely independent.
Our EM approach allows a unique opportunity to investigate long-range patterns of gene activity in the rDNA array. To study the distribution of active 5S and 35S genes, we mapped a particularly favorable spread from a control yeast strain with ∼143 rDNA repeats (NOY1051) (Table 1). We were able to map 120 repeats, mostly contiguous, representing 84% of the rRNA genes from a single nucleolus (Fig. 3A). For each repeat, the activity of the 5S and 35S rRNA genes was determined, except for a few regions where the 5S genes were ambiguous due to overlying chromatin. Of the 120 traceable rDNA repeat units, 71, or 59% of the 35S genes, were active. Of the 110 traceable 5S genes, 37, or 34%, displayed one or more polymerases and were designated as active. Figure 3B shows a linear representation of 113 rDNA repeats shown in Fig. 3A. Active 35S genes are shown above the line; active 5S genes are shown below the line. Statistical tests were used to analyze the activity pattern (see File S1 in the supplemental material). Dammann et al. (9) showed that the activity of 35S genes was random when they were considered as neighboring gene pairs, and we obtained the same result for both 35S-35S gene pairs and 5S-5S gene pairs (see Table S1 in the supplemental material). The data also allowed the first test of the activity status of neighboring 35S and 5S genes in single rDNA repeats, which were found to occur in proportions predicted by a random pattern (see Table S1 in the supplemental material), indicating that the activity of the 5S gene was largely independent of the activity of the neighboring 35S gene and vice versa. The results of analysis of longer-range activity patterns (i.e., across multiple neighboring rDNA repeats), however, revealed a moderate inverse correlation (P = 0.01) between regions of the array that had 35S-on/5S-off repeats versus 35S-off/5S-on repeats and also between regions of the array that were more or less active (P = 0.02) (see Table S4 in the supplemental material). Clearly there was no strict constraint on the coexpression of both genes in a repeat unit, though the results of the longer-range analysis suggest position effects that may be due, e.g., to chromatin structure or subnucleolar localization (see File S1 in the supplemental material).
FIG. 3.
Activity of 5S genes and 35S genes in the linear array. (A) A dispersed nucleolus from NOY1051, in which 120 of the ∼143 genes were traceable, shown as EM image only, trace only, and superimposed EM image and trace. Repeats (with one 35S and one 5S gene) are numbered. Underlining of the repeat number indicates that the 35S gene is inactive. Two breaks in the array (between 70 and 71 and 78 and 79) are presumed to be contiguous for panel B since they can be unambiguously aligned due to gene polarity. (B) Linear schematic of 113 repeats, with active 35S genes shown as red bars above the line and active 5S genes as blue bars below the line. Question marks indicate 5S genes that were somewhat ambiguous due to overlying chromatin, but which are more likely to be inactive than active (see File S1 in the supplemental material).
5S genes were regulated at two levels: on versus off and polymerase loading.
The EM approach allowed us to obtain semiquantitative results regarding the number of polymerases per 5S gene and the fraction of 5S genes with bound polymerases. (For simplicity, we will refer to genes with bound polymerases as “active” genes, keeping in mind the caveat mentioned above.) We asked how these parameters vary as 5S RNA transcription varies. In considering the number of active 5S genes, one must take into account the fact that the total rDNA repeat number (and thus, 5S gene number) varies between 140 and 200 copies for control strains and can be reduced by experimental manipulation (32). Four control strains were studied, with rDNA repeat numbers of ∼190, ∼177, ∼143, and ∼42 (Tables 1 and 2; see Table S5 in the supplemental material). (Strains with as few as 42 rDNA copies synthesize rRNA and grow as well as control strains; further reduction in gene number is deleterious to growth rate [32].)
TABLE 2.
Activity of 5S genes in yeast strains with different gene copy numbers, strains at different growth stages, and strains with mutations
| Strain (genotype) | Growth stage | Approximate no. of rDNA repeats/cell | No. of 5S genes tallied | Active 5S genes/cell (%) | Approximate no. of active 5S genes/cellc | Mean no. of pols/active gene | SD | 95% CI | Approximate no. of pols on 5S genes/celld |
|---|---|---|---|---|---|---|---|---|---|
| NOY886 | Log | 42b | 464 | 69 | 29 | 1.96 | 0.74 | 0.08 | 57 |
| 30 min of rapamycin | 42 | 103 | 46 | 19 | 1.87 | 0.82 | 0.24 | 36 | |
| Post-log | 42 | 122 | 34 | 14 | 1.39 | 0.59 | 0.18 | 19 | |
| NOY1051a | Log | 143 | 347 | 30 | 43 | 1.73 | 0.71 | 0.14 | 74 |
| NOY388 (wt SPT4) | Log | 177 | 227 | 21 | 37 | 1.81 | 0.88 | 0.25 | 67 |
| NOY2167 (spt4Δ) | Log | 64 | 234 | 35 | 22 | 1.77 | 0.79 | 0.17 | 39 |
| CY1 (wt LHP1) | Log | 190 | 249 | 26 | 49 | 1.54 | 0.66 | 0.13 | 75 |
| CY4 (lhp1Δ) | Log | 140 | 160 | 34 | 48 | 2.12 | 0.67 | 0.14 | 102 |
The NOY1051 data include the results for the single nucleolus shown in Fig. 3, as well as additional data representing smaller-rDNA-repeat multimers from this strain, similar to the data used for the other strains shown here. See breakdown in Table S5 in the supplemental material.
The four bold rows show data for four logarithmically growing control strains with different gene copy numbers. These data were used for the graphs in Fig. 4A to C.
Derived from rDNA repeat number and estimated percentage of active genes per cell.
Derived from approximate number of active 5S genes/cell and mean number of pols/active gene.
As 5S gene copy number increased in the control strains over a 4.5-fold range (42 to 190), there was a decrease in the percentage of active genes (from 69% to ∼25%) (Fig. 4A), accompanied by an increase in the number of active genes/cell (from 29 to 49) (Table 2). There was also a modest decrease in the mean number of polymerases/active gene (from ∼2 to ∼1.5) (Fig. 4B). (Although the range of possible numbers for polymerases/active gene is very small, from 1 to 3, we were able to obtain reasonable 95% confidence intervals for the means by tallying large numbers of genes, as shown by the error bars in Fig. 4B). These estimates for number of active genes/cell and mean number of polymerases/active gene were used to approximate the total number of polymerases engaged on 5S genes/cell (Fig. 4C), which ranged from 57 to 75 in log-phase cells and averaged 68.3 ± 8.3 (mean ± standard deviation) for the four control strains or 72 ± 4.4 for the three normal-copy-number strains. In the latter, the polymerases were distributed on ∼40 to 50 active genes/cell, with an average gene having one to two polymerases. In the 42-copy strain, ∼30 active genes were engaged by an average of two polymerases.
FIG. 4.
Regulation of 5S transcription by variation of active gene number and by polymerase loading per gene. (A) Correlation between percent active 5S genes and gene copy number for four control strains in log-phase growth (bold rows in Table 2). Correlation coefficient (r) and trend line are shown (y = −0.32x + 80.60). (B) Correlation between mean number of polymerases/active gene and gene copy number for control strains as described for panel A (trend line, y = −0.002x + 2.06). Error bars represent 95% confidence intervals for the mean. (C) Correlation between number of engaged polymerases/cell and gene copy number for control strains as described for panel A (trendline, y = 0.10x + 53.79). (D to F) For four experimental situations (post-log phase, 30-min rapamycin treatment, spt4Δ, and lhp1Δ), data points are displayed for the same parameters plotted for control strains in panels A to C. For comparison, the control trend lines from panels A to C are also shown. (G) Percentage of genes with zero, one, two, or three polymerases in the 42-copy yeast strain (NOY886) in log phase, post-log phase, and after 30-min rapamycin treatment.
In a previous study of cells lacking elongation factor Spt4, we found by pulse-labeling that the synthesis of both 35S rRNA and 5S rRNA was decreased by ∼30% (55). We asked if and how the decrease in 5S RNA synthesis manifested at the individual gene level. Our previous finding (55) that the spt4 deletion strain had a smaller rDNA array size than its control strain (∼64 versus ∼177) was taken into account. Based on correlations determined from the results shown in Fig. 4A and B (shown by the trend lines in Fig. 4D and E), it can be predicted that a strain with ∼64 genes would have about ∼60% of them active and that these genes would average 1.92 polymerases/gene. In fact, only 35% of the 5S genes were active in the spt4Δ strain (Fig. 4D), while the observed number of polymerases/gene (1.77) was closer to the predicted value (shown by proximity to the trend line, Fig. 4E). These data yield an average of 39 engaged polymerases/cell on 5S genes in the spt4Δ strain, corresponding to a decrease in engaged polymerases/cell in the range from 23% (compared to the Spt4 control strain) to 43% (compared to the average of 68 for the four control strains), indicating that the ∼30% decrease in transcription correlated with a corresponding decrease in polymerases transcribing the 5S gene. We have not investigated the mechanism of this effect, nor whether it is direct or indirect via Spt4/Spt5's role in the regulation of Pol I and Pol II genes (55, 69). However, these results allowed us to conclude that in log-phase cells, there was a direct correlation between the 5S RNA synthesis rate and the number of polymerases engaged on 5S genes and, also, that a decrease in the number of active genes appeared to be in large part responsible for the ∼30% decrease in 5S RNA synthesis in the absence of Spt4.
Next we examined the downregulation of 5S RNA synthesis (Fig. 4G). We applied either a brief repressive treatment (30-min treatment with rapamycin, which inhibits the growth-regulatory TOR pathway) or we allowed cells to leave exponential growth, representing a more extensive growth repression. The 42-copy strain was used for this analysis because 69% of its 5S genes are active during log-phase growth and, thus, it is a more sensitive indicator of changes. A 30-min rapamycin treatment resulted in a 37% decrease in engaged polymerases/cell (from 57 to 36), with the majority of the decrease being due to fewer active genes/cell (33% decrease, from 69% to 46%) rather than to fewer polymerases/gene (5% decrease, from 1.96 to 1.87) (Fig. 4D to F). With the more extensive downregulation that occurs in the shift to diauxic growth, the number of engaged polymerases/cell decreased by 67% (from 57 to 19), with a 51% decrease in the percentage of active 5S genes (from 69% to 34%) and a 29% decrease in the mean number of polymerases/gene (1.96 to 1.39), indicating that both parameters were downregulated as the growth rate slowed (Fig. 4D to F).
A nonessential cotranscriptional role for Lhp1, the yeast La protein.
To investigate whether Lhp1 had any discernible cotranscriptional role in 5S RNA synthesis in yeast, we analyzed 5S genes in strains CY4 (lhp1Δ) and CY1 (control). The results of EM analysis revealed that 5S genes from lhp1Δ cells had more polymerases/gene on average than the control strain (2.12 versus 1.54), as shown in images of representative genes in Fig. 5A. In fact, they had a higher average polymerase density than any other strain analyzed. To determine a possible contribution from a change in rDNA repeat number, we used Southern hybridization analysis (55) and found that the lhp1Δ strain had ∼140 rDNA repeats, in comparison to its control strain which has ∼190 repeats (not shown). The percentage of active repeats for the lhp1Δ strain (34%) (Fig. 4D) was very close to the expected value for a 140-copy strain (36%), while the number of polymerases/gene (mean 2.12) was significantly different (P < 0.001) from both its control strain and from NOY1051 with a similar rDNA copy number, resulting in an unusually high estimate of 102 total engaged polymerases/cell (Fig. 4E and F) Thus, the percentage of active genes was consistent with the gene copy number for the lhp1Δ strain (P = 0.42), but the number of polymerases/gene was higher than expected, leading to more polymerases engaged on 5S genes/cell. We obtained a second lhp1Δ strain for EM analysis to see if this result was reproducible (see Table S5 in the supplemental material). The 5S genes of this strain (BY4741 Δlhp1) displayed an average of 1.91 polymerases/gene (n = 152), while its control strain (BY4741) had 1.53 polymerases/gene on average (n = 156), supporting the conclusion that deletion of Lhp1 results in more polymerases/gene (P < 0.001).
FIG. 5.
The yeast La protein, Lhp1, and 5S transcription. (A) Representative EM images of 5S genes from wt (CY1) and lhp1Δ (CY4) cells. Transcription is from left to right. The 5S promoter is to the left in all EM images. (B) ChIP analysis of Lhp1 association with 5S rDNA and with the 35S rDNA promoter. After formaldehyde cross-linking, cell extracts were split in half. One half was left untreated (No RNase) while the other half was treated with 3 mg/ml RNase A (+ RNase). Quantification was by real-time PCR as described in Materials and Methods. Error bars equal ±1 standard deviation of data. (C) wt and lhp1Δ cells were pulse-labeled with [3H]uracil for 2 min and chased for the indicated times with excess unlabeled uracil. Total RNA was extracted and fractionated in a 5% polyacrylamide-8.3 M urea gel. (D) Total RNA, extracted from the indicated strains, was subjected to Northern analysis and probed with an oligonucleotide complementary to the 3′ end of pre-5S RNA. Because this oligonucleotide includes 13 nt of complementarity to mature 5S RNA, both precursor and mature 5S RNAs are detected. Primary transcripts are indicated by arrow. As controls, the blot was reprobed to detect mature 5S RNA (middle panel) and signal recognition protein (SRP) RNA (bottom panel). In rex1Δ strains, 5S RNAs accumulate that are ∼3 nt longer than the wt RNA (46, 61).
To determine whether Lhp1 can directly affect Pol III transcription, we measured the association of hemagglutinin-tagged Lhp1 with 5S rDNA by using ChIP assays. We observed a clear association of Lhp1 with the 5S coding region compared to that in the negative-control strain (Fig. 5B; untagged control). Additionally, we observed a somewhat lower, but significant, association of Lhp1 with multiple regions of the rDNA (e.g., the 35S promoter region) (Fig. 5B). These observations are consistent with previous observations made in mammalian studies (17, 28). We note that the degree of association of Lhp1 with 5S rDNA is more than sixfold greater than the background signal, but the signal is still rather weak. One potential explanation for low ChIP signal is indirect association of Lhp1 with DNA, by association for example, with nascent RNA transcripts. To determine if Lhp1 binds primarily to RNA, we repeated the ChIP analysis after treating cell extracts with RNase A. All ChIP signaling was reduced in the rDNA, supporting the hypothesis that Lhp1 associates with RNA. There was some remnant signal in the 5S coding region after RNase treatment (ca. twofold greater than the background). This may reflect incomplete digestion of 5S RNA or affinity of Lhp1 for non-RNA members of the Pol III transcription complex, as was reported for human cells (17). These data demonstrate that Lhp1 associates with the 5S rRNA coding region, at least in part by direct association with nascent RNA, and this association renders the protein poised to directly modulate Pol III transcription.
Previous results with other yeast Pol III genes suggest that the absence of Lhp1 will not affect 5S RNA synthesis (44, 73). However, knowing that Lhp1 is present on 5S genes in vivo and that its absence resulted in an increase in the expected number of polymerases/gene, we asked if the rate of synthesis or size of 5S RNA changed in lhp1Δ cells. By labeling for 2 min with [3H]uracil, followed by a 3- to 30-min chase, we compared the appearance of newly synthesized 5S RNA in wild-type (wt) and lhp1Δ cells. The pattern of 5S RNA accumulation was essentially identical in the two strains (Fig. 5C), indicating that there is no significant increase or decrease in the rate of 5S RNA synthesis in the absence of Lhp1.
To determine if the length of 5S RNA changed in lhp1Δ cells, we probed total RNA from wt and mutant strains by performing Northern blotting with an oligonucleotide that overlaps the 3′ end of mature 5S RNA, including 10 nt complementary to an 11- to 13-nt 3′ extension present in the primary 5S RNA transcript (46). This pre-5S RNA, containing a U-rich tail, is the 5S species bound by Lhp1 (72). Rex1, a 3′ exonuclease, is required for its normal processing (61). As shown previously (46, 61), the very-short-lived primary transcript is not easily detectable in normal cells but can be detected in rex1Δ cells (Fig. 5D, lane 3, pre-5S band), in addition to a 3-nt extended product that accumulates in the absence of Rex1, due to partial processing of pre-5S by an unidentified nuclease. In lhp1Δ cells, 5S RNA was produced at its normal size and amount (Fig. 5D, lane 2), indicating that Lhp1 was not required for 5S RNA synthesis or processing, similar to previous findings with yeast tRNA and U6 snRNA (44, 73). However, as previously reported for pre-tRNAs (73), Lhp1 likely protects the 3′ end of pre-5S RNAs from exonucleases, as the primary transcripts that accumulated in rex1Δ cells were undetectable in the rex1Δ lhp1Δ double mutant (Fig. 5D, lanes 3 to 4). Due to the speed of 3′-end processing (which was not detectably slowed in lhp1Δ cells) (Fig. 5D, lane 2) and the inability to detect the primary transcript except in the absence of Rex1 and the presence of Lhp1 (Fig. 5D, lane 3), it was not possible to determine whether Pol III terminates 5S RNA transcription at the same position in lhp1Δ cells as in control cells. However, since 5S genes in lhp1Δ cells had no more than three polymerases, as did normal 5S genes, the position of termination is either the same or very close to the normal position. Additional controls with rrp6Δ cells and rrp6Δ lhp1Δ cells showed that the deletion of Rrp6, a nuclear-exosome-associated exo-RNase, did not affect the production of mature 5S RNA, as expected (62). The combined results show that in the absence of Lhp1, 5S RNA is made at its normal rate and size (Fig. 5C and D) and on the expected number of genes (Fig. 4D) but that more polymerases are engaged on 5S genes than for control cells on a per-gene and per-cell basis (Fig. 4D to F and 5A).
DISCUSSION
Pol III genes can be densely packed with polymerases, explaining RNA production rates for known genes.
It is generally assumed that the very short Pol III genes are occupied by a single polymerase, influencing, for example, models of Pol III transcription factories (49) and leading to the proposal that 100 yeast 5S genes must be active to accommodate known ribosome synthesis rates (15). However, Costanzo et al. (8), using various footprinting techniques, found that about a third to a half that many 5S genes are bound by Pol III or TFIIIB. The results of EM visualization clarify the situation by agreeing reasonably well with the latter estimate for active gene number while showing that these genes can be engaged by up to three polymerases. Three polymerases/gene corresponds to the highest density allowed on these 132-nt genes, based on the elongating Pol III footprint of 37 nt (3). The finding is not surprising given the known high transcription rate from Pol III genes, which in turn is due to nonlimiting concentrations of TFIIIB (58), the remarkable stability of TFIIIB-DNA complexes during multiple rounds of transcription (7, 30, 31), and facilitated reinitiation of Pol III (14, 15). It also implies that a proposed “hand-back” mechanism for facilitated recycling (18) should be able to accommodate multiple polymerases per gene.
Strains with typical rDNA array sizes (140 to 190 5S genes) have an average of ∼1.5 polymerases on ∼45 active 5S genes, while the 42-copy strain has ∼2 polymerases/gene on ∼30 active genes/cell. This increase in polymerase density as gene number decreases was previously seen on Pol I genes in the 42-copy strain (19) and indicates that polymerase loading per gene can be regulated. The estimated number of total polymerases engaged on 5S genes/cell was quite similar for the three normal-copy-number strains (i.e., 67, 74, and 75; mean, 72 ± 4.4) and ∼20% lower (i.e., 57) for the 42-copy strain (Table 2). Since these four strains grow with similar doubling times and since Pol III transcripts typically are not limiting for cell growth under normal conditions (57), it is possible that 5S RNA may be somewhat overproduced in the normal-copy-number strains, with its steady-state amount determined by protein association or incorporation into ribosomes (33). Another possibility is that the 5S RNA elongation rate or half-life may vary somewhat in the different strains.
Using the average of 72 Pol IIIs engaged on 5S genes per cell, as well as the estimate that log-phase yeast cells make ∼2,000 ribosomes/minute (65) (and assuming that all 5S RNAs made are incorporated into ribosomes), it is possible to estimate the transcription rate. To make 2,000 5S RNAs/min, each of the 72 Pol IIIs would need to make ∼28 transcripts/min or ∼1 transcript/2.16 s. 5S genes are 132 nt long, so the transcription elongation rate would be 132 nt/2.16 s or 61 nt/s, similar to the 54 nt/s determined for yeast Pol I using similar reasoning (19). (If the four control strains are considered separately, a range in estimated elongation rates of 58 nt/s to 76 nt/s for Pol III is obtained.) In comparison, elongation rates for mammalian RNA polymerases have been estimated at 95 nt/s for Pol I (16) and 72 nt/s for Pol II (10). The yeast rates are expected to be somewhat lower than mammalian rates based on the differential in growth temperature (53).
The average polymerase density in the three normal-copy-number strains studied is 1.69 polymerases/gene or 1 polymerase/78 nt, while in the 42-copy strain, it is 1.96 polymerases/gene or 1 polymerase/67 nt. Based on these numbers, the Pol III reinitiation interval at the promoter would be 1.1 to 1.3 s, similar to reinitiation rates for yeast and mammalian Pol I (16, 19). These estimates can accommodate a previous prediction that the yeast SCR1 gene (a single-copy Pol III gene encoding 7SL RNA) needs to make ∼50 RNAs/min (13). That is, a 61 nt/s elongation rate and 1.2-s reinitiation rate would result in the production of 1 RNA/1.2 s or 50 7SL RNAs/min with a polymerase density of one polymerase/73 nt. This would correspond to seven polymerases total on this 522-bp gene, the longest class III gene known.
Repression of 5S gene transcription in log-phase and post-log-phase cultures.
Given the high polymerase occupancy found for essentially all other class III genes (26), it is interesting that the majority of 5S genes are inactive in control strains and that ∼12 of the 5S genes in the 42-copy strain remain unengaged by Pol III in spite of the fact that control strains have more than 42 active genes/cell. 5S gene activity can be controlled independently of other Pol III genes via the 5S gene-specific factor, TFIIIA. The inactive genes may be maintained by a negative feedback loop that limits the amount of TFIIIA available to nucleate the initiation complex on 5S genes, which involves 5S RNA, 5S DNA, TFIIIA, and a ribosomal protein that binds 5S RNA (6, 47).
We have considered the possibility that the genes that appear inactive are in fact active, in the sense that they are open complexes with TFIIIB bound, but given their short length and the mere 2 s required for transcription, our snapshot approach has caught a split second with no engaged polymerases. This may account for some missed active genes, but we consider it unlikely to account for the majority for the following reasons: (i) the facilitated recycling mechanism, which is supported by results herein, predicts that polymerases do not disengage from active genes (14, 15); (ii) our result for active gene number in normal strains agrees very well with the results of a previous study of yeast 5S genes using footprinting methods (8); and (iii) the numbers make sense in accounting for known transcription output and in agreeing with typical elongation rates. Thus, we are assuming that most genes that appear inactive are indeed so. (If we are mistaken, one can apply a Poisson distribution to estimate that five 5S genes would be expected in the active but unengaged category in the 42-copy strain, thus predicting a population of ∼7 rather than 12 inactive genes in this strain.)
As cells left the exponential growth phase, there was a substantial decrease in the number of active 5S genes, as well as in polymerases/gene, with total engaged polymerases/cell decreasing by 66% (Fig. 4F). However, a shorter repressive treatment (30 min of rapamycin treatment) resulted in a decrease of 37% in engaged polymerases/cell, primarily due to fewer active genes/cell rather than to fewer polymerases/gene. This finding that downregulation first manifested as an ∼35% decrease in 5S genes with engaged polymerases agrees reasonably well with the results of a previous ChIP analysis showing an ∼50% decrease in yeast 5S genes occupied by Pol III after a short treatment with rapamycin (33). In addition, the distribution of monomers, dimers, and trimers on 5S genes after 30 min of rapamycin treatment (Fig. 4G) shows that the polymerase density/gene is not significantly affected on those genes remaining active, e.g., the proportion of trimers is as high as before repression.
Recent results with the Maf1 protein, which receives input from multiple signaling pathways (including the Tor pathway studied here) and conveys that information to repress Pol III transcription (42, 48, 52, 60), have led to various models for its action that differ in whether Pol III (either free or engaged) or TFIIIB is the primary Maf1 target (discussed in references 21 and 68). Maf1 has been located on mammalian 5S genes (24), and the concentration of Maf1 is inversely correlated with 5S RNA synthesis in both yeast (48) and mammals (24). Our finding that downregulation of 5S genes first manifests as fewer active genes rather than fewer polymerases/gene is most easily reconcilable with an early Maf1 response mediated by TFIIIB to turn genes off (11, 24), but they do not necessarily rule out a response mediated by Maf1 binding to Pol III. For example, genes displaying normal polymerase density under short-term repressive conditions may not be transcribing at normal rates (due to Maf1 binding) but rather have polymerases poised for transcription should nutrients be provided (52).
Cotranscriptional action of the yeast La protein, Lhp1.
In the absence of the yeast La homolog, Lhp1, 5S RNA is made at its normal rate and size (Fig. 5C and D). However, Lhp1 associates with the nascent RNA on 5S genes (Fig. 5B), and in lhp1Δ strains, there is an unexpected increase in polymerase density on 5S genes (Fig. 4E and F and 5A), corresponding to an extra polymerase on ∼30 to 40% of the active genes. Several explanations can be proposed for this increase in polymerase density. Lhp1 might increase either the synthesis rate or half-life of 5S RNA, such that in its absence more polymerases are needed to make the same amount of RNA. However, given that there is no detectable change in RNA synthesis, the simplest explanation is one that would reconcile two previous conclusions that were not obviously compatible—first, that La is completely dispensable for Pol III transcription (27, 34, 44, 66, 73), and second, that La plays a role in transcript release and polymerase recycling at the 3′ end of Pol III genes (25, 36, 38). In this scenario, the extra polymerases seen by EM would be those transiently detained at the 3′ end of the gene due to the absence of Lhp1 in its role as a release factor. Considering that the Lhp1 binding site resides in the 3′ extension of 5S RNA, which is very short lived (Fig. 5C and D), and that this sequence is not accessible for binding until the 3′ end is extruded from the exit channel of Pol III, it is reasonable to propose a role at the 3′ end of the gene. For example, as proposed from in vitro studies (25, 38), Lhp1 may be involved in the release of the completed pre-5S transcript and dissociation of the ternary complex. Structural studies of Pol III (29) reveal binding surfaces for structured RNA in two proteins near the RNA exit channel. Lhp1 may help displace a transient association of this nature, leading to transcript and polymerase release. Whatever its role, it is clearly not a rate-limiting step in vivo, perhaps due to an excess of Pol III available for initiation. The combined results show that Lhp1 plays a cotranscriptional but nonessential and non-rate-limiting role in 5S RNA synthesis in yeast, with one possibility being a role in transcript release and polymerase recycling upon transcription termination.
Supplementary Material
Acknowledgments
The work was supported by NIH Public Health Service grants GM-63952 (to A.L.B.), GM-35949 (to M.N.), and GM-48410 (to S.L.W.) and a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research (to D.A.S.).
We thank A. van Hoof and R. Parker for yeast strains.
Footnotes
Published ahead of print on 12 May 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 2.Bai, C., and P. P. Tolias. 2000. Genetic analysis of a La homolog in Drosophila melanogaster. Nucleic Acids Res. 281078-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartholomew, B., D. Durkovich, G. A. Kassavetis, and E. P. Geiduschek. 1993. Orientation and topography of RNA polymerase III in transcription complexes. Mol. Cell. Biol. 13942-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, B. R., B. Bartholomew, G. A. Kassavetis, and E. P. Geiduschek. 1992. Topography of transcription factor complexes on Saccharomyces cerevisiae 5S RNA genes. J. Mol. Biol. 2281063-1077. [DOI] [PubMed] [Google Scholar]
- 5.Braun, B. R., G. A. Kassavetis, and E. P. Geiduschek. 1992. Bending of the Saccharomyces cerevisiae 5S rRNA gene in transcription factor complexes. J. Biol. Chem. 26722562-22569. [PubMed] [Google Scholar]
- 6.Brow, D. A., and E. P. Geiduschek. 1987. Modulation of yeast 5S rRNA synthesis in vitro by ribosomal protein YL3. J. Biol. Chem. 26213953-13958. [PubMed] [Google Scholar]
- 7.Cloutier, T. E., M. D. Librizzi, A. K. Mollah, M. Brenowitz, and I. M. Willis. 2001. Kinetic trapping of DNA by transcription factor IIIB. Proc. Natl. Acad. Sci. USA 989581-9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costanzo, G., S. Camier, P. Carlucci, L. Burderi, and R. Negri. 2001. RNA polymerase III transcription complexes on chromosomal 5S rRNA genes in vivo: TFIIIB occupancy and promoter opening. Mol. Cell. Biol. 213166-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dammann, R., R. Lucchini, T. Koller, and J. M. Sogo. 1995. Transcription in the yeast rRNA gene locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol. Cell. Biol. 155294-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darzacq, X., Y. Shav-Tal, V. de Turris, Y. Brody, S. M. Shenoy, R. D. Phair, and R. H. Singer. 2007. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 14796-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai, N., J. Lee, R. Upadhya, Y. Chu, R. D. Moir, and I. M. Willis. 2005. Two steps in Maf1-dependent repression of transcription by RNA polymerase III. J. Biol. Chem. 86455-6462. [DOI] [PubMed] [Google Scholar]
- 12.Dieci, G., G. Fiorino, M. Castelnuovo, M. Teichmann, and A. Pagano. 2007. The expanding RNA polymerase III transcriptome. Trends Genet. 23614-622. [DOI] [PubMed] [Google Scholar]
- 13.Dieci, G., S. Giuliodori, M. Catellani, R. Percudani, and S. Ottonello. 2002. Intragenic promoter adaptation and facilitated RNA polymerase III recycling in the transcription of SCR1, the 7SL RNA gene of Saccharomyces cerevisiae. J. Biol. Chem. 2776903-6914. [DOI] [PubMed] [Google Scholar]
- 14.Dieci, G., and A. Sentenac. 1996. Facilitated recycling pathway for RNA polymerase III. Cell 84245-252. [DOI] [PubMed] [Google Scholar]
- 15.Dieci, G., and A. Sentenac. 2003. Detours and shortcuts to transcription reinitiation. Trends Biochem. Sci. 28202-209. [DOI] [PubMed] [Google Scholar]
- 16.Dundr, M., U. Hoffmann-Rohrer, Q. Hu, I. Grummt, L. I. Rothblum, R. D. Phair, and T. Misteli. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science 2981623-1626. [DOI] [PubMed] [Google Scholar]
- 17.Fairley, J. A., T. Kantidakis, N. S. Kenneth, R. V. Intive, R. J. Maraia, and R. J. White. 2005. Human La is found at RNA polymerase III-transcribed genes in vivo. Proc. Natl. Acad. Sci. USA 10218350-18355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrari, R., C. Rivetti, J. Acker, and G. Dieci. 2004. Distinct roles of transcription factors TFIIIB and TFIIIC in RNA polymerase III transcription reinitiation. Proc. Natl. Acad. Sci. USA 10113442-13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.French, S. L., Y. N. Osheim, F. Cioci, M. Nomura, and A. L. Beyer. 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 231558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geiduschek, E. P., and G. A. Kassavetis. 2001. The RNA polymerase III transcription apparatus. J. Mol. Biol. 3101-26. [DOI] [PubMed] [Google Scholar]
- 21.Geiduschek, E. P., and G. A. Kassavetis. 2006. Transcription: adjusting to adversity by regulating RNA polymerase. Curr. Biol. 16R849-R851. [DOI] [PubMed] [Google Scholar]
- 22.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74527-534. [DOI] [PubMed] [Google Scholar]
- 23.Goodfellow, S. J., and R. J. White. 2007. Regulation of RNA polymerase III transcription during mammalian cell growth. Cell Cycle 62323-2326. [DOI] [PubMed] [Google Scholar]
- 24.Goodfellow, S. J., E. L. Graham, T. Kantidakis, L. Marshall, B. A. Coppins, D. Oficjalska-Pham, M. Gérard, O. Lefebvre, and R. J. White. 4 March 2008. Regulation of RNA polymerase III transcription by Maf1 in mammalian cells. J. Mol. Biol. doi: 10.1016/j.jmb. 2008.02.060. [DOI] [PubMed]
- 25.Gottlieb, E., and J. A. Steitz. 1989. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J. 8851-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harismendy, O., C. G. Gendrel, P. Soularue, X. Gidrol, A. Sentenac, M. Werner, and O. Lefebvre. 2003. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 224738-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu, P., S. Wu, and N. Hernandez. 2003. A minimal RNA polymerase III transcription system from human cells reveals positive and negative roles for CK2. Mol. Cell 12699-709. [DOI] [PubMed] [Google Scholar]
- 28.Intine, R. V., M. Dundr, A. Vassilev, E. Schwartz, Y. Zhao, Y. Zhao, M. L. Depamphilis, and R. J. Maraia. 2004. Nonphosphorylated human La antigen interacts with nucleolin at nucleolar sites involved in rRNA biogenesis. Mol. Cell. Biol. 2410894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jasiak, A. J., K. J. Armache, B. Martens, R. P. Jansen, and P. Cramer. 2006. Structural biology of RNA polymerase III: subcomplex C17/25 X-ray structure and 11 subunit model. Mol. Cell 2371-81. [DOI] [PubMed] [Google Scholar]
- 30.Kassavetis, G. A., B. R. Braun, L. H. Nguyen, and E. P. Geiduschek. 1990. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 60235-245. [DOI] [PubMed] [Google Scholar]
- 31.Kassavetis, G. A., and E. P. Geiduschek. 2006. Transcription factor TFIIIB and transcription by RNA polymerase III. Biochem. Soc. Trans. 341082-1087. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi, T., D. J. Heck, M. Nomura, and T. Horiuchi. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 123821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laferté, A., E. Favry, A. Sentenac, M. Riva, C. Carles, and S. Chedin. 2006. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 202030-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin-Marq, N., and S. G. Clarkson. 1998. Efficient synthesis, termination and release of RNA polymerase III transcripts in Xenopus extracts depleted of La protein. EMBO J. 172033-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 36.Maraia, R. J. 1996. Transcription termination factor La is also an initiation factor for RNA polymerase III. Proc. Natl. Acad. Sci. USA 933383-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maraia, R. J., and R. V. Intine. 2001. Recognition of nascent RNA by the human La antigen: conserved and diverged features of structure and function. Mol. Cell. Biol. 21367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maraia, R. J., D. J. Kenan, and J. D. Keene. 1994. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol. Cell. Biol. 142147-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, O. L., Jr. 1981. The nucleolus, chromosomes, and visualization of genetic activity. J. Cell Biol. 9115s-27s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moqtaderi, Z., and K. Struhl. 2004. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol. Cell. Biol. 244118-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller, M., R. Lucchini, and J. M. Sogo. 2000. Replication of yeast rDNA initiates downstream of transcriptionally active genes. Mol. Cell 5767-777. [DOI] [PubMed] [Google Scholar]
- 42.Oficjalska-Pham, D., O. Harismendy, W. J. Smagowicz, A. Gonzalez de Peredo, M. Boguta, A. Sentenac, and O. Lefebvre. 2006. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol. Cell 22623-632. [DOI] [PubMed] [Google Scholar]
- 43.Osheim, Y. N., S. L. French, K. M. Keck, E. A. Champion, K. Spasov, F. Dragon, S. J. Baserga, and A. L. Beyer. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell 16943-954. [DOI] [PubMed] [Google Scholar]
- 44.Pannone, B. K., D. H. Xue, and S. L. Wolin. 1998. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 177442-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park, J. M., M. J. Kohn, M. W. Bruinsma, C. Vech, R. V. Intine, S. Fuhrmann, A. Grinberg, I. Mukherjee, P. E. Love, M. S. Ko, M. L. DePamphilis, and R. J. Maraia. 2006. The multifunctional RNA-binding protein La is required for mouse development and for the establishment of embryonic stem cells. Mol. Cell. Biol. 261445-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piper, P. W., J. W. Bellatin, and A. Lockheart. 1983. Altered maturation of sequences at the 3′ terminus of 5S gene transcripts in a Saccharomyces cerevisiae mutant that lacks an RNA processing endonuclease. EMBO J. 2353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pittman, R. H., M. T. Andrews, and D. R. Setzer. 1999. A feedback loop coupling 5S rRNA synthesis to accumulation of a ribosomal protein. J. Biol. Chem. 27433198-33201. [DOI] [PubMed] [Google Scholar]
- 48.Pluta, K., O. Lefebvre, N. C. Martin, W. J. Smagowicz, D. R. Stanford, S. R. Ellis, A. K. Hopper, A. Sentenac, and M. Boguta. 2001. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol. Cell. Biol. 215031-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pombo, A., D. A. Jackson, M. Hollinshead, Z. Wang, R. G. Roeder, and P. R. Cook. 1999. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 182241-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rivetti, C., S. Codeluppi, G. Dieci, and C. Bustamante. 2003. Visualizing RNA extrusion and DNA wrapping in transcription elongation complexes of bacterial and eukaryotic RNA polymerases. J. Mol. Biol. 3261413-1426. [DOI] [PubMed] [Google Scholar]
- 51.Roberts, D. N., A. J. Stewart, J. T. Huff, and B. R. Cairns. 2003. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl. Acad. Sci. USA 10014695-14700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts, D. N., B. Wilson, J. T. Huff, A. J. Stewart, and B. R. Cairns. 2006. Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol. Cell 22633-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryals, J., R. Little, and H. Bremer. 1982. Temperature dependence of RNA synthesis parameters in Escherichia coli. J. Bacteriol. 151879-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheer, U. 1982. A novel type of chromatin organization in lampbrush chromosomes of Pleurodeles waltlii: visualization of clusters of tandemly repeated, very short transcriptional units. Biol. Cell 44213-220. [Google Scholar]
- 55.Schneider, D. A., S. L. French, Y. N. Osheim, A. O. Bailey, L. Vu, J. Dodd, J. R. Yates, A. L. Beyer, and M. Nomura. 2006. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc. Natl. Acad. Sci. USA 10312707-12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schramm, L., and N. Hernandez. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 162593-2620. [DOI] [PubMed] [Google Scholar]
- 57.Sethy, I., R. D. Moir, M. Librizzi, and I. M. Willis. 1995. In vitro evidence for growth regulation of tRNA gene transcription in yeast. J. Biol. Chem. 27028463-28470. [DOI] [PubMed] [Google Scholar]
- 58.Sethy-Coraci, I., R. D. Moir, A. Lopez-de-Leon, and I. M. Willis. 1998. A differential response of wild type and mutant promoters to TFIIIB70 overexpression in vivo and in vitro. Nucleic Acids Res. 262344-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tarn, W. Y., T. A. Yario, and J. A. Steitz. 1995. U12 snRNA in vertebrates: evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of intron. RNA 1644-656. [PMC free article] [PubMed] [Google Scholar]
- 60.Upadhya, R., J. Lee, and I. M. Willis. 2002. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol. Cell 101489-1494. [DOI] [PubMed] [Google Scholar]
- 61.van Hoof, A., P. Lennertz, and R. Parker. 2000. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 191357-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Horn, D. J., C. J. Yoo, D. Xue, H. Shi, and S. L. Wolin. 1997. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA 31434-1443. [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, Z., T. Luo, and R. G. Roeder. 1997. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 112371-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24437-440. [DOI] [PubMed] [Google Scholar]
- 66.Weser, S., M. Bachmann, K. H. Seifart, and W. Meissner. 2000. Transcription efficiency of human polymerase III genes in vitro does not depend on the RNP-forming autoantigen La. Nucleic Acids Res. 283935-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White, R. J. 2004. RNA polymerase III transcription and cancer. Oncogene 233208-3216. [DOI] [PubMed] [Google Scholar]
- 68.Willis, I. M., and R. D. Moir. 2007. Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem. Sci. 3251-53. [DOI] [PubMed] [Google Scholar]
- 69.Winston, F. 2001. Control of eukaryotic transcription elongation. Genome Biol. 21006.1-1006.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Véronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285901-906. [DOI] [PubMed] [Google Scholar]
- 71.Wolin, S. L., and T. Cedervall. 2002. The La protein. Annu. Rev. Biochem. 71375-403. [DOI] [PubMed] [Google Scholar]
- 72.Yoo, C. J., and S. L. Wolin. 1994. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol. Cell. Biol. 145412-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoo, C. J., and S. L. Wolin. 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89393-402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.