Abstract
APLF is a novel protein of unknown function that accumulates at sites of chromosomal DNA strand breakage via forkhead-associated (FHA) domain-mediated interactions with XRCC1 and XRCC4. APLF can also accumulate at sites of chromosomal DNA strand breaks independently of the FHA domain via an unidentified mechanism that requires a highly conserved C-terminal tandem zinc finger domain. Here, we show that the zinc finger domain binds tightly to poly(ADP-ribose), a polymeric posttranslational modification synthesized transiently at sites of chromosomal damage to accelerate DNA strand break repair reactions. Protein poly(ADP-ribosyl)ation is tightly regulated and defects in either its synthesis or degradation slow global rates of chromosomal single-strand break repair. Interestingly, APLF negatively affects poly(ADP-ribosyl)ation in vitro, and this activity is dependent on its capacity to bind the polymer. In addition, transient overexpression in human A549 cells of full-length APLF or a C-terminal fragment encoding the tandem zinc finger domain greatly suppresses the appearance of poly(ADP-ribose), in a zinc finger-dependent manner. We conclude that APLF can accumulate at sites of chromosomal damage via zinc finger-mediated binding to poly(ADP-ribose) and is a novel component of poly(ADP-ribose) signaling in mammalian cells.
The rapid repair of chromosomal DNA single- and double-strand breaks is critical for genome integrity, and defects in this process result in a variety of hereditary genetic diseases (21). Recently, we and others identified the human protein APLF (aka C2orf13, PALF, and XIP1) as a novel component of the DNA single-strand break repair (SSBR) and double-strand break repair (DSBR) machinery (4, 14, 15, 19). The amino terminus of APLF contains a highly conserved forkhead-associated (FHA) domain that mediates interaction with the SSBR and DSBR factors XRCC1 and XRCC4, respectively. In addition, APLF interacts with Ku80 in an FHA domain-independent manner. The C terminus of APLF contains a second highly conserved region that encodes two tandem zinc fingers (designated ZNF1 and ZNF2) and a highly acidic tail. Both the FHA domain and the tandem ZNFs can facilitate, by independent mechanisms, the accumulation of APLF at sites of DNA strand breakage (4, 14, 15). Whereas the FHA domain facilitates APLF accumulation via interaction with CK2-phosphorylated XRCC1, the mechanism by which the ZNFs achieve this is unclear.
The ZNFs in APLF most closely resemble the tandem zinc fingers present in tristetraprolin. Tristetraprolin binds specific mRNA species, with each of two ZNFs targeting a separate 5′-UAUU-3′ subsite located within a larger, AU-rich recognition sequence (5, 18). Although APLF does not bind this mRNA species (unpublished observations), it is possible that the APLF ZNFs might interact with some other type of adenine-rich structure. One such structure that arises during DNA strand break repair is poly(ADP-ribose) (pADPr), a branched nucleic acid-like polymer synthesized rapidly at DNA strand breaks by pADPr polymerase-1 (PARP-1) and linked covalently to specific protein acceptors to signal the presence of chromosome damage (reviewed in references 6 and 17). Protein poly(ADP-ribosyl)ation can affect protein function in a variety of ways, such as by regulating the enzymatic activity of its protein acceptor and/or by facilitating ionic interactions with other proteins. The major targets of poly(ADP-ribosyl)ation at chromosomal DNA strand breaks are histone H1, histone H2B, and PARP-1 itself. The poly(ADP-ribosyl)ation of these proteins at chromosomal DNA breaks appears to regulate chromatin structure and compaction and, also, the accumulation of DNA repair protein complexes at sites of chromosome damage (30-32). Importantly, both pADPr synthesis and its subsequent catabolism by pADPr glycohydrolase (PARG) (reviewed in reference 11) are critical for rapid rates of chromosomal SSBR, indicating that the control of pADPr levels at sites of DNA strand breakage is a dynamic and regulated process (10, 12). Here, we show that APLF binds tightly to pADPr via its tandem zinc finger domain and that this binding can suppress protein poly(ADP-ribosyl)ation in vitro and in cells. These data provide a mechanism for the zinc finger-dependent recruitment of APLF to chromosome damage and suggest that APLF is a novel component of pADPr signaling/metabolism in mammalian cells.
MATERIALS AND METHODS
Expression constructs.
pET16b-APLF encoding full-length His-APLF has been described previously (14). pET16b encoding His-APLF360-511, His-APLF360-511zfm1 (harboring a mutated ZNF1 domain), and APLF360-511 zfm2 (harboring a mutated ZNF2 domain) were created by subcloning the 0.4-kb SphI/BamHI fragment from the construct pEYFP-C1-APLF, pEYFP-C1-APLFzfm1, or pEYFP-C1-APLFzfm2 (14), respectively, into NdeI (blunted)/BamHI sites of pET16b. The mutations introduced into the ZNF1 and ZNF2 domains were C379A/C385A (ZNF1) and C421A/C427A (ZNF2). pET16b-APLF1-469 was created by subcloning an NdeI/XbaI (blunted) restriction fragment from pET16b-APLF into the NdeI/BamHI (blunted) sites of pET16b. The small interfering RNA targeting-resistant (TR) His-Myc-APLF expression construct pcD2E-His-Myc-APLFTR was generated by site-directed mutagenesis (Quickchange; Stratagene) of pcD2E-His-Myc-APLF (14) using appropriate oligonucleotides (5′-GAAAAGAAGAAATCTGTAAGGACAAATCCCAGCTAAAC-3′ and 5′-GTTTAGCTGGGATTTGTCCTTACAGATTTCTTCTTTTC-3′) to introduce three silent mutations (underlined). The targeting-resistant open reading frame was then subcloned from pcD2E-His-Myc-APLFTR into the EcoR1 sites of pCI-puro, creating the puromycin-selectable construct pCI-puro-His-Myc-APLFTR. C379/C385 and C421/C427 were each then changed to alanine in pCI-puro-His-Myc-APLFTR by site-directed mutagenesis, creating targeting-resistant expression constructs for His-Myc-APLFzfm1 and His-Myc-APLFzfm2, respectively. A targeting-resistant expression construct for His-Myc-APLF1-469 was generated by subcloning a 1.4-kb EcoRI/XbaI restriction fragment from pCI-puro-His-Myc-APLFTR into the EcoRI/XbaI sites of pCI-puro. pCI-puro-APLF360-511 was generated by subcloning an NcoI (blunted)/BamHI (blunted) fragment from pET16b-APLF360-511 (14) into the NheI (blunted) site of pCI-puro. The truncation in APLF360-511 renders this protein targeting resistant.
Proteins.
Histidine-tagged XRCC1 and APLF proteins were expressed from pET16b constructs in Escherichia coli BL21(DE3) cells and purified by immobilized-metal chelate chromatography and, for some experiments, gel filtration. Recombinant human PARP-1 and PARG were purchased from Trevigen or, where indicated in the case of PARP-1, expressed in insect cells (13) and purified on 3-aminobenzamide-conjugated ECH Sepharose 4B as previously described (The PARP Link Web resource [http://parplink.u-strasbg.fr/protocols/tools/parp_purification.html]). Calf thymus histone H1 (mixed isoforms) was from Sigma, and recombinant human histone H1.2 was from Axxora. Recombinant human histone H2B was from New England Biolabs.
A549 transfection.
A549 cell lines stably transfected with pSUPER-APLF, designated A549-APLFKD (14), were maintained in minimal essential medium supplemented with 15% fetal calf serum, 1.5 mg/ml G418, 2 mM l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin. For subsequent transient transfections, A549-APLFKD cells were seeded onto coverslips (1 × 104 per cm2) and, 48 h later, transfected with the pCI-puro construct encoding the APLF proteins indicated in Fig. 5 using GeneJuice (Novagen) according to the manufacturer's instructions. Twenty-four hours after transfection, the transfected cells were treated with H2O2 and examined by immunofluorescence as described below.
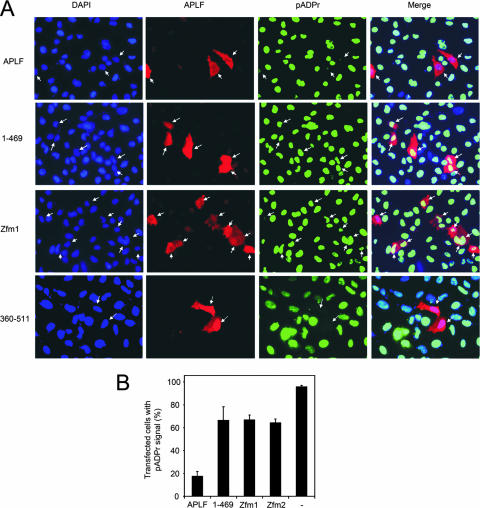
FIG. 5.
Suppression of pADPr signaling by APLF overexpression. (A) A549-APLFKD cells were transiently transfected with expression constructs encoding targeting-resistant His-Myc-APLF, His-Myc-APLF1-469, His-Myc-APLFzfm1, and APLF360-511, and 48 h later, treated with H2O2 (20 min on ice) followed by 1 min in drug-free medium. Cells were then immunostained with anti-APLF (red) and anti-pADPr (green) antibodies and analyzed by immunofluorescence microscopy. Transfected cells expressing detectable levels of recombinant APLF are highlighted by white arrows. Cell nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI; blue). Images are representative of multiple experiments and were captured at ×40 magnification. (B) The fraction of A549-APLFKD cells with detectable pADPr signal was quantified from the APLF-expressing (red) subset of transiently transfected cells in panel A or from total populations of untransfected cells (−). Data are the means (± 1 standard error of the mean) of the results from three independent experiments.
Immunofluorescence.
For laser-induced UVA damage, wild-type A549 cells or A549-PARP-1KD cells stably depleted of PARP-1 (10) were seeded onto gridded coverslips (MatTek). After preincubation for 10 min with 10 μg/ml Hoechst dye 33258 at 37°C, selected cells were irradiated by using a 351-nm UVA laser (approximately 0.35 J/m2). Where indicated (see Fig. 1A), cells were preincubated with the PARP inhibitor KU58948 (500 nM). After exposure, coverslips were incubated at 37°C for 4 min and washed in ice-cold phosphate-buffered saline (PBS). For H2O2 treatment, cells were treated with 10 mM H2O2 in PBS on ice for 20 min, rinsed in PBS, and incubated for 1 min in drug-free medium at 37°C. Following DNA damage and repair incubation, all cells were rinsed in ice-cold PBS and fixed in PBS-4% paraformaldehyde (5 min at room temperature), followed by methanol (20 min at −20°C). Cells were then permeabilized with PBS-1% Triton X-100 (5 min), blocked in PBS-5% nonfat dried milk (NFDM) for 30 min, and incubated in 1% NFDM-PBS-TS (0.1% Tween-20, 0.02% sodium dodecyl sulfate [SDS] in PBS) containing anti-pADPr monoclonal antibody (MAb) (10H; Alexis) and anti-APLF polyclonal antibody (SK3595 [14]) at 1/200 dilution for 2 h at room temperature. After being rinsed (three times) with PBS-TS, cells were incubated in Alexa Fluor 488 goat anti-mouse immunoglobulin G and Alexa Fluor 555 anti-rabbit immunoglobulin G (Invitrogen) at 1/200 dilution in 1% NFDM-PBS-TS for 1 h at room temperature. Cells were then rinsed in PBS-TS (five times) and mounted in Vectashield (Vector Laboratories).
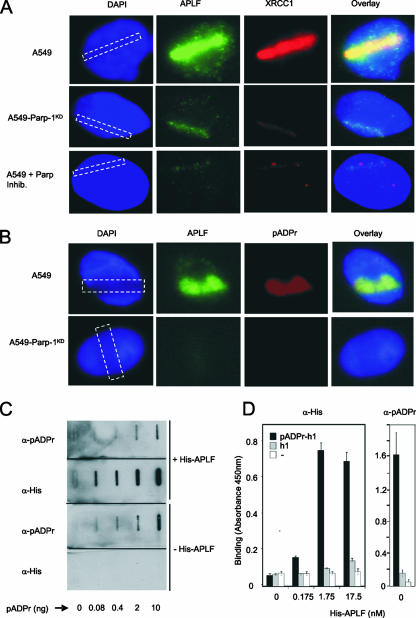
FIG. 1.
APLF colocalizes with and binds pADPr. (A) Human A549 cells, A549 cells stably depleted of PARP-1 (A549-PARP-1KD) (10), or A549 cells incubated with PARP inhibitor (A549 + Parp Inhib.) were damaged by UVA laser in the indicated area (dashed rectangle) and, 4 min later, fixed and immunostained with anti-APLF or anti-XRCC1 MAb. Nuclear DNA was counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). Representative images are shown. (B) Human A549 cells or A549 cells stably depleted of PARP-1 (A549-PARP-1KD) (10) were damaged by UVA laser, and 4 min later, fixed and immunostained with anti-APLF and anti-pADPr MAb. Other details are as described for panel A. (C) The indicated amounts of pADPr were slot blotted, and the filters incubated in the presence or absence of His-APLF (88 nM). Washed filters were then incubated with anti-His-tag or anti-pADPr MAb antibodies as indicated. (D) Multiwell dishes coated with calf thymus histone H1 (mixed isoforms) were mock ribosylated (h1 and −) or poly(ADP-ribosyl)ated (pADPr-h1) and, after being washed, quantified for levels of immobilized pADPr (right) or incubated with the indicated amounts of recombinant full-length His-APLF for 30 min on ice (left). Immobilized His-APLF or pADPr was detected with anti-His-tag (left) or anti-pADPr primary antibody (right), followed by anti-HRP-conjugated secondary antibody, and quantified colorimetrically (450 nm). Where indicated (−), histone-coated wells were pretreated with trypsin to remove all protein prior to poly(ADP-ribosyl)ation. Error bars show standard errors of the mean. α, anti; +, present; −, absent.
Slot blot analysis of His-APLF binding to free pADPr and poly(ADP-ribosyl)ated proteins in vitro.
Different amounts of pADPr (Trevigen) were slot blotted onto Hybond-N+ (Amersham Bioscience) under vacuum and cross-linked with UV. For the analysis of poly(ADP-ribosyl)ated proteins, 1 μg of each of the proteins indicated in Fig. 2A was slot blotted onto Hybond C-extra membrane (Amersham Bioscience) and blocked in 5% NFDM, and filter-bound proteins mock ribosylated or poly(ADP-ribosyl)ated with 4 U/ml recombinant human PARP-1 (Trevigen) in PARP buffer (25 mM Tris, pH 8, 10 mM MgCl2, 100 mM NaCl, 0.5 mM dithiothreitol), 340 μM NAD+ (absent from mock ribosylation reactions), and 4 μg/ml of activated DNA. Membranes were blocked in binding buffer (PBS plus 0.05% Tween 20) containing 5% NFDM for 30 min, washed, and incubated in binding buffer in the presence or absence of 88 nM His-APLF for 30 min at room temperature. APLF and pADPr were detected with anti-His tag (Sigma) and anti-pADPr (10H) MAb, respectively.
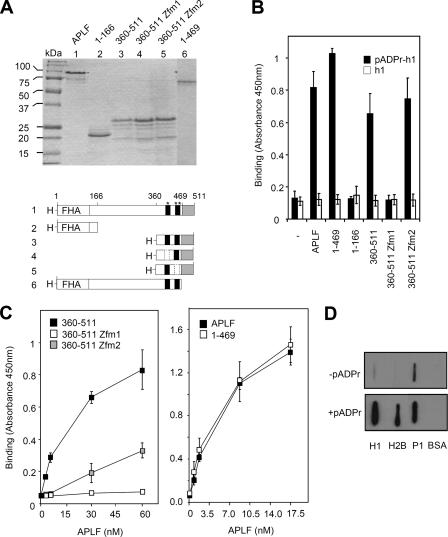
FIG. 2.
The tandem zinc finger domain of APLF binds poly(ADP-ribosyl)ated proteins in vitro. (A) Recombinant human His-APLF proteins. One microgram of recombinant His-APLF and the indicated mutated derivatives were fractionated by SDS-PAGE and stained with Coomassie blue. A schematic of the recombinant His-APLF proteins is shown (bottom). “H-” and “FHA” denote the decahistidine tag and FHA domain, respectively. The C-terminal acidic tail (gray box) and tandem ZNF domains (black boxes: *, ZNF1; **, ZNF2) are also indicated. Dotted boxes denote a mutated ZNF. Amino acid positions are indicated at the top. (B) Multiwell dishes coated with calf thymus histone H1 were poly(ADP-ribosyl)ated (pADPr-h1) or not (h1) as described above and incubated in the absence (−) or presence of 17.5 nM of the indicated His-APLF protein for 30 min on ice. Bound His-APLF was quantified as described above. (C) Multiwell dishes coated with calf thymus histone H1 were poly(ADP-ribosyl)ated and incubated with the indicated concentration of the indicated His-APLF protein for 30 min at 37°C. Bound His-APLF was quantified as described above. (D) Two micrograms of BSA, calf thymus histone H1, recombinant human histone H2B, or recombinant human PARP-1 was slot blotted onto nitrocellulose and mock treated (−pADPr) or poly(ADP-ribosyl)ated (+pADPr) with recombinant human PARP-1 (P1). Filters were incubated with 35 nM His-APLF, and after extensive washing, filter-bound APLF was detected with anti-His-Tag MAb and appropriate secondary antibody. Error bars show standard errors of the mean.
Colorimetric analysis of His-APLF binding to poly(ADP-ribosyl)ated proteins.
Multiwell dishes coated with unfractionated calf thymus histone H1 (Trevigen) were either mock treated or poly(ADP-ribosyl)ated with recombinant PARP enzyme as recommended by the manufacturer (Trevigen). After being rinsed (four times with PBS-0.1% Triton X-100), wells were blocked in PBS-0.05% Tween 20-5% bovine serum albumin (BSA) for 30 min on ice and rinsed four times with PBS-0.05% Tween 20. The wells were then either incubated or not as indicated with wild-type or mutant APLF protein for 30 min on ice (unless otherwise indicated). His-APLF was detected with anti-His-tag MAb (Sigma) (30 min on ice) followed by anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (30 min on ice). pADPr was detected with anti-pADPr MAb (10H), followed by the secondary antibody described above. Rinsed immunocomplexes were detected colorimetrically by incubation with the HRP substrate TACS-Sapphire (Trevigen) as directed by the manufacturer.
Polyacrylamide gel electrophoresis (PAGE) analysis of His-APLF binding and PARG activity on pADPr-histone H1.2 and pADPr-PARP-1.
Flat-bottomed, 96-well polystyrene plates (Greiner) were incubated with 0.1 mg/ml recombinant human H1.2 (Axxora) or recombinant human PARP-1 (Trevigen) in PBS overnight at 4°C. Bound histone was then poly(ADP-ribosyl)ated in the presence of 4 μg/ml activated DNA (Trevigen), 4 U/ml recombinant human PARP-1 (Trevigen), and 100 nM 32P-NAD+ (GE Healthcare) for 1 h at room temperature. The wells containing pADPr-H1.2 were then rinsed and incubated in the absence or presence of the indicated amounts of APLF (see Fig. 3) in 20 mM potassium phosphate (pH 7.2), 50 mM KCl, 0.1 mg/ml BSA, 0.1% Triton X-100 (PARG buffer; Trevigen) at 30°C or 15 min. The wells were then washed and incubated with 10 ng/ml human PARG (Trevigen) for 15 min, after which released material was collected. Released and residually bound materials were incubated with 2.5 mg/ml trypsin for 30 min at room temperature to cleave the ester linkage between pADPr and protein acceptor, and equivalent aliquots of each fraction separated on polyacrylamide sequencing gels (20% in Tris-borate-EDTA) and detected by phosphorimager.
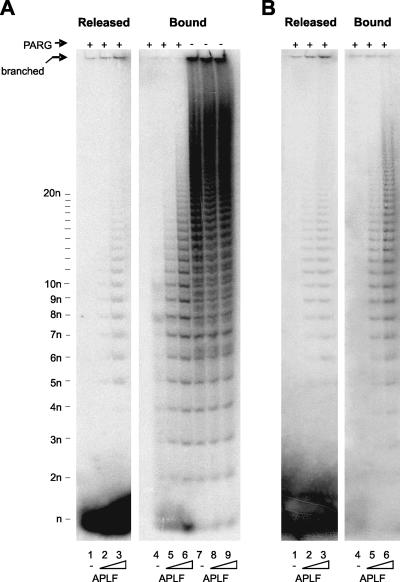
FIG. 3.
APLF binding protects pADPr from degradation by PARG. (A) Multiwell dishes coated with recombinant human histone H1.2 were poly(ADP-ribosyl)ated in the presence of 32P-NAD+ and incubated with 0 nM (lanes 1, 4, and 7), 1.75 nM (lanes 2, 5, and 8), or 17.5 nM (lanes 3, 6, and 9) of His-APLF for 15 min. The histone H1.2-coated dishes were then incubated in the presence (+) or absence (−) of PARG protein for 15 min. Released and bound materials were analyzed by native PAGE and detected by phosphorimager. The position of mADPr (“n”), pADPr of two or more ADPr units (2n, 3n, and 4n, etc.), and branched pADPr molecules remaining at the origin (16, 27) are indicated. (B) Multiwell dishes were coated with recombinant human PARP-1 (purified from insect cells). Other details are as described for panel (A).
Analysis of His-APLF activity on ribosylated histone H1.2.
Histone-coated polystyrene plates were prepared as described above. The histones were ribosylated for 30 min at room temperature in the presence of 68 μM cold NAD+, 40 nM 32P-NAD+, 2 μg/ml activated DNA, and 4 U/ml recombinant human PARP-1. After being rinsed, the ribosylated histones were incubated in 2 μg/ml recombinant His-APLF or mutated His-APLF at 30°C for the times indicated in the figures, after which the released material was collected. After trypsination of the bound fraction, bound and released materials were quantified by phosphorimaging.
Analysis of PARP-1 autoribosylation.
All reactions were carried out in PARP buffer containing 20 U/ml (0.6 μg/ml) recombinant human PARP-1 in the presence of 68 μM cold NAD+, 40 nM 32P-NAD+, and 4 μg/ml activated DNA. The His-APLF protein indicated in the figures (88 nM), His-XRCC1 (88 nM), or 5 μg/ml BSA (negative control) was added to the reaction mixture. After incubation at 30°C for the indicated time, the reaction was stopped by 1 h incubation on ice in acid loading buffer (30 mM phosphoric acid-Tris, pH 6, 4.5 M urea, 0.2% SDS, 5 mM dithiothreitol, 0.1% bromophenol blue) to stabilize the pADPr-protein ester link. Samples were then subjected to acid PAGE analysis (7.5% in 30 mM phosphoric acid-Tris, pH 6, 0.1% SDS) and quantification by phosphorimaging.
RESULTS
Yellow fluorescent protein- or green fluorescent protein-tagged APLF can accumulate at sites of chromosomal breakage by a mechanism that is dependent on a highly conserved C-terminal tandem zinc finger motif (4, 14, 15). Given the similarity between the tandem zinc finger domain of APLF and that of tristetraprolin, which binds an adenine-rich RNA motif, we examined the relationship between endogenous APLF and sites of pADPr synthesis following laser-induced UVA damage. Both APLF and its interacting protein XRCC1 rapidly accumulated at sites of UVA damage in human A549 cells and did so in a manner that was dependent on the presence and activity of PARP-1 (Fig. 1A). Moreover, the sites of UVA laser damage and APLF accumulation were associated with high levels of PARP-1-dependent pADPr synthesis (Fig. 1B). These data confirm that endogenous APLF accumulates at sites of pADPr synthesis, in a PARP-1 dependent manner, and are consistent with a functional relationship between APLF and pADPr.
Next, we employed slot blot analyses to determine whether APLF interacts with pADPr. Full-length recombinant human His-APLF bound to nitrocellulose filter in a manner that was dependent on the amount of slot-blotted polymer (Fig. 1C). Because cellular pADPr is a protein posttranslational modification, we also examined whether His-APLF interacted with poly(ADP-ribosyl)ated proteins. Indeed, His-APLF bound much more efficiently to dishes coated with poly(ADP-ribosyl)-ated histone H1 (pADPr-H1) than to unmodified histone H1-coated dishes (Fig. 1D). To identify the region(s) of APLF responsible for pADPr binding, we employed a number of mutated derivatives lacking or mutated in specific domains (Fig. 2A). A C-terminal fragment of His-APLF (His-APLF360-511) containing the highly conserved tandem ZNFs and acidic tail bound efficiently to pADPr-H1, but an amino-terminal fragment encoding the FHA domain (His-APLF1-166) did not (Fig. 2B). Whereas deletion of the acidic C-terminal tail (His-APLF1-469) did not affect binding to pADPr-H1, mutation of the ZNF1 domain (His-APLF360-511zfm1) greatly reduced or ablated it (Fig. 2B). Mutation of ZNF2 (His-APLF360-511zfm2) did not reduce pADPr-H1 binding under the same conditions, but did reduce binding by approximately half at physiological temperature (Fig. 2C, left panel). Binding by His-APLF360-511zfm2 may thus be temperature sensitive. His-APLF also bound poly(ADP-ribosyl)ated histone H2B and PARP-1, indicating that binding is not restricted to poly(ADP-ribosyl)ated histone H1 (Fig. 2D). We conclude that APLF binds to pADPr and to poly(ADP-ribosyl)ated proteins and that this activity is mediated by the tandem ZNFs.
To examine His-APLF binding to pADPr-H1 in more detail, we examined the impact of this protein on the sensitivity of pADPr to PARG, the cellular enzyme that catabolizes pADPr (7, 33). For this purpose, we employed multiwell dishes coated with [32P]poly(ADP-ribosyl)ated recombinant human histone H1.2 (pADPr-H1.2). Incubation with PARG released almost all pADPr from histone H1 as mono(ADP-ribose) (mADPr) within 30 min, consistent with the robust exoglycosidic activity of this protein (Fig. 3A, lanes 1 and 4). In contrast, preincubation of pADPr-H1.2 with 17.5 nM His-APLF resulted in the appearance of a PARG-resistant “footprint” of ∼1 to 20 ADPr units in the residual histone H1-bound material (Fig. 3A, lane 5). This suggests that His-APLF binds tightly to pADPr-H1.2, close to the junction between pADPr and histone. Similar results were observed with 10-fold-higher concentrations of His-APLF, although in this case the footprint was larger and thus extended further from the polymer-histone junction (Fig. 3A, lane 6). A small amount of protected pADPr was also present in the released material (see Fig. 3A, lanes 2 and 3), which contains polymer released “spontaneously” due to the intrinsic instability of some of the ester bonds linking pADPr to the protein acceptor (22-24, 26). Similar results were observed for poly(ADP-ribosyl)ated PARP-1, suggesting that His-APLF also binds tightly to pADPr-PARP-1 (Fig. 3B).
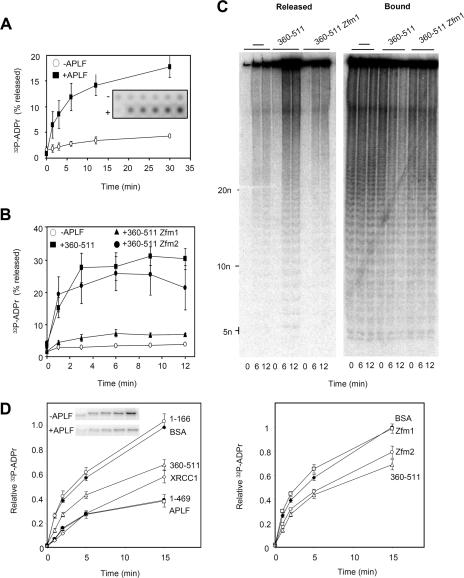
The ability to bind tightly to pADPr provides a mechanism for the ZNF-dependent accumulation of APLF into subnuclear foci at sites of chromosomal DNA strand breakage (4, 14, 15). However, we wondered whether interaction with pADPr may also impact on pADPr metabolism, especially since we noticed in experiments similar to those described above, but in described above which we omitted PARG, that incubation with His-APLF increased the amount of pADPr released from histone H1.2 (data not shown). Quantitative analysis revealed that both His-APLF and His-APLF360-511 increased pADPr release ∼threefold above the amount released spontaneously and that this was dependent on an intact ZNF1 domain (Fig. 4A and B). Separation of the reaction products on a sequencing gel revealed that the pADPr released by APLF was comprised of branched and linear polymers of all sizes, similar to that released “spontaneously,” suggesting that APLF binding decreased the stability of some of the labile ester linkages between pADPr and histone H1.2 (Fig. 4C). In contrast to histone H1.2, His-APLF did not measurably promote the release of pADPr from autoribosylated PARP-1 (data not shown).
FIG. 4.
Impact of APLF on pADPr metabolism in vitro. (A) Release of [32P]pADPr from ribosylated histone H1.2 by APLF. Multiwell dishes coated with recombinant human histone H1.2 were poly(ADP-ribosyl)ated in the presence of NAD+ that was spiked with 32P-NAD+ and then washed and incubated with His-APLF (+APLF) or buffer alone (−APLF) for the times indicated at 30°C. Released material and bound material were detected by phosphorimager, and the released material plotted as a percentage of total 32P signal (released plus bound) in each reaction mixture. Inset shows a representative phosphorimage. (B) Release of [32P]pADPr from ribosylated histone H1.2 by the C-terminal domain of APLF. Ribosylated histone H1.2 was incubated as described above with buffer alone, His-APLF360-511, His-APLF360-511zfm1, or His-APLF360-511zfm2 for the times indicated. Other details are as described for panel A. (C) Analysis by native PAGE of material generated in one of the experiment whose results are shown in panel B. −, buffer alone. (D) The results in the left panel show the impact of APLF on PARP-1 autoribosylation. PARP-1 autoribosylation was conducted in the presence of NAD+ that was spiked with 32P-NAD+, and 32P-labeled PARP-1 was quantified on acid PAGE gels by phosphorimager. Reaction mixtures included BSA, His-APLF, His-APLF360-511, His-APLF1-166, His-APLF1-469, or His-XRCC1. A representative phosphorimage showing PARP-1 ribosylation is included (inset). Data are plotted relative to the signal obtained after 15 min of incubation in the presence of BSA (negative control). The results in the right panel show the impact of the C-terminal domain of APLF on PARP-1 autoribosylation. Reactions were conducted as described for the left panel, and the reaction mixtures included BSA, His-APLF360-511, His-APLF360-511zfm1, or His-APLF360-511zfm2. Error bars show standard errors of the mean.
We also considered the possibility that APLF binding might affect pADPr synthesis, since negative regulation of pADPr synthesis by PARP-1 has been reported for XRCC1, which also binds to pADPr (20, 29). Notably, full-length His-APLF slowed the rate of PARP-1 autoribosylation in vitro to an extent similar to that observed with the protein XRCC1 (Fig. 4D, left panel). The His-APLF360-511 protein, containing the highly conserved tandem ZNFs and acidic tail, also slowed the rate of PARP-1 autoribosylation, whereas His-APLF1-166, encoding just the FHA domain, did not. As observed for pADPr binding, deletion of the acidic C-terminal tail (His-APLF1-469) did not affect the impact of APLF on pADPr synthesis (Fig. 4D, left panel), whereas mutation of the ZNF1 domain (His-APLF360-511zfm1) greatly reduced or ablated it (Fig. 4D, right panel). Mutation of the ZNF2 domain, which under the conditions employed does not measurably affect pADPr binding, had little or no impact on the ability of APLF to inhibit pADPr synthesis. We thus conclude that APLF binding can affect the rate of pADPr synthesis in vitro.
We next examined whether His-APLF might affect cellular pADPr metabolism by examining the effect of transiently expressing targeting-resistant derivatives of His-Myc-APLF protein in A549 cells stably depleted of APLF (A549-APLFKD) (14). Surprisingly, transient overexpression of targeting-resistant full-length protein (His-Myc-APLFTR) in A549-APLFKD cells greatly reduced or ablated the appearance of pADPr, even at very early times (1 min) after H2O2 treatment (Fig. 5A, top panels, and B). To examine whether the suppression of pADPr signaling by APLF involved polymer binding, we examined whether the ZNFs were sufficient and/or required for this effect. Indeed, the overexpression of APLF360-511, encoding just the C-terminal tandem ZNFs and acidic tail, was sufficient for the suppression of pADPr following H2O2 treatment (Fig. 5A, bottom panels, and B). Moreover, the overexpression of His-Myc-APLF harboring a mutant of either ZNF1 (“zfm1”) or ZNF2 (“zfm2”) failed to suppress pADPr efficiently, confirming the importance of the tandem ZNF domain for this activity (Fig. 5A and B). We noted that deletion of the acidic C-terminal tail (APLF1-469), which is dispensable for pADPr binding in vitro, also suppressed the effect of APLF on pADPr accumulation after H2O2 treatment, suggesting that while polymer binding is required for this purpose, it is not sufficient (Fig. 5A and B).
In summary, we show that APLF binds tightly to pADPr via its tandem zinc finger domain and that this binding can suppress protein poly(ADP-ribosyl)ation in vitro and in cells. These data provide a mechanism for the zinc finger-dependent recruitment of APLF to chromosome damage and suggest that APLF is a novel component of pADPr signaling/metabolism in mammalian cells.
DISCUSSION
APLF is comprised of highly conserved amino- and carboxyl-terminal domains separated by a relatively nonconserved central region and was recently identified as a novel component of DNA SSBR and DSBR (4, 14, 15, 19). The amino-terminal domain encodes an FHA domain that interacts with CK2-phosphorylated XRCC1 and XRCC4 and facilitates the accumulation of APLF at sites of chromosomal DNA damage in an XRCC1-dependent manner. In addition, the accumulation of APLF can occur independently of the FHA domain via a mechanism that utilizes the C-terminal tandem zinc finger domain. The finding reported here that the tandem ZNFs interact with pADPr provides a likely mechanism for the ZNF-dependent accumulation of APLF at chromosome breaks. Consistent with this idea, mutation of the ZNF1 domain ablated both pADPr binding and APLF accumulation, whereas mutation of the ZNF2 domain allowed residual levels of both (Fig. 2C) (13). These data are in agreement with the results in a recent report suggesting that the ability to bind free pADPr polymer is a common function of the type of zinc finger present in APLF and in several other proteins (3). In the current experiments, APLF also bound polymer that was conjugated to histone H1, histone H2B, and PARP-1 (Fig. 2D). This is important because cellular pADPr arises at sites of chromosomal damage largely as a posttranslational modification of these proteins rather than as free polymer.
It is possible that pADPr binding by APLF fulfils another role in addition to targeting this protein to chromosomal DNA strand breaks. Consistent with this idea, APLF overexpression in APLF-depleted A549 cells reduced the steady-state level of pADPr at early times following H2O2 treatment, in a zinc finger-dependent manner, raising the possibility that APLF might play a role in regulating cellular pADPr levels. Interestingly, the highly conserved acidic C-terminal tail was also required for the ability of APLF to suppress cellular pADPr, despite the dispensability of this domain for pADPr binding. Consequently, while pADPr binding by APLF is most likely required for the suppression of cellular pADPr, it is unlikely to be sufficient for this activity. APLF promoted the release of up to 30% of pADPr from histone H1.2 in vitro (Fig. 4A), suggesting that polymer binding may destabilize some of the labile ester linkages that connect pADPr to histone (1, 2, 24, 26). The limited release of pADPr exhibited by His-APLF is reminiscent of the activity of mADPr lyase, an activity that cleaves a subset of ester bonds linking mADPr to protein (24, 26). However, we failed to detect the release of significant amounts of mADPr by APLF (unpublished observations). The limited activity of APLF in vitro, and particularly the absence of activity on PARP-1, which is the major poly(ADP-ribosyl)ated protein in cells, suggests that pADPr release is unlikely to account for the impact of APLF on cellular pADPr levels, at least by itself. Another contributing factor may be the ability of APLF to inhibit pADPr synthesis by PARP-1 that was observed in this study. This activity was similarly dependent on an intact ZNF1 domain, suggesting that it too requires polymer binding. It is possible that tight binding by APLF to nascent pADPr chains affects access to the polymer terminus by PARP-1, thereby negatively regulating polymer extension.
It remains to be determined whether or not regulating the cellular levels of pADPr is a physiological role of APLF. However, such a role would be an important component of chromosomal DNA strand break repair, because pADPr levels are highly dynamic following DNA breakage and greatly affect the accumulation and/or stability of DNA repair protein complexes at chromosomal DNA strand breaks (9, 10, 25). This is illustrated by the observation that depletion of PARP-1 and/or PARG reduces global rates of SSBR to a similar extent (10). In addition, the poly(ADP-ribosyl)ation of histones can relax higher-order chromatin folding and facilitate histone shuttling (8, 28, 30, 31), suggesting that the proper regulation of pADPr levels may be critical for controlling chromatin structure during cellular SSBR reactions.
Footnotes
Published ahead of print on 12 May 2008.
REFERENCES
- 1.Adamietz, P., R. Brodehorst, and H. Hilz. 1978. ADP-ribosylated histone H1 from HeLa cultures. Eur. J. Biochem. 92317-326. [DOI] [PubMed] [Google Scholar]
- 2.Adamietz, P., and H. Hilz. 1976. Poly(adenosine diphosphate ribose) is covalently linked to nuclear proteins by two types of bonds. Hoppe-Seylers Z. Physiol. Chem. 357527-534. [DOI] [PubMed] [Google Scholar]
- 3.Ahel, I., D. Ahel, T. Matsusaka, A. J. Clark, J. Pines, S. J. Boulton, and S. C. West. 2008. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 45181-85. [DOI] [PubMed] [Google Scholar]
- 4.Bekker-Jensen, S., K. Fugger, J. R. Danielsen, I. Gromova, M. Sehested, J. Celis, J. Bartek, J. Lukas, and N. Mailand. 2007. Human Xip1 (C2orf13) is a novel regulator of cellular responses to DNA strand breaks. J. Biol. Chem. 28219638-19643. [DOI] [PubMed] [Google Scholar]
- 5.Carballo, E., W. S. Lai, and P. J. Blackshear. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 2811001-1005. [DOI] [PubMed] [Google Scholar]
- 6.D'Amours, D., S. Desnoyers, I. D'Silva, and G. G. Poirier. 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342249-268. [PMC free article] [PubMed] [Google Scholar]
- 7.Davidovic, L., M. Vodenicharov, E. B. Affar, and G. G. Poirier. 2001. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp. Cell Res. 2687-13. [DOI] [PubMed] [Google Scholar]
- 8.de Murcia, G., A. Huletsky, D. Lamarre, A. Gaudreau, J. Pouyet, M. Daune, and G. G. Poirier. 1986. Modulation of chromatin superstructure induced by poly(ADP-ribose) synthesis and degradation. J. Biol. Chem. 2617011-7017. [PubMed] [Google Scholar]
- 9.El-Khamisy, S. F., M. Masutani, H. Suzuki, and K. W. Caldecott. 2003. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 315526-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher, A. E. O., H. Hochegger, S. Takeda, and K. W. Caldecott. 2007. Poly (ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell. Biol. 275597-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagne, J. P., M. J. Hendzel, A. Droit, and G. G. Poirier. 2006. The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr. Opin. Cell Biol. 18145-151. [DOI] [PubMed] [Google Scholar]
- 12.Gao, H., D. L. Coyle, M. L. Meyer-Ficca, R. G. Meyer, E. L. Jacobson, Z. Q. Wang, and M. K. Jacobson. 2007. Altered poly(ADP-ribose) metabolism impairs cellular responses to genotoxic stress in a hypomorphic mutant of poly(ADP-ribose) glycohydrolase. Exp. Cell Res. 313984-996. [DOI] [PubMed] [Google Scholar]
- 13.Giner, H., F. Simonin, G. de Murcia, and J. Menissier-de Murcia. 1992. Overproduction and large-scale purification of the human poly(ADP-ribose) polymerase using a baculovirus expression system. Gene 114279-283. [DOI] [PubMed] [Google Scholar]
- 14.Iles, N., S. Rulten, S. F. El-Khamisy, and K. W. Caldecott. 2007. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell. Biol. 273793-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanno, S., H. Kuzuoka, S. Sasao, Z. Hong, L. Lan, S. Nakajima, and A. Yasui. 2007. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J. 262094-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiehlbauch, C. C., N. Aboul-Ela, E. L. Jacobson, D. P. Ringer, and M. K. Jacobson. 1993. High resolution fractionation and characterization of ADP-ribose polymers. Anal. Biochem. 20826-34. [DOI] [PubMed] [Google Scholar]
- 17.Kim, M. Y., T. Zhang, and W. L. Kraus. 2005. Poly(ADP-ribosyl)ation by PARP-1: “PAR-laying” NAD+ into a nuclear signal. Genes Dev. 191951-1967. [DOI] [PubMed] [Google Scholar]
- 18.Lai, W. S., E. Carballo, J. M. Thorn, E. A. Kennington, and P. J. Blackshear. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J. Biol. Chem. 27517827-17837. [DOI] [PubMed] [Google Scholar]
- 19.Macrae, C. J., R. D. McCulloch, J. Ylanko, D. Durocher, and C. A. Koch. 2008. APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair (Amsterdam) 7292-302. [DOI] [PubMed] [Google Scholar]
- 20.Masson, M., C. Niedergang, V. Schreiber, S. Muller, J. Menissier-de Murcia, and G. de Murcia. 1998. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 183563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinnon, P. J., and K. W. Caldecott. 2007. DNA strand break repair and human genetic disease. Annu. Rev. Genomics Hum. Genet. 837-55. [DOI] [PubMed] [Google Scholar]
- 22.Ogata, N., K. Ueda, and O. Hayaishi. 1980. ADP-ribosylation of histone H2B. Identification of glutamic acid residue 2 as the modification site. J. Biol. Chem. 2557610-7615. [PubMed] [Google Scholar]
- 23.Ogata, N., K. Ueda, H. Kagamiyama, and O. Hayaishi. 1980. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J. Biol. Chem. 2557616-7620. [PubMed] [Google Scholar]
- 24.Oka, J., K. Ueda, O. Hayaishi, H. Komura, and K. Nakanishi. 1984. ADP-ribosyl protein lyase. Purification, properties, and identification of the product. J. Biol. Chem. 259986-995. [PubMed] [Google Scholar]
- 25.Okano, S., L. Lan, K. W. Caldecott, T. Mori, and A. Yasui. 2003. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 233974-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okayama, H., M. Honda, and O. Hayaishi. 1978. Novel enzyme from rat liver that cleaves an ADP-ribosyl histone linkage. Proc. Natl. Acad. Sci. USA 752254-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panzeter, P. L., and F. R. Althaus. 1990. High resolution size analysis of ADP-ribose polymers using modified DNA sequencing gels. Nucleic Acids Res. 182194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panzeter, P. L., C. A. Realini, and F. R. Althaus. 1992. Noncovalent interactions of poly(adenosine diphosphate ribose) with histones. Biochemistry 311379-1385. [DOI] [PubMed] [Google Scholar]
- 29.Pleschke, J. M., H. E. Kleczkowska, M. Strohm, and F. R. Althaus. 2000. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 27540974-40980. [DOI] [PubMed] [Google Scholar]
- 30.Poirier, G. G., G. de Murcia, J. Jongstra-Bilen, C. Niedergang, and P. Mandel. 1982. Poly(ADP-ribosy)lation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl. Acad. Sci. USA 793423-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Realini, C. A., and F. R. Althaus. 1992. Histone shuttling by poly(ADP-ribosylation). J. Biol. Chem. 26718858-18865. [PubMed] [Google Scholar]
- 32.Rouleau, M., R. A. Aubin, and G. G. Poirier. 2004. Poly(ADP-ribosyl)ated chromatin domains: access granted. J. Cell Sci. 117815-825. [DOI] [PubMed] [Google Scholar]
- 33.Ueda, K., J. Oka, S. Naruniya, N. Miyakawa, and O. Hayaishi. 1972. Poly ADP-ribose glycohydrolase from rat liver nuclei, a novel enzyme degrading the polymer. Biochem. Biophys. Res. Commun. 46516-523. [DOI] [PubMed] [Google Scholar]