Abstract
Rpd3(L) and Rpd3(S) are distinct multisubunit complexes containing the Rpd3 histone deacetylase. Disruption of the GCN5 histone acetyltransferase gene shows a strong synthetic phenotype when combined with either an sds3 mutation affecting only the Rpd3(L) complex or an rco1 mutation affecting only Rpd3(S). However, these synthetic growth defects are not seen in a gcn5 sds3 rco1 triple mutant, suggesting that the balance between Rpd3(L) and Rpd3(S) is critical in cells lacking Gcn5. Different genetic interactions are seen with mutations affecting the FACT chromatin reorganizing complex. An sds3 mutation affecting only Rpd3(L) has a synthetic defect with FACT mutants, while rco1 and eaf3 mutations affecting Rpd3(S) suppress FACT mutant phenotypes. Rpd3(L) therefore acts in concert with FACT, but Rpd3(S) opposes it. Combining FACT mutations with mutations in the Esa1 subunit of the NuA4 histone acetyltransferase results in synthetic growth defects, and these can be suppressed by an rco1 or set2 mutation. An rco1 mutation suppresses phenotypes caused by mutations in the ESA1 and ARP4 subunits of NuA4, while Rco1 overexpression exacerbates these defects. These results suggest a model in which NuA4 and Rpd3(S) compete. Chromatin immunoprecipitation experiments show that eliminating Rpd3(S) increases the amount of NuA4 binding to the ARG3 promoter during transcriptional activation and to the sites of DNA repair induced by a double-strand break. Our results suggest that the Rpd3(L) and Rpd3(S) complexes have distinct functions in vivo and that the relative amounts of the two forms alter the effectiveness of other chromatin-altering complexes, such as FACT and NuA4.
The structure of chromatin plays an important role in regulating gene expression, DNA replication, and repair of DNA damage. Posttranslational modification of histones can create sites recognized by other chromatin factors (2). Histone K acetyltransferase (KAT) enzymes create acetyl-lysine residues that can be recognized by proteins containing bromo domains (2), and these marks can be removed by histone deacetylase (HDAC) enzymes (60). Histone K methyltransferase (KMT) enzymes methylate lysines, and methyl-lysine residues can be recognized by proteins with chromo domains or PHD domains (9, 63). The recruitment of these chromatin factors that bind to modified nucleosomes can have profound effects on transcription, replication, and DNA repair.
There are multiple KAT complexes (46). SAGA and NuA4 are two well-studied KATs that modify the tail regions of histones H3 and H4, respectively. SAGA contains at least 16 subunits, with the GCN5 gene encoding the catalytic KAT subunit of SAGA. gcn5 null mutants are viable and display defects in transcriptional regulation and DNA repair (1). The essential ESA1 gene encodes the catalytic subunit of NuA4, and viable esa1 mutants affect both transcription and DNA repair (17). NuA4 has 13 subunits, including Eaf3, a chromodomain protein, which recognizes methylated H3(K36) (12, 27, 28), Arp4, an actin-related protein that binds histones and is required for the binding of NuA4 to sites of DNA damage (15), and Yng2, a PHD domain protein that binds methylated H3(K4) (32). These subunits are not unique to NuA4; Eaf3 is also present in Rpd3(S), Arp4 is in the Ino80 and Swr1 complexes, and Yng2 is in the Piccolo/NuA4 complex. It is believed that these histone-binding subunits of NuA4 determine its ability to bind to nucleosomes with specific modifications.
RPD3 encodes the catalytic subunit present in two HDAC complexes, Rpd3(L) and Rpd3(S) (60). The larger Rpd3(L) complex contains at least nine subunits, of which three are also present in the smaller Rpd3(S) complex. Disruption of genes encoding subunits specific to one complex disrupts only that complex. For example, an rco1 mutation eliminates the Rpd3(S) complex but does not affect Rpd3(L), and deletion of the Rpd3(L)-specific gene SDS3 does not affect Rpd3(S) (12, 31). Rpd3(L) and Rpd3(S) appear to have different functions, with Rpd3(L) localized primarily to promoter regions and Rpd3(S) at transcribed regions (27). It is believed that active RNA polymerase II brings the Set2 methyltransferase to transcribed regions; thus, Set2 can methylate H3(K36) in the transcribed regions, and the Eaf3 subunit of Rpd3(S) that binds methylated K36 (Me-K36) can recruit Rpd3(S) to the 3′ region of actively transcribed genes (12, 27, 28). In contrast, Rpd3(L) appears to function at promoters to repress transcription.
We have been studying the FACT complex, which reorganizes chromatin structure (19). In contrast to the Swi/Snf family of ATP-dependent chromatin remodeling factors, which reposition nucleosomes (10), FACT changes the accessibility of DNA within nucleosomes without hydrolyzing ATP and without repositioning the histone octamer core relative to the DNA (7, 20, 41). Yeast FACT is encoded by two essential genes, SPT16 and POB3, and viable spt16 and pob3 mutations have been isolated with defects in both transcription and DNA replication (20, 35, 43, 45). We have shown that the effects of FACT mutations on transcription and replication can be suppressed by mutations in SET2, a KMT that acts on histone H3(K36), and by a mutation in CHD1, an ATP-dependent chromatin remodeler with a chromodomain (4-6). Here we show that an rco1 mutation affecting the Rpd3(S) complex also suppresses FACT mutants; this is not surprising given the described relationship between Set2 and Rpd3(S). However, in the course of these studies, we made a number of surprising findings. While eliminating Rpd3(S) suppresses FACT defects, an sds3 mutation eliminating Rpd3(L) exacerbates FACT mutant phenotypes, suggesting that the relative levels of the two HDAC complexes, Rpd3(L) and Rpd3(S), become very important in FACT mutants. The importance of the relative levels of Rpd3(L) and Rpd3(S) is also seen in gcn5 mutants, affecting the catalytic subunit of the SAGA KAT, where eliminating either of the Rpd3 complexes is toxic but eliminating both is tolerated well. Finally, we also show that mutating the Eaf3 subunit present in both Rpd3(S) and NuA4 suppresses FACT mutants. These results led to a model where Rpd3(S) and NuA4, both containing the methyl-lysine binding chromodomain subunit Eaf3, compete for binding to chromatin sites. Consistent with this hypothesis, an rco1 mutation suppresses phenotypes associated with mutation in subunits of NuA4. Our chromatin immunoprecipitation (ChIP) data show that an rco1 mutation that eliminates the Rpd3(S) complex results in increased binding of NuA4 to specific sites.
MATERIALS AND METHODS
Standard genetic methods (48) were used for constructing strains (see Table S1 at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). The Rco1(ΔPHD) and Rco1(Yng2-PHD) alleles were PCR amplified from strains YBL632 and YBL648 (32), provided by Jerry Workman, and transformed into W303 strain DY11327 (rco1:URA3MX), selecting for gene replacement on fluoroorotic acid (FOA) medium. The hta2(S129A)::TRP1 allele from strain SKY2939 (26), provided by Steve Kron, was PCR amplified and transferred into a fresh W303 strain, selecting for the TRP1 marker, and a W303 strain with hta1(S129A)::HIS3MX was constructed by a two-step PCR procedure (52). These strains were crossed to isolate the hta1(S129A) hta2(S129A) double mutant.
Cells were grown in yeast extract-peptone-dextrose medium with 2% glucose (48) or on medium containing an appropriate amount of hydroxyurea (HU) or methylmethanesulfonate (MMS) at various temperatures. Strains with plasmids were grown on synthetic complete medium lacking uracil with 2% glucose at 30°C. For amino acid starvation experiments, cells were grown at 25°C in yeast extract-peptone-dextrose medium with 2% glucose to early log phase and then filtered and transferred to synthetic complete medium lacking isoleucine and valine and containing 0.2 μg/ml sulfometuron-methyl (SM). For GAL1::HO induction experiments, cells were grown at 30°C in synthetic complete medium lacking uracil with 2% raffinose, and galactose was added to a final concentration of 2%.
Plasmids are listed in Table S2 at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement. The RCO1 gene was PCR amplified from genomic DNA and inserted into the EcoRI site of pRS426 (8) to construct plasmid M5316. A similar strategy was used to clone the RCO1(ΔPHD), RCO1(Yng2- PHD), and RCO1-Myc::TRP1 alleles in constructing plasmids M5322, M5327, and M5340. A 3.9-kb KpnI-SphI fragment with the POB3 gene was cloned into YCplac33 (22) to construct plasmid M4211.
Sin3-hemagglutinin (HA) immunoprecipitation was performed as described previously (14) using anti-HA antibody (12CA5; University of Utah Bioprocessing Resource) and Pan Mouse Dynabeads, except that 25 mM HEPES-KOH (pH 7.6)-150 mM KOAc-1 mM EDTA-1 mM EGTA-2 mM MgSO4-0.1% NP-40-10% glycerol was used as the extraction buffer. Blots were probed with anti-HA and anti-Myc (9E10; Covance) antibodies and scanned and quantitated using a Li-Cor infrared scanner. Western immunoblots to examine Esa1-Myc levels were performed with anti-Myc antibodies, along with anti-Pgk1 antibodies as a loading control. ARG3 mRNA levels were measured by reverse transcription followed by quantitative PCR as described previously (56). ChIPs were performed as described previously (3) using 4A6 (Upstate) monoclonal antibody to the Myc epitope, anti-histone H3 (07-690; Upstate), anti-histone H3(Ac-Lys14) (07-353; Upstate), anti-acetyl-histone H4 (06-598; Upstate), anti-histone H3(Me-Lys36) (ab9050; Abcam), anti-Spt16 (provided by Tim Formosa), and antibody-coated magnetic beads (rabbit and pan-mouse immunoglobulin G beads; Dynal Biotech). ChIP assays were analyzed as described previously by real-time PCR (18), and standard deviations for normalized PCR replicates were calculated using equation 7 from the work of van Kempen and van Vliet (55). Oligonucleotides used for PCR, ChIP, or RT-PCR are listed in Table S3 at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement.
RESULTS
gcn5 shows growth defects when combined with rco1 or sds3 mutations.
Disruption of the GCN5 gene encoding a histone acetyltransferase causes transcriptional defects that can be suppressed by mutations in either RPD3 or SIN3 (39, 61). There are two Rpd3 complexes, Rpd3(L) and Rpd3(S), and we were interested in whether mutations affecting only one complex could suppress gcn5 phenotypes. We constructed gcn5 sds3 and gcn5 rco1 double-mutant strains, since the Sds3 and Rco1 subunits are specific to either Rpd3(L) or Rpd3(S), respectively. The results were rather surprising: the gcn5 sds3 and gcn5 rco1 double mutants both showed marked growth defects compared to the single mutants (Fig. 1A). In contrast, the gcn5 rco1 sds3 triple mutant grew reasonably well (Fig. 1A). Thus, eliminating Gcn5, Rpd3(L), or Rpd3(S) singly is tolerated well, but eliminating Gcn5 along with either Rpd3(L) or Rpd3(S) makes cell very sick. However, cells lacking Gcn5 along with both Rpd3(L) and Rpd3(S) are reasonably healthy. Disruption of the RPD3 gene eliminates both Rpd3(L) and Rpd3(S), and one might expect the gcn5 rpd3 double mutant to grow normally since Gcn5 and Rpd3 are thought to act in opposition. However, there is a modest growth defect in the gcn5 rpd3 double mutant, and importantly, this defect is similar to that seen for the gcn5 rco1 sds3 triple mutant (Fig. 1B). These results suggest there is a balance between the two types of HDAC complexes, Rpd3(L) and Rpd3(S), and that eliminating one or both of these complexes in a GCN5 strain is tolerated well. In contrast, eliminating only one of the complexes with an sds3 or an rco1 mutation in a gcn5 mutant is extremely toxic, but eliminating both Rpd3 complexes is less of a problem. Thus, the balance between Rpd3(L) and Rpd3(S) is extremely important in cells lacking Gcn5.
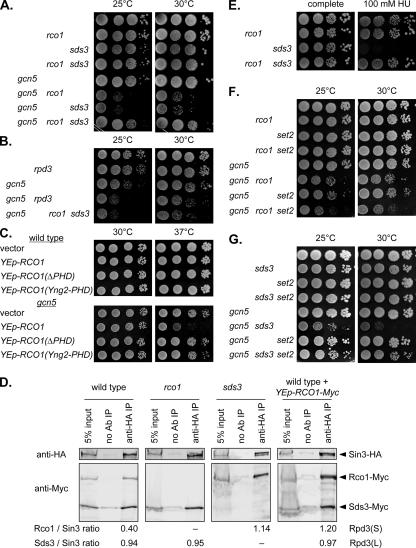
FIG. 1.
gcn5 mutants lacking either Rpd3(L) or Rpd3(S) are sick. (A) Tenfold dilutions of strains DY5699 (wild type), DY10398 (rco1), DY11148 (sds3), DY10460 (rco1 sds3), DY11354 (gcn5), DY11355 (gcn5 rco1), DY11357 (gcn5 sds3), and DY11359 (gcn5 rco1 sds3) were plated on complete medium for 3 days at either 25°C or 30°C. (B) Tenfold dilutions of strains DY5699 (wild type), DY4895 (rpd3), DY11354 (gcn5), DY5169 (gcn5 rpd3), and DY11359 (gcn5 rco1 sds3) were plated on complete medium for 2 days at either 25°C or 30°C. (C) Strains DY150 (wild type) and DY11087 (gcn5) were transformed with the indicated multicopy plasmid, and 10 dilutions were plated on selective medium lacking uracil for 3 days at 30°C or for 3 days at 37°C. (D) Strains DY13050 (Sin3-HA Rco1-Myc Sds3-Myc), DY13056 (Sin3-HA rco1Δ Sds3-Myc), and DY13082 (Sin3-HA Rco1-Myc sds3Δ) were grown in synthetic complete medium and strain DY13050 (Sin3-HA Rco1-Myc Sds3-Myc) transformed with plasmid M5340 (YEp-RCO1-Myc) was grown in selective medium lacking uracil. Extracts were prepared and immunoprecipitated with anti-HA antibody (anti-HA IP) or with no antibody as a control (no Ab IP). The immunoprecipitated proteins were electrophoresed and transferred to Western blots, along with a control corresponding to 5% of the input before immunoprecipitation (5% input), and the blots were probed with anti-HA and anti-Myc antibodies. Protein bands in the immunoblots were quantitated, with Rpd3(S) levels corresponding to the ratio of Rco1-Myc to Sin3-HA and Rpd3(L) levels corresponding to the ratio of Sds3-Myc to Sin3-HA. (E) Tenfold dilutions of strains DY5699 (wild type), DY10398 (rco1), DY11148 (sds3), and DY10460 (rco1 sds3) were plated on complete medium for 3 days at 30°C or on medium containing 100 mM HU for 5 days at 30°C. (F) Tenfold dilutions of strains DY5699 (wild type), DY10398 (rco1), DY8825 (set2), DY10402 (set2 rco1), DY11087 (gcn5), DY11504 (gcn5 rco1), DY11509 (gcn5 set2), and DY11505 (gcn5 rco1 set2) were plated on complete medium for 3 days at 25°C or for 2 days at 30°C. (G) Tenfold dilutions of strains DY5699 (wild type), DY11148 (sds3), DY8825 (set2), DY10430 (set2 sds3), DY11087 (gcn5), DY11357 (gcn5 sds3), DY11509 (gcn5 set2), and DY11513 (gcn5 sds3 set2) were plated on complete medium for 4 days at either 25°C or 30°C.
To test this idea of the relative levels of Rpd3(L) and Rpd3(S), we constructed a multicopy plasmid with the RCO1 gene, which could increase the amount of Rpd3(S). The YEp-RCO1 plasmid showed toxicity in a gcn5 mutant but not in wild-type cells (Fig. 1C). Rco1 contains a PHD domain that may recognize methylated lysine residues (32), and we examined the role of this PHD domain in promoting this toxic effect. A multicopy plasmid with an RCO1 allele lacking the PHD domain (32) did not affect growth of the gcn5 strain (see Fig. S1A at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). Immunoblots show that the Rco1 and Rco1(ΔPHD) proteins are expressed at similar levels (data not shown). Li et al. (32) also constructed an RCO1 allele with the endogenous PHD domain replaced with the PHD domain from Yng2, and overexpression of this Rco1(Yng2-PHD) protein also does not affect growth of the gcn5 mutant (see Fig. S1A at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). These results support the idea that gcn5 mutants are sensitive to the relative amounts of Rpd3(L) and Rpd3(S) and that the PHD domain of Rco1 is required for Rpd3(S) to affect this balance.
Immunoprecipitation experiments were performed to measure the relative amounts of Rpd3(S) and Rpd3(L) (Fig. 1D). A strain was constructed with three epitope-tagged proteins, Sin3-HA, Rco1-Myc, and Sds3-Myc. Sin3, like Rpd3, is present in both the Rpd3(S) and Rpd3(L) complexes. Rco1-Myc and Sds3-Myc, present in Rpd3(S) and Rpd3(L), respectively, differ in size and are easily separable on sodium dodecyl sulfate (SDS) gels. Sin3-HA was immunoprecipitated, and the immunoprecipitated proteins were separated on SDS gels, transferred to nitrocellulose, and probed with antibodies to the HA and Myc epitopes. The bands in the Western blots were quantitated, and the amounts of Rpd3(S) and Rpd3(L) were defined as the ratio of Rco1-Myc to Sin3-HA and the ratio of Sds3-Myc to Sin3-HA, respectively. In the rco1 mutant, the amount of Rpd3(L) does not increase compared to that for the wild type. In contrast, the amount of the Rpd3(S) complex is much greater in the sds3 mutant than in the wild type. Finally, the wild-type strain with the three-epitope tagged proteins was transformed with a multicopy plasmid that overexpresses Rco1-Myc. The YEp-Rco1-Myc plasmid caused a major increase in the amount of Rpd3(S), while Rpd3(L) levels were unaffected. These experiments showed that genetic effects of the rco1 mutation are due to the absence of Rpd3(S), since the amount of Rpd3(L) does not change. In contrast, the toxic effects of either an sds3 mutation or Rco1 overexpression are due to an increase in Rpd3(S) levels.
We also found that an sds3 mutant was sensitive to the DNA replication inhibitor HU (Fig. 1E). Significantly, the HU sensitivity of an sds3 mutant was lost by introducing an rco1 mutation. Thus, while a cell lacking Rpd3(L) was HU sensitive, a cell lacking both Rpd3(L) and Rpd3(S) had normal HU sensitivity, supporting the importance of balance between the two types of Rpd3 complexes.
Rpd3(S) is recruited to chromatin containing methylated H3(K36) (12, 27, 28), and K36 is methylated by Set2 (50). A set2 mutation should reduce the association of Rpd3(S) with chromatin. If the synthetic defect caused by combining gcn5 with rco1 is solely due to a failure to recruit Rpd3(S) to sites with methylated H3(K36), then one might expect growth defects in both the gcn5 rco1 and gcn5 set2 double-mutant strains. In fact, the growth defect in the gcn5 rco1 strain was worse than that in the gcn5 set2 strain (Fig. 1F). Additionally, the slightly poorer growth of the gcn5 rco1 set2 triple mutant than that of the double mutants suggests that Rco1 and Set2 have independent functions. In contrast, the gcn5 sds3 double mutant was strongly suppressed by a set2 mutation (Fig. 1G). Here, the problem caused by a deficiency of Rpd3(L) in the gcn5 mutant can be ameliorated by preventing methylation of the histone residue that recruits binding of Rpd3(S), just as removing the PHD domain from Rpd3(S) made it ineffective in balancing Rpd3(L).
In summary, gcn5 mutants displayed a marked growth defect when only one of the two Rpd3 complexes, Rpd3(L) and Rpd3(S), was absent. The gcn5 mutants grew well when both complexes were absent or when the Rpd3(L) complex was mutated and the activity of the Rpd3(S) was restricted by preventing H3(K36) methylation or by preventing Rpd3(S) from binding to this modification. These results indicate that imbalanced HDAC activity is detrimental in the absence of normal Gcn5 KAT activity.
rco1 mutation suppresses FACT mutant phenotypes.
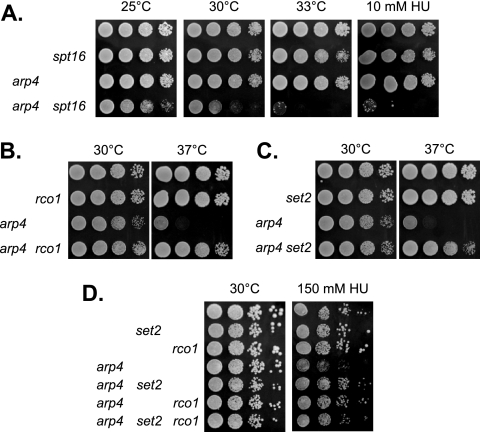
Mutations affecting the FACT complex result in a number of phenotypes, including temperature-sensitive growth, sensitivity to the DNA replication inhibitor HU, and synthetic lethality with a variety of mutations in replication or transcription factors (20, 21, 49). An rpd3 mutation can suppress some growth and transcriptional defects caused by FACT mutations (20), and we now examined the effect of mutations affecting only Rpd3(L) or Rpd3(S) on FACT mutants. An rco1 mutation suppressed both the temperature-sensitive and HU-sensitive phenotypes of three FACT mutant alleles, spt16-11, pob3(L78R), and pob3(Q308K) (Fig. 2A; also see Fig. S1 at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). This suppression by rco1 is consistent with the similar suppression of FACT mutants by a set2 deletion (4) and suggests that Rpd3(S) recruitment may be the principle function of Set2 in opposing FACT activity. Additivity of set2 and rco1 mutations in suppressing pob3(L78R) was not seen (see Fig. S2A at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement), consistent with the idea that Set2 and Rco1 also function in the same pathway. We note that while set2 and rco1 did not cause additive effects in the pob3(L78R) mutant, additivity was seen in the gcn5 mutant, suggesting that Rco1 and Set2 may have independent functions as well.
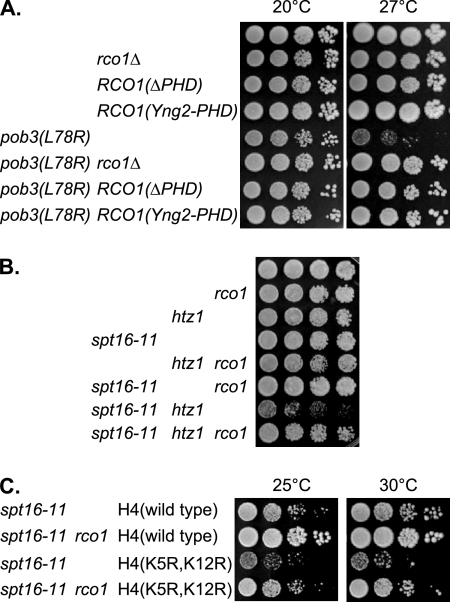
FIG. 2.
The Rco1 PHD domain affects pob3(L78R), and rco1 suppresses the spt16-11 htz1 and spt16-11 H4(K5R,K12R) synthetic lethalities. (A) Tenfold dilutions of strains DY150 (wild type), DY10398 (rco1), DY12800 [RCO1(ΔPHD)], DY12881 [RCO1(Yng2-PHD)], DY7379 [pob3(L78R)], DY10406 [pob3(L78R) rco1], DY12802 [RCO1(ΔPHD) pob3(L78R)], and DY12883 [pob3(L78R) RCO1(Yng2-PHD)] were plated on complete medium for 4 days at either 20°C or 27°C. (B) Tenfold dilutions of strains DY150 (wild type), DY10398 (rco1), DY7835 (htz1), DY8107 (spt16-11), DY11612 (htz1 rco1), DY11373 (spt16-11 rco1), DY9807 (spt16-11 htz1), and DY11606 (spt16-11 htz1 rco1) were plated on complete medium for 3 days at 30°C. (C) Tenfold dilutions of strains DY11848 (spt16-11), DY11850 (spt16-11 rco1), DY11852 [spt16-11 HHF2(K5, K12R)], and DY11854 [spt16-11 rco1 HHF2(K5, K12R)] were plated on complete medium for 4 days at either 25°C or 30°C.
HTZ1 encodes the yeast H2A.Z histone variant of H2A, which localizes preferentially to promoter regions (34, 40, 62). An htz1 mutation shows synthetic growth defects when combined with spt16-11 (4), and this growth defect was suppressed by an rco1 mutation (Fig. 2B). Histone H4 is acetylated at K5 and K12 during DNA replication (24), and consistent with a FACT role in DNA replication, an H4(K5R,K12R) mutant shows a strong synthetic phenotype in combination with spt16-11 (21). An rco1 mutation suppressed this spt16-11 H4(K5R,K12R) synthetic defect (Fig. 2C). Thus, rco1 suppresses FACT mutant phenotypes associated with both replication and transcription.
We examined the role of the Rco1 PHD domain in suppression of pob3(L78R). RCO1 alleles either lacking the PHD domain or with a replacement of the Yng2 PHD2 domain also suppressed the pob3(L78R) defects as did an rco1 gene deletion (Fig. 2A). Although Rco1(ΔPHD) was expressed at levels similar to those of native Rco1 (see Fig. S1A at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement), Rco1(ΔPHD) did not cause toxicity in the pob3(L78R) mutant. These results suggest that the Rco1(ΔPHD) mutant suppresses pob3(L78R) because it fails to target Rpd3(S) to specific genomic locations.
Synthetic defects in sds3 FACT double mutants.
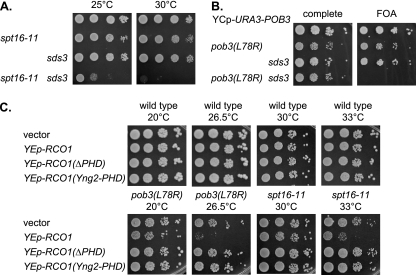
Since mutation of the rco1 subunit of Rpd3(S) suppresses FACT defects, we constructed strains where disruption of the SDS3 gene encoding an Rpd3(L) subunit was combined with FACT mutations. Instead of suppression, we observed more severe phenotypes in the double mutant. The spt16-11 sds3 double mutant strain was very sick at 25°C and lethal at 30°C (Fig. 3A). Since the gcn5 sds3 synthetic phenotypes can be suppressed by mutations in RCO1 or SET2, we looked for similar suppression and found that the spt16-11 sds3 growth defect can be suppressed by set2 or rco1 (see Fig. S2 at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). Although rco1 and set2 mutations each suppressed the spt16-11 sds3 growth defect, we did not see additivity with multiple suppressor mutations (see Fig. S2D at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). We were unable to recover any viable pob3(L78R) sds3 spores in crosses. We constructed a pob3(L78R) sds3 strain with a YCp-URA3-POB3 plasmid; this strain was unable to lose the plasmid and grow on 5-FOA medium (Fig. 3B), demonstrating the synthetic lethality between pob3(L78R) and sds3. We crossed this pob3(L78R) sds3 strain with the YCp-URA3-POB3 plasmid to strains with an rco1 or set2 mutation, and we were able to recover viable strains without the plasmid (see Fig. S3A at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). Thus, rco1 and set2 mutations each can suppress the pob3(L78R) sds3 synthetic lethality. Finally, a multicopy plasmid with the RCO1 gene inhibited growth of pob3(L78R) and spt16-11 mutants, an effect that requires the Rco1 PHD domain (Fig. 3C). These experiments suggest that Rpd3(L) supports FACT function while Rpd3(S) with the ability to bind methylated H3(K36) inhibits it.
FIG. 3.
FACT mutants are lethal with an sds3 mutation or RCO1 overexpression. (A) Tenfold dilutions of strains DY150 (wild type), DY8107 (spt16-11), DY2413 (sds3), and DY10482 (spt16-11 sds3) were plated on complete medium for 2 days at either 25°C or 30°C. (B) Tenfold dilutions of strains DY150 (wild type), DY7378 [pob3(L78R)], DY2413 (sds3), and DY10507 [pob3(L78R) sds3] with a YCp-URA3-POB3 plasmid were plated on either complete or FOA-containing medium for 3 days at 20°C. (C) Strains DY150 (wild type), DY7379 [pob3(L78R)], and DY8107 (spt16-11) were transformed with the indicated multicopy plasmid, and 10-fold dilutions were plated on selective medium lacking uracil for 4 days at 20°C or 26.5°C or for 2 days at 30°C or 33°C.
eaf3 mutation suppresses certain FACT phenotypes.
We also determined the effect of an eaf3 mutation, since Eaf3 is a subunit of Rpd3(S) but not Rpd3(L); Eaf3 is also present in the NuA4 complex. We found that eaf3 suppressed both the temperature- and HU-sensitive phenotypes for two FACT alleles, spt16-11 (Fig. 4A) and pob3(L78R) (Fig. 4B). Additionally, eaf3 suppressed the lethality of pob3(L78R) sds3 (see Fig. S3A at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement) and also suppressed the growth defect of the spt16-11 sds3 double mutant (see Fig. S3B at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). Since both set2 and eaf3 mutations suppress the pob3(L78R) temperature sensitivity, we constructed strains with multiple mutations to look for additive effects. The pob3(L78R) set2 eaf3 triple mutant grew no better than the pob3(L78R) set2 or pob3(L78R) eaf3 double mutant (see Fig. S3C at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). Thus, no additive effects were seen from combining set2 and eaf3 mutations, suggesting that Set2 and Eaf3 function in the same pathway in regulating FACT functions in vivo.
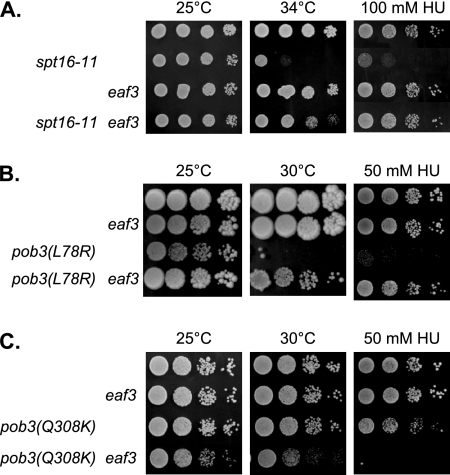
FIG. 4.
eaf3 suppresses FACT mutants. (A) Tenfold dilutions of strains DY150 (wild type), DY8117 (spt16-11), DY10382 (eaf3), and DY10479 (spt16-11 eaf3) were plated on complete medium for 2 days at 25°C or at 34°C or on medium containing 100 mM HU for 4 days at 25°C. (B) Tenfold dilutions of strains DY150 (wild type), DY10382 (eaf3), DY7379 [pob3(L78R)], and DY10390 [pob3(L78R) eaf3] were plated on complete medium for 3 days at 25°C or for 5 days at 30°C or on medium containing 50 mM HU for 4 days at 25°C. (C) Tenfold dilutions of strains DY2860 (wild type), DY10880 (eaf3), DY10722 [pob3(Q308K)], and DY10883 [pob3(Q308K) eaf3] were plated on complete medium for 2 days at 25°C or 30°C or on medium containing 50 mM HU for 4 days at 25°C.
However, in contrast to the suppression seen with spt16-11 and pob3(L78R), a pob3(Q308K) eaf3 double mutant showed increased sensitivity either to elevated temperature or to HU (Fig. 4C). This experiment provided two important results. First, it pointed out an important difference between the pob3(Q308K) allele and the spt16-11 and pob3(L78R) alleles. Second, while rco1 and set2 each suppressed all three FACT alleles tested, eaf3 was different because it failed to suppress pob3(Q308K) and instead enhanced the pob3(Q308K) defect. This difference could be because Eaf3 is present in NuA4, in addition to being a subunit of Rpd3(S).
NuA4 histone acetyltransferase may be involved in DNA replication.
Because Eaf3 is also a subunit of the NuA4 histone acetyltransferase complex, the genetic effects of eaf3 mutants cannot be ascribed simply to Rpd3(S). We therefore examined genetic interactions between mutations specifically affecting NuA4 and FACT. ESA1 is an essential gene encoding the histone acetyltransferase subunit of NuA4, and there are conditional esa1 alleles, such as esa1(L254P) and esa1-Δ414 (13). We previously showed that a spt16-11 esa1(L254P) double mutant shows a synthetic growth defect at elevated temperature (21), and here we showed that the same is true for the combination of the spt16-11 and esa1-Δ414 mutations (see Fig. S4 at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). In addition to the synthetic growth defect at 33°C, the spt16-11 esa1 double mutants also showed increased sensitivity to HU (see Fig. S4 at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). Mutations in SET2 and RCO1 suppressed both the temperature and HU sensitivity of the spt16-11 esa1 double mutants, although suppression by rco1 was less pronounced (see Fig. S4 at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). We also found that esa1 mutations cause mild sensitivity to HU, and this can be suppressed by rco1 (see Fig. S5A at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). Nonetheless, combining an esa1 mutation with either rco1 or set2 resulted in an additive growth defect at 35°C (see Fig. S5B at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). In examining the ability of eaf3 to suppress FACT defects, it is significant that a mutation in the ESA1 subunit of NuA4 caused synthetic defects when combined with spt16-11. This suggests that the suppressive effect of the eaf3 mutation on FACT mutants is due to the loss of Rpd3(S).
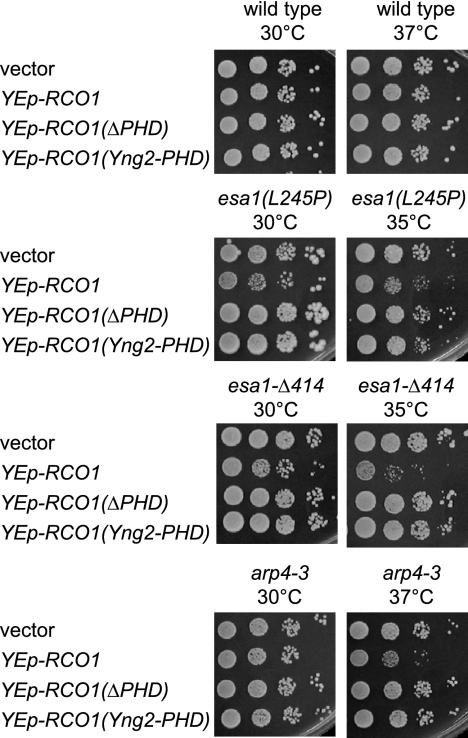
Like Rco1, Eaf3 binds to methylated H3(K36) (12, 27, 28). To explain our observations, we therefore considered a model in which NuA4 and Rpd3(S) compete with one another for binding to methylated H3(K36). In this model, NuA4 binding promotes FACT function and Rpd3(S) binding inhibits it. Thus, the synthetic growth defects of spt16-11 esa1 double mutants could be suppressed by rco1 because the mutation prevents Rpd3(S) from binding to methylated H3(K36), promoting the NuA4-based enhancement of FACT action. Eliminating the competitor for binding could also suppress the NuA4 defect caused by the esa1 mutation. Suppression of the HU sensitivity caused by esa1 mutations by rco1 supports this idea (see Fig. S5A at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). In this view, overexpression of Rco1 could increase the amount of Rpd3(S), resulting in toxicity in NuA4 mutant strains. Consistent with this, a multicopy plasmid with the RCO1 gene was toxic in esa1 mutants, and the Rco1 PHD domains were required for toxicity (Fig. 5). A set2 mutation would eliminate methylation of H3(K36), ending the competition for binding to Me-K36 between Rpd3(S) and NuA4. However, NuA4 has additional subunits that bind histones. Thus, the set2 mutation would provide a competitive advantage for NuA4 binding compared to Rpd3(S), explaining the suppression of FACT defects by a set2 mutation.
FIG. 5.
RCO1 overexpression is toxic in esa1 and arp4 mutants. Strains DY150 (wild type), DY7560 [esa1(L254P)], DY7558 (esa1-Δ414), and DY5856 (arp4-3) were transformed with the indicated multicopy plasmid, and 10-fold dilutions were plated on selective medium lacking uracil. All plates were incubated for 3 days at the indicated temperature for 2 days, except for the esa1-Δ414 cells at 35°C, which were incubated for 4 days.
The Arp4 subunit of NuA4 is encoded by an essential gene, but a strain with the arp4-3 allele was unable to grow at 37°C. A strain with both the arp4-3 and spt16-11 mutations showed a growth defect at 30°C and synthetic lethality either at 33°C or in the presence of a low concentration of HU (Fig. 6A). The synthetic phenotypes of the arp4-3 spt16-11 double mutant could not be suppressed by rco1 or set2 (data not shown). However, the arp4-3 temperature sensitivity could be suppressed by both rco1 (Fig. 6B) and set2 (Fig. 6C). The arp4-3 mutant is sensitive to 150 mM HU (23), and this could be suppressed by either rco1 or set2 (Fig. 6D). The arp4-3 sds3 double mutant was extremely sensitive to HU, much more sensitive that either single mutant; the HU sensitivity of the arp4-3 sds3 strain could be suppressed by an rco1 mutation (see Fig. S6 at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). Finally, like esa1 mutants, the arp4-3 strain is sensitive to overexpression of Rco1 containing its PHD domain (Fig. 5). Of course, interpreting results with the arp4 mutant is complicated by the fact that Arp4 is also present in the Ino80 and Swr1 complexes (29, 30, 37, 47).
FIG. 6.
arp4 mutant phenotypes are suppressed by rco1 and set2. (A) Tenfold dilutions of strains DY150 (wild type), DY8107 (spt16-11), DY5856 (arp4-3), and DY11778 (arp4-3 spt16-11) were plated on complete medium for 3 days at 25°C, for 2 days at 30°C, or for 2 days at 33°C or on medium containing 10 mM HU for 3 days at 30°C. (B) Tenfold dilutions of strains DY150 (wild type), DY10398 (rco1), DY4136 (arp4-3), and DY11119 (arp4-3 rco1) were plated on complete medium for 2 days at 30°C or for 3 days at 37°C. (C) Tenfold dilutions of strains DY150 (wild type), DY8690 (set2), DY4136 (arp4-3), and DY11133 (arp4-3 set2) were plated on complete medium for 2 days at 30°C or for 3 days at 37°C. (D) Tenfold dilutions of strains DY150 (wild type), DY8782 (set2), DY10398 (rco1), DY5856 (arp4-3), DY11133 (arp4-3 set2), DY11119 (arp4-3 rco1), and DY11136 (arp4-3 rco1 set2) were plated on complete medium for 2 days at 30°C or on medium containing 150 mM HU for 4 days at 30°C.
The esa1 and arp4 mutations affecting NuA4 were sensitive to changes in the relative levels of Rpd3(L) and Rpd3(S). An sds3 mutation eliminating Rpd3(L) showed synthetic defects when combined with either an esa1 or an arp4 mutation, and overexpression of Rco1 was toxic in these NuA4 mutants. Mutations in either RCO1 or SET2, which resulted in either eliminating Rpd3(S) or the methyl mark at H3(K36) that Rpd3(S) binds, suppressed the NuA4 mutations. These results are consistent with the idea that NuA4 and Rpd3(S) act in opposition, possibly by competing for binding to methylated H3(K36).
NuA4 and Rpd3(S) compete for binding to ARG3.
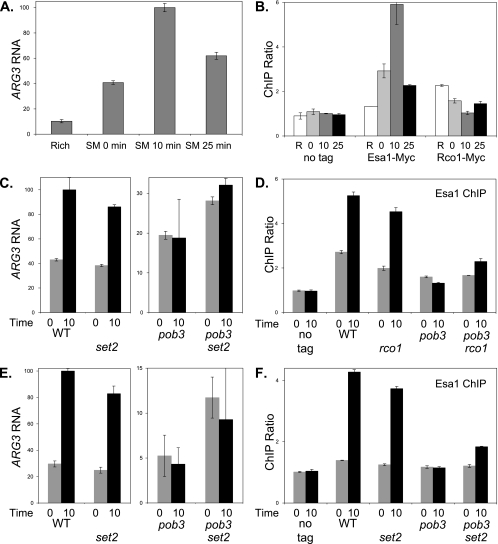
To test for competition between NuA4 and Rpd3(S) directly, we examined binding at the ARG3 promoter during transcriptional induction. Swanson et al. (51) have shown that amino acid starvation results in NuA4 recruitment to promoters of arginine biosynthesis genes, including ARG3 (42). Following their protocol, we added SM to the medium, causing inhibition of isoleucine and valine biosynthesis that resulted in activation of Gcn4 target genes, including ARG3. RNA measurements showed ARG3 mRNA levels were maximal 10 min after addition of SM (Fig. 7A), and ChIP analysis showed Esa1-Myc binding at the ARG3 promoter also peaked 10 min after SM addition (Fig. 7B). Although Esa1-Myc bound to the ARG3 promoter, it did not bind to the ARG3 open reading frame (see Fig. S7A at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). We also examined binding of Rco1-Myc to ARG3, and the results were reciprocal to those of Esa1: Rco1 bound to ARG3 in the absence of expression, and binding disappeared when ARG3 was highly expressed (Fig. 7B). Additionally, Rco1 binding was not detected at the ARG3 open reading frame (see Fig. S7B at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). Previous work suggested that Rco1 and Rpd3(S) function primarily at open reading frames and not promoters (12, 27), and thus, our results are surprising. At 1 kb in size, ARG3 is a rather small gene, and larger genes have greater dependence on Rpd3(S) (33). To verify the quality of our Rco1 ChIPs, we examined STE11, since previous work found Eaf3 [present in Rpd3(S), as well as NuA4 and other complexes] binds to the 3′ region of the STE11 open reading frame (12). We found Rco1 binding to the STE11 open reading frame with an efficiency similar to that seen at the ARG3 promoter (see Fig. S8A at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). The Rco1(ΔPHD) mutant does not bind to nucleosomes in vitro (32), and we examined binding of Rco1(ΔPHD) in vivo with ChIP assays. Rco1(ΔPHD) did not bind to either the STE11 open reading frame or the ARG3 promoter (see Fig. S8B at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). Although Rpd3(S) may function primarily at open reading frames, we conclude that Rpd3(S) is bound to the ARG3 promoter and its pattern of binding at ARG3 is opposite to that of NuA4.
FIG. 7.
pob3 and rco1 affect ARG3 induction and NuA4 binding. (A) Strain DY150 (wild type) was grown at 25°C in rich medium, and then cells were transferred to minimal medium containing SM to starve cells for amino acids. Samples were taken before the shift and at 0, 10, and 25 min following the shift. RNA was isolated, and mRNA levels were determined by reverse transcription (RT)-PCR for ARG3 and ACT1 (internal control). The results are given as the ratio of ARG3 to the ACT1 internal control, with the error bars showing the standard deviation of the triplicate PCRs. (B) Strains DY150 (no tag), DY12268 (Esa1-Myc), and DY11045 (Rco1-Myc) were starved for amino acids as in panel A, and ChIP assays were performed to measure Esa1-Myc and Rco1-Myc binding to the ARG3 promoter. “R” indicates Rich medium, and the numbers indicate the time after transfer to starvation medium. PCR assays were also performed to measure protein occupancy at a control locus on chromosome I. The results give the ratio of the ChIP signals at ARG3 to the control interval, and error bars show the standard deviations of the ChIP PCRs performed in triplicate. (C) Strains DY150 (wild type [WT]), DY10398 (rco1), DY8881 [pob3(L78R)], and DY10406 [pob3(L78R) rco1] were starved for amino acids as in panel A. RNA was isolated, and mRNA levels were determined by RT-PCR for ARG3 and ACT1 (internal control). “0” and “10” indicate time after the shift to starvation conditions. The results are given as the ratio of ARG3 to the ACT1 internal control, with the error bars showing the standard deviations of the triplicate PCRs. Note that the scales for the left and right panels are different. (D) Strains DY150 (no tag), DY12268 (Esa1-Myc), DY12343 (Esa1-Myc rco1), DY12270 [Esa1-Myc pob3(L78R)], and DY12342 [Esa1-Myc pob3(L78R) rco1] were starved for amino acids as in panel A, and ChIP assays were performed to measure Esa1-Myc binding to the ARG3 promoter. “0” and “10” indicate time after the shift to starvation conditions. The results give the ratio of the ChIP signals at ARG3 to the control interval, and error bars show the standard deviation of the ChIP PCRs performed in triplicate. (E) Strains DY150 (wild type), DY8780 (set2), DY8881 [pob3(L78R)], and DY8877 [pob3(L78R) set2] were starved for amino acids as in panel A. RNA was isolated, and mRNA levels were determined by RT-PCR for ARG3 and ACT1 (internal control). “0” and “10” indicate time after the shift to starvation conditions. The results are given as the ratio of ARG3 to the ACT1 internal control, with the error bars showing the standard deviation of the triplicate PCRs. Note that the scales for the left and right panels are different. (F) Strains DY150 (no tag), DY12268 (Esa1-Myc), DY12339 (Esa1-Myc set2), DY12270 [Esa1-Myc pob3(L78R)], and DY12337 [Esa1-Myc pob3(L78R) set2] were starved for amino acids as in panel A, and ChIP assays were performed to measure Esa1-Myc binding to the ARG3 promoter. “0” and “10” indicate time after the shift to starvation conditions. The results give the ratio of the ChIP signals at ARG3 to the control interval, and error bars show the standard deviations of the ChIP PCRs performed in triplicate.
We next examined the effect of pob3(L78R) and rco1 mutations on ARG3 induction and NuA4 binding to ARG3. The pob3 mutation eliminated ARG3 induction at the 10-min time point (Fig. 7C) and sharply reduced binding of Esa1-Myc at this time (Fig. 7D). This suggests that the pob3 mutation affects the recruitment of NuA4 to the ARG3 promoter. Interestingly, the rco1 mutation partially suppressed the pob3 defect in both ARG3 induction and NuA4 recruitment.
We also examined changes in histone occupancy and modification during induction of ARG3 (see Fig. S9 at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). Eviction of nucleosomes can accompany gene activation (59). We used antibody to histone H3 in ChIP assays to measure loss of nucleosomes, and we found ARG3 induction was accompanied by nucleosome loss in the promoter and throughout the open reading frame. A pob3 mutation substantially reduced nucleosome loss through the open reading frame, as expected, since pob3 reduces ARG3 expression. However, a pob3 mutation also reduced nucleosome loss at the ARG3 promoter. We next performed ChIP assays to measure H3(K36) methylation, H3(K14) acetylation, and H4 acetylation with the modification-specific ChIP signal normalized to H3 occupancy to account for the nucleosome eviction. No increase in methylation of H3(K36) occurs when ARG3 is induced, possibly because ARG3 is a small gene (33). We detected strong H3(K36) methylation at the PMA1 open reading frame, which served as a positive control for these ChIP assays. There was only a modest increase in H3 acetylation at the ARG3 promoter, while the sharp increase in H4 acetylation was consistent with binding of the NuA4 complex, which specifically acetylates histone H4. Less histone acetylation was seen at the ARG3 open reading frame. There was less H4 acetylation at the promoter in the pob3 mutant, as expected with less NuA4 bound, and a similar level of H4 acetylation was seen in the pob3 rco1 double mutant. The ratio before and after induction of H4 acetylation was not markedly different in the wild-type and pob3 strains, however.
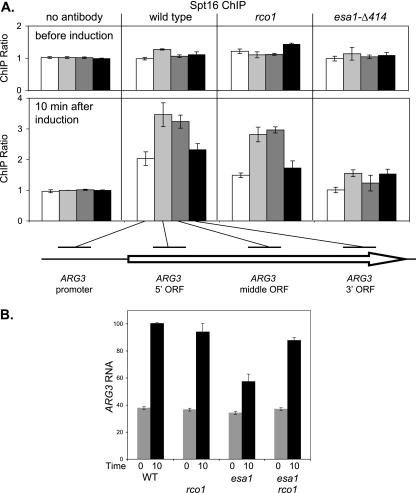
We also examined binding of FACT to ARG3 using ChIP assays. Strong binding of FACT was seen at the ARG3 open reading frame following induction with SM (Fig. 8A), consistent with previous reports that FACT travels with elongating RNA polymerase II (36, 44). Only modest FACT binding to the ARG3 promoter was seen, and this binding could reflect its role in stimulating initiation at ARG3, or the ChIP signal could be due to the proximity of the promoter probe to the open reading frame. An esa1 mutation resulted in markedly decreased FACT binding, while an rco1 mutation had little effect. Thus, FACT and NuA4 are both required for ARG3 induction, and a mutation of either chromatin factor affects binding of the other. RNA analysis showed that the esa1 mutation markedly decreased induction of ARG3 (Fig. 8B). Importantly, the defect in ARG3 induction caused by the esa1 mutation was suppressed by rco1, further supporting the role of Rpd3(S) in negatively regulating the ARG3 promoter.
FIG. 8.
An esa1 mutation affects FACT binding and ARG3 expression. (A) FACT binding to the ARG3 open reading frame is reduced in an esa1 mutant. Strains DY150 (wild type), DY10398 (rco1), and DY7558 (esa1-Δ414) were grown at 25°C in rich medium and transferred to minimal medium containing SM to starve cells for amino acids, and samples were taken for ChIP before and 10 min after induction. ChIP assays were performed with cross-linked extracts, and a mock precipitation without antibody was performed with extracts from wild-type cells. PCRs measured binding to the ARG3 promoter (−295 to −53) (white), the ARG3 5′ open reading frame (−34 to +169) (light gray), the ARG3 middle open reading frame (+214 to +415) (dark gray), and the ARG3 3′ open reading frame (+707 to +926) (black). The results give the ratio of the ChIP signals at the ARG3 region to the control interval, and error bars show the standard deviation of the ChIP PCRs performed in triplicate. (B) An esa1 mutation reduces ARG3 expression. Strains DY150 (wild type [WT]), DY10398 (rco1), DY7558 (esa1-Δ414), and DY11116 (esa1-Δ414 rco1) were starved for amino acids as in panel A. RNA was isolated, and mRNA levels were determined by RT-PCR for ARG3 and ACT1 (internal control). “0” and “10” indicate time after the shift to starvation conditions. The results are given as the ratio of ARG3 to the ACT1 internal control, with the error bars showing the standard deviations of the triplicate PCRs.
Since rco1 and set2 mutations both suppressed the HU- and temperature-sensitive phenotypes caused by FACT mutations, we also examined whether set2 could suppress the effects of a pob3 mutation at ARG3. A set2 mutation showed some ability to suppresses the pob3 defect in both induction of ARG3 RNA and binding of NuA4 to the ARG3 promoter (Fig. 7E and F). We were concerned that a pob3 mutation might affect accumulation of the Esa1 protein and that mutations in RCO1 or SET2 might reverse this. Immunoblot experiments show that Esa1 protein levels are not affected by pob3, rco1, or set2 mutations (see Fig. S8C at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement).
In summary, a pob3 mutation affects ARG3, reducing both NuA4 binding and the level of mRNA induction. These defects can be suppressed by either an rco1 mutation or a set2 mutation. The suppression by set2 suggests that methylation of H3(K36) is important for recruitment of both Rpd3(S) and NuA4, and there is a slight increase in H3(K36) methylation at the ARG3 promoter following induction. Additionally, the suppression by rco1 is consistent with competition for binding to Me-K36 by NuA4 and Rpd3(S). Disruption of the EAF3 gene encoding a NuA4 subunit required for binding of Me-K36 did not affect ARG3 mRNA induction (data not shown), but Eaf3 is also present in Rpd3(S). A number of other transcriptional coactivators are recruited to ARG3 in addition to NuA4, and it is possible that interactions with these other coactivators facilitate NuA4 binding despite the absence of K36 methylation or Eaf3, the Me-K36 binding subunit.
rco1 mutation increases NuA4 binding to sites of DNA damage.
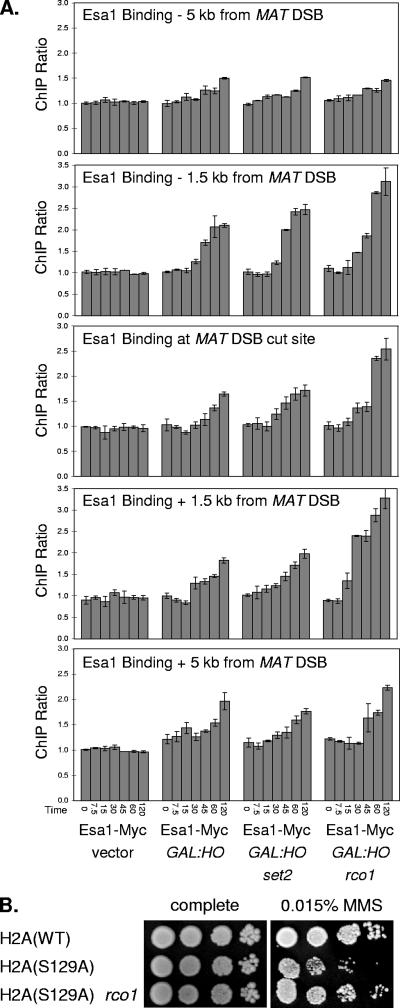
We next examined other genomic locations where NuA4 binds but other transcriptional coactivators may not be present. NuA4 is recruited to sites of DNA damage (15), such as double-strand DNA breaks created by the HO endonuclease. Double-strand breaks were created at the MAT locus by inducing HO expression from the GAL1 promoter, and we used an Esa1-Myc strain for ChIP assays with seven sets of PCR primers to monitor binding of NuA4 both to the cleavage site region and to regions 1.5, 5, and 10 kb on either side of the breakpoint (Fig. 9A). NuA4 binding was seen at 30 min, after HO induction, and persisted up to 120 min in this strain, where deletion of the HML and HMR loci prevents repair of the double-strand break. NuA4 binding was strong in the 3-kb region centered on the break site and weaker 5 kb away (Fig. 9A); binding was not detected at a region 10 kb away from the double-strand break (data not shown). ChIP experiments did not detect Rco1 binding in the region of the HO cleavage site, but significant Rco1 binding was seen at the kb −5, −1.5, and +1.5 locations, which are present within open reading frames (see Fig. S10A at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement). H3(K36) methylation is also seen at these regions (see Fig. S10B at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement), although the level of H3(K36) methylation does not correlate with the level of Rco1 binding. The levels of Rco1 binding and H3(K36) methylation are not significantly affected by the double-strand break. Importantly, an rco1 mutation results in a significant increase in NuA4 binding (Fig. 9A), consistent with the idea that Rpd3(S) competes with NuA4 for binding at sites of DNA damage. However, an increase in NuA4 binding was not seen in set2 mutants. Thus, there is specificity in that disruption of RCO1 results in increased NuA4 binding to sites of DNA damage while disruption of SET2 does not.
FIG. 9.
rco1 affects Esa1 binding to MAT double-strand break and suppresses H2A(S129A). (A) Strains DY12561 (Esa1-Myc hmlΔ hmrΔ), with either the empty vector or the GAL1::HO plasmid, and strains DY12784 (Esa1-Myc hmlΔ hmrΔ set2) and DY12553 (Esa1-Myc hmlΔ hmrΔ set2), both with the GAL1::HO plasmid, were grown on selective medium lacking uracil at 25°C, and galactose was added to induce expression of the HO endonuclease. Samples were taken at various time points after induction for ChIP assays to measure Esa1-Myc binding at the indicated regions near the MAT locus and at the chromosome I control region. The times after galactose addition are given. The results give the ratio of the ChIP signals at the specific MAT region to the control interval, and error bars show the standard deviation of the ChIP PCRs performed in triplicate. (B) Tenfold dilutions of strains DY5699 (wild type [WT]), DY12351 [hta1(S129A) hta2(S129A)], and DY12404 [hta1(S129A) hta2(S129A) rco1] were plated on complete medium or medium containing 0.015% MMS for 3 days at 30°C.
DNA damage also results in activation of the ATM family Mec1 kinase, which localizes to sites of DNA damage and phosphorylates histone H2A on S129 (16). It has been shown that Nu4A binds to the chromatin with phosphorylated H2A(S129) at sites of DNA damage via the Arp4 subunit (15). Thus, NuA4 can bind chromatin via the Arp4/phosphorylated H2A(S129) interaction and via the Eaf3/methylated H3(K36) interaction; other NuA4 subunits can also mediate chromatin interactions. Substitution of serine 129 with an alanine results in chromatin which cannot be phosphorylated in response to DNA damage, and the H2A(S129A) mutant shows increased sensitivity to the DNA-damaging agent MMS (25). We show that the MMS sensitivity of the H2A(S129A) mutant can be partially suppressed by an rco1 mutation (Fig. 9B). Mutations in SET2 do not suppress the sensitivity of the H2A(S129A) mutant to MMS, however (data not shown). The MMS sensitivity of the H2A(S129A) mutant is interpreted as being due to reduced binding of NuA4 to chromatin at sites of DNA damage (15). An rco1 mutation eliminates the competition of Rpd3(S) for binding to methylated H3(K36) and thereby improves the chromatin binding ability of NuA4, resulting in suppression of the MMS sensitivity caused by the H2A(S129A) mutation.
DISCUSSION
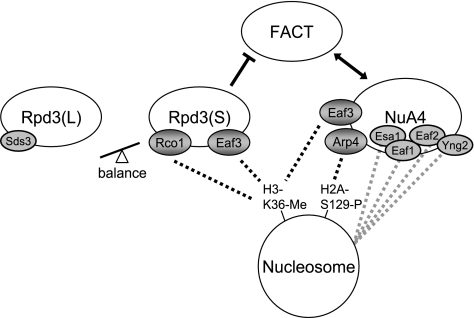
The factors that modify chromatin structure play important roles in regulating transcription, DNA replication, and repair of DNA damage. In this work we have identified important functional interactions between the FACT chromatin reorganizing factor, the NuA4 histone acetyltransferase, and the two Rpd3 histone deacetylases, Rpd3(L) and Rpd3(S). Our genetic studies show that FACT and NuA4 mutually reinforce one another and that FACT activity is opposed by Rpd3(S) (Fig. 10). NuA4 and Rpd3(S) each has at least two subunits involved in binding nucleosomes, and the nucleosome binding subunits of Rpd3(S), Eaf3 and Rco1, recognize methylated K36 of histone H3. Our genetic experiments led to a model where NuA4 and Rpd3(S) compete for binding to nucleosomes (Fig. 10), with one consequence of the outcome of the competition being the level of FACT activity required for growth. The model is supported by ChIP experiments showing that mutations eliminating Rpd3(S) result in increased NuA4 binding in vivo and by reciprocal binding of NuA4 and Rpd3(S) at an inducible promoter.
FIG. 10.
Model of competition between Rpd3(S) and NuA4. Rpd3(S) and NuA4 each have at least two subunits that mediate association with nucleosomes. Rco1 and Eaf3 each recognize methylated K36 of histone H3. Arp4 is thought to recognize histones in several ways, one of which is dependent on phosphorylation of H2A(S129). FACT NuA4 double mutants show synthetic phenotypes, and a mutation in one reduces binding of the other factor, and thus, NuA4 and FACT each reinforce the function of the other factor. Mutations which decrease Rpd3(S) binding to nucleosomes increase NuA4 binding and also stimulate FACT activity. The balance between the two types of Rpd3 HDAC complexes is important, and changing this balance can either suppress or exacerbate FACT mutant phenotypes.
Our results are consistent with those in previous reports showing that Rpd3(L) and Rpd3(S) have different roles (11, 12), but it is a surprise to find that they can have opposing functions. Conditions that decrease the ratio of Rpd3(S) to Rpd3(L), such as elimination of the Rpd3(S)-specific subunit Rco1, suppress defects in either FACT or NuA4, and mutations that increase this ratio, such as deletion of SDS3 or overexpression of Rco1, enhance defects in FACT or NuA4. The appropriate distribution of common subunits between Rpd3(S) and Rpd3(L) complexes is therefore important at least partly because these complexes have opposing effects on FACT and NuA4 and therefore the ratio of the two HDAC complexes regulates the activity of these two essential factors.
The need to balance Rpd3(L) and Rpd3(S) is also evident in gcn5 mutants. Here, mutations that favor either Rpd3(L) or Rpd3(S) complex formation are detrimental. Only balanced levels or, more surprisingly, the absence of any Rpd3 complexes is compatible with robust growth. The growth defects of a gcn5 mutant lacking Rpd3(L) can be suppressed by disruption of the SET2 gene encoding a KMT that modifies H3(K36). Rpd3(S) binds to methylated H3(K36) (12, 27, 28), and preventing this modification and thus making a change in where Rpd3(S) acts is sufficient to overcome the problem caused by the absence of Rpd3(L) in the gcn5 mutant.
NuA4 and Rpd3(S) bind nucleosomes differently despite the presence in both complexes of the Eaf3 subunit, which binds methylated H3(K36). NuA4 has not been shown to recognize methylated H3(K36) nucleosomes in vitro, while Rpd3(S) does bind such modified nucleosomes in vitro because of the combined action of the chromodomain of Eaf3 and the PHD domain of Rco1 (32). Thus, Rpd3(S) contains two subunits, Rco1 and Eaf3, that bind to modified histone residues (Fig. 10). Rco1 contains a PHD domain that binds methyl-lysine (32), and the Rco1 PHD domain is required for Rco1 activity in vivo. An rco1 gene disruption suppresses growth defects caused by a FACT mutation, as do rco1 mutants either lacking the PHD domain or with the native PHD domain replaced by a Yng2 PHD domain. Additionally, while overexpression of Rco1 is toxic in certain mutants, overexpression of Rco1(ΔPHD) or Rco1(Yng2-PHD) does not inhibit growth. An EAF3 gene disruption also suppresses the growth defects of FACT mutants. Importantly, NuA4 lacking the Eaf3 subunit displays altered histone acetyltransferase activity in vitro (J. Cote, personal communication). Eaf3 is present in both Rpd3(S) and NuA4, raising the question of how the eaf3 mutation suppresses. However, point mutations in two subunits of NuA4, Esa1 and Arp4, show synthetic defects when combined with FACT mutations, suggesting that the suppressive effect of the eaf3 mutation on FACT mutants is due to the absence of Eaf3 from Rpd3(S).
In addition to Eaf3, NuA4 contains other subunits that may bind histones, including Eaf1 and Eaf2, with SANT domains: Esa1 with a chromodomain and Yng2 with a PHD domain (17). Whereas the two nucleosome binding subunits in Rpd3(S) are dependent on methylation of histone H3(K36), nucleosome binding by NuA4 is largely independent of K36 methylation (Fig. 10). The Set2 enzyme methylates H3(K36), and a set2 mutation eliminates binding of Rpd3(S) to nucleosomes (12, 27, 28). Importantly, a set2 mutation does not significantly affect binding of NuA4 to nucleosomes in vivo (Fig. 7F and 9A), presumably due to NuA4 subunits other than Eaf3 that promote association with nucleosomes. While a set2 mutation robustly suppresses both FACT mutants (4) and FACT NuA4 double mutants (see Fig. S4 at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement), set2 shows either weak suppression or synthetic defects when combined with mutations in the ARP4 (Fig. 6C) or ESA1 (see Fig. S4 at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement) subunit of NuA4. Thus, FACT and NuA4 are differently affected by a set2 mutation. The suppression of FACT mutants by a set2 mutation could happen because the absence of methylated H3(K36) prevents Rpd3(S) binding. This idea is consistent with the lack of additivity in suppression by the combination of set2 and rco1 mutations. Finally, although rco1 and set2 mutations are both robust suppressors of a variety of FACT defects, including those of FACT NuA4 double mutants, rco1 and set2 both show mild synthetic defects in combination with esa1 mutations (see Fig. S5B at http://www.path.utah.edu/research/labs/david-stillman-lab/supplement), and thus, rco1 and set2 do not suppress all defects in these pathways.
Although our work shows effects of mutations affecting Rpd3(S), Set2, and FACT on transcriptional initiation, previous work has provided functions for these factors in transcriptional elongation. In contrast, it has been shown that Sds3 and Htz1 both localize primarily to promoter elements (27, 34, 40, 62), and it is possible that sds3 and htz1 mutations show synthetic defects when combined with FACT mutants because of a linkage between transcriptional initiation and elongation. Further work will be needed to understand whether a defect in transcriptional initiation could affect elongation.
Based on our findings, we developed a model of competition between NuA4 and Rpd3(S) (Fig. 10). NuA4 and Rpd3(S) act in opposition, and both complexes contain a common Me-K36 binding subunit, Eaf3. The similar genetic effects of mutating the Set2 methyltransferase or the Rco1 subunit of Rpd3(S) on yFACT and NuA4 mutants suggested there may be competition for binding to methylated H3(K36). To address the question of competition between NuA4 and Rpd3(S), we examined factor binding to the ARG3 promoter, where transcriptional induction leads to NuA4 binding. Induction of arginine biosynthesis genes starts with binding of the Gcn4 activator, which then recruits NuA4 along with other coactivators (51). Our ChIP assays show NuA4 binding concurrent with transcriptional activation. Interestingly, Rco1 is present at the promoter before induction but disappears as the gene is activated, as if NuA4 displaces Rpd3(S) from the promoter, thereby supporting our hypothesis of competition. We did not observe an increase in NuA4 binding at ARG3 in an rco1 mutant, possibly because NuA4 binding is dependent on other coactivators (51). However, the defect in NuA4 binding at ARG3 in a pob3 mutant is partially suppressed deletion of the RCO1 gene. Also, of note, the presence of Rpd3(S) at the promoter of an uninduced gene is rather surprising, since previous work suggested Rpd3(S) is present only at the 3′ portion of actively transcribed regions (12, 27).
DNA damage results in the phosphorylation of the C-terminal tail of H2AX (or H2A in yeast), and this phosphorylation leads to the recruitment of multiple chromatin-modifying complexes, including Ino80, Swr1, and NuA4 (15, 38, 53). The Arp4 subunit is required for efficient binding of NuA4 to nucleosomes with phosphorylated H2A(S129) (15). We expressed the HO endonuclease to induce double-strand breaks and found that NuA4 binding in the vicinity of the DNA breaks is significantly increased in an rco1 mutant lacking Rpd3(S). This increased NuA4 binding to double-strand breaks in an rco1 strain compared to that of the wild type strongly supports the idea of competition between NuA4 and Rpd3(S).
In response to DNA damage, the Mec1 and Tel1 kinases phosphorylate serine 129 of histone H2A (16). An S129A mutation in histone H2A prevents this phosphorylation, and yeast strains with H2A(S129A) are sensitive to DNA-damaging agents (15, 25), possibly because NuA4 binds less efficiently to the regions of DNA damage. The fact that an rco1 mutation can suppress sensitivity of the H2A(S129A) mutant to DNA damage suggests that Rpd3(S) directly or indirectly inhibits binding of factors such as NuA4 that are important for repairing DNA damage.
The FACT complex plays an important role in DNA replication (6, 54, 57, 58), and our results suggest that the NuA4 KAT complex is also involved in DNA replication. A mutation in the Esa1 catalytic subunit results in mild sensitivity to HU, and an spt16 esa1 double mutant shows additivity in HU sensitivity. Importantly, the HU sensitivities of spt16 and esa1 single mutants and the spt16 esa1 double mutant can be suppressed by disruption of RCO1.
Our results suggest that NuA4 and FACT work together in promoting both transcription and DNA replication. FACT NuA4 double mutants show synthetic phenotypes, and a mutation in one reduces binding of the other factor. The two Rpd3 HDAC complexes differently affect this pathway, with Rpd3(L) acting in support and Rpd3(S) opposing the pathway. The synthetic defects seen in the Rpd3(L) FACT double mutants lead to the genetic argument that Rpd3(L) supports FACT; however, these synthetic defects can also be explained by increased levels of the Rpd3(S) complex in the sds3 mutant. This idea is supported by the observation that overexpression of the Rpd3(S)-specific Rco1 subunit is toxic in FACT mutants.
The competition between NuA4 and Rpd3(S) is apparent in cells with limited FACT activity, where optimal function becomes crucial for growth. FACT is stimulated by NuA4 and opposed by Rpd3(S), and further work is needed to understand how these enzymes that affect histone acetylation affect FACT activity.
Acknowledgments
We thank Steve Kron, Lorraine Pillus, and Jerry Workman for providing strains, Jacques Cote for communicating unpublished results, and Tim Formosa for many helpful discussions.
This work was supported by grants from the National Institutes of Health.
Footnotes
Published ahead of print on 19 May 2008.
REFERENCES
- 1.Baker, S. P., and P. A. Grant. 2007. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene 265329-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. [DOI] [PubMed] [Google Scholar]
- 3.Bhoite, L. T., Y. Yu, and D. J. Stillman. 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 152457-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas, D., R. Dutta-Biswas, D. Mitra, Y. Shibata, B. D. Strahl, T. Formosa, and D. J. Stillman. 2006. Opposing roles for Set2 and yFACT in regulating TBP binding at promoters. EMBO J. 254479-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas, D., R. Dutta-Biswas, and D. J. Stillman. 2007. Chd1 and yFACT act in opposition in regulating transcription. Mol. Cell. Biol. 276279-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas, D., S. Takahata, H. Xin, R. Dutta-Biswas, Y. Yu, T. Formosa, and D. J. Stillman. 2008. A role for Chd1 and Set2 in negatively regulating DNA replication in Saccharomyces cerevisiae. Genetics 178649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas, D., Y. Yu, M. Prall, T. Formosa, and D. J. Stillman. 2005. The Yeast FACT complex has a role in transcriptional initiation. Mol. Cell. Biol. 255812-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14115-132. [DOI] [PubMed] [Google Scholar]
- 9.Brehm, A., K. R. Tufteland, R. Aasland, and P. B. Becker. 2004. The many colours of chromodomains. Bioessays 26133-140. [DOI] [PubMed] [Google Scholar]
- 10.Cairns, B. R. 2005. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 15185-190. [DOI] [PubMed] [Google Scholar]
- 11.Carrozza, M. J., L. Florens, S. K. Swanson, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 173177-87. [DOI] [PubMed] [Google Scholar]
- 12.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123581-592. [DOI] [PubMed] [Google Scholar]
- 13.Clarke, A. S., J. E. Lowell, S. J. Jacobson, and L. Pillus. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 192515-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colina, A. R., and D. Young. 2005. Raf60, a novel component of the Rpd3 histone deacetylase complex required for Rpd3 activity in Saccharomyces cerevisiae. J. Biol. Chem. 28042552-42556. [DOI] [PubMed] [Google Scholar]
- 15.Downs, J. A., S. Allard, O. Jobin-Robitaille, A. Javaheri, A. Auger, N. Bouchard, S. J. Kron, S. P. Jackson, and J. Cote. 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16979-990. [DOI] [PubMed] [Google Scholar]
- 16.Downs, J. A., N. F. Lowndes, and S. P. Jackson. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 4081001-1004. [DOI] [PubMed] [Google Scholar]
- 17.Doyon, Y., and J. Cote. 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14147-154. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson, P., D. Biswas, Y. Yu, J. M. Stewart, and D. J. Stillman. 2004. TATA-binding protein mutants that are lethal in the absence of the Nhp6 high-mobility-group protein. Mol. Cell. Biol. 246419-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Formosa, T. 2003. Changing the DNA landscape: putting a SPN on chromatin. Curr. Top. Microbiol. Immunol. 274171-201. [DOI] [PubMed] [Google Scholar]
- 20.Formosa, T., P. Eriksson, J. Wittmeyer, J. Ginn, Y. Yu, and D. J. Stillman. 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 203506-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Formosa, T., S. Ruone, M. D. Adams, A. E. Olsen, P. Eriksson, Y. Yu, A. R. Rhoades, P. D. Kaufman, and D. J. Stillman. 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway. Polymerase passage may degrade chromatin structure. Genetics 1621557-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74527-534. [DOI] [PubMed] [Google Scholar]
- 23.Gorzer, I., C. Schuller, E. Heidenreich, L. Krupanska, K. Kuchler, and U. Wintersberger. 2003. The nuclear actin-related protein Act3p/Arp4p of Saccharomyces cerevisiae is involved in transcription regulation of stress genes. Mol. Microbiol. 501155-1171. [DOI] [PubMed] [Google Scholar]
- 24.Gunjan, A., J. Paik, and A. Verreault. 2005. Regulation of histone synthesis and nucleosome assembly. Biochimie 87625-635. [DOI] [PubMed] [Google Scholar]
- 25.Harvey, A. C., S. P. Jackson, and J. A. Downs. 2005. Saccharomyces cerevisiae histone H2A Ser122 facilitates DNA repair. Genetics 170543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Javaheri, A., R. Wysocki, O. Jobin-Robitaille, M. Altaf, J. Cote, and S. J. Kron. 2006. Yeast G1 DNA damage checkpoint regulation by H2A phosphorylation is independent of chromatin remodeling. Proc. Natl. Acad. Sci. USA 10313771-13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joshi, A. A., and K. Struhl. 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20971-978. [DOI] [PubMed] [Google Scholar]
- 28.Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny, S. R. Collins, M. Schuldiner, K. Chin, T. Punna, N. J. Thompson, C. Boone, A. Emili, J. S. Weissman, T. R. Hughes, B. D. Strahl, M. Grunstein, J. F. Greenblatt, S. Buratowski, and N. J. Krogan. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123593-605. [DOI] [PubMed] [Google Scholar]
- 29.Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings, A. J. Link, H. D. Madhani, and J. Rine. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 121565-1576. [DOI] [PubMed] [Google Scholar]
- 31.Lechner, T., M. J. Carrozza, Y. Yu, P. A. Grant, A. Eberharter, D. Vannier, G. Brosch, D. J. Stillman, D. Shore, and J. L. Workman. 2000. Sds3 (suppressor of defective silencing 3) is an integral component of the yeast Sin3·Rpd3 histone deacetylase complex and is required for histone deacetylase activity. J. Biol. Chem. 27540961-40966. [DOI] [PubMed] [Google Scholar]
- 32.Li, B., M. Gogol, M. Carey, D. Lee, C. Seidel, and J. L. Workman. 2007. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 3161050-1054. [DOI] [PubMed] [Google Scholar]
- 33.Li, B., M. Gogol, M. Carey, S. G. Pattenden, C. Seidel, and J. L. Workman. 2007. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 211422-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, B., S. G. Pattenden, D. Lee, J. Gutierrez, J. Chen, C. Seidel, J. Gerton, and J. L. Workman. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 10218385-18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malone, E. A., C. D. Clark, A. Chiang, and F. Winston. 1991. Mutations in SPT68/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 115710-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mason, P. B., and K. Struhl. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 238323-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303343-348. [DOI] [PubMed] [Google Scholar]
- 38.Morrison, A. J., J. Highland, N. J. Krogan, A. Arbel-Eden, J. F. Greenblatt, J. E. Haber, and X. Shen. 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119767-775. [DOI] [PubMed] [Google Scholar]
- 39.Perez-Martin, J., and A. D. Johnson. 1998. Mutations in chromatin components suppress a defect of Gcn5 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 181049-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu, S. L. Schreiber, O. J. Rando, and H. D. Madhani. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhoades, A. R., S. Ruone, and T. Formosa. 2004. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell. Biol. 243907-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robert, F., D. K. Pokholok, N. M. Hannett, N. J. Rinaldi, M. Chandy, A. Rolfe, J. L. Workman, D. K. Gifford, and R. A. Young. 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowley, A., R. A. Singer, and J. C. Johnston. 1991. CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol. Cell. Biol. 115718-5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saunders, A., J. Werner, E. D. Andrulis, T. Nakayama, S. Hirose, D. Reinberg, and J. T. Lis. 2003. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 3011094-1096. [DOI] [PubMed] [Google Scholar]
- 45.Schlesinger, M. B., and T. Formosa. 2000. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics 1551593-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shahbazian, M. D., and M. Grunstein. 2007. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 7675-100. [DOI] [PubMed] [Google Scholar]
- 47.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406541-544. [DOI] [PubMed] [Google Scholar]
- 48.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 1941-21. [DOI] [PubMed] [Google Scholar]
- 49.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 211764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strahl, B. D., P. A. Grant, S. D. Briggs, Z. W. Sun, J. R. Bone, J. A. Caldwell, S. Mollah, R. G. Cook, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 221298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swanson, M. J., H. Qiu, L. Sumibcay, A. Krueger, S. J. Kim, K. Natarajan, S. Yoon, and A. G. Hinnebusch. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 232800-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toulmay, A., and R. Schneiter. 2006. A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae. Yeast 23825-831. [DOI] [PubMed] [Google Scholar]
- 53.van Attikum, H., and S. M. Gasser. 2005. ATP-dependent chromatin remodeling and DNA double-strand break repair. Cell Cycle 41011-1014. [DOI] [PubMed] [Google Scholar]
- 54.VanDemark, A. P., M. Blanksma, E. Ferris, A. Heroux, C. P. Hill, and T. Formosa. 2006. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol. Cell 22363-374. [DOI] [PubMed] [Google Scholar]
- 55.van Kempen, G. M., and L. J. van Vliet. 2000. Mean and variance of ratio estimators used in fluorescence ratio imaging. Cytometry 39300-305. [DOI] [PubMed] [Google Scholar]
- 56.Voth, W. P., Y. Yu, S. Takahata, K. L. Kretschmann, J. D. Lieb, R. L. Parker, B. Milash, and D. J. Stillman. 2007. Forkhead proteins control the outcome of transcription factor binding by antiactivation. EMBO J. 264324-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wittmeyer, J., and T. Formosa. 1997. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 174178-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wittmeyer, J., L. Joss, and T. Formosa. 1999. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry 388961-8971. [DOI] [PubMed] [Google Scholar]
- 59.Workman, J. L. 2006. Nucleosome displacement in transcription. Genes Dev. 202009-2017. [DOI] [PubMed] [Google Scholar]
- 60.Yang, X. J., and E. Seto. 2008. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell. Biol. 9206-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu, Y., P. Eriksson, and D. J. Stillman. 2000. Architectural transcription factors and the SAGA complex function in parallel pathways to activate transcription. Mol. Cell. Biol. 202350-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, H., D. N. Roberts, and B. R. Cairns. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y. 2006. It takes a PHD to interpret histone methylation. Nat. Struct. Mol. Biol. 13572-574. [DOI] [PubMed] [Google Scholar]