Abstract
We previously created a knock-in mutant mouse harboring a dominantly negative mutant thyroid hormone receptor β (TRβPV/PV mouse) that spontaneously develops a follicular thyroid carcinoma similar to human thyroid cancer. We found that β-catenin, which plays a critical role in oncogenesis, was highly elevated in thyroid tumors of TRβPV/PV mice. We sought to understand the molecular basis underlying aberrant accumulation of β-catenin by mutations of TRβ in vivo. Cell-based studies showed that thyroid hormone (T3) induced the degradation of β-catenin in cells expressing TRβ via proteasomal pathways. In contrast, no T3-induced degradation occurred in cells expressing the mutant receptor (TRβPV). In vitro binding studies and cell-based analyses revealed that β-catenin physically associated with unliganded TRβ or TRβPV. However, in the presence of T3, β-catenin was dissociated from TRβ-β-catenin complexes but not from TRβPV-β-catenin complexes. β-Catenin signaling was repressed by T3 in TRβ-expressing cells through decreasing β-catenin-mediated transcription activity and target gene expression, whereas sustained β-catenin signaling was observed in TRβPV-expressing cells. The stabilization of β-catenin, via association with a mutated TRβ, represents a novel activating mechanism of the oncogenic protein β-catenin that could contribute to thyroid carcinogenesis in TRβPV/PV mice.
β-Catenin, a structural component of cell adhesion complexes, interacts with the transmembrane protein E-cadherin to regulate actin filament assembly to regulate cellular functions (9). In addition, β-catenin also functions as a coactivator for a family of transcription factors known as T-cell factor/lymphoid enhancer factor (TCF/LEF). Upon increased cellular levels and nuclear accumulation, β-catenin-TCF complexes bind to the promoters of downstream target genes involved in cell proliferation, survival, and migration (23). Induction of these genes has significant effects on tissue development and oncogenesis; abnormal subcellular localization and aberrant accumulation of β-catenin have been reported for human cancers of the colon, prostate, uterus, liver, and thyroid (3, 6, 8, 22, 24, 32). The cellular levels of β-catenin protein are tightly regulated by two distinct adenomatous polyposis coli (APC)-dependent proteasomal degradation pathways, namely, a glycogen synthase kinase 3β (GSK3β)-regulated pathway involving the APC-axin complex (27) and a p53-inducible pathway involving Siah-1 (20). Recently, nuclear receptors such as the peroxisome proliferator-activated receptor γ (PPARγ) and the retinoid X receptor (RXR) have been reported to regulate the cellular levels of β-catenin via APC/GSK3β/p53-independent mechanisms (31, 36). Both PPARγ2 and RXRα have increased physical interaction with β-catenin following agonist binding, and they are degraded together with β-catenin via proteasomal pathways (21, 31, 36). These findings have added a new dimension to the understanding of the regulation of β-catenin in cells. However, it is not known whether this mode of regulation is shared by other nuclear receptors.
We have created a knock-in mutant mouse that harbors a mutated thyroid hormone β receptor (TRβPV). TRβPV was identified from a patient with a genetic disorder, i.e., resistance to thyroid hormone. TRβPV has a frameshift mutation in the C-terminal 14 amino acids (25), resulting in complete loss of thyroid hormone (T3) binding and transcription activity (15). As the homozygous mice (TRβPV/PV mice) age, they spontaneously develop follicular thyroid carcinoma with a pathological progression similar to that of human thyroid cancer (33, 39). This mouse model of thyroid cancer provides us with an unusual opportunity to understand gene alterations and aberrant signaling during thyroid carcinogenesis (39). Several oncogenes, including the β-catenin gene, were found to be activated during thyroid carcinogenesis (39). Importantly, we discovered that the cellular abundance of β-catenin was aberrantly elevated in thyroid tumors of TRβPV/PV mice. Therefore, TRβPV/PV mice provided us with a tool to understand how TRβ and its mutants regulate the cellular levels of β-catenin in vivo and, furthermore, whether the APC/GSK3β-independent regulatory mechanisms of β-catenin reported for PPARγ2 and RXRα (31, 36) could be extended to TRβ. Indeed, we found that similar to PPARγ2 and RXRα, TRβ could also regulate the β-catenin protein level via APC-independent proteasome pathways. However, in contrast to PPARγ2 and RXRα, where the interaction of β-catenin with PPARγ2 or RXRα is strengthened by ligand, the physical interaction of β-catenin with TRβ was favored in the unliganded state. TRβPV, which has lost T3 binding, constitutively bound to β-catenin to block the proteasomal degradation of β-catenin, thereby leading to sustained activation of β-catenin-mediated downstream target gene expression to contribute to thyroid carcinogenesis in TRβPV/PV mice.
MATERIALS AND METHODS
Mouse strains and cell lines.
All aspects of the care and handling of animals used in this study were approved by the National Cancer Institute Animal Care and Use Committee. Generation and genotyping of TRβPV mice were described previously (15). The establishment of HeLa cells stably expressing Flag-hemagglutinin-tagged TRβ1 (FH-TRβ), FH-TRβPV, and control cells (FH) was reported previously (38).
Plasmids and antibodies.
The pGEX4T1-GST-β-catenin vector was generously provided by Beatrice Darimont (University of Oregon, Eugene, OR). The plasmids used for in vitro transcription/translation were pcDNA3.1 (TRβ1 and TRβPV), pJL08 (ED41), pJL06 (MD32), pCJ4 (KD25), and pCJ7 (KP24), which have been described previously (18). The plasmids used for transfection were pcDNA-hTCF4 (generously provided by Bert Vogelstein, Howard Hughes Medical Institute, Baltimore, MD), pcDNA3-β-catenin (generously provided by Stephen Byers, Georgetown University, Washington, DC), pcDNA3 (Invitrogen Corp.), TOP-Flash, and FOP-Flash (Upstate, Lake Placid, NY). The primary antibodies used were mouse anti-TR (18), rabbit anti-β-catenin (1:2,000 dilution) (9562; Cell Signaling Technology), rabbit anti-phospho-β-catenin (Ser33/37/Thr41; 1:1,000 dilution) (9561; Cell Signaling Technology), rabbit anti-phospho-β-catenin (Ser552; 1:2,000 dilution) (generously provided by Linheng Li, Stowers Institute for Medical Research, Kansas City, MO), mouse anti-MT1-MMP (1:1,000 dilution) (MAB3317; Chemicon International), rabbit anti-GSK3β (1:1,000 dilution) (9315; Cell Signaling Technology), rabbit anti-phospho-GSK3α/β (Ser21/9; 1:1,000 dilution) (9331; Cell Signaling Technology), goat anti-Siah-1 (1:1,000 dilution) (ab2237-100; Abcam), rabbit anti-p53 (1:200 dilution) (6243; Santa Cruz), rabbit anti-c-Myc (1:1,000 dilution) (Cell Signaling Technology), mouse anti-cyclin D1 (1:200) (450; Santa Cruz), rabbit anti-protein disulfide isomerase (anti-PDI; 3632) (2), rabbit anti-PARP (1:200 dilution) (7150; Santa Cruz), and mouse anti-α-tubulin (1:6,000 dilution) (T6199; Sigma).
Immunohistochemistry of β-catenin.
Immunohistochemistry on thyroid tissue sections for β-catenin was performed as previously described (40). The primary antibody used was purchased from Cell Signaling Technology (9562).
Western blot analysis.
The levels of cellular proteins in FH-TRβ and FH-TRβPV cells were determined via Western blotting. Cells were plated at a density of 9 × 105 cells/60-mm dish and cultured for 24 h in Dulbecco's modified Eagle's medium (DMEM) containing 10% calf serum, antibiotic cocktail (PSN; Gibco), and 0.25 mg/ml Geneticin (Gibco). Cells were then incubated for 48 h in DMEM with 10% charcoal-dextran-stripped calf serum (T3-depleted, or Td, serum). They were then incubated with or without 100 nM of T3 for the indicated duration and lysed. The proteasome inhibitor MG132 (25 μM; dissolved in dimethyl sulfoxide [DMSO] [Calbiochem]) or the vehicle DMSO was added to the Td medium in the presence or absence of 100 nM T3, and cells were lysed after 17 h. Whole-cell lysates were prepared using a buffer containing 50 mM Tris, 250 mM NaCl, 5 mM EDTA, 0.5% NP-40, a cocktail of protease inhibitors (Roche Diagnostics), 5 mM N-ethylmaleimide, 10 nM ubiquitinase aldehyde, 0.2 μM okadaic acid, 10 mM NaF, and 1 mM Na3VO4. After a 10-minute incubation on ice, cell lysates were centrifuged at maximal speed for 10 min, and the supernatants were collected. Nuclear and cytoplasmic fractions were separated by using NE-PER nuclear and cytoplasmic extraction reagents (Pierce Biotechnology) following the manufacturer's instructions. Following determination of protein concentration, 30 μg was heat denatured in a sodium dodecyl sulfate (SDS) denaturing buffer (2× SDS protein gel loading solution; Quality Biological, Inc.) before being loaded into a 4% to 20% Tris-glycine gel (Invitrogen).
Thyroid lysates were prepared from wild-type and TRβPV/PV mice in a manner similar to that described previously (7, 17). Thirty micrograms of lysate was used for the determination of c-Myc, cyclin D1, p-β-catenin (S552), total β-catenin, and the membrane type matrix metalloproteinase (MT1-MMP) by Western blot analysis as described above. The antibodies for c-Myc (1:1,000 dilution), total β-catenin (1:1,000 dilution), and p-β-catenin (S552) (1:2,000 dilution) were purchased from Cell Signaling; that for cyclin D1 (1:200 dilution) was purchased from Santa Cruz Biotech. The concentration of rabbit anti-PDI antibodies used for loading controls was 1 μg/ml.
In vitro and in vivo studies of TR interaction with β-catenin.
Binding of 35S-labeled TRβ or TRβPV to glutathione S-transferase (GST)-β-catenin was carried out as described previously (37). In vitro-translated 35S-labeled TRβ and TRβPV proteins were synthesized by using a TNT kit (Promega, Inc.) and incubated with GST-β-catenin, with or without 1 μM T3, at 4°C for 12 h with constant shaking. The translated 35S-labeled proteins were quantified by the intensities of bands after SDS gel electrophoresis and autoradiography, and equal amounts of 35S-labeled proteins (TRβ, TRβPV, or truncated TRβ proteins) were used in the binding to GST-β-catenin. The beads were washed three times, and the bound proteins were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Coimmunoprecipitation studies were carried out using cytoplasmic and nuclear extracts of FH-TRβ, FH-TRβPV, or FH cells. Cells were cultured for 72 h in Td medium in 15-cm dishes. They were then incubated with or without T3 for 30 min. Cell lysis was performed for 15 min on ice with a buffer containing 10 mM HEPES, 0.1 mM EDTA, 10 mM KCl, 1× protease inhibitor cocktail (Roche), 10 mM dithiothreitol, and 0.6% NP-40, supplemented or not with T3 (1 μM). Lysates were centrifuged at 3,000 rpm for 10 min, and the supernatant containing the cytoplasmic extracts was immediately transferred to a new tube and kept on ice, whereas the pellets containing the nuclear extracts were subsequently incubated with buffer C (20 mM HEPES, 450 mM NaCl, 1 mM EDTA, 1× protease inhibitor cocktail [Roche], 10 mM dithiothreitol), with or without T3 (1 μM), for 15 min on ice. Nuclear fraction samples were vortexed every minute and then centrifuged at 14,000 rpm for 5 min. Nuclear and cytoplasmic extract concentrations were determined by spectrophotometry, and the concentration of each sample was adjusted using lysis buffer. Samples were then diluted three times in buffer D (20 mM HEPES, 1 mM EDTA, 1× protease inhibitor cocktail, 0.1% NP-40), with or without T3 (1 μM). Samples were incubated with rabbit anti-β-catenin (4 μl) (9562; Cell Signaling Technology) or rabbit immunoglobulin G (IgG) (NIO1; Calbiochem) overnight at 4°C with constant rotation and then with 15 μl protein A agarose (Roche) for 3 h at 4°C with constant rotation. Beads were washed three times with BC100 buffer (20 mM HEPES, 1 mM EDTA, 100 mM KCl, 0.1% NP-40), with or without T3 (1 μM). Bound proteins were analyzed by SDS-PAGE.
Nuclear extracts from TRβPV/PV mouse thyroids were prepared using an NE-PER nuclear and cytoplasmic extraction reagent kit (Pierce Biotechnology, Rockford, IL). Briefly, frozen thyroids were ground in liquid nitrogen and homogenized in CER I buffer with a Dounce homogenizer. Immunoprecipitation was performed using 450 μg proteins incubated overnight with 5 μg of a mouse monoclonal anti-TR antibody (302) or mouse IgG (I8765; Sigma) in Tris-buffered saline-0.1% NP-40 with protease inhibitors (Roche) at 4°C. The samples were then mixed with 20 μl protein G-agarose (Roche) at 4°C for 2 h, and beads were washed five times with TBS-0.1% NP-40 containing protease inhibitors. Bound proteins were analyzed by SDS-PAGE, and β-catenin was detected by using a rabbit anti-β-catenin antibody (1:1,000 dilution) (9562; Cell Signaling Technology).
Transient transfection assays.
FH-TRβ, FH-TRβPV, and FH cells were plated at a density of 3 × 105 cells/well in six-well plates and transfected the next day, using Fugene6 (Boehringer Mannheim, Indianapolis, IN) according to the manufacturer's protocol. The reporter plasmids TOP-Flash and FOP-Flash (1 μg) were transfected with pcDNA-hTCF4 (0.1 μg; generously provided by Bert Vogelstein, Howard Hughes Medical Institute, Baltimore, MD) and pcDNA3-β-catenin (0.1 μg; generously provided by Stephen Byers, Georgetown University, Washington, DC). The empty vector pcDNA3.1 (0.2 μg; Invitrogen) was used to supplement an equal amount of DNA. Briefly, 3.6 μl of Fugene6 was mixed with 1.2 μg of DNA (1 μg of reporter with 0.1 μg of pcDNA-hTCF4 and 0.1 μg pcDNA3-β-catenin or 0.2 μg empty vector per well) in 150 μl of OptiMEM1 (Gibco-BRL, Rockville, MD) and added to cells cultured in 2 ml OptiMEM1. After a 7-hour incubation, transfection medium was replaced by fresh DMEM containing 10% Td calf serum. Eighteen hours later, cells were cultured with or without 100 nM T3 for an additional 24 h. Cells were lysed in reporter lysis buffer (3× cell lysis buffer; BD Bioscience Pharmingen). Lysates were assayed for luciferase activity and normalized to total protein concentration.
Determination of c-Myc mRNA expression by real-time RT-PCR.
FH-TRβ, FH-TRβPV, and FH cells were preincubated with Td medium for 24 h and then incubated with T3 (100 nM) for 24 h. Total RNA was isolated from three to six independent samples. To determine the effect of T3 on the expression of c-Myc mRNA, real-time reverse transcription-PCR (RT-PCR) was carried out using a QuantiTect SYBR green RT-PCR kit from Qiagen following the manufacturer's instructions. The primers used to amplify c-Myc mRNA were previously published (26) and were as follows: A227, TGGTGCTCCATGAGGAGAC; and A228, CCACAGAAACAACATCGATTTC. The primers used to amplify the endogenous control gene 18S were as follows: forward, ACCGCAGCTAGGAATAATGGA; and reverse, CAAATGCTTTCGCTCTGGTC. All PCR products were analyzed by agarose gel electrophoresis (2% agarose) followed by ethidium bromide staining to ensure amplification of the appropriately sized products.
Statistical analysis.
All statistical analyses were carried out using StatView 5.0 (SAS Institute, Inc.) as described previously (16). Statistical analysis was performed with the use of analysis of variance, and P values of <0.05 were considered significant. All data are expressed as means ± standard errors of the means.
RESULTS
Aberrant accumulation of β-catenin in thyroid tumors of TRβPV/PV mice.
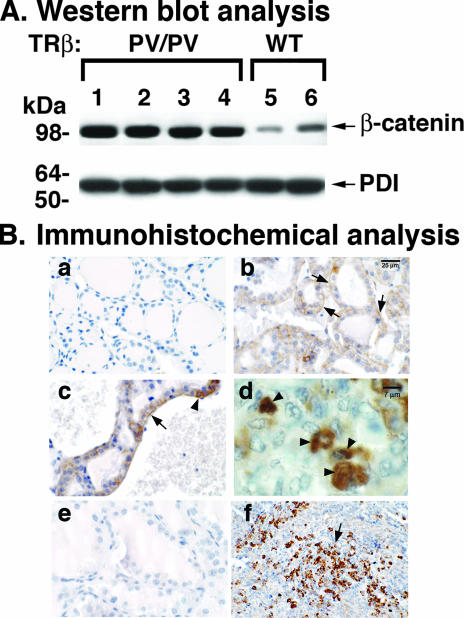
Using cDNA microarray analysis followed by quantitative real-time PCR, we previously found that the expression of β-catenin mRNA in the thyroid is significantly elevated during thyroid carcinogenesis (39). We therefore ascertained whether β-catenin protein levels were altered during tumor progression by Western blot analysis (Fig. 1A). Compared with the case for wild-type mice (Fig. 1A, lanes 5 and 6), lanes 1 to 4 show an approximately fourfold increase in β-catenin protein abundance in thyroid tumors of TRβPV/PV mice. PDI was used as a loading control (Fig. 1A, bottom panel). These results indicate that during thyroid carcinogenesis, β-catenin protein levels are elevated.
FIG. 1.
Aberrantly elevated β-catenin protein in thyroid tumors of TRβPV/PV mice. (A) Thyroid lysates from TRβPV/PV (lanes 1 to 4) and wild-type (lanes 5 and 6) mice aged 8 to 9 months were analyzed by Western blotting for β-catenin protein abundance as described in Materials and Methods. PDI was used as a loading control. Results are representative of three independent experiments. (B) Immunohistochemistry analysis of β-catenin on wild-type (a) or hyperplastic (b to d) thyroids and anaplastic tumors of TRβPV/PV mice (f). No staining could be detected on hyperplastic thyroids (e) when the primary antibody was omitted, indicating the specificity of the signals detected in panels b to d and f.
The increased abundance of β-catenin protein was further confirmed by immunohistochemical analysis. Compared with normal thyroids of wild-type mice (Fig. 1B, panel a), intensive staining of β-catenin protein was observed in the hyperplastic thyroid epithelia (arrows in panels b and c). The major distribution was at the basolateral surface, which is more clearly evident in panel c (shown by an arrow). Higher magnification of thyroid sections showed intense β-catenin staining in the nuclei of epithelial cells (arrowheads in panel d). The signals shown in panels b to d and f were specific because when the primary anti-β-catenin antibodies were omitted, no staining was detected (panel e). Interestingly, in advanced lesions of spindle cell anaplasia, foci of intensive β-catenin labeling were detected (panel f), indicating that the sustained elevation of β-catenin protein is associated with tumor progression.
The increased expression of β-catenin mRNA shown in our previous studies (39) could possibly contribute to the elevated β-catenin protein levels demonstrated in the present study (Fig. 1). However, it is known that β-catenin protein is degraded via the proteasomal pathways, and its degradation is accelerated when it is bound to liganded RXRα or PPARγ2 (21, 31, 36). These observations prompted us to test the hypothesis that TR could play a regulatory role in the stability of β-catenin protein via the proteasomal machinery.
Liganded TRβ regulates the stability of β-catenin.
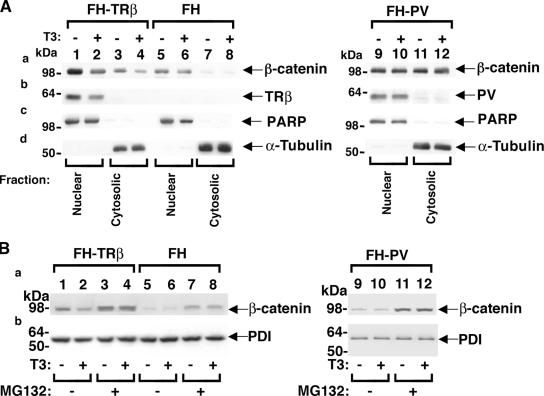
To evaluate whether TRβ plays a role in the stability of β-catenin, we used HeLa cell lines stably expressing FH-TRβ or FH-TRβPV and cells containing only a parental expression vector for controls (FH) (38). Cellular extracts were fractionated into nuclear and cytoplasmic extracts, as indicated by the markers poly(ADP-ribose) polymerase (Fig. 2A, panel c) and α-tubulin, respectively (Fig. 2A, panel d). As shown in Fig. 2A, T3 treatment of FH-TRβ cells decreased protein levels of endogenous β-catenin in the nuclear fraction (Fig. 2A, panel a, compare lane 2 with lane 1) as well as in the cytoplasmic fraction (Fig. 2A, panel a, compare lane 4 with lane 3). The ratio of nuclear to cytoplasmic distribution of β-catenin protein was 4.6 and 3.9 in the presence and absence of T3, respectively, suggesting that T3 facilitates the nuclear translocation of β-catenin. In contrast, in TRβPV-expressing cells (FH-TRβPV), an approximately equal distribution of β-catenin protein between the nuclear (lanes 9 and 10) and the cytoplasmic compartments (lanes 11 and 12) was observed. Furthermore, T3 treatment had no effect on the levels of endogenous β-catenin in the nuclear (Fig. 2A, panel a, compare lane 10 with lane 9) and cytoplasmic (Fig. 2A, panel a, compare lane 12 with lane 11) compartments.
FIG. 2.
Regulation of β-catenin protein levels by liganded TRβ through proteasomal pathways in cells. (A) FH-TRβ (lanes 1 to 4), FH (controls) (lanes 5 to 8), and FH-TRβPV (lanes 9 to 12) cells were cultured with (+) or without (−) T3 for 24 h. Nuclear and cytoplasmic fractions were analyzed by Western blotting for the expression of β-catenin (a), TRβ and TRβPV (b), and the markers for nuclear (PARP) (c) and cytoplasmic (α-tubulin) (d) fractions. (B) FH-TRβ, FH, and FH-TRβPV cells were cultured with (+) or without (−) T3 and with the vehicle control DMSO or the proteasome inhibitor MG132 (25 μM). Whole-cell lysates were analyzed by Western blotting for the expression of β-catenin (a) and the loading control, PDI (b), as described in Materials and Methods. Lanes are as marked.
To determine whether T3-induced down-regulation of β-catenin was mediated via proteasomal degradation pathways, FH-TRβ cells were treated with a proteasome-specific inhibitor, MG132 (Fig. 2B). Compared with untreated cells (Fig. 2B, panel a, lanes 1 and 2), MG132 treatment blocked T3-induced degradation (Fig. 2B, panel a, compare lane 4 with lane 3). In FH cells in which there was no TRβ, MG132 treatment led to T3-independent stabilization of β-catenin, which is consistent with the known degradation of β-catenin by the proteasomal machinery (19). Since TRβPV does not bind T3 (25), inhibition of degradation by MG132 was not affected by T3 (Fig. 2B, panel a, lanes 11 and 12). The loading controls using PDI are shown in Fig. 2B, panel b. These data indicate that liganded TRβ facilitates the proteasome-mediated degradation of β-catenin but that unliganded TR or TRβPV that does not bind T3 does not.
β-Catenin protein physically interacts with TRβ and TRβPV in vitro and in cells.
To understand how liganded TRβ facilitated the degradation of β-catenin, we considered the possibility that the physical interaction of TR with β-catenin could affect the stability of β-catenin. To test this hypothesis, we used GST pulldown assays to assess whether β-catenin interacted with TRβ or TRβPV (Fig. 3A). When increasing amounts of 35S-labeled TRβ (lanes 1 to 3) or 35S-labeled TRβPV (lanes 4 to 6) were incubated with a constant amount of GST-β-catenin, 35S-labeled TRβ bound to GST-β-catenin in a concentration-dependent manner in the absence of T3 (lanes 7 to 9). However, T3 was found to significantly weaken the interaction of TRβ with GST-β-catenin (compare lane 10 with lane 9). TRβPV also bound to GST-β-catenin in a concentration-dependent manner (lanes 11 to 13). Still, at each corresponding input concentration, TRβPV bound with GST-β-catenin more strongly than TRβ did (compare lane 11 with lane 7, lane 12 with lane 8, and lane 13 with lane 9). A quantitative comparison is shown in Fig. 3B. Moreover, T3 had no effect on the interaction of TRβPV with β-catenin, because similar amounts of TRβPV were bound to GST-β-catenin whether T3 was present or not (Fig. 3A, compare lane 13 with lane 14). Lanes 15 and 16 (Fig. 3A) show the negative controls, for which no binding to TRβ or TRβPV was detected, respectively, when GST alone was used.
FIG. 3.
Physical interaction of β-catenin with TRβ or TRβPV in vitro. (A) Increasing concentrations of 35S-labeled TRβ (lanes 7 to 9) or 35S-labeled TRβPV (lanes 11 to 13) were incubated with constant amounts of GST-β-catenin, with T3 (lanes 10 and 14) or without T3 (lanes 7 to 9 and 11 to 13), or with agarose-GST only (lanes 15 and 16), as described in Materials and Methods. The input of 35S-labeled TRβ was identical to that of 35S-labeled TRβPV at each corresponding concentration (lanes 1 and 4, lanes 2 and 5, and lanes 3 and 6). (B) The bands of TRβ or TRβPV bound to GST-β-catenin were scanned, and their intensities were quantified by using ImageJ software (ImageJ 1.34s [http://rsb.info.nih.gov/ij]) and then graphed. (C) Schematic representation of full-length and truncated TRβ proteins, with domains and boundaries indicated. (D) Study of the binding of 35S-labeled full-length and truncated TRβ to GST-β-catenin. Lanes 1 to 5 show the 10% input of full-length and truncated TR proteins, as marked; lanes 6 and 7 show the binding of TRβ and the truncated TRβ protein named ED41 (domains C, D, and E) with GST-β-catenin; and lanes 8 to 10 indicate that MD32 (domains D and E), KD25, and KP24 (domain E) do not interact with GST-β-catenin.
We further mapped the region of TRβ that interacted with β-catenin by using a series of truncated TRβ proteins (Fig. 3C). Deletion of the A/B domain had no effect on the interaction of TRβ with GST-β-catenin (Fig. 3D, compare lane 7 with the full-length TRβ in lane 6). But when the DNA binding domain was deleted, the binding with β-catenin was virtually lost (lanes 8, 9, and 10). No signals were detected when GST alone was used (lanes 11 to 15 are negative controls for lanes 6 to 10, respectively). These results show that the DNA binding domain of TRβ is critical in the interaction with β-catenin.
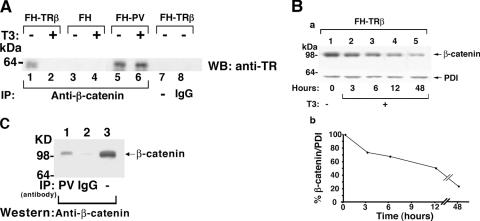
We further demonstrated the interaction of endogenous β-catenin with TRβ or TRβPV in cells by coimmunoprecipitation assays. Nuclear extracts of FH-TRβ, FH-TRβPV, or FH cells were immunoprecipitated with anti-β-catenin antibodies, followed by Western blot analysis using a monoclonal antibody that recognizes the common amino-terminal epitope of TRβ and TRβPV. As shown in Fig. 4A, lanes 1 and 5, in the absence of T3, TRβ and TRβPV were coimmunoprecipitated with endogenous β-catenin, indicating their physical interaction in cells. In FH cells in which no TRβ or TRβPV was expressed, no detectable signals were observed (lane 3). The negative controls—using IgG (lane 8) or no antibodies (lane 7)—further confirmed the specificity of the TRβ and TRβPV detected in lanes 1 and 5. However, due to significantly small amounts of TRβ and TRβPV localized in the cytoplasmic fraction, we could not detect the β-catenin-associated TRβ and TRβPV (data not shown).
FIG. 4.
β-Catenin interacts with TRβ and TRβPV in cells and with TRβPV in the thyroids of TRβPV/PV mice. (A) FH-TRβ, FH-TRβPV, or FH cells (control) were cultured with (+) or without (−) T3 for 30 min, and nuclear extracts were immunoprecipitated using a polyclonal anti-β-catenin antibody (lanes 1 to 6) or rabbit IgG (lane 8). An additional control in which the primary antibody was omitted is shown in lane 7. Analysis was performed by Western blotting using mouse anti-TR antibody (J52) to detect TR. (B) Time-dependent degradation of β-catenin in FH-TRβ cells treated with T3, determined by Western blot analysis (a). The band intensities of β-catenin (upper panel) and PDI (lower panel) were quantified by using ImageJ software (ImageJ 1.34s [http://rsb.info.nih.gov/ij]), and the percentage of β-catenin to PDI protein levels at different time points after T3 treatment is shown in panel b. (C) Thyroid nuclear extracts of TRβPV/PV mice were immunoprecipitated with a specific mouse monoclonal anti-TRβPV antibody (302) (lane 1; TRβPV) or mouse IgG (lane 2; IgG) as described in Materials and Methods. Nuclear extracts (20 μg; lane 3) and immunoprecipitates were analyzed by Western blotting using a rabbit polyclonal anti-β-catenin antibody.
In vitro GST pull-down analysis indicated that T3 weakened the interaction of TRβ with β-catenin (Fig. 3A, compare lane 10 with lane 9). To determine whether this T3-induced dissociation of TRβ-β-catenin complexes also occurred in cells, we carried out coimmunoprecipitation assays in the presence of T3. As indicated in Fig. 2A, T3 induced the degradation of β-catenin. Therefore, whether T3 induced the dissociation of TRβ-β-catenin complexes would have to be determined under the conditions in which the T3-induced degradation had not yet occurred. As further assessed by Western blot analysis (Fig. 4B), T3-induced degradation occurred in a time-dependent manner (Fig. 4B, panel a), and the half-life was determined to be about 12 h (Fig. 4B, panel b).
On the basis of kinetic data shown in Fig. 4B, we carried out coimmunoprecipitation after cells were treated with T3 for 30 min, at which time β-catenin remained relatively stable. Compared with cells without T3 treatment (Fig. 4A, lane 1), no TRβ was detectable under the experimental conditions (Fig. 4A, lane 2), indicating that T3 treatment led to dissociation of the TRβ-β-catenin complexes. However, no T3-induced dissociation of TRβPV from TRβPV-β-catenin complexes was observed (lanes 5 and 6). Consistent with the in vitro GST pull-down assays (Fig. 3A and B), in cells TRβPV interacted more strongly with β-catenin than TRβ did (Fig. 4A, compare lane 5 with lane 1). Furthermore, T3 induced the release of TRβ from the TRβ-β-catenin complexes but did not release TRβPV from TRβPV-β-catenin complexes (Fig. 4A, compare lane 6 with lane 2). These results suggest that β-catenin is protected from proteasome degradation when it is associated with unliganded TRβ or TRβPV in cells.
An interaction between endogenous β-catenin and TRβPV was also demonstrated in vivo in the thyroids of TRβPV/PV mice by coimmunoprecipitation assays. Nuclear thyroid extracts were immunoprecipitated by specific monoclonal anti-TRβPV antibodies, followed by Western blot analysis using anti-β-catenin antibody (Fig. 4C, lane 1). Lane 2 shows the negative control, using an irrelevant mouse IgG antibody in the immunoprecipitation, and lane 3 shows the β-catenin protein marker of the TRβPV/PV thyroid by direct Western blot analysis. Thus, these data indicate that the association of β-catenin and TRβPV occurs not only in exogenously TRβPV-expressing cells but also in tissues.
Liganded TRβ does not activate GSK3β- and Siah-1-regulated APC pathways to mediate β-catenin degradation.
Two APC-dependent proteasomal degradation pathways are known to regulate the cellular levels of β-catenin, namely, a GSK3β-regulated pathway involving the APC-axin complex (27) and a p53-inducible pathway involving Siah-1 (20). GSK3β phosphorylates β-catenin and targets it to ubiquitylation and degradation by the proteasome pathways (1, 10, 12, 13, 29, 30, 35). In the second pathway, p53 up-regulates Siah-1, which interacts with APC, thereby recruiting a ubiquitylation complex to β-catenin and targeting β-catenin for degradation. To evaluate whether T3-induced degradation of β-catenin could be mediated through changes in these mechanisms, we examined the effect of T3 on the cellular levels of several key regulators in these two pathways. As shown in Fig. 5A, panel a, in the absence of T3, the steady-state level of phosphorylated β-catenin in FH-TRβ cells was higher than that in FH-TRβPV cells, while the T3-induced degradation of β-catenin had occurred (Fig. 5A, panel b, lanes 1 and 2). We further determined the ratios of p-β-catenin versus β-catenin levels by quantification of the intensities of the bands. In FH-TRβ cells, the ratio was higher in the presence of T3 than in the absence of T3 (1.55 and 3.64, respectively), suggesting that more β-catenin was targeted for degradation. In FH-TRβPV cells, the ratio of p-β-catenin versus β-catenin levels was not significantly affected by T3 (1.34 and 1.24 in the absence and presence of T3, respectively).
FIG. 5.
T3-induced degradation of β-catenin occurs via APC/GSK3β or APC/p53/Siah-1-independent mechanisms. FH-TRβ (lanes 1 and 2), FH-TRβPV (lanes 3 and 4), or FH control (lanes 5 and 6) cells were treated with (+) or without (−) T3 for 24 h, and whole-cell lysates were analyzed by Western blotting for the expression of components of the classical β-catenin degradation pathways. (A) Expression levels of phosphorylated β-catenin (p-β-catenin) and total β-catenin. (B) Expression levels of phosphorylated GSK3β (p-GSK-3β) and total GSK3β. (C) Expression levels of p53 and Siah-1.
However, we found that the ratio of phosphorylated GSK3β (panel b) to total GSK3β (panel a) did not show any significant alterations upon T3 treatment (Fig. 5B). Figure 5C shows that T3 treatment of FH-TRβ cells increased the cellular abundance of p53 (Fig. 5C, panel a, compare lane 2 with lane 1), but such treatment did not have that effect in either FH-TRβPV (Fig. 5C, panel a, lanes 3 and 4) or FH (Fig. 5C, panel a, lanes 5 and 6) cells. However, despite the T3-induced up-regulated p53, the Siah-1 protein levels were not affected by T3 treatment (Fig. 5C, panel b, lanes 1 and 2) and were similar to those in FH-TRβPV and FH cells (Fig. 5C, panel b, lanes 3 to 6). Figure 5C, panel c, shows the loading controls using PDI. Thus, the cellular levels of these two key regulators in the APC-dependent proteasomal degradation pathways were not affected by the liganded TRβ. These findings suggest that proteasomal degradation of β-catenin per se is not affected by liganded TRβ; instead, more β-catenin is available to be degraded by proteasome degradation once T3 induces its release from the TRβ-β-catenin complexes (see the proposed molecular model in Fig. 9).
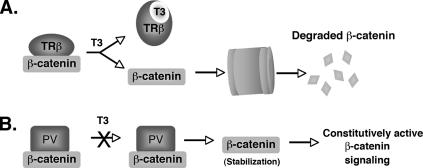
FIG. 9.
Proposed molecular model for the nongenomic action of thyroid hormone receptor in regulating cellular levels of β-catenin. (A) Binding of T3 to TRβ weakens the physical interaction between β-catenin and TRβ and allows the uncomplexed β-catenin to be targeted for degradation via proteasomal pathways. (B) TRβPV also forms complexes with β-catenin, but since this mutant does not bind T3, β-catenin remains associated with TRβPV and is not degraded. This leads to β-catenin stabilization and results in the constitutive activation of the β-catenin signaling pathway.
β-Catenin signaling is repressed by liganded TRβ but not by TRβPV.
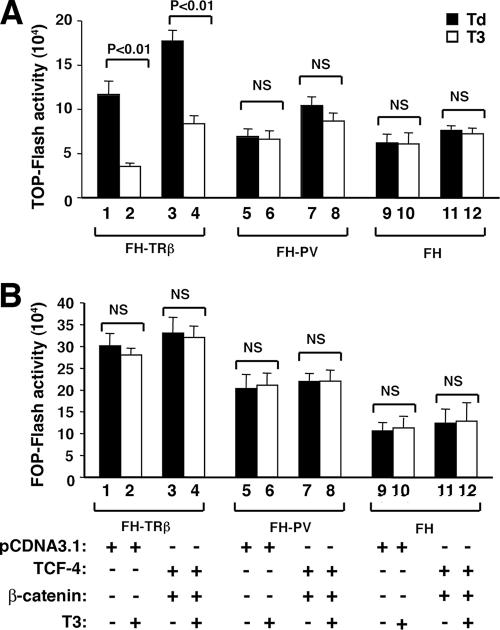
The above findings would predict that in the presence of T3, the β-catenin-mediated transcription activity would be decreased due to a lower cellular level of β-catenin. We tested this possibility by using a luciferase reporter system containing TCF binding sites of the c-Myc promoter (TOP-Flash reporter) or mutated TCF binding sites (FOP-Flash reporter). As shown in Fig. 6A, a 75% reduction in the reporter activity was observed in FH-TRβ cells treated with T3 for 24 h (Fig. 6A, compare bars 1 and 2). In the absence of T3, the exogenously transfected β-catenin further enhanced the TOP-Flash reporter activity (Fig. 6A, bar 3). In the presence of T3, TOP-Flash-mediated activity was also repressed (bar 4). In contrast, no effect of T3 was observed in the TOP-Flash reporter activity mediated by the endogenous β-catenin in FH-TRβPV cells (bars 5 and 6) or in FH cells that had no TRβ (bars 9 and 10). While the enhanced TOP-Flash reporter activity was detected due to exogenously transfected β-catenin (Fig. 6A, compare bar 7 to bar 5), no repression of the TOP-Flash reporter activity was observed in the presence of T3 (compare bar 8 to bar 7). Figure 6B shows the results of negative controls in which a FOP-Flash reporter containing mutated TCF binding sites was used. No detectable reporter activity was detected either in the presence of endogenous β-catenin (Fig. 6B, bars 1 and 2) or in the presence of transfected β-catenin (Fig. 6B, bars 3 and 4), indicating the specificity of the reporter activity shown in Fig. 6A. These results indicate that in cells with T3-bound TRβ, β-catenin-mediated transcription was decreased, whereas in cells expressing TRβPV, no repression of the β-catenin-mediated transcription was detected, resulting in constitutive activation.
FIG. 6.
Analysis of regulation of β-catenin signaling activity by wild-type and mutant TRβ by luciferase reporters. FH-TRβ, FH-TRβPV, and FH cells were transfected with either the β-catenin-TCF4-responsive TOP-Flash luciferase reporter or a FOP-Flash reporter that harbors mutations in the TCF4 DNA binding sequences, along with the expression vectors pcDNA-hTCF-4 (TCF4) and pcDNA3-β-catenin or the empty vector pcDNA3.1, as indicated in Materials and Methods. Results of three independent experiments for TOP-Flash activity (A) and FOP-Flash activity (B) are shown. Bars are means ± SEM. NS, not significant.
We further determined the expression of the endogenous c-Myc gene, known to be regulated by β-catenin (11). c-Myc is a master regulator of genes involved in diverse cellular processes, such as cell apoptosis and proliferation. In FH-TRβ cells, the expression of the c-Myc mRNA was repressed ∼50% in the presence of T3 (Fig. 7A, compare bar 2 with bar 1). In contrast, no changes in the expression of c-Myc mRNA levels by T3 were observed in FH-TRβPV or FH cells (Fig. 7A, bar 4 versus bar 3 and bar 6 versus bar 5). Consistent with reduced expression of mRNA, an approximately 70% reduction in cellular protein levels of c-Myc was detected in T3-treated FH-TRβ cells (Fig. 7B, panel a, compare lane 2 with lane 1). In contrast, T3 had no effects on the cellular levels of c-Myc in FH-TRβPV cells (lanes 3 and 4) or in FH cells that had no TRβ (lanes 5 and 6). Figure 7B, panel b, shows the loading controls using PDI.
FIG. 7.
Repression of the c-Myc oncogene, a downstream target of the β-catenin signaling pathway, by T3-bound TRβ but not by TRβPV. (A) Total RNA was prepared from FH-TRβ (bars 1 and 2), FH-TRβPV (bars 3 and 4), and FH (bar 5 and 6) cells cultured with (+) or without (−) T3 for 24 h and analyzed by real-time RT-PCR for c-Myc mRNA levels as described in Materials and Methods. (B) Whole-cell lysates from FH-TRβ cells (lanes 1 and 2), FH-TRβPV cells (lanes 3 and 4), or FH cells (control; lanes 5 and 6) cultured with (+) or without (−) T3 for 24 h were analyzed by Western blotting for c-Myc and PDI protein levels. In panel A, bars show means ± SEM for three to six samples. NS, not significant.
Aberrant accumulation of β-catenin in the thyroids of TRβPV/PV mice leads to constitutive β-catenin signaling.
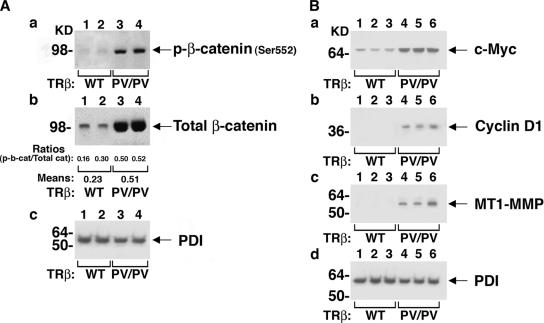
Phosphorylation of β-catenin at serine residue 552 (p-β-catenin-Ser552) by AKT has been reported to increase β-catenin transcriptional activity (4). To determine whether the accumulation of β-catenin observed in the thyroids of TRβPV/PV mice was associated with increased levels of p-β-catenin-S552, thereby contributing to constitutive β-catenin signaling, Western blotting was performed using thyroid lysates from wild-type and TRβPV/PV mice. p-β-Catenin-Ser552 protein levels were dramatically increased in TRβPV/PV thyroids compared with those in wild-type thyroids (Fig. 8A, panel a, compare lanes 1 and 2 with lanes 3 and lane 4), along with the increase in total β-catenin protein levels (Fig. 8A, panel b, compare lanes 1 and 2 with lanes 3 and 4). The ratios of p-β-catenin-Ser552 protein and total β-catenin were determined by quantification of band intensities. The increased ratios in the thyroid extracts of TRβPV/PV mice indicate an increased phosphorylation of p-β-catenin-Ser552. Panel c shows the PDI loading control for the thyroid extracts in corresponding lanes.
FIG. 8.
Constitutively active β-catenin signaling in TRβPV/PV thyroids. Thyroid extracts of wild-type and TRβPV/PV mice were analyzed by Western blotting for protein abundance of several markers of β-catenin signaling activation. (A) Blots of β-catenin phosphorylated at serine residue 552 (p-β-catenin-S552) (a), total β-catenin (b), and the loading control PDI (c). (B) Protein levels of c-Myc (a), cyclin D1 (b), MT1-MMP (c), and PDI (d) in the thyroid extracts of wild-type and TRβPV/PV mice. Lanes are marked.
The expression levels of several β-catenin signaling downstream targets were also evaluated. c-Myc protein levels were dramatically increased in the thyroids of TRβPV/PV mice compared with those in wild-type mice (Fig. 8B, panel a, compare lanes 1 to 3 with lanes 4 to 6), consistent with our cell-based findings. Similarly, the protein levels of cyclin D1, which is a critical cell cycle regulator (5), were increased in the mutant thyroids (Fig. 8B, panel b, compare lanes 1 to 3 with lanes 4 to 6). Our study of MT1-MMP, which is associated with tumor invasion (34), showed a marked elevation of its protein levels in the thyroids of TRβPV/PV mice compared with those in wild-type mice (Fig. 8B, panel c, compare lanes 1 to 3 with lanes 4 to 6). Panel d shows the PDI loading control for the thyroid extracts in the corresponding lanes. Altogether, our data suggest that β-catenin signaling is constitutively activated in the thyroids of TRβPV/PV mice and thereby contributes to the development of follicular thyroid carcinoma.
DISCUSSION
β-Catenin is the central mediator of the Wnt signaling pathway, which is critical for various cellular processes, including oncogenesis (28). The cellular level of β-catenin is tightly controlled by two APC-dependent proteasomal pathways, including a GSK3β-regulated pathway and a p53-Siah-1 pathway (27). Recently, however, APC-independent pathways were discovered to participate in the regulation of the cellular level of β-catenin. These newly identified pathways are mediated via physical interaction of β-catenin with PPARγ2 or RXRα, and these ligand receptor-β-catenin complexes are degraded together through proteasomal pathways (21, 31, 36). These observations prompted the question of whether this mode of regulation of the cellular level of β-catenin is general to all members of the nuclear receptor family or limited to just a few. The findings that the β-catenin cellular level was highly elevated in the thyroid tumors of TRβPV/PV mice provided an opportunity to address this question in vivo. We found that, indeed, similar to PPARγ2 or RXRα, TRβ and its mutant TRβPV physically interacted with β-catenin in vitro and in cells. However, in contrast to PPARγ2 or RXRα, binding of T3 to TRβ weakened such an interaction that would allow more uncomplexed β-catenin to be degraded via the proteasomal pathway (Fig. 9A). In contrast, the interaction of β-catenin with TRβPV is not affected by T3, because TRβPV does not bind T3, and therefore the constitutive association of TRβPV with β-catenin would lead to the stabilization of β-catenin (Fig. 9B). Thus, the present study has uncovered a novel nongenomic mechanism by which TR could regulate the cellular stability of β-catenin.
The present study shows that there is a commonality in the early steps of the APC-independent regulation of β-catenin mediated by TRβ and by PPARγ2 and RXRα. Each of these nuclear receptors first physically interacts with β-catenin. However, the effect of ligand binding distinguishes the subsequent mode of actions between TRβ and PPARγ2 or RXRα. The present study shows that the DNA binding domain of TRβ is important in its interaction with β-catenin. In contrast, the A/B domain of RXRα (36) and the ligand binding domain of PPARγ2 (21) were reported to be necessary in mediating the effective degradation of β-catenin. These different regions of TRβ, RXRα, and PPARγ2 that are critical in the interaction with β-catenin could differently affect the avidity of the interaction of liganded receptors with β-catenin. It is well known that ligand binding to TRβ, RXRα, and PPARγ induces significant structural alterations compared with binding to aporeceptors (14). It is entirely possible that the liganded TRβ assumes a conformation that is not favorable in its interaction with β-catenin, whereas the liganded RXRα and PPARγ2 adopt a structure that strengthens their interaction with β-catenin. By doing so, the effect of ligand binding would dictate the receptor specificity in the APC-independent regulation of cellular β-catenin to affect Wnt signaling. It would be important to study other nuclear receptors to determine whether they could also mediate the APC-independent regulation of β-catenin. If they do, it would be of interest to assess which mode of regulation they belong to (TRβ mode, RXR mode, or another, novel mode of regulation).
The present study uncovered a novel action of TRβ, i.e., to cross talk with Wnt/β-catenin signaling via regulating the cellular level of β-catenin. Indeed, we have shown that c-Myc, one of the downstream target genes, was repressed in the presence of T3 at both the mRNA and protein levels (Fig. 7). These findings suggest that other Wnt/β-catenin-regulated downstream target genes could also be modulated similarly to maintain normal cellular functions. In cells in which TRβ is mutated such that T3 binding is lost, this regulatory pathway would be perturbed by sustained activation of β-catenin-regulated downstream target genes due to stabilization of β-catenin (Fig. 9B). This notion is supported by the findings that, similar to the case for human cancers (3, 32), thyroid carcinogenesis is associated with elevation of cellular β-catenin in TRβPV/PV mice (Fig. 1). One of the Wnt/β-catenin downstream target genes, cyclin D1, is highly activated and has been shown in our previous studies to contribute to thyroid carcinogenesis (39). In addition to cyclin D1, the present study also found that c-Myc and MT1-MMP were activated in the thyroid tumors of TRβPV/PV mice (Fig. 8B). Future studies would be required to identify other target genes that are affected by the cross talk of TR with Wnt/β-catenin signaling to ascertain its effects on cellular functions and in disease.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print on 12 May 2008.
REFERENCES
- 1.Behrens, J., B. A. Jerchow, M. Wurtele, J. Grimm, C. Asbrand, R. Wirtz, M. Kuhl, D. Wedlich, and W. Birchmeier. 1998. Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science 280596-599. [DOI] [PubMed] [Google Scholar]
- 2.Cheng, S. Y., S. Hasumura, M. C. Willingham, and I. Pastan. 1986. Purification and characterization of a membrane-associated 3,3′,5-triiodo-l-thyronine binding protein from a human carcinoma cell line. Proc. Natl. Acad. Sci. USA 83947-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chesire, D. R., and W. B. Isaacs. 2003. Beta-catenin signaling in prostate cancer: an early perspective. Endocr. Relat. Cancer 10537-560. [DOI] [PubMed] [Google Scholar]
- 4.Fang, D., D. Hawke, Y. Zheng, Y. Xia, J. Meisenhelder, H. Nika, G. B. Mills, R. Kobayashi, T. Hunter, and Z. Lu. 2007. Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. J. Biol. Chem. 28211221-11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu, M., C. Wang, Z. Li, T. Sakamaki, and R. G. Pestell. 2004. Cyclin D1: normal and abnormal functions. Endocrinology 1455439-5447. [DOI] [PubMed] [Google Scholar]
- 6.Fukuchi, T., M. Sakamoto, H. Tsuda, K. Maruyama, S. Nozawa, and S. Hirohashi. 1998. Beta-catenin mutation in carcinoma of the uterine endometrium. Cancer Res. 583526-3528. [PubMed] [Google Scholar]
- 7.Furumoto, H., H. Ying, G. V. Chandramouli, L. Zhao, R. L. Walker, P. S. Meltzer, M. C. Willingham, and S. Y. Cheng. 2005. An unliganded thyroid hormone beta receptor activates the cyclin D1/cyclin-dependent kinase/retinoblastoma/E2F pathway and induces pituitary tumorigenesis. Mol. Cell. Biol. 25124-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Rostan, G., G. Tallini, A. Herrero, T. G. D'Aquila, M. L. Carcangiu, and D. L. Rimm. 1999. Frequent mutation and nuclear localization of beta-catenin in anaplastic thyroid carcinoma. Cancer Res. 591811-1815. [PubMed] [Google Scholar]
- 9.Gottardi, C. J., and B. M. Gumbiner. 2001. Adhesion signaling: how beta-catenin interacts with its partners. Curr. Biol. 11R792-R794. [DOI] [PubMed] [Google Scholar]
- 10.Hart, M. J., R. de los Santos, I. N. Albert, B. Rubinfeld, and P. Polakis. 1998. Downregulation of beta-catenin by human axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr. Biol. 8573-581. [DOI] [PubMed] [Google Scholar]
- 11.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-Myc as a target of the APC pathway. Science 2811509-1512. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda, S., S. Kishida, H. Yamamoto, H. Murai, S. Koyama, and A. Kikuchi. 1998. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 171371-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh, K., V. E. Krupnik, and S. Y. Sokol. 1998. Axis determination in Xenopus involves biochemical interactions of axin, glycogen synthase kinase 3 and beta-catenin. Curr. Biol. 8591-594. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, B. A., E. M. Wilson, Y. Li, D. E. Moller, R. G. Smith, and G. Zhou. 2000. Ligand-induced stabilization of PPARgamma monitored by NMR spectroscopy: implications for nuclear receptor activation. J. Mol. Biol. 298187-194. [DOI] [PubMed] [Google Scholar]
- 15.Kaneshige, M., K. Kaneshige, X. Zhu, A. Dace, L. Garrett, T. A. Carter, R. Kazlauskaite, D. G. Pankratz, A. Wynshaw-Boris, S. Refetoff, B. Weintraub, M. C. Willingham, C. Barlow, and S. Cheng. 2000. Mice with a targeted mutation in the thyroid hormone beta receptor gene exhibit impaired growth and resistance to thyroid hormone. Proc. Natl. Acad. Sci. USA 9713209-13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato, Y., H. Ying, L. Zhao, F. Furuya, O. Araki, M. C. Willingham, and S. Y. Cheng. 2006. PPARgamma insufficiency promotes follicular thyroid carcinogenesis via activation of the nuclear factor-kappaB signaling pathway. Oncogene 252736-2747. [DOI] [PubMed] [Google Scholar]
- 17.Kim, C. S., H. Ying, M. C. Willingham, and S. Y. Cheng. 2007. The pituitary tumor-transforming gene promotes angiogenesis in a mouse model of follicular thyroid cancer. Carcinogenesis 28932-939. [DOI] [PubMed] [Google Scholar]
- 18.Lin, K. H., M. C. Willingham, C. M. Liang, and S. Y. Cheng. 1991. Intracellular distribution of the endogenous and transfected beta form of thyroid hormone nuclear receptor visualized by the use of domain-specific monoclonal antibodies. Endocrinology 1282601-2609. [DOI] [PubMed] [Google Scholar]
- 19.Liu, C., Y. Li, M. Semenov, C. Han, G. H. Baeg, Y. Tan, Z. Zhang, X. Lin, and X. He. 2002. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108837-847. [DOI] [PubMed] [Google Scholar]
- 20.Liu, J., J. Stevens, C. A. Rote, H. J. Yost, Y. Hu, K. L. Neufeld, R. L. White, and N. Matsunami. 2001. Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol. Cell 7927-936. [DOI] [PubMed] [Google Scholar]
- 21.Liu, J., H. Wang, Y. Zuo, and S. R. Farmer. 2006. Functional interaction between peroxisome proliferator-activated receptor gamma and beta-catenin. Mol. Cell. Biol. 265827-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyoshi, Y., K. Iwao, Y. Nagasawa, T. Aihara, Y. Sasaki, S. Imaoka, M. Murata, T. Shimano, and Y. Nakamura. 1998. Activation of the beta-catenin gene in primary hepatocellular carcinomas by somatic alterations involving exon 3. Cancer Res. 582524-2527. [PubMed] [Google Scholar]
- 23.Moon, R. T., B. Bowerman, M. Boutros, and N. Perrimon. 2002. The promise and perils of Wnt signaling through beta-catenin. Science 2961644-1646. [DOI] [PubMed] [Google Scholar]
- 24.Ougolkov, A. V., K. Yamashita, M. Mai, and T. Minamoto. 2002. Oncogenic beta-catenin and MMP-7 (matrilysin) cosegregate in late-stage clinical colon cancer. Gastroenterology 12260-71. [DOI] [PubMed] [Google Scholar]
- 25.Parrilla, R., A. J. Mixson, J. A. McPherson, J. H. McClaskey, and B. D. Weintraub. 1991. Characterization of seven novel mutations of the c-erbA beta gene in unrelated kindreds with generalized thyroid hormone resistance. Evidence for two “hot spot” regions of the ligand binding domain. J. Clin. Investig. 882123-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peter, M., C. Rosty, J. Couturier, F. Radvanyi, H. Teshima, and X. Sastre-Garau. 2006. Myc activation associated with the integration of HPV DNA at the Myc locus in genital tumors. Oncogene 255985-5993. [DOI] [PubMed] [Google Scholar]
- 27.Polakis, P. 2002. Casein kinase 1: a Wnt'er of disconnect. Curr. Biol. 12R499-R501. [DOI] [PubMed] [Google Scholar]
- 28.Polakis, P. 2007. The many ways of Wnt in cancer. Curr. Opin. Genet. Dev. 1745-51. [DOI] [PubMed] [Google Scholar]
- 29.Sakanaka, C., J. B. Weiss, and L. T. Williams. 1998. Bridging of beta-catenin and glycogen synthase kinase-3beta by axin and inhibition of beta-catenin-mediated transcription. Proc. Natl. Acad. Sci. USA 953020-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salic, A., E. Lee, L. Mayer, and M. W. Kirschner. 2000. Control of beta-catenin stability: reconstitution of the cytoplasmic steps of the wnt pathway in Xenopus egg extracts. Mol. Cell 5523-532. [DOI] [PubMed] [Google Scholar]
- 31.Sharma, C., A. Pradeep, L. Wong, A. Rana, and B. Rana. 2004. Peroxisome proliferator-activated receptor gamma activation can regulate beta-catenin levels via a proteasome-mediated and adenomatous polyposis coli-independent pathway. J. Biol. Chem. 27935583-35594. [DOI] [PubMed] [Google Scholar]
- 32.Sparks, A. B., P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 581130-1134. [PubMed] [Google Scholar]
- 33.Suzuki, H., M. C. Willingham, and S. Y. Cheng. 2002. Mice with a mutation in the thyroid hormone receptor beta gene spontaneously develop thyroid carcinoma: a mouse model of thyroid carcinogenesis. Thyroid 12963-969. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi, M., T. Tsunoda, M. Seiki, Y. Nakamura, and Y. Furukawa. 2002. Identification of membrane-type matrix metalloproteinase-1 as a target of the beta-catenin/Tcf4 complex in human colorectal cancers. Oncogene 215861-5867. [DOI] [PubMed] [Google Scholar]
- 35.Wu, G., and X. He. 2006. Threonine 41 in beta-catenin serves as a key phosphorylation relay residue in beta-catenin degradation. Biochemistry 455319-5323. [DOI] [PubMed] [Google Scholar]
- 36.Xiao, J. H., C. Ghosn, C. Hinchman, C. Forbes, J. Wang, N. Snider, A. Cordrey, Y. Zhao, and R. A. Chandraratna. 2003. Adenomatous polyposis coli (APC)-independent regulation of beta-catenin degradation via a retinoid X receptor-mediated pathway. J. Biol. Chem. 27829954-29962. [DOI] [PubMed] [Google Scholar]
- 37.Yap, N., C. L. Yu, and S. Y. Cheng. 1996. Modulation of the transcriptional activity of thyroid hormone receptors by the tumor suppressor p53. Proc. Natl. Acad. Sci. USA 934273-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ying, H., F. Furuya, L. Zhao, O. Araki, B. L. West, J. A. Hanover, M. C. Willingham, and S. Y. Cheng. 2006. Aberrant accumulation of PTTG1 induced by a mutated thyroid hormone beta receptor inhibits mitotic progression. J. Clin. Investig. 1162972-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ying, H., H. Suzuki, H. Furumoto, R. Walker, P. Meltzer, M. C. Willingham, and S. Y. Cheng. 2003. Alterations in genomic profiles during tumor progression in a mouse model of follicular thyroid carcinoma. Carcinogenesis 241467-1479. [DOI] [PubMed] [Google Scholar]
- 40.Ying, H., H. Suzuki, L. Zhao, M. C. Willingham, P. Meltzer, and S. Y. Cheng. 2003. Mutant thyroid hormone receptor beta represses the expression and transcriptional activity of peroxisome proliferator-activated receptor gamma during thyroid carcinogenesis. Cancer Res. 635274-5280. [PubMed] [Google Scholar]