Abstract
RASSF1A is a tumor suppressor protein involved in death receptor-dependent apoptosis utilizing the Bax-interacting protein MOAP-1 (previously referred to as MAP-1). However, the dynamics of death receptor recruitment of RASSF1A and MOAP-1 are still not understood. We have now detailed recruitment to death receptors (tumor necrosis factor receptor 1 [TNF-R1] and TRAIL-R1/DR4) and identified domains of RASSF1A and MOAP-1 that are required for death receptor interaction. Upon TNF-α stimulation, the C-terminal region of MOAP-1 associated with the death domain of TNF-R1; subsequently, RASSF1A was recruited to MOAP-1/TNF-R1 complexes. Prior to recruitment to TNF-R1/MOAP-1 complexes, RASSF1A homodimerization was lost. RASSF1A associated with the TNF-R1/MOAP-1 or TRAIL-R1/MOAP-1 complex via its N-terminal cysteine-rich (C1) domain containing a potential zinc finger binding motif. Importantly, TNF-R1 association domains on both MOAP-1 and RASSF1A were essential for death receptor-dependent apoptosis. The association of RASSF1A and MOAP-1 with death receptors involves an ordered recruitment to receptor complexes to promote cell death and inhibit tumor formation.
Allelic loss within the short arm of human chromosome 3 is an early event that occurs frequently in numerous human cancers (12, 48). One gene of interest in this region is RASSF1 (Ras association domain family protein 1) (11, 48). The RASSF1 locus encodes two major splice variants (A and C) that predominantly characterize this gene family (12). The longer RASSF1A isoform consists of 340 amino acids and a unique 119-amino-acid amino-terminal (N-terminal) region encoded by exon 1α. Methylation of the promoter for exon 1α of RASSF1A occurs without epigenetic loss of the other RASSF1 isoforms, suggesting that RASSF1A serves an important in vivo function. There are nine related RASSF genes with different chromosomal locations and uncertain in vivo function (12, 22, 48). Similar to RASSF1A, the expression of RASSF2 and RASSF4 was lost in most lung tumor cell lines (51) and, recently, RASSF6 was found to be downregulated in 30 to 60% of tumor-derived tissues of several primary tumors (such as those of the breast, kidney, and liver) (1). RASSF7 was demonstrated to be localized to mitotic spindles and centrosomes and to be an important component of neural tube mitosis in Xenopus (41). All RASSF proteins contain a Ras binding domain (RBD) within their primary sequence, but direct association with Ras (mainly K-Ras) has been observed only for RASSF2, RASSF4, and RASSF5 (also known as Nore1 [novel ras effector 1]; with two isoforms, Nore1A or Nore1B [also known as RapL, regulator for cell adhesion and polarization enriched in lymphoid tissues]) (16, 25). K-Ras-associated RASSF6 has also been reported and was found to augment cell death in 293 T cells (1), but not in HeLa cells (22). An association of RASSF1A with K-Ras has also been reported but is considered to be weak and indirect through RASSF5/Nore1A (52).
We and others have demonstrated that RASSF1A is a cytoskeletal protein that colocalizes with microtubules. Song et al. demonstrated a role for RASSF1A in mitosis dependent upon its localization on microtubules (42). In addition, Liu et al. identified RASSF1A protein complexes in the mitochondria and in the nucleus (30). It has been suggested that the cytoskeletal localization of RASSF1A may play an important role in regulating mitotic stability and ensure that abnormal cells do not arise. However, Rassf1a−/− mice revealed no differences in cell cycle profiles or in the proliferative rate of fibroblasts or splenocytes, nor were there any differences in mitotic instability (47, 49). Rassf1a−/− mice do, however, show increased tumor incidence in the breast, lung, gastrointestinal tract, and immune system (mainly B-cell-related lymphomas) and have decreased death receptor-dependent apoptosis (2). Taken together, these data lend support to the tumor suppressor function for RASSF1A and for its role in apoptosis.
Apoptosis is essential for normal development, maintenance of cellular homeostasis, and tumor survival (3, 7). Two types of signaling pathways promote apoptosis by utilizing the mitochondrion, the central organelle involved in cell death. The “intrinsic” pathway is activated by noxious factors, such as DNA damage. In contrast, specific death receptors (e.g., tumor necrosis factor receptor 1 [TNF-R1], TNF-α-related apoptosis-inducing ligand receptor [TRAIL-R1 or DR4], or Fas [CD95]) stimulate the “extrinsic” pathway (13, 46, 53). Surface activation results in death receptor trimerization, movement into lipid rafts, and formation of receptor complexes stimulating apoptosis, such as the death-inducing signaling complex (DISC) (46, 53). DISC assembly and the subsequent activation of initiator caspases (mainly caspase-8) convey signals to the mitochondria to promote the release of small apoptogenic factors (such as cytochrome c) from the mitochondrial matrix into the cytosol. Released cytochrome c (together with caspase-9 and Apaf-1) assembles into a multiprotein complex, the apoptosome, that activates downstream effector caspases (such as caspase-3) (5, 21), cleaves several nuclear proteins [such as poly(ADP-ribose) polymerase, or PARP], activates DNA endonucleases, and ultimately results in nuclear/cytoplasmic breakdown and cell death (15).
Many, if not all, of these events are regulated by Bcl-2 family proteins, defined by the presence of one or more Bcl-2 homology (BH) domains. Multi-BH-domain proapoptotic proteins (such as Bax, Bak, and Bok) normally exist as monomers, but upon activation by upstream signals, they are thought to oligomerize and either directly form a pore releasing inner mitochondrial membrane proteins or interact with intrinsic mitochondrial proteins to form such a pore (28, 33, 40). BH3-only pro-apoptotic proteins (e.g., Bid, Puma, and Bim) promote Bak/Bax oligomerization and pore formation, resulting in a conformational change in Bax prior to or coincident with its insertion into the mitochondrial membrane (13). Apoptosis-inhibiting (antiapoptotic) proteins, such as Bcl-2 and Bcl-xL, function to sequester BH3-only proteins, thereby preventing Bax activation (28). Although these outcomes are known, the mechanisms that specifically activate Bax, a key premitochondrial regulator of apoptosis, have only recently been suggested by our observations from investigations of death receptor signaling (2) and by the observations of Tan et al. demonstrating the direct role of MOAP-1 in the regulation of Bax at the level of the mitochondrial membrane (44, 45).
We and others have recently defined some of the molecular mechanisms of apoptotic regulation by RASSF1A. RASSF1A has been recently demonstrated to modulate the activation of the MST2/LATS1/YAP pathway by controlling p73 transcriptional activity linked to expression of PUMA (a key proapoptotic effector) (18, 31). RASSF1A has been shown to associate with the proapoptotic kinase, MST1, and function with K-Ras to promote apoptosis (34, 36). The connector enhancer of KST (CNK1) has also been shown to associate with RASSF1A and modulate the activity of MST1 (37). We have demonstrated that ectopic expression of RASSF1A enhanced death receptor-evoked apoptosis stimulated by TNF-α (2). RASSF1A and MOAP-1 knockdown cells (generated by RNA interference) and Rassf1a−/− knockout mouse embryonic fibroblasts have defective cytochrome c release and Bax translocation and impaired death receptor-dependent apoptosis (2). TNF-α stimulation resulted in the formation of RASSF1A/TNF-R1, MOAP-1/TNF-R1, and RASSF1A/MOAP-1 complexes and allowed for MOAP-1 regulation of Bax activity (2). However, TNF-α-stimulated recruitment of RASSF1A and MOAP-1 to death receptor complexes followed quite different kinetics resulting in the association of RASSF1A with TNF-R1 at 3 h of TNF-α stimulation, whereas MOAP-1 associated with TNF-R1 within 2 h of TNF-α stimulation (2). Most DISC components, such as TRADD, FADD, and caspase-8, are recruited to death receptor complexes within 1 h of TNF-α stimulation (29, 32, 39). It has been demonstrated that shortly after internalization, death receptors (such as TNF-R1) encounter an “apoptotic checkpoint” whereby complexes are brought together to continue to promote survival or stimulate apoptosis (23, 29).
In this study, we further investigated the involvement of RASSF1A and MOAP-1 in death receptor-dependent apoptosis. We now demonstrate that MOAP-1 was specifically required for RASSF1A association with TNF-R1 and that MOAP-1 residues important for TNF-R1 and TRAIL-R1 association were distinct from MOAP-1 residues required for RASSF1A association. Furthermore, MOAP-1 associated with the death domain of TNF-R1 upon TNF-α stimulation. We propose that RASSF1A (and MOAP-1) residues are important components that modulate the direction of TNF-α signaling and influence an important step in the decision to promote cell death.
MATERIALS AND METHODS
Antibodies and reagents.
Anti-mouse Alexa Fluor 546, propidium iodide, and annexin V Alexa Fluor 647 were obtained from Molecular Probes. Rabbit anti-Erk1/Erk2 (sc-93/sc-154), mouse anti-green fluorescent protein (GFP) (sc-9996) and glutathione S-transferase (GST) (sc-138), rabbit anti-TNF-R1 (sc-7895), anti-FADD (rabbit [sc-5559] and goat [sc-6035] antibodies), rabbit anti-TRADD (sc-7868), rabbit anti-caspase-8 (sc-7890), rabbit anti-RIP1 (sc-7881) (Santa Cruz Biotechnology), goat anti-TRAIL R1 (AF-347; R&D), rabbit anti-MOAP-1 (QED), mouse anti-RASSF1A (EBiosciences), and human TNF-α and TRAIL (300-014 and 310-04, respectively, from Peprotech) were purchased from the indicated commercial sources. Murine monoclonal anti-hemagglutinin (HA) (12CA5) and anti-Myc (9E10) were purified from their corresponding in-house hybridomas. A rabbit anti-RASSF1A was generated in house and also used for immunoblotting of endogenous RASSF1A in U2OS cells. Both the rabbit anti-RASSF1A and mouse anti-RASSF1A were validated as described by Baksh et al. (2). In-house enhanced chemiluminescence (ECL) detection was used for all immunostaining analysis.
Cell lines and transfection.
COS-1 and U2OS cells were maintained in Dulbecco's modified Eagle's medium (DMEM) plus 10% bovine growth serum. H1299 cells were maintained in RPMI medium plus 10% bovine growth serum. All cells were maintained in a 37°C, 5% CO2 incubator. To generate stable H1299 cells, transfections were carried out using the linear 25-kDa polymer polyethyleneimine (PEI) obtained from Polysciences (catalog no. 23966-2). PEI transfections were carried out by mixing PEI with DNA in a ratio of 3 μl PEI to 1 μg DNA (COS-1 and U2OS cells) or 4 μl PEI to 1 μg DNA (H1299 cells) in 400 μl of serum-free DMEM (for transfection in a six-well dish). The mixture was allowed to incubate for 15 min. While incubating, cells were washed three times with serum-free medium and 2 ml of complete medium was added after the washes. The PEI-DNA mixture was added to cells, mixed gently by swirling, and incubated overnight. The medium was changed at 12 to 16 h, and lysates were harvested at 48 h posttransfection. Further details are available from S.B. upon request.
Cell lysis and immunoprecipitations.
Unless otherwise indicated, cells were stimulated with 50 ng/ml TNF-α or TRAIL, followed by lysis in radioimmunoprecipitation assay (RIPA) buffer as previously described (2). TNF-R1 immunoprecipitations were carried out using 1.5 μg of rabbit anti-TNF-R1 antibody, TRAIL-R1 immunoprecipitations were carried out with 1.0 μg of goat anti-TRAIL R1, and immunoprecipitations for HA- and Myc-tagged proteins were carried out with 20 μl of our in-house hybridoma supernatant. All immunoprecipitations were carried out overnight as previously described (2). For all whole-cell lysate (WCL) immunoblots, 10% of input was used (∼70 μg of protein/lane).
Immunofluorescence.
Cells were plated onto 18- by 18-mm coverslips in six-well dishes (Corning) and fixed in 3.7% paraformaldehyde-phosphate-buffered saline(PBS) for 15 min at room temperature, followed by two 1× PBS washes and permeabilization with 0.1% Triton X-100 in 1× PBS. Cells were washed twice with 1× PBS and blocked with 0.2% fish gelatin (Sigma) for 10 min, followed by incubation for 1 h with monoclonal mouse anti-HA (1:200 dilution in 0.2% fish gelatin). Following primary antibody incubation, coverslips were washed twice with 1× PBS and incubated for 30 min with secondary antibody (1:500 of goat anti-mouse Alexa Flour 546 in 0.2% fish gelatin). Confocal images were acquired using a Zeiss laser-scanning microscope and analyzed with LSM510 software.
Apoptosis assays.
Human TNF-α or TRAIL was added together with 10 μg/ml cycloheximide (CHX) for the indicated times, and annexin V staining analysis was carried out as previously described (2). Annexin V staining by fluorescence-activated cell sorting analysis is a standard assay utilized to detect the early to late apoptotic event of exposure of phosphatidylserine residues. The annexin V antibody will recognize phosphatidylserine residues on the surface of cells that correlate well with the degree of cell death occurring inside the cell. All apoptosis assays were performed at least six times. Data for all immunofluorescence and apoptosis assays were evaluated by Student's t test (two tailed), unless otherwise stated.
Expression vectors.
Expression vectors for HA-RASSF1A and Myc-MOAP-1 were generated as previously described (2). All RASSF1A, MOAP-1, and TNF-R1 deletion and point mutation expression constructs were generated by PCR using the QuikChange site-directed mutagenesis kit (Stratagene). All HA- and Myc-tagged proteins contained single tags at their amino termini. For GFP-RASSF1A used in this study, GFP was at the N terminus of RASSF1A and was PCR cloned in at EcoRI/BamHI of the pEGFP-C2 vector using the forward primer 5′-TGCGAATTCACTAGTATGTCGGGGGAGCCT-3′ and the reverse primer 5′-GGTGGATCCATATCACCCAAGGGGGCAGGC-3′. HA-RASSF1A deletion constructs (amino acids 82 to 340, 128 to 340, 139 to 340, 143 to 340, 158 to 340, and 175 to 340) were subcloned using BamHI/NotI into pCDNA3-HA. The following primers were used. The reverse primer was 5′-TATGCGGCCGCTATTCACCCAAGGGGGCAGGC-3′, and the forward primers were 5′-CAGTGCGGATCCATCGATTGCGCGCATTGCAAG-3′, 5′-CAGTGCGGATCCATCGATGACCTTTCTCAAGCT-3′, and 5′-CAGTGCGGATCCATCGATATCAAGGAGTACAAT-3′, 5′-CAGTGCGGATCCATCGATAATGCCCAGATCAAC-3′, 5′-CAGTGCGGATCCATCGATGGTTCTTACACAGGC-3′, and 5′-CAGTGCGGATCCATCGATGTGCCCTCCAGCAAG-3′, respectively. For RASSF1A R1 to R6, the primers were R1 101CLDCCGPR→CLDAAGPR (CTCGTCTGCCTGGACGCTGCCGGGCCCCGGGACCTG), R2 84HCKFTCHYRC→HCKFTAAYRA (CATTGCAAGTTCACCGCCGCCTACCGCGCCCGCGCGCTCGTC), R3 79GLQCAHCK→GLQAAAAK (AAAGGCCTGCAGGCCGCGGCTGCCAAGTTCACCTGC), R4 84HCKFTCHYRC→HCAFTCHYRC (CAGTGCGCGCATTGCGCGTTCACCTGCCACTAC), R5 76VRKGLQCAHCK→VRKAAQCAHCK (GGCGTCGTGCGCAAAGCCGCGCAGTGCGCGCATTGC), and R6 69GDFIWGVVRK→GDFAAAAVRK (TGTGGCGACTTCGCCGCGGCCGCCGTGCGCAAAGGC). For TNF-R1 mutants T1 and T2, the primers were T1 408TWRRR→TWAAA (CTGGCGACCTGGGCGGCGGCCACGCCGCGGCGC) and T2 435CLEDIWEEA→CLAAIWAAA (CTGGGCTGCCTGGCGGCCATCGCGGCGGCGCTTTGCGGC). All Myc-MOAP-1 mutants (M1, M2, and M3) were generated as previously described (2), and deletion constructs to MOAP-1 were generated by Fu Naiyang (Institute of Molecular and Cell Biology). Further details on the generation of other constructs are available from S.B. upon request. All expression constructs were confirmed by sequencing.
Computational methods.
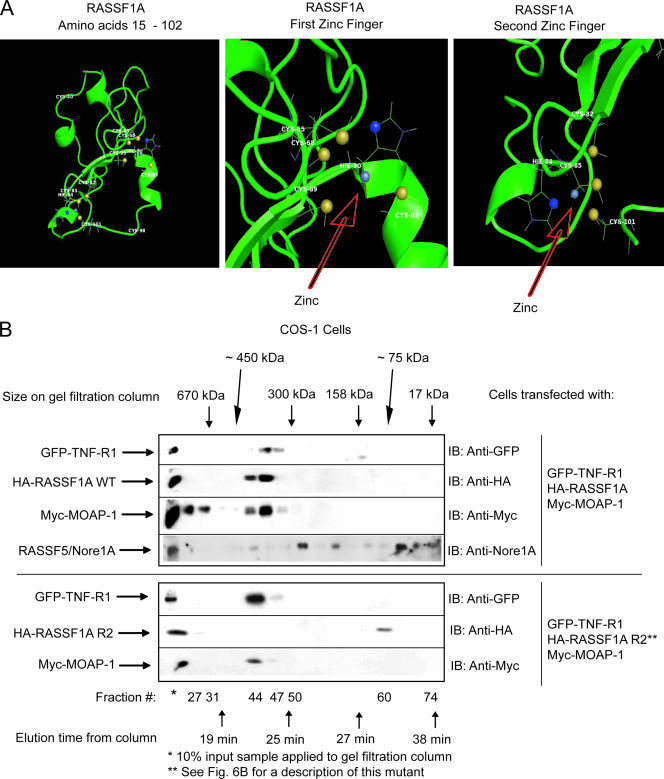
The MODELLER (38) program was utilized to align the domain of RASSF1A consisting of residues 15 to 102 with that of RASSF5/Nore1A. This was used to thread the RASSF1A sequence onto the RASSF5/Nore1A backbone to produce an initial coordinate file. The Rosetta comparative modeling protocol (8, 9, 27) was then used to build the missing loops, consisting of residues 34 through 59 and 70 through 72. Full-atom refinement was performed on the whole model to give a total of 8,000 models. Selecting models with scores representing full-atom energies less than a value of −110 resulted in 35 structures, which, when clustered at a root-mean-square deviation of 5 Å, gave three major groups of structures, with 9, 11, and 15 elements, respectively. The lowest-scoring Rosetta structure (indicative of the most stable structure) belonged to the cluster of size 11, and was chosen for the representation shown in Fig. 7A; the other two clusters are shown in Fig. S3 in the supplemental material. The likely sets of cysteine and histidine residues comprising the two zinc fingers could be identified from these structures. Zinc atoms were added into the centers of these groups of sites at distances similar to those observed in the RASSF5/Nore1A structure, and the system was minimized for 100 steps with distance restraints on the zinc in AMBER8 (6). Water was then added, and the solvated model was subjected to another 600 steps of conjugate gradient minimization with distance restraints on the zinc atoms. Finally, the figures were created using the PyMOL visualization program (W. L. DeLano, The PyMOL Molecular Graphics System [2002]; http://www.pymol.org).
FIG. 7.
Ternary complex formation between RASSF1A and TNF-R1 requires an intact C1 domain of RASSF1A. (A) Computer-generated model of residues 15 to 102 of RASSF1A. This model was generated by sequence and structural comparison with the NMR structure of RASSF5/Nore1A using the MODELLER program (38) and ROSETTA comparative modeling protocol (9, 27). The left panel shows the structure of residues 15 to 102 of RASSF1A, revealing two zinc finger motifs as indicated. The middle panel shows the first zinc finger motif with the zinc coordinated to four residues (C-68, C-89, H-90, and C-93). The right panel shows the second zinc finger motif with the zinc coordinated to four residues (C-82, H-84, C-85, and C-101). The zinc atom is gray, the histidine ND1 atoms are blue, and the cysteine SG atoms are yellow. (B) Transfection was carried out (as indicated) on a 4- by 10-cm2 dish using 5 μg of each component. (See Fig. 6B for a description of RASSF1A R2, a RASSF1A mutant that lacks the ability to associate with TNF-R1.) Plate samples were pooled by scraping into 1 ml of 1× PBS, followed by lysis in 800 μl of RIPA buffer. Pooled samples were then analyzed by gel filtration analysis as described in the legend to Fig. 2. Fractions were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. IB, immunoblotting. This experiment was carried out two times with similar outcomes. A representative experiment is shown.
RESULTS
MOAP-1 is required for RASSF1A association with TNF-R1.
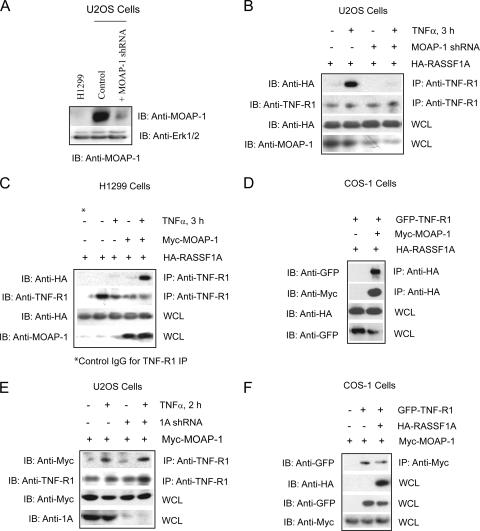
Stable expression of a MOAP-1 short hairpin RNA (shRNA) in U2OS cells resulted in >80% decrease in MOAP-1 expression (Fig. 1A). The loss of MOAP-1 expression inhibited the ability of RASSF1A to associate with TNF-R1 (Fig. 1B). Furthermore, in a non-small-cell lung cancer cell lacking detectable expression of MOAP-1 (Fig. 1A, H1299 cells) and RASSF1A (4), the ability of ectopically expressed RASSF1A to associate with TNF-R1 was also lost (Fig. 1C). This suggested that MOAP-1 was required for RASSF1A to associate with TNF-R1. In contrast to the observations by Vos et al. (50), we have observed robust associations between RASSF1A and MOAP-1 and between RASSF1A and TNF-R1 independent of the presence of activated K-Ras. We further confirmed the requirement for MOAP-1 by the stable reconstitution of MOAP-1 in H1299, which restored the ability of ectopically expressed RASSF1A to associate with TNF-R1 upon TNF-α stimulation (Fig. 1C, last two lanes). In support of these data, RASSF1A association with TNF-R1 was not observed unless MOAP-1 was coexpressed in COS-1 cells (Fig. 1D). In contrast, knockdown of RASSF1A did not impede MOAP-1 association with TNF-R1 (Fig. 1E) and complex formation between ectopically expressed MOAP-1 and TNF-R1 was readily detected in COS-1 cells in the absence or presence of overexpressed HA-RASSF1A (Fig. 1F). This suggested that MOAP-1 does not require the presence of RASSF1A in order to form a complex with TNF-R1.
FIG. 1.
MOAP-1 is required for the association of RASSF1A with TNF-R1. (A) Expression of endogenous MOAP-1 in H1299 cells, control U2OS cells, or U2OS cells containing MOAP-1 shRNA. (B) Pooled U20S cells expressing control (−) or MOAP-1 shRNA (+) were used to determine RASSF1A association with TNF-R1. The association of ectopically expressed HA-RASSF1A with endogenous TNF-R1 was recovered by immunoprecipitation (IP) with an anti-TNF-R1 antibody followed by immunoblotting (IB) with the indicated antibodies. (C) HA-RASSF1A association with endogenous TNF-R1 was carried out as in panel B but in H1299 cells without (−) or with (+) ectopically expressed Myc-MOAP-1. Control antibodies (control immunoglobulin G) did not reveal immunoprecipitation of TNF-R1 or HA-RASSF1A (data not shown, but see Fig. 6C for an example). (D) RASSF1A, MOAP-1, and TNF-R1 were ectopically expressed in COS-1 cells, and proteins associated with RASSF1A were recovered by immunoprecipitation with an anti-HA antibody followed by immunoblotting with the indicated antibodies. (E) Pooled U20S cells expressing control (−) or RASSF1A (1A) shRNA (+) as previously described (2) were used to determine MOAP-1 association with endogenous TNF-R1 as outlined in panel B. (F) MOAP-1, RASSF1A, and TNF-R1 were ectopically expressed in COS-1 cells, and proteins associated with MOAP-1 were recovered by immunoprecipitation with an anti-Myc antibody followed by immunoblotting with the indicated antibodies.
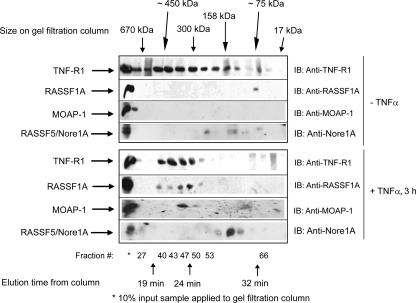
These data suggested that an ordered series of events was required for RASSF1A to associate with TNF-R1, most likely subsequent to association of MOAP-1 with TNF-R1. To support our observations of a ternary complex between MOAP-1, TNF-R1, and RASSF1A, we used high-performance liquid chromatography (HPLC) analytical gel filtration chromatography to analyze endogenous complexes formed upon TNF-α stimulation in U2OS cells (Fig. 2). This technique separates protein complexes based on size, with the larger complexes eluting earlier and the smaller ones eluting later. It has been reported that RASSF5/Nore1A can form heterodimers with RASSF1A under basal conditions (35). In Fig. 2, with or without TNF-α stimulation, the majority of RASSF5/Nore1A migrates within fractions that do not contain RASSF1A, MOAP-1, and TNF-R1, suggesting that RASSF5/Nore1A is not part of the complex containing RASSF1A, MOAP-1, and TNF-R1 that functions to activate Bax and promote cell death. We can, however, detect a minor fraction of endogenous RASSF5/Nore1A basally in association with endogenous RASSF1A (Fig. 2, upper panel). It is therefore feasible that a portion of the RASSF1A pool may undergo heterodimerization with RASSF1A/Nore1A; however, RASSF5/Nore1A is not in a complex with TNF-R1 (see Fig. S5B in the supplemental material) or MOAP-1 (data not shown). Gel filtration chromatography revealed the presence of an ∼75-kDa complex of endogenous RASSF1A in U2OS cells not stimulated with TNF-α, suggesting that RASSF1A (normally 39 kDa) may basally exist as a homodimer. Surprisingly, endogenous MOAP-1 eluted with high-molecular-weight aggregrates in unstimulated cells (Fig. 2, third panel from the top, fraction 27, and see Fig. 7B, third panel from the top, fractions 27 and 31). However, upon TNF-α stimulation, an ∼300- to 350-kDa complex composed of a substantial amount of endogenous TNF-R1, MOAP-1, and RASSF1A was observed (Fig. 2, plus TNF-α, elution time of 23 to 25 min, fractions 47 to 50). We speculate that this complex may be composed of trimeric TNF-R1 (55 kDa × 3 = 165 kDa), monomeric RASSF1A (39 kDa), and MOAP-1 (41 kDa) (total, ∼245 kDa). We have also detected endogenous TRAF2 comigrating within fractions containing TNF-R1, MOAP-1, and RASSF1A (R. Chow and S. Baksh, unpublished observations). Preliminary evidence suggests that MOAP-1 becomes ubiquitinlyated upon TNF-α stimulation and TRAF2 is the E3 ligase for MOAP-1 ubiquitinylation (Chow and Baksh, unpublished). We hypothesize that TRAF2 (∼57 kDa) may account for the remaining component of this complex to allow the complex to migrate at ∼300 kDa. However, further analysis will be required, such as protein cross-linking experiments or native gel electrophoresis in order to confirm the correct size of the RASSF1A/MOAP-1/TNF-R1 complex.
FIG. 2.
Gel filtration analysis of complex formation of endogenous TNF-R1, MOAP-1, and RASSF1A. U2OS cells were grown on 4- by 10-cm2 dishes and left unstimulated or stimulated with TNF-α. Cells were lysed by scraping into 1 ml of 1× PBS, combined, and lysed in 800 μl of RIPA buffer. After lysate was clarified by centrifugation, supernatant was dialyzed against HPLC washing buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 2 mM EGTA) with 1 liter of buffer overnight and then 1 liter of buffer for 8 h the next day. The dialyzed sample was then concentrated to ∼200 μl, and 150 μl was applied to an analytical HPLC gel filtration Superose 200 column. The flow rate applied was 400 μl/min, and 200-μl fractions were collected in a 96-well plate. The column was standardized using Bio-Rad's gel filtration standard mixture of thryoglobulin (∼670 kDa, with an elution time of 19.5 min); gamma globulin (∼158 kDa, with an elution time of 27 min), ovalbumin (∼44 kDa, with an elution time of 35 min), myoglobin (∼17 kDa, with a retention time of 38 min), and vitamin B12 (∼1.3 kDa, with an elution time of 47 min). Fractions were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. This experiment was carried out three times with similar outcomes. A representative experiment is shown for complex formation between TNF-R1, MOAP-1, and RASSF1A, with additional immunoblotting (IB) for RASSF5/Nore1A. Approximate locations of gel filtration standards, fraction numbers, and elution times are indicated.
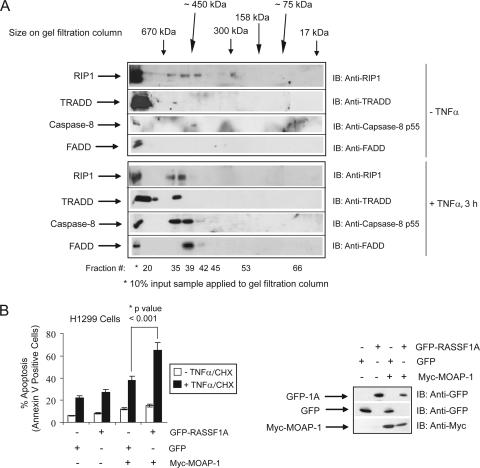
Analysis of DISC components (containing RIP1, TRADD, FADD, and caspase-8) within the fractions used in Fig. 2, revealed the presence of a protein complex of >400 kDa that eluted before fraction 40 upon TNF-α stimulation (Fig. 3A). (RASSF1A and MOAP-1 eluted after fraction 40 upon TNF-α stimulation in Fig. 2.) We have previously demonstrated that Bax conformational change and mitochondrial insertion were inhibited in RASSF1A shRNA knockdown cells, whereas Bid cleavage and translocation were not (2). Combined with our results in Fig. 3A, these observations suggest that the MOAP-1/TNF-R1/RASSF1A complex (which functions to activate Bax and promote Bax insertion into the mitochondrial membrane) (2) may migrate as a distinct complex from the DISC, a complex that functions to activate caspase-8 in order to promote Bid cleavage and insertion into the mitochondrial membrane. In support of this data, the association of DISC components (FADD, TRADD, and caspase-8) was not lost in RASSF1A or MOAP-1 shRNA knockdown cells (see Fig. S1 in the supplemental material). The presence of a signaling complex composed of MOAP-1, TNF-R1, and RASSF1A may thus be required to activate Bax and not Bid. Furthermore, the ability of H1299 cells to respond to death receptor stimuli was greatly enhanced in the presence of both MOAP-1 and RASSF1A (Fig. 3B), suggesting that the RASSF1A/MOAP-1 pathway plays a critical role in death receptor-dependent apoptosis in H1299 cells.
FIG. 3.
The RASSF1A/TNF-R1/MOAP-1 complex migrates to distinct regions within a gel filtration (away from the DISC complex) and requires the presence of both MOAP-1 and RASSF1A. (A) U2OS cells were grown on 4- by 10-cm2 dishes and left unstimulated or stimulated with TNF-α. Samples for gel filtration were treated as described in the legend to Fig. 2. Fractions were immunoblotted (IB) for endogenous DISC components: RIP1, FADD, TRADD, and caspase-8. These analyses were carried out two times. (B) The left panel shows annexin V staining of TNF-α-CHX-treated H1299 cells stably expressing control vector (−) or stably expressing Myc-MOAP-1 (+). The transfection efficiency of H1299 cells was between 30 and 50% GFP-positive cells. The experiment was repeated six times, and significance was evaluated by Student's t test (two tailed) with the indicated P value (*). The right panel shows expression of proteins used in annexin V staining.
Electrostatic interactions determine MOAP-1 association with TNF-R1.
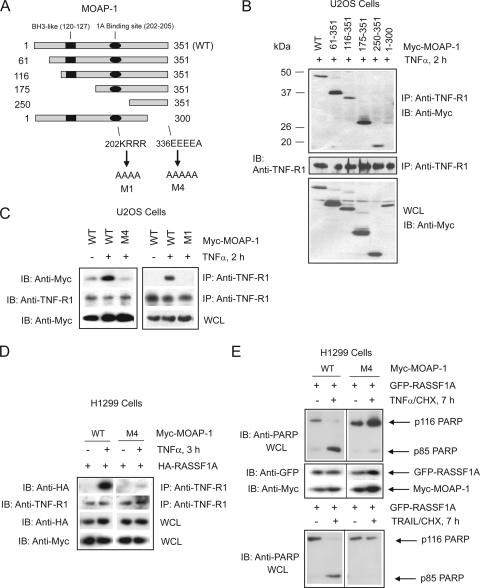
Next, we proceeded with analysis of the regions of MOAP-1 required for mediating association with TNF-R1. Using a series of deletion mutants, we identified the C-terminal 51 amino acid residues of MOAP-1 as important for TNF-R1 association (Fig. 4A and B, comparing TNF-R1 association of wild-type amino acids 1 to 351 with amino acids 1 to 300 of MOAP-1). Further deletion analysis within the last 51 amino acids revealed an acidic stretch within MOAP-1 that was required for association with TNF-R1 (Fig. 4A, 336EEEEA, and C, mutant M4 containing the mutation 336EEEEA→336AAAAA) and TRAIL-R1 (also known as DR4) (see Fig. S2A in the supplemental material). These data suggest that MOAP-1/TNF-R1 and MOAP-1/TRAIL-R1 association may involve an electrostatic association between 336EEEEA of MOAP-1 and, most likely, a basic region within TNF-R1 and TRAIL-R1. As we have demonstrated previously that MOAP-1 can associate with an acidic region within RASSF1A via amino acids 202 to 205 of MOAP-1 (202KRRR) (2), our results suggest that MOAP-1 might associate simultaneously with RASSF1A and TNF-R1 by utilizing different charged regions.
FIG. 4.
Location of MOAP-1 residues important for TNF-R1 association. (A) Schematic of MOAP-1 mutants used in panel B indicating the locations of critical residues and domains. Numbers indicate amino acid location, and arrows denote amino acid changes. MOAP-1 deletion mutants (B) or point mutants (C) were ectopically expressed, and proteins associated with endogenous TNF-R1 were recovered by immunoprecipitation (IP) with anti-TNF-R1 antibodies followed by immunoblotting (IB) with the indicated antibodies. All lanes were stimulated with TNF-α for 2 h. WT, wild type. (D) H1299 stable cells were generated containing either wild-type MOAP-1 or MOAP-1 mutant M4, which lacked the ability to associate with endogenous TNF-R1. HA-RASSF1A was ectopically expressed, and association with endogenous TNF-R1 was recovered by immunoprecipitation with anti-TNF-R1 antibodies followed by immunoblotting with the indicated antibodies. (E) H1299 stable cells were generated as in panel D, and GFP-RASSF1A was ectopically expressed, stimulated with TNF-α (top) or TRAIL (bottom), lysed in RIPA buffer, and immunoblotted with the indicated antibodies.
We next investigated the ability of the “open” mutant of MOAP-1 to associate with death receptors. Previously, we demonstrated that the Bax-interacting domain on MOAP-1, the BH3-like domain, is “covered” by an intraelectrostatic association involving regions 202KRRR and 178EEEF of MOAP-1 (2). The association of RASSF1A with 202KRRR resulted in the loss of this intraelectrostatic association and “opening” up of MOAP-1 (2). Once MOAP-1 has acquired the “open” conformation (as generated by mutations in mutant M1 [Fig. 4A]), RASSF1A was not required for MOAP-1 association with Bax (2). Based on these observations, we would predict that, once in the “open” conformation, MOAP-1 may not require association with death receptors (since it now has the ability to activate Bax). We, therefore, utilized MOAP-1 mutant M1 to test our hypothesis for association with TNF-R1 and TRAIL-R1. As previously demonstrated, we observed an inducible association of the MOAP-1 wild type with TNF-R1 (Fig. 4C) and TRAIL-R1 (see Fig. S2A in the supplemental material). In support of our prediction, MOAP-1 mutant M1 lost its ability to associate with both TNF-R1 (Fig. 4C) and TRAIL-R1 (see Fig. S2A in the supplemental material) following death receptor stimulation.
We further assessed the importance of the TNF-R1 association site on MOAP-1 for interaction of RASSF1A with the TNF-R1 complex (residues 336EEEE, this is mutated in mutant M4). The ability of RASSF1A to associate with TNF-R1 was lost in the presence of this mutant of MOAP-1 (Fig. 4D); again suggesting that formation of a MOAP-1/TNF-R1 complex was critical for RASSF1A association with TNF-R1. Furthermore, the ability of MOAP-1 to promote RASSF1A-mediated PARP cleavage was greatly impeded by mutation of the TNF-R1 association site on MOAP-1 (Fig. 4E). The cleavage of PARP from a 116-kDa protein to an 85-kDa protein is a marker of the later stages of apoptosis and is dependent on the generation of effector caspase activity originating from Bax activity and mitochondrial perturbation. H1299 cells stably expressing MOAP-1 wild type and transiently transfected with GFP-RASSF1A resulted in the appearance of the cleaved 85-kDa fragment of PARP (p85 PARP) upon TNF-α stimulation (Fig. 4E, left panel). However, when stable cells containing MOAP-1 mutant M4 were transiently transfected with GFP-RASSF1A, the cleavage of PARP was almost completely lost (Fig. 4E, right panel), suggesting an inhibitory action of the presence of the MOAP-1 mutant M4. These data are in support of an important role for MOAP-1 in TNF-α-induced, RASSF1A-mediated apoptosis that is dependent upon the presence of MOAP-1. MOAP-1 is recruited to TNF-R1 receptor complexes and functions to promote the association of RASSF1A with TNF-R1.
MOAP-1 influences RASSF1A self-association.
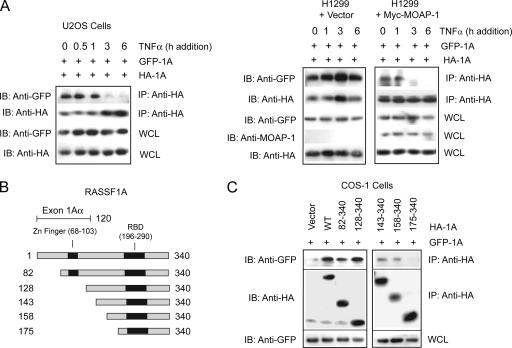
In most of the cellular settings investigated, DISC components are assembled within 0.5 h to 1 h following TNF-α stimulation. MOAP-1 association occurs approximately 2 h following TNF-α stimulation and is subsequently followed by RASSF1A recruitment to TNF-R1 approximately 1 h later (2). Since RASSF1A has been observed to undergo self-association and associations with RASSF5/Nore1A (35), we explored the possibility that the dissolution of these complexes of RASSF1A may explain the delay in RASSF1A association with TNF-R1 upon TNF-α stimulation. In Fig. 2, we demonstrate that only a minor portion of endogenous RASSF5/Nore1A comigrates with endogenous RASSF1A, suggesting that RASSF1A/RASSF5 complexes may not play a role in RASSF1A-mediated cell death. We, therefore, explored the possibility that the dissolution of RASSF1A self-associations may explain the delay in RASSF1A association with TNF-R1 upon TNF-α stimulation. Ectopic expression of HA-RASSF1A and GFP-RASSF1A resulted in RASSF1A self-association (Fig. 5A, no TNF-α addition). However, upon TNF-α stimulation, self-association was lost and was almost completely undetectable by 3 h of TNF-α stimulation (Fig. 5A, left panel). Loss of RASSF1A self-association coincided with the association of RASSF1A with both MOAP-1 and TNF-R1—that is, by 3 h of TNF-α addition (2). Interestingly, in H1299 cells lacking detectable levels of MOAP-1 (Fig. 1A), self-association of RASSF1A was not lost upon TNF-α stimulation (Fig. 5A, right panel) but was reestablished once MOAP-1 was ectopically expressed (Fig. 5A, right panel, plus Myc-MOAP-1). This suggested that MOAP-1 may be required to promote RASSF1A monomerization and aid in association of RASSF1A with TNF-R1. We speculate that only a portion of the RASSF1A pool may undergo monomerization to allow MOAP-1/TNF-R1 association to mediate cell death since RASSF1A may serve other important in vivo functions in response to TNF-α stimulation. Further experiments will be required in order to address the stoichiometry of RASSF1A association with itself and with TNF-R1, but this analysis is beyond the scope of this study. We propose that TNF-α stimulation may function at many levels to (i) recruit MOAP-1 to TNF-R1 (this study); (ii) promote the dissociation of the RASSF1A self-association, allowing for recruitment to TNF-R1 (this study); and, lastly, (iii) to promote MOAP-1 conformational change by the association of RASSF1A with MOAP-1 (2).
FIG. 5.
Regulation of RASSF1A self-association. (A) Coimmunoprecipitation (IP) of GFP-RASSF1A and HA-RASSF1A with anti-HA antibodies following TNF-α stimulation in U2OS cells (left panel) or H1299 cells (right panel) as indicated. IB, immunoblotting. (B) Schematic of RASSF1A deletion mutants used in panel C indicating the location of the C1 domain containing a potential zinc finger binding motif. Numbers indicate amino acid locations. (C) Coimmunoprecipitation of GFP-RASSF1A (GFP-1A) and HA-RASSF1A (HA-1A) deletion mutants in U2OS cells using anti-HA antibodies.
Since RASSF1A self-association was lost upon TNF-α stimulation, we explored the requirements for RASSF1A self-association using deletion mutants of HA-RASSF1A (as shown in Fig. 5B and 5C). Both wild-type and HA-RASSF1A expression constructs containing amino acids 158 to 340 resulted in association with GFP-RASSF1A, whereas an HA-RASSF1A expression construct containing amino acids 175 to 340 did not. The loss of amino acids between 158 and 175 resulted in the loss of RASSF1A self-association (Fig. 5C), suggesting that RASSF1A self-association was localized within this region.
The C1 domain of RASSF1A governs TNF-R1 association.
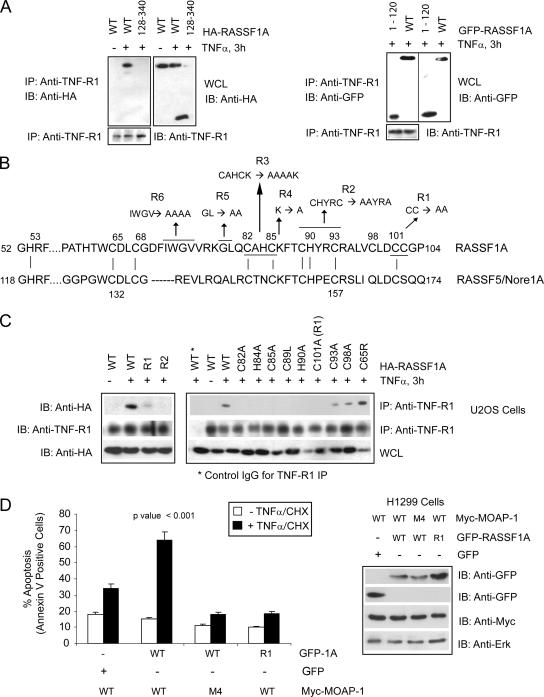
The results described above indicate the presence of a distinct region within RASSF1A for self-association that may influence how RASSF1A associates with the TNF-R1/MOAP-1 complex. We next addressed the determinants on RASSF1A for association with the TNF-R1/MOAP-1 complex. Utilizing the deletion mutants from Fig. 4C, we observed the importance of the first 128 amino acids of RASSF1A for TNF-R1 association (Fig. 6A) and TRAIL-R1 association (see Fig. S2B in the supplemental material). Within this region is a C1 domain (Fig. 5B and 6B) similar to one found in protein kinase C that influences association with membrane lipids and modulates the binding of zinc (12, 26). RASSF5/Nore1A has a similar C1 domain to RASSF1A (Fig. 6B) and was recently shown by nuclear magnetic resonance (NMR) to contain a zinc finger binding motif coordinating two molecules of zinc (19). Mutation of cysteine (C) and histidine (H) residues within the C1 domain of RASSF1A resulted in the loss of RASSF1A association with TNF-R1 (Fig. 6C and see Fig. S2C in the supplemental material). A more detailed analysis revealed the importance of C-82, H-84, C-85, C-89, H-90, and C-100/C-102 (mutant R1) (Fig. 6B) to the formation of the zinc finger domain within RASSF1A (Fig. 6C [all mutations to alanine unless otherwise stated]). Mutations at C-93 and C-98 also interfered with TNF-R1 association, but a mutation at C-65 (the C65R polymorphism of RASSF1A [12]) revealed not a loss but a reproducible enhancement of TNF-R1 association (Fig. 6C). Mutation of noncysteine or -histidine residues within this region did not result in the loss of association with TNF-R1 (see Fig. S2C, mutants R4 to R6, in the supplemental material), suggesting the importance of the core cysteine and histidine residues involved in forming the zinc finger binding domain. RASSF1A association with TRAIL-R1 was also significantly inhibited by mutations at C-82, C-85, H-84 (see Fig. S2D in the supplemental material), and C-101/C-102 (mutant R1; data not shown), but, again a mutation at C-65 did not result in the loss of TRAIL-R1 association with RASSF1A (see Fig. S2D in the supplemental material). Interestingly, some of the cysteine and histidine mutations also interfered with the association of RASSF1A with MOAP-1 (see Fig. S2E in the supplemental material). However, the presence of the C65R mutation did not result in the loss of MOAP-1 association (see Fig. S2E in the supplemental material), in contrast to observations by Vos et al. (50). Interestingly, stable U2OS cells containing this mutant exhibited reduced RASSF1A-mediated cell death, even though this mutant demonstrated a reproducible enhancement of TNF-R1 association (M. El-Kalla et al., unpublished observations). Preliminary data suggest a gain of function for this mutant in modulating survival (non-cell death) pathways linked to TNF-α signaling (and to TNF-R1) in U2OS and HCT116 cells (El-Kalla et al., unpublished).
FIG. 6.
Location of RASSF1A residues important for TNF-R1 association. (A) Coimmunoprecipitation (IP) of endogenous TNF-R1 with wild-type (WT) HA-RASSF1A and a deletion mutant lacking the N terminus of RASSF1A (left panel, 128-340) or C terminus (right panel, 1-120). Numbers indicate amino acid locations. (B) Amino acid sequence of the C1 domain of RASSF1A and RASSF5/Nore1A with indicated mutants of RASSF1A (R1, R2, etc.). Vertical lines between the two protein sequences indicate locations of conserved cysteine and histidine residues involved in the formation of the coordination spheres for zinc (as characterized for RASSF5/Nore1A by Harjes et al. [19]) Numbers indicate amino acid locations, and underlined residues and arrows indicate amino acid changes to alanine with the RASSF1A sequence. (C) Coimmunoprecipitation of endogenous TNF-R1 with HA-RASSF1A wild type or point mutants within the C1 domain of RASSF1A (R1 and R2 and indicated mutations). Numbers indicate amino acid location changes to alanine within the RASSF1A sequence. IgG, immunoglobulin G; L, leucine; R, arginine; A, alanine. (D) The left panel shows annexin V staining of TNF-α-CHX-treated H1299 cells stably expressing the Myc-MOAP-1 wild type or MOAP-1 mutant M4 (Fig. 4A). GFP constructs were transiently transfected to the level of 30 to 40%. Experiments were repeated six times, and significance was evaluated by one-way analysis of variance (P < 0.001). The right panel shows expression of proteins used in the annexin V staining experiment.
We next explored how these cysteine and histidine mutations may affect RASSF1A self-association. Unlike association with TNF-R1, RASSF1A self-association was not lost in the presence of cysteine and histidine mutations within the C1 domain of RASSF1A (see Fig. S2F in the supplemental material), nor was microtubule attachment lost (data not shown). These data suggest that disruption of the C1 domain does not interfere with the three-dimensional structure of RASSF1A required for self-association and localization to microtubules, but it did interfere with a RASSF1A three-dimensional structure required for association with MOAP-1/death receptor complexes.
We proceeded to test the importance of both RASSF1A and MOAP-1 TNF-R1 interaction sites in TNF-α-evoked cell death. Analysis was carried out in H1299 cells stably expressing Myc-tagged wild-type MOAP-1 or Myc-MOAP-1 mutant M4 (a mutant that lacked the ability to associate with TNF-R1 [Fig. 4C]). H1299 cells do not have detectable levels of endogenous MOAP-1 (Fig. 1A) and thus do not have a significant amount of inducible TNF-α-evoked cell death, even in the presence of overexpressed RASSF1A (Fig. 6D). However, stable expression of Myc-MOAP-1 in H1299 cells demonstrated increased sensitivity to TNF-α-evoked cell death in the presence of overexpressed GFP-RASSF1A, but not with GFP. In the presence of the MOAP-1 mutant M4, ectopically expressed GFP-RASSF1A did not augment TNF-α-evoked cell death (Fig. 6D and see Fig. S3A in the supplemental material) or Bax activation (see Fig. S3D in the supplemental material). Similarly, in the presence of RASSF1A that lacked receptor association (GFP-1A mutant R1 [Fig. 6C]), TNF-α-evoked cell death was compromised in Myc-MOAP-1 wild-type stable cells (Fig. 6D). Similar observations were also made for TRAIL-evoked apoptosis in H1299 cells (see Fig. S3B and C in the supplemental material).
To support our observations of the importance of the C1 domain of RASSF1A in TNF-R1 association, we carried out computational structural analysis on the C1 domain of RASSF1A based on the established NMR structure of RASSF5/Nore1A. This methodology produced three stable structures that confirmed that the C1 domain of RASSF1A can form two zinc finger domains coordinating two molecules of zinc. (Fig. 7A shows the most stable structure; see Fig. S4A and S4B in the supplemental material for the other two stable structures.) This analysis revealed that the first 102 amino acids of RASSF1A incorporated two zinc finger motifs composed of the C and H residues, as described in Fig. 7A. This structure confirmed our empirical data in Fig. 6C, with the exception of C-65. C-65 appears to be in the coordination sphere of the first zinc finger motif, but mutation of C-65 did not interfere with TNF-R1 association (Fig. 6C). We speculate that C-68 may be more significant to the coordination of the zinc molecule in this area than C-65. Based on our result in Fig. 6C, C-65 appears to be inessential to the stability of the zinc finger and it is not uncommon for zinc to be tetrahedrally coordinated by only four C or H residues.
To further add support for the importance of the zinc finger motif for complex formation with TNF-R1 and MOAP-1, we carried out HPLC analytical gel filtration chromatography to analyze complexes formed upon coexpression of Myc-MOAP-1 (∼41-kDa protein), GFP-TNF-R1 (∼81-kDa protein), and the HA-RASSF1A wild type (∼39-kDa protein) (Fig. 7B, upper panel) or RASSF1A R2 mutant (Fig. 7B, lower panel). (Note that R2 is a mutant that does not associate with TNF-R1.) Gel filtration chromatography revealed the presence of a 300- to 450-kDa complex composed of a substantial amount of GFP-TNF-R1, Myc-MOAP-1, and HA-RASSF1A (Fig. 7B, upper panel, elution time of 23 min, fractions 44 to 49). We speculate that this complex may be composed of GFP-TNF-R1 trimer (as TNF-R1 trimerization follows TNF-α stimulation; 81 kDa × 3 = 243 kDa) plus Myc-MOAP-1 (41 kDa) plus HA-RASSF1A (39 kDa), for a total of 323 kDa. However, further experimentation is required to confirm the size of this complex.
Similar to Fig. 2 (upper panel, no TNF-α stimulation), we can detect a minor fraction of endogenous RASSF5/Nore1A comigrating with GFP-TNFR1/RASSF1A/MOAP-1 in COS-1 cells as a result of forced expression of GFP-TNF-R1, HA-RASSF1A, and Myc-MOAP-1 (Fig. 7B, top panel, fraction 44). However, under endogenous conditions, we do not think that RASSF5/Nore1A is part of the RASSF1A/MOAP-1/TNF-R1 complex that activates Bax activity. In support of the formation of a complex of RASSF1A, MOAP-1, and TNF-R1, RASSF1A mutant R2 (which lacked TNF-R1 association) failed to elute with times comparable to those of wild-type RASSF1A on an analytical gel filtration column (compare the results for anti-HA immunoblotting in lanes 44 to 49 with those of lane 60 in the upper panel versus lower panel of Fig. 7B). A predicted molecular mass, based on elution times off the analytical gel filtration column, is ∼75 kDa for the RASSF1A mutant R2 in the lower panel (most likely the RASSF1A dimer) (Fig. 7B). The fractions containing RASSF1A mutant R2 do not have detectable MOAP-1 or TNF-R1. However, fractions containing MOAP-1 also contained TNF-R1, suggesting that MOAP-1 retained its ability to associate with TNF-R1 even in the presence of a RASSF1A mutant R2 that lacked TNF-R1 association. These observations suggest that distinct domains exist on RASSF1A for self-association (between amino acids 158 and 175), microtubule attachment (N- and C-terminal residues), and TNF-R1 association (the zinc finger domain between amino acids 80 and 105).
To summarize these data, the presence of MOAP-1 mutant M4 (i) inhibited the ability of RASSF1A to associate with TNF-R1 and promote apoptosis (Fig. 4D and 6D), (ii) inhibited the ability of PARP to be cleaved by caspases (Fig. 4E), and (iii) interfered with the ability of TNF-α to promote Bax conformational change (see Fig. S3D in the supplemental material) and apoptosis (Fig. 6D and see Fig. S3A to C in the supplemental material). Taken together, these data lend support to the importance of the death receptor interaction site on MOAP-1 (336EEEE) for RASSF1A- and MOAP-1-mediated apoptosis.
The death domain of TNF-R1 associates with MOAP-1.
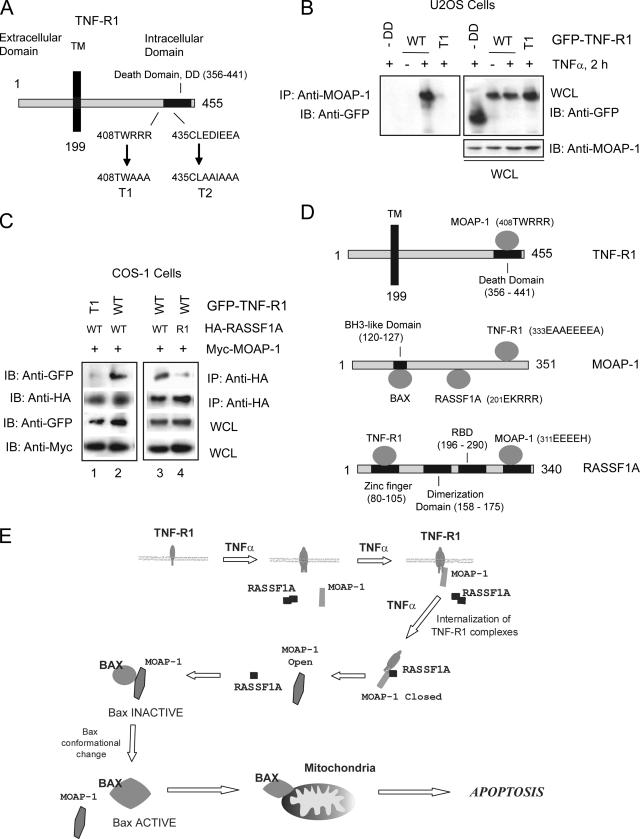
TNF-R1 and TRAIL-R1 undergo receptor trimerization following ligand binding (14). Members of the TNF-R1 superfamily of receptors contain a death domain in their cytoplasmic tail responsible for the recruitment of adaptor molecules essential in carrying out death receptor-dependent apoptosis (Fig. 8A). In this study, we have focused on the dynamics of association of both RASSF1A and MOAP-1 with TNF-R1. Without MOAP-1, RASSF1A does not find its target (i.e., TNF-R1) (Fig. 1B and C). Therefore, we investigated the binding requirements on TNF-R1 for MOAP-1 association. Deletion of the death domain of TNF-R1 resulted in the loss of MOAP-1 association (Fig. 8B), suggesting that MOAP-1 associated with TNF-R1 via its death domain. Since the acidic region of MOAP-1 (336EEEE) was required for TNF-R1 association (Fig. 4C), we investigated the role of two charged regions within the death domain of TNF-R1 (Fig. 8A, TNF-R1 mutants T1 and T2). Mutation of the basic stretch (T1) (Fig. 8B) and not the acidic stretch (T2) (see Fig. S5A in the supplemental material) resulted in the loss of MOAP-1 association with TNF-R1, supporting the importance of an interelectrostatic interaction between MOAP-1 and TNF-R1. In addition, as both TNF-R1 and TRAIL-1 have death domain regions, we speculate that MOAP-1 may also interact with a basic region within the death domain of TRAIL-R1. A sequence, 347SQRRRL, can be found within the death domain of TRAIL-1 that may coordinate MOAP-1 association, but this remains to be tested. Furthermore, the ability of ectopically expressed wild-type RASSF1A to associate with GFP-TNF-R1 in COS-1 cells was dependent upon the ability of TNF-R1 to associate with MOAP-1 (compare lanes 1 and 2 in Fig. 8C). Similarly, the ability of RASSF1A to associate with wild-type TNF-R1 and MOAP-1 was dependent upon a functional RASSF1A C1 domain (compare lanes 3 and 4 in Fig. 8C). These observations support a model proposing an ordered set of dynamic protein associations between MOAP-1, TNF-R1, and RASSF1A in order to promote apoptosis (Fig. 8E).
FIG. 8.
Location of TNF-R1 residues important for RASSF1A and MOAP-1 association. (A) Schematic of TNF-R1 indicating the locations of binding domains. Numbers indicate amino acid locations. TM, transmembrane domain. (B) Coimmunoprecipitation (IP) in U2OS cells of endogenous MOAP-1 with ectopically expressed GFP-TNF-R1 wild type (WT) or the death domian (DD) T1 mutant of TNF-R1. Associated proteins with MOAP-1 were recovered by immunoprecipitation with a rabbit anti-MOAP-1 antibody followed by immunoblotting (IB) as indicated. (C) Coimmunoprecipitation in COS-1 cells of ectopically expressed HA-RASSF1A or R1 mutant (Fig. 6C) with the GFP-TNF-R1 wild type or GFP-TNF-R1 T1 mutant (Fig. 8A). (D) Schematic summary of binding regions on TNFR1, MOAP-1, and RASSF1A. The information provided in parentheses indicates amino acid residues required for association with the respective binding partner. For example, in the upper panel for TNF-R1, 408TWRRR is the region within TNF-R1 that associates with MOAP-1. (E) Model for RASSF1A and MOAP-1 association with TNF-R1. TNF-α stimulation results in membrane recruitment of MOAP-1 to receptor complexes, followed by receptor internalization, association of monomeric RASSF1A, and promotion of the “open” conformation of MOAP-1 (2). Subsequent activation of Bax results in stimulation of apoptosis. See text for details.
DISCUSSION
RASSF1A is a proapoptotic modulator of death receptor signaling.
In this study, we detail the formation of death receptor complexes with RASSF1A and MOAP-1 (Fig. 8E). MOAP-1 is recruited to TNF-R1 followed by the recruitment of RASSF1A. Without MOAP-1, RASSF1A does not undergo loss of self-association (Fig. 5A), does not associate with TNF-R1 (Fig. 4D), and does not have the ability to evoke TNF-α-dependent apoptosis (Fig. 6D). We propose that MOAP-1 modulates RASSF1A recruitment to TNF-R1. Once monomeric RASSF1A has associated with TNF-R1, it can then modulate MOAP-1 conformational change to produce a more “open” form of MOAP-1 with its BH3-like domain exposed for association with Bax (as described previously [2]). This results in Bax conformational change, insertion into the mitochondrial membrane, and stimulation of apoptosis (Fig. 8E).
RASSF1A/MOAP-1/TNF-R1 associations are predominantly governed by electrostatic interactions.
The open/closed form of MOAP-1 is regulated by an intraelectrostatic interaction within MOAP-1 (involving regions 202KRRR and 178EEEF of MOAP-1) and by an interelectrostatic interaction between MOAP-1 and RASSF1A (involving region 202KRRR of MOAP-1 and region 311EEEEH of RASSF1A) (Fig. 8D) (2). Results in this study continue to support a role for electrostatic associations governing MOAP-1 associations and subsequent activation of apoptosis. In this study, we now identify the C-terminal sequence of MOAP-1 (336EEEEA) as important for both TNF-R1 and TRAIL-R1 association and, correspondingly, identify a basic stretch within the death domain of TNF-R1 (region 408TWRRR) as an important site for MOAP-1 association. MOAP-1 is a highly charged protein with a pI of −9.0 at pH 7.0 and contains 45% polar or charged amino acids. Similarly, RASSF1A has a charge of +13 at pH 7.0 and 50% polar or charged amino acids. Therefore, electrostatic associations between these two proteins were not entirely surprising. Electrostatic associations have been demonstrated to be strong stabilizing forces for protein-protein and protein-DNA associations (24, 54) and they appear to be important for the biology of RASSF1A and MOAP-1. Conceivably, additional contact regions may be required to aid in complex formation between MOAP-1, RASSF1A, and TNF-R1, but electrostatic associations predominate. MOAP-1 can, therefore, potentially associate simultaneously with both TNF-R1 and RASSF1A following TNF-α stimulation, and these electrostatic pairings allow for the exposure of the BH3-like domain and subsequent Bax association.
RASSF1A, MOAP-1, and TNF-R1 associations are assembled as an ordered complex that functions to activate Bax and promote cell death.
In this study, we demonstrate that RASSF1A requires MOAP-1 in order to associate with death receptors following TNF-α stimulation. If RASSF1A cannot bind to MOAP-1 (as demonstrated by our 311EEEEH→311AAAAH mutant of RASSF1A [2]), it loses its ability to associate with TNF-R1 (data not shown). A dynamic order of complex formation with TNF-R1 and apoptotic proteins has been observed for the majority of TNF-R1 complexes, such as TNF-R1/TRAF2/RIP1/IKK and TNF-R1/TRADD/FADD/caspase-8 (20, 32, 39). It is not surprising that this also occurs with the RASSF1A/MOAP-1 proapoptotic pathway, whereby the activation of Bax is controlled by the TNF-R1/MOAP-1/RASSF1A complex (Fig. 8E). Furthermore, without a death receptor signal, RASSF1A most likely exists as a homodimer (as observed in Fig. 2, 5A, and 7B) and MOAP-1 may direct the loss of RASSF1A self-association following TNF-α stimulation (Fig. 5A). The factors that basally maintain RASSF1A self-association are not understood, but preliminary data suggest that RASSF1A self-association is maintained by association between specific 14-3-3 family members and the 14-3-3 binding site on RASSF1A (Chow and Baksh, unpublished data).
In most human cancers, the promoter for exon 1Aα is silenced by epigenetic mechanisms resulting in the lack of expression of RASSF1A (11, 12). We have demonstrated that the C1 domain of RASSF1A (containing the zinc finger binding motifs encoded by exon 1Aα) is required for death receptor association. Most of the cysteine and histidine residues within the C1 domain of RASSF1A are highly conserved in human RASSF5/Nore1A (Fig. 6B) and human RASSF3A (GenBank accession no. AY062002), but not within the other RASSF family members. RASSF5/Nore1A did not associate with TNF-R1 (see Fig. S5B in the supplemental material) due to lack of association with MOAP-1 (data not shown). Sequence analysis revealed that RASSF5/Nore1A contained a potentially disrupted MOAP-1 binding site (compare 310REEEEH of RASSF1A to 385EKEEQDK of RASSF5/Nore1A). We have not investigated the association of RASSF3A with TNF-R1, but we speculate it may also lack TNF-R1 association due to a disrupted MOAP-1 binding site (compare 310REEEEH of RASSF1A to 386KEEQDK of RASSF3A). The other RASSF proteins do not have a conserved C1 domain, but RASSF2, RASSF4, and RASSF6 have potential MOAP-1 binding sites (compare 310REEEEH of RASSF1A to 291QEEEDRE of RASSF2, 289KEEEERE of RASSF4, and 300NEEEKRE of RASSF6). The association of MOAP-1 with these RASSF family members remains to be determined. The tumor suppressor property of RASSF1A is thus mainly governed by its N-terminal region and would suggest a unique role for MOAP-1 in TNF-R1 association and subsequent activation of cell death.
MOAP-1 and RASSF1A associate with defined subpopulations of TNF-R1.
We have observed in U2OS cells that following 1 to 2 h of TNF-α stimulation, 60% of TNF-R1 receptors are on the surface; however, by 3 h only 20% of TNF-R1 receptors remain surface bound, with the remaining 80% internalized (data not shown). Once internalized, TNF-R1 relocalizes to the endosomal vesicles, requiring an intact microtubular network (10, 17, 43). Upon TNF-α stimulation, RASSF1A does not associate with death receptors until approximately 3 h of receptor stimulation (2), suggesting that RASSF1A may associate only with the internalized receptors (treatment at 4°C does not disrupt RASSF1A microtubule localization; data not shown). Analysis of MOAP-1/TNF-R1 and RASSF1A/TNF-R1 receptor complexes at 4°C revealed that MOAP-1 still associated with TNF-R1, whereas RASSF1A did not (see Fig. S5C in the supplemental material). Treatment at 4°C interferes with receptor-mediated endocytosis and thus would prevent internalization of TNF-R1. We speculate that MOAP-1 associated with surface-bound TNF-R1 and RASSF1A associated with the internalized complex of MOAP-1 and TNF-R1.
Once internalized, TNF-R1 appears in endosomes, where it may encounter active associations that will determine the outcome of TNF-α signaling (23, 39), such as the association with RASSF1A. TNF-R1 internalization may be dependent on a stable microtubular network influenced by RASSF1A. The requirement for MOAP-1 association with TNF-R1 and the requirement for internalization of TNF-R1 complexes may explain why RASSF1A does not associate with TNF-R1 until approximately 3 h following TNF-α stimulation. However, if analysis is carried out with TNF-α and CHX stimulation (in order to inhibit protein synthesis and inhibit the NF-κB pathway), we observed a much earlier association of MOAP-1 and RASSF1A with TNF-R1 (see Fig. S5D and E in the supplemental material). Under such conditions, we still observed an initial recruitment of MOAP-1 to TNF-R1 followed by RASSF1A recruitment. We propose that in the presence of CHX treatment, TNF-α signaling will predominately drive apoptosis and not NF-κB activation, resulting in an earlier association with proapoptotic elements, such as MOAP-1 and RASSF1A. Furthermore, the kinetics of complex formation of MOAP-1, RASSF1A, and TNF-R1 is now similar to the kinetics of DISC assembly leading to caspase-8 activation and Bid cleavage.
MOAP-1 and RASSF1A potentially function as key elements in an apoptotic checkpoint in death receptor-dependent cell death.
As mentioned earlier, it has been proposed that shortly after internalization, death receptors (such as TNF-R1) encounter an “apoptotic checkpoint” bringing together complexes to promote survival or stimulate apoptosis. Here we demonstrate that RASSF1A and MOAP-1 may be important components of this “apoptotic checkpoint” and directly influence the direction of TNF-R1 (and most likely TRAIL-R1) signaling. Death receptor stimulation functions to bring together RASSF1A and MOAP-1 to receptor complexes in order for RASSF1A to regulate the conformation of MOAP-1. This is an important step in the activation of Bax and subsequent stimulation of apoptosis. Conceivably, other cell death pathways can promote Bax activation and cell death independent of the RASSF1A/MOAP-1 pathway. These pathways may cooperate with the RASSF1A/MOAP-1 to ensure maximal activation of Bax and promotion of cell death. In this study, we have gained further insights into the role played by RASSF1A in MOAP-1 death receptor-dependent apoptosis. The findings reported here and in our previous study highlight the importance of the RASSF1A/MOAP-1 pathway to death receptor signaling and possibly to the prevention of carcinogenesis.
Supplementary Material
Acknowledgments
We thank Benjamin G. Neel for taking the time to critically read the manuscript. We also thank Timothy Hemesath and the members of the Benjamin G. Neel and Victor C. Yu laboratories for helpful discussions and use of reagents and Geoffrey Clark for the kind gift of a rabbit anti-RASSF5/Nore1A antibody. We thank Christina Onyskiw, Karen Levi, and Paul Semchuk for excellent technical assistance. We thank Bin Qian for help with the Rosetta comparative modeling protocol.
Support for this study was provided by a University of Alberta, Faculty of Medicine and Department of Pediatrics research grant; by a CIHR (MOP-79494) and Alberta Cancer Board (ACB-22107) research grant (S.B.), by NIH R3Z CA49152 (providing initial support for this work in the laboratory of Benjamin G. Neel); by the Office of the Provost and VP Academic Award (S.C.); by the Franklin W. Olin College of Engineering (C.J.F. and J.C.P); and by the Agency of Science, Technology and Research (N.Y.F. and V.C.Y).
Footnotes
Published ahead of print on 12 May 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allen, N. P., H. Donninger, M. D. Vos, K. Eckfeld, L. Hesson, L. Gordon, M. J. Birrer, F. Latif, and G. J. Clark. 2007. RASSF6 is a novel member of the RASSF family of tumor suppressors. Oncogene 266203-6211. [DOI] [PubMed] [Google Scholar]
- 2.Baksh, S., S. Tommasi, S. Fenton, V. C. Yu, L. M. Martins, G. P. Pfeifer, F. Latif, J. Downward, and B. G. Neel. 2005. The tumor suppressor RASSF1A and MAP-1 link death receptor signaling to Bax conformational change and cell death. Mol. Cell 18637-650. [DOI] [PubMed] [Google Scholar]
- 3.Blank, M., and Y. Shiloh. 2007. Programs for cell death: apoptosis is only one way to go. Cell Cycle 6686-695. [DOI] [PubMed] [Google Scholar]
- 4.Burbee, D. G., E. Forgacs, S. Zochbauer-Muller, L. Shivakumar, K. Fong, B. Gao, D. Randle, M. Kondo, A. Virmani, S. Bader, Y. Sekido, F. Latif, S. Milchgrub, S. Toyooka, A. F. Gazdar, M. I. Lerman, E. Zabarovsky, M. White, and J. D. Minna. 2001. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J. Natl. Cancer Inst. 93691-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cain, K., S. B. Bratton, and G. M. Cohen. 2002. The Apaf-1 apoptosome: a large caspase-activating complex. Biochimie 84203-214. [DOI] [PubMed] [Google Scholar]
- 6.Case, D. A., T. E. Cheatham III, T. Darden, H. Gohlke, R. Luo, K. M. Merz, Jr., A. Onufriev, C. Simmerling, B. Wang, and R. J. Woods. 2005. The Amber biomolecular simulation programs. J. Comput. Chem. 261668-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao, D. T., and S. J. Korsmeyer. 1998. BCL-2 family: regulators of cell death. Annu. Rev. Immunol. 16395-419. [DOI] [PubMed] [Google Scholar]
- 8.Chivian, D., D. E. Kim, L. Malmstrom, P. Bradley, T. Robertson, P. Murphy, C. E. Strauss, R. Bonneau, C. A. Rohl, and D. Baker. 2003. Automated prediction of CASP-5 structures using the Robetta server. Proteins 53(Suppl. 6)524-533. [DOI] [PubMed] [Google Scholar]
- 9.Chivian, D., D. E. Kim, L. Malmstrom, J. Schonbrun, C. A. Rohl, and D. Baker. 2005. Prediction of CASP6 structures using automated Robetta protocols. Proteins 61(Suppl. 7)157-166. [DOI] [PubMed] [Google Scholar]
- 10.D'Alessio, A., R. S. Al-Lamki, J. R. Bradley, and J. S. Pober. 2005. Caveolae participate in tumor necrosis factor receptor 1 signaling and internalization in a human endothelial cell line. Am. J. Pathol. 1661273-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dammann, R., C. Li, J. H. Yoon, P. L. Chin, S. Bates, and G. P. Pfeifer. 2000. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 25315-319. [DOI] [PubMed] [Google Scholar]
- 12.Dammann, R., U. Schagdarsurengin, C. Seidel, M. Strunnikova, M. Rastetter, K. Baier, and G. P. Pfeifer. 2005. The tumor suppressor RASSF1A in human carcinogenesis: an update. Histol. Histopathol. 20645-663. [DOI] [PubMed] [Google Scholar]
- 13.Danial, N. N., and S. J. Korsmeyer. 2004. Cell death: critical control points. Cell 116205-219. [DOI] [PubMed] [Google Scholar]
- 14.Dempsey, P. W., S. E. Doyle, J. Q. He, and G. Cheng. 2003. The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev. 14193-209. [DOI] [PubMed] [Google Scholar]
- 15.Denecker, G., D. Vercammen, M. Steemans, T. Vanden Berghe, G. Brouckaert, G. Van Loo, B. Zhivotovsky, W. Fiers, J. Grooten, W. Declercq, and P. Vandenabeele. 2001. Death receptor-induced apoptotic and necrotic cell death: differential role of caspases and mitochondria. Cell Death Differ. 8829-840. [DOI] [PubMed] [Google Scholar]
- 16.Fujita, H., S. Fukuhara, A. Sakurai, A. Yamagishi, Y. Kamioka, Y. Nakaoka, M. Masuda, and N. Mochizuki. 2005. Local activation of Rap1 contributes to directional vascular endothelial cell migration accompanied by extension of microtubules on which RAPL, a Rap1-associating molecule, localizes. J. Biol. Chem. 2805022-5031. [DOI] [PubMed] [Google Scholar]
- 17.Gao, Y., M. Hansson, J. Calafat, H. Tapper, and I. Olsson. 2004. Sorting soluble tumor necrosis factor (TNF) receptor for storage and regulated secretion in hematopoietic cells. J. Leukoc. Biol. 76876-885. [DOI] [PubMed] [Google Scholar]
- 18.Guo, C., S. Tommasi, L. Liu, J. K. Yee, R. Dammann, and G. P. Pfeifer. 2007. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr. Biol. 17700-705. [DOI] [PubMed] [Google Scholar]
- 19.Harjes, E., S. Harjes, S. Wohlgemuth, K. H. Muller, E. Krieger, C. Herrmann, and P. Bayer. 2006. GTP-Ras disrupts the intramolecular complex of C1 and RA domains of Nore1. Structure 14881-888. [DOI] [PubMed] [Google Scholar]
- 20.Harper, N., M. Hughes, M. MacFarlane, and G. M. Cohen. 2003. Fas-associated death domain protein and caspase-8 are not recruited to the tumor necrosis factor receptor 1 signaling complex during tumor necrosis factor-induced apoptosis. J. Biol. Chem. 27825534-25541. [DOI] [PubMed] [Google Scholar]
- 21.Ho, A. T., and E. Zacksenhaus. 2004. Splitting the apoptosome. Cell Cycle 3446-448. [PubMed] [Google Scholar]
- 22.Ikeda, M., S. Hirabayashi, N. Fujiwara, H. Mori, A. Kawata, J. Iida, Y. Bao, Y. Sato, T. Iida, H. Sugimura, and Y. Hata. 2007. Ras-association domain family protein 6 induces apoptosis via both caspase-dependent and caspase-independent pathways. Exp. Cell Res. 3131484-1495. [DOI] [PubMed] [Google Scholar]
- 23.Jones, S. J., E. C. Ledgerwood, J. B. Prins, J. Galbraith, D. R. Johnson, J. S. Pober, and J. R. Bradley. 1999. TNF recruits TRADD to the plasma membrane but not the trans-Golgi network, the principal subcellular location of TNF-R1. J. Immunol. 1621042-1048. [PubMed] [Google Scholar]
- 24.Karshikov, A., and W. Bode. 1993. Electrostatic properties of thrombin: importance for structural stabilization and ligand binding. Semin. Thromb. Hemost. 19334-343. [DOI] [PubMed] [Google Scholar]
- 25.Katagiri, K., A. Maeda, M. Shimonaka, and T. Kinashi. 2003. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat. Immunol. 4741-748. [DOI] [PubMed] [Google Scholar]
- 26.Kazanietz, M. G. 2000. Eyes wide shut: protein kinase C isozymes are not the only receptors for the phorbol ester tumor promoters. Mol. Carcinog. 285-11. [DOI] [PubMed] [Google Scholar]
- 27.Kim, D. E., D. Chivian, and D. Baker. 2004. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32W526-W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuwana, T., M. R. Mackey, G. Perkins, M. H. Ellisman, M. Latterich, R. Schneiter, D. R. Green, and D. D. Newmeyer. 2002. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111331-342. [DOI] [PubMed] [Google Scholar]
- 29.Legler, D. F., O. Micheau, M. A. Doucey, J. Tschopp, and C. Bron. 2003. Recruitment of TNF receptor 1 to lipid rafts is essential for TNFalpha-mediated NF-kappaB activation. Immunity 18655-664. [DOI] [PubMed] [Google Scholar]
- 30.Liu, L., A. Vo, and W. L. McKeehan. 2005. Specificity of the methylation-suppressed A isoform of candidate tumor suppressor RASSF1 for microtubule hyperstabilization is determined by cell death inducer C19ORF5. Cancer Res. 651830-1838. [DOI] [PubMed] [Google Scholar]
- 31.Matallanas, D., D. Romano, K. Yee, K. Meissl, L. Kucerova, D. Piazzolla, M. Baccarini, J. K. Vass, W. Kolch, and E. O'Neill. 2007. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol. Cell 27962-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Micheau, O., and J. Tschopp. 2003. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114181-190. [DOI] [PubMed] [Google Scholar]
- 33.Mikhailov, V., M. Mikhailova, K. Degenhardt, M. A. Venkatachalam, E. White, and P. Saikumar. 2003. Association of Bax and Bak homo-oligomers in mitochondria. Bax requirement for Bak reorganization and cytochrome c release. J. Biol. Chem. 2785367-5376. [DOI] [PubMed] [Google Scholar]
- 34.Oh, H. J., K. K. Lee, S. J. Song, M. S. Jin, M. S. Song, J. H. Lee, C. R. Im, J. O. Lee, S. Yonehara, and D. S. Lim. 2006. Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res. 662562-2569. [DOI] [PubMed] [Google Scholar]
- 35.Ortiz-Vega, S., A. Khokhlatchev, M. Nedwidek, X. F. Zhang, R. Dammann, G. P. Pfeifer, and J. Avruch. 2002. The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene 211381-1390. [DOI] [PubMed] [Google Scholar]
- 36.Praskova, M., A. Khoklatchev, S. Ortiz-Vega, and J. Avruch. 2004. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem. J. 381453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabizadeh, S., R. J. Xavier, K. Ishiguro, J. Bernabeortiz, M. Lopez-Ilasaca, A. Khokhlatchev, P. Mollahan, G. P. Pfeifer, J. Avruch, and B. Seed. 2004. The scaffold protein CNK1 interacts with the tumor suppressor RASSF1A and augments RASSF1A-induced cell death. J. Biol. Chem. 27929247-29254. [DOI] [PubMed] [Google Scholar]
- 38.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234779-815. [DOI] [PubMed] [Google Scholar]
- 39.Schneider-Brachert, W., V. Tchikov, J. Neumeyer, M. Jakob, S. Winoto-Morbach, J. Held-Feindt, M. Heinrich, O. Merkel, M. Ehrenschwender, D. Adam, R. Mentlein, D. Kabelitz, and S. Schutze. 2004. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity 21415-428. [DOI] [PubMed] [Google Scholar]
- 40.Scorrano, L., and S. J. Korsmeyer. 2003. Mechanisms of cytochrome c release by proapoptotic BCL-2 family members. Biochem. Biophys. Res. Commun. 304437-444. [DOI] [PubMed] [Google Scholar]
- 41.Sherwood, V., R. Manbodh, C. Sheppard, and A. D. Chalmers. 2008. RASSF7 is a member of a new family of RAS association domain-containing proteins and is required for completing mitosis. Mol. Biol. Cell 191772-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song, M. S., J. S. Chang, S. J. Song, T. H. Yang, H. Lee, and D. S. Lim. 2005. The centrosomal protein RAS association domain family protein 1A (RASSF1A)-binding protein 1 regulates mitotic progression by recruiting RASSF1A to spindle poles. J. Biol. Chem. 2803920-3927. [DOI] [PubMed] [Google Scholar]
- 43.Storey, H., A. Stewart, P. Vandenabeele, and J. P. Luzio. 2002. The p55 tumour necrosis factor receptor TNFR1 contains a trans-Golgi network localization signal in the C-terminal region of its cytoplasmic tail. Biochem. J. 36615-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan, K. O., N. Y. Fu, S. K. Sukumaran, S. L. Chan, J. H. Kang, K. L. Poon, B. S. Chen, and V. C. Yu. 2005. MAP-1 is a mitochondrial effector of Bax. Proc. Natl. Acad. Sci. USA 10214623-14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan, K. O., K. M. Tan, S. L. Chan, K. S. Yee, M. Bevort, K. C. Ang, and V. C. Yu. 2001. MAP-1, a novel proapoptotic protein containing a BH3-like motif that associates with Bax through its Bcl-2 homology domains. J. Biol. Chem. 2762802-2807. [DOI] [PubMed] [Google Scholar]
- 46.Thorburn, A. 2004. Death receptor-induced cell killing. Cell. Signal. 16139-144. [DOI] [PubMed] [Google Scholar]
- 47.Tommasi, S., R. Dammann, Z. Zhang, Y. Wang, L. Liu, W. M. Tsark, S. P. Wilczynski, J. Li, M. You, and G. P. Pfeifer. 2005. Tumor susceptibility of Rassf1a knockout mice. Cancer Res. 6592-98. [PubMed] [Google Scholar]
- 48.van der Weyden, L., and D. J. Adams. 2007. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim. Biophys. Acta 177658-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Weyden, L., K. K. Tachibana, M. A. Gonzalez, D. J. Adams, B. L. Ng, R. Petty, A. R. Venkitaraman, M. J. Arends, and A. Bradley. 2005. The RASSF1A isoform of RASSF1 promotes microtubule stability and suppresses tumorigenesis. Mol. Cell. Biol. 258356-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vos, M. D., A. Dallol, K. Eckfeld, N. P. Allen, H. Donninger, L. B. Hesson, D. Calvisi, F. Latif, and G. J. Clark. 2006. The RASSF1A tumor suppressor activates Bax via MOAP-1. J. Biol. Chem. 2814557-4563. [DOI] [PubMed] [Google Scholar]
- 51.Vos, M. D., C. A. Ellis, C. Elam, A. S. Ulku, B. J. Taylor, and G. J. Clark. 2003. RASSF2 is a novel K-Ras-specific effector and potential tumor suppressor. J. Biol. Chem. 27828045-28051. [DOI] [PubMed] [Google Scholar]
- 52.Vos, M. D., A. Martinez, C. A. Ellis, T. Vallecorsa, and G. J. Clark. 2003. The pro-apoptotic Ras effector Nore1 may serve as a Ras-regulated tumor suppressor in the lung. J. Biol. Chem. 27821938-21943. [DOI] [PubMed] [Google Scholar]
- 53.Wajant, H., K. Pfizenmaier, and P. Scheurich. 2003. Tumor necrosis factor signaling. Cell Death Differ. 1045-65. [DOI] [PubMed] [Google Scholar]
- 54.Xu, D., S. L. Lin, and R. Nussinov. 1997. Protein binding versus protein folding: the role of hydrophilic bridges in protein associations. J. Mol. Biol. 26568-84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.