Abstract
Translocation of proteins from the cytosol across the mitochondrial inner membrane is driven by the action of the import motor, which is associated with the translocon on the matrix side of the membrane. It is well established that an essential peripheral membrane protein, Tim44, tethers mitochondrial Hsp70 (mtHsp70), the core of the import motor, to the translocon. This Tim44-mtHsp70 interaction, which can be recapitulated in vitro, is destabilized by binding of mtHsp70 to a substrate polypeptide. Here we report that the N-terminal 167-amino-acid segment of mature Tim44 is sufficient for both interaction with mtHsp70 and destabilization of a Tim44-mtHsp70 complex caused by client protein binding. Amino acid alterations within a 30-amino-acid segment affected both the release of mtHsp70 upon peptide binding and the interaction of Tim44 with the translocon. Our results support the idea that Tim44 plays multiple roles in mitochondrial protein import by recruiting Ssc1 and its J protein cochaperone to the translocon and coordinating their interactions to promote efficient protein translocation in vivo.
On the order of 99% of proteins residing in mitochondria are encoded by nuclear DNA and synthesized on cytosolic ribosomes (38). Thus, mitochondrial biogenesis and function are dependent on efficient protein import mechanisms. Proteins destined for the mitochondrial matrix are first recognized by receptors on the outer mitochondrial membrane and then imported and sorted by the action of multiprotein complexes, the TOM and TIM23 complexes, in the inner and outer membranes, respectively (3, 6, 13, 14, 19, 28, 30, 33, 36). Most matrix-bound proteins are synthesized as preproteins with positively charged N-terminal targeting sequences, whose insertion into the TIM23 complex is strictly dependent on a membrane potential across the inner membrane (2, 26). Translocation of the remainder of the preprotein requires the presequence translocase-associated motor complex (PAM) (32, 34).
PAM acts as a functional unit to couple the action of mitochondrial Hsp70 (mtHsp70) chaperone (Ssc1 in Saccharomyces cerevisiae) to the movement of preproteins through the Tim23 translocon into the mitochondrial matrix. The core of the import motor is mtHsp70 (12, 18). The essential, 44-kDa peripheral membrane protein Tim44 recruits Ssc1 to the translocon, where Ssc1's interaction with incoming preproteins is initiated (15, 22, 25, 35, 37, 43). Like other Hsp70s, Ssc1 rapidly interacts with client proteins when it is in the ATP-bound state. The interaction is stabilized by ATP hydrolysis, triggered by interaction with a J protein partner. Pam18 (also known as Tim14), which forms a stable heterodimer with the related protein Pam16 (also known as Tim16), serves this function in the case of import (8, 10, 31, 41). In vitro, the Tim44-Ssc1 complex is destabilized upon binding of peptide substrate, leading to the idea that release of preprotein-bound Ssc1 allows another Ssc1(ATP) to be recruited to the import channel, facilitating further import (7, 23, 40). In addition, Tim44 is involved in tethering the Pam18-Pam16 heterodimer to the translocon, as depletion leads to reduced association (20, 31).
Tim44 plays a central role in protein translocation. Besides acting as a tether that regulates the interaction of Ssc1 with the translocon (7), it is also important for the association of the Pam16-Pam18 heterodimer with the TIM23 translocase (20, 29, 31). Despite these critical roles, little functional information about Tim44 is available. We undertook a comprehensive mutational analysis of the portion of TIM44 encoding the N-terminal 18 kDa of the mature protein, a segment we found to be sufficient for interaction with Ssc1 and for which there is no structural information available (17). Residues critical for association of Tim44 with the TIM23 translocase and for regulation of the stability of the interaction with Ssc1 were identified. Our results are consistent with the existence of a dynamic interaction between Tim44 and both Ssc1 and the translocon, mediated by a small segment of Tim44.
MATERIALS AND METHODS
Site-directed mutagenesis of TIM44.
TIM44 truncations were constructed using a two-step PCR procedure. In the case of tim4443-209, the DNA sequence encoding amino acids 1 to 209 and the 3′-untranslated region of TIM44 were amplified first and then fused in a second round of PCR. For creation of tim44210-431, DNAs encoding amino acids 210 to 431 and 1 to 48, which contain the cleavable mitochondrial targeting sequence (residues 1 to 42) and the processing site (C terminus of amino acid 42), were fused by PCR and cloned into pRS314-TIM44 (23) by replacing the open reading frame of TIM44. TIM44 mutants encoding internal deletions within the N-terminal segment of Tim44 were constructed using a two-step PCR procedure. DNA sequences carrying the 5′-untranslated region and 3′-untranslated region of TIM44, as well as the coding sequences flanking the desired deletions, were amplified first and then fused in a second-round PCR, resulting in an EcoRI restriction site replacing the deleted DNA stretch. The following primers were used: tim44(Δ68-82) sense, 5′ TGG GAG AAG TCT CAG GAA GAA TTC GGC GAG TCT; tim44(Δ68-82) antisense, 5′ AGA GTC CTT GAA TTC CCG CTC AGA CTC CGG ATG; tim44(Δ85-99) sense, 5′ TTA GGC GAG GAA TTC AGA GGC TCC ACA ATT GTG; tim44(Δ85-99) antisense, 5′ GTG TTA ACA CCT CGG AGA CTT AAG GAG CGG ATT; tim44(Δ103-118) sense, 5′ AGT GGC TAT GAA TTC GGA GCC TCT CTG AGC TTT; tim44(Δ103-118) antisense, 5′ AGA GGC TCC GAA TTC ATA GCC ACT AAG GCC TGG; tim44(Δ115-127) sense, 5′ ACC GGT GAG GAA TTC CTC GGT AAG AAC ACA AGA; tim44(Δ115-127) antisense, 5′ CTT ACC GAG GAA TTC CTC ACC GGT CTT TTT CAA; tim44(Δ131-147) sense, 5′ CAC TGG CTC GAA TTC CTT ACC GAG CTC GGA C; tim44(Δ131-147) antisense, 5′ CTC GGT AAG GAA TTC GAG CCA GTG AGA CAG AC; tim44(Δ150-164) sense, 5′ TTT GAG CCA GAA TTC GAT GGG GAG AGT TCC C; tim44(Δ150-164) antisense, 5′ TCT CCC CAT CGA ATT CTG GCT CAA AAC TCT CAT C; tim44(Δ170-185) sense, 5′ GAG AGT TCC GAA TTC AGA GAC CTG GCC TCT GG; tim44(Δ170-185) antisense, 5′ CCA GGT CTC TGA ATT CGG AAC TCT CCC CAT CGT C; tim44(Δ185-202) sense, 5′ GAG ACT TAA ACG TGC AGG AAC AGC AGT GG; and tim44(Δ185-202) antisense, 5′ CTG CTG TTC CTG CAC GTT TAA GTC TCC TTT GC. For alanine scanning mutagenesis of the segment of full-length TIM44 encoding amino acids 126 to 210, a site-directed mutagenesis kit (Stratagene, Heidelberg, Germany) was used, starting with pRS314-TIM44 as a template. Primers were designed and mutagenesis was carried out according to the manufacturer's instructions. For creation of tim44 mutants, the following oligonucleotides were used: tim44(150AAAA) sense, 5′ GAG AGT TTT GAG CCA GCG GCA GCG GCG AAG ATT TAC AAG GAA; tim44(150AAAA) antisense, 5′ TTC CTT GTA AAT CTT CGC CGC TGC CGC TGG CTC AAA ACT CTC; tim44(154AAAA) sense, 5′ CCA GTG AGA CAG ACG GCG GCT GCC GCG GAA GTC TCA GAA GTC; tim44(154AAAA) antisense, 5′ GAC TTC TGA GAC TTC CGC GGC AGC CGC CGT CTG TCT CAC TGG; tim44(170AAAA) sense, 5′ GAT GGG GAG AGT TCC GCA GCC GCT GCG TTT ATC ACG AAA GAG; tim44(170AAAA) antisense, 5′ CTC TTT CGT GAT AAA CGC AGC GGC TGC GGA ACT CTC CCC ATC; tim44(174AAAA) sense, 5′ TCC CGA TAC GGT GGG GCT GCC GCG GCA GAG CAA AGG AGA CTT; tim44(174AAAA) antisense, 5′ AAG TCT CCT TTG CTC TGC CGC GGC AGC CCC ACC GTA TCG GGA; tim44(178AAAA) sense, 5′ GGG TTT ATC ACG AAA GCG GCA GCG GCA CTT AAA CGT GAG AGA; tim44(178AAAA) antisense, 5′ TCT CTC ACG TTT AAG TGC CGC TGC CGC TTT CGT GAT AAA CCC; tim44(R180A) sense, 5′ ATC ACG AAA GAG CAA GCG AGA CTT AAA CGT GAG; tim44(R180A) antisense, 5′ CTC ACG TTT AAG TCT CGC TTG CTC TTT CGT GAT; tim44(R180K) sense, 5′ ATC ACG AAA GAG CAA AAG AGA CTT AAA CGT GAG; and tim44(R180K) antisense, 5′ CTC ACG TTT AAG TCT CTT TTG CTC TTT CGT GAT. The mutated TIM44 DNA sequences were then subcloned into pRS314-TIM44, using intrinsic restriction sites for PstI and BglII within TIM44. The presence of the expected mutations was verified by sequencing of the respective mutagenized open reading frames of TIM44.
Protein purification and size-exclusion chromatography.
N-terminally six-His-tagged wild-type Tim44, Tim44 variants, and terminally truncated Tim44 proteins were purified from the yeast strain BJ3497 (pep4::HIS3 ura3-52 his3Δ200), using the expression plasmid pYES 2.0 (Invitrogen Corp., Carlsbad, CA), as described previously (16, 23). Purification of His-tagged Ssc1 was carried out as described previously (24).
To assess the protein-protein interaction in vitro, purified Tim44 and Ssc1 were incubated at a molar ratio of 2:1 (Tim44 to Ssc1) in the presence of ATP for 60 min at 23°C. The reaction mixture was subjected to chromatography on a Superdex HiLoad 16/60 column (GE Healthcare BioSciences AB, Uppsala, Sweden) run at 1 ml/min with column buffer (25 mM HEPES-KOH, pH 7.4, 80 mM KCl, 10 mM MgCl2, 10% glycerol, and 0.05% Triton X-100) at 4°C. Five-hundred-microliter fractions were collected, and aliquots from alternate fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting using Ssc1- and Tim44-specific antibodies. Signals were quantitated by densitometry using MacBAS 2.0 (Fuji) software.
Testing of in vivo phenotypes and Tim44 expression.
To test the ability of TIM44 mutants to rescue the lethal phenotype of a chromosomal deletion mutant of TIM44, yeast strain Y1182 (his3 leu2 lys2-801 trp1 ura3-1 ade2-1 met2 tim44::LYS2) harboring plasmid ycplac33-TIM44 (35) was transformed with pRS314 plasmids encoding wild-type or the indicated mutant TIM44 under the control of the TIM44 promoter (see Table 1 for all plasmids used). Cells having lost ycplac33-TIM44 were selected on 5-fluoroorotic acid (5-FOA), which inhibits growth of cells retaining the URA3-containing plasmid ycplac33-TIM44, since the functional expression of the URA3 gene (encoding orotidine-5′-monophosphate decarboxylase and critical for a step in the biosynthesis of uracil) converts the otherwise nontoxic form of 5-FOA to 5-fluorouracil, which is toxic (4). For testing of the level of expression of Tim44 variants that could not maintain viability, cell extracts were prepared from initial transformants harboring both the plasmid encoding wild-type Tim44 and a plasmid encoding a Tim44 variant. Cell extracts were prepared by incubating a cell pellet equivalent to an optical density at 600 nm of 0.15 in 1 M NaOH for 5 min at 23°C. Cells were sedimented and resuspended in 2× SDS-PAGE sample buffer containing 1 mM phenylmethylsulfonyl fluoride (PMSF). After being mixed vigorously for 5 min at 23°C, the cells were boiled for 3 min. After a clarifying spin, the supernatant was subjected to SDS-PAGE and immunoblot analysis using Tim44-specific antibodies.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| YCplac33-TIM44 | YCplac33(URA3) carrying complete TIM44 gene | 35 |
| pRS314 | pBluescript; TRP1 CEN6 ARSH4 yeast shuttle vector | 39 |
| pRS314-Tim44 | Encodes full-length Tim44 under control of native TIM44 promoter | 23 |
| pRS314-Tim4443-209 | Encodes N-terminal 209 amino acids under control of native TIM44 promoter | This study |
| pRS314-Tim44210-431 | Encodes Tim441-48 and Tim44210-431 under control of native TIM44 promoter | This study |
| pRS314-Tim44Δ68-82 | Tim44 with internal deletion of codons 68 to 82 | This study |
| pRS314-Tim44Δ85-99 | Tim44 with internal deletion of codons 85 to 99 | This study |
| pRS314-Tim44Δ103-118 | Tim44 with internal deletion of codons 103 to 118 | This study |
| pRS314-Tim44Δ115-127 | Tim44 with internal deletion of codons 115 to 127 | This study |
| pRS314-Tim44Δ131-147 | Tim44 with internal deletion of codons 131 to 147 | This study |
| pRS314-Tim44Δ150-164 | Tim44 with internal deletion of codons 150 to 164 | This study |
| pRS314-Tim44Δ170-185 | Tim44 with internal deletion of codons 170 to 185 | This study |
| pRS314-Tim44Δ185-202 | Tim44 with internal deletion of codons 185 to 202 | This study |
| pRS314-Tim44150AAAA | Tim44 with V150A, R151A, Q152A, and T153A substitutions | This study |
| pRS314-Tim44154AAAA | Tim44 with K154A, I155A, Y156A, and K157A substitutions | This study |
| pRS314-Tim44170AAAA | Tim44 with R170A, Y171A, G172A, and G173A substitutions | This study |
| pRS314-Tim44174AAAA | Tim44 with F174A, I175A, T176A, and K177A substitutions | This study |
| pRS314-Tim44178AAAA | Tim44 with E178A, Q179A, R180A, and R181A substitutions | This study |
| pRS314-Tim44R180A | Tim44 with R180A substitution | This study |
| pRS314-Tim44R180K | Tim44 with R180K substitution | This study |
| pYES | Yeast protein expression vector with the GAL1 promoter | Invitrogen |
| pYES-Tim44-His tag | Tim44 with C-terminal His tag | 23 |
| pYES-Tim4443-209-His tag | Tim4443-209 with C-terminal His tag | This study |
| pYES-Tim44210-431-His tag | Tim44210-431 with C-terminal His tag | This study |
| pYES-Tim44174AAAA-His tag | Tim44174AAAA with C-terminal His tag | This study |
| pYES-Tim44R180A-His tag | Tim44R180A with C-terminal His tag | This study |
To test for the accumulation of precursors in vivo, tim44 strains were grown in minimal medium at 23°C to mid-logarithmic phase and then shifted to 37°C for 6 h. Cell extracts (optical density at 600 nm, 0.2) were prepared and analyzed by SDS-PAGE and immunoblotting using antibodies specific for the mitochondrial matrix protein Hsp60.
Isolation and analysis of mitochondria.
Mitochondria were isolated as described previously (24). For submitochondrial protein localization studies, mitochondria were diluted to 0.1 mg/ml in sonication buffer (25 mM HEPES-KOH, pH 7.4, 50 mM KCl, 10 mM magnesium acetate, and 1 mM PMSF) and subjected to sonication five times for 5 seconds each on ice at output level 4 (W-220 sonicator; Heat Systems-Ultrasonics, Farmingdale, NY). After a clarifying spin (3,000 × g for 5 min), mitochondria were separated into membrane and soluble fractions at 250,000 × g for 20 min. The membrane fraction was washed once with sonication buffer. Proteins in the soluble fraction were precipitated with trichloroacetic acid, which was added to a final concentration of 15% (wt/vol). After 30 min on ice, the supernatant was centrifuged at 16,000 × g at 4°C for 15 min. The sediment was washed twice with ice-cold acetone. The dried pellet was resuspended in SDS-PAGE sample buffer (soluble fraction). Total, soluble, and membrane fractions were analyzed by SDS-PAGE and immunoblotting using antibodies specific for Tim44, Mge1, and Tim23. To test whether Tim44 variants were prone to aggregation at elevated temperatures, energized mitochondria were incubated at 37°C or, as a control, on ice for 30 min. Subsequently, mitochondria were lysed with Triton X-100 for 10 min at 4°C. Lysates were subjected to ultracentrifugation at 106,000 × g for 1 h at 4°C, as described previously (5). Solubilized and insoluble fractions were analyzed by SDS-PAGE and immunoblotting.
In vitro import of precursor proteins was carried out as described previously (21, 42). Radiolabeled cytochrome b2(220)Δ19-dihydrofolate reductase [cytochrome b2(220)Δ19-DHFR] was synthesized by in vitro transcription and translation of the precursor from an SP6 promoter in rabbit reticulocyte lysate in the presence of [35S]methionine, using a TNT coupled reticulocyte lysate system (Promega, Madison, WI). The precursor was partially purified by precipitation with ammonium sulfate and ultracentrifugation and incubated with energized mitochondria. Import reactions were stopped by the addition of 1 μM valinomycin and cooling on ice. Following protease treatment with proteinase K (ratio of mitochondrial protein to protease mass, 10:1) for 20 min on ice to remove unimported precursor, protease-resistant Cytb2(220)Δ19-DHFR was detected by SDS-PAGE and autoradiography.
Coimmunoprecipitation in mitochondrial lysates.
To analyze the interaction of Tim44 with Ssc1 in mitochondrial lysates, mitochondria (100 μg protein) were lysed in lysis buffer (20 mM morpholinepropanesulfonic acid [MOPS]-KOH, pH 7.2, 250 mM sucrose, 80 mM KCl, 0.2% [vol/vol] Triton X-100, and 1 mM PMSF) in the presence of either 1 mM ATP and 10 mM magnesium acetate or 5 mM EDTA for 20 min on ice. Nonsolubilized material was sedimented at 20,000 × g for 15 min. The supernatant was incubated with antibodies against Tim44 cross-linked to protein A beads for 1 h at 4°C, as described previously (23). Tim44, Mge1, and Ssc1 bound to the beads were detected by immunoblot analysis using Tim44-, Ssc1-, and Mge1-specific antibodies.
For analysis of the association of Tim44, Pam16, and Pam18 with the TIM23 translocase, mitochondria were lysed in lysis buffer (25 mM Tris-Cl, pH 7.5, 80 mM KCl, 5 mM EDTA, 10% [vol/vol] glycerol, 1% [wt/vol] digitonin, 1 mM PMSF) for 1 h at 4°C with gentle agitation. Insoluble material was sedimented at 20,000 × g for 20 min. The supernatant was incubated with affinity-purified Tim23-specific antibodies cross-linked to protein A beads for 1.5 h at 4°C. The beads were sedimented and washed three times with lysis buffer. Proteins bound to the beads were eluted with SDS-PAGE sample buffer and detected by immunoblot analysis. As a control for unspecific binding of PAM components, preimmune serum and purified Pam16, Pam18, or Tim44 were added to anti-Tim23 beads. No unspecific binding of these proteins to antibodies against Tim23 cross-linked to protein A beads was detected.
Miscellaneous.
Affinity purification of antibodies against Tim23, Tim44, and Ssc1 and cross-linking of the purified antibodies to protein A beads by use of dimethylpimelimidate dihydrochloride were carried out as described previously (9, 23). Immunoblot analysis was carried out using an ECL system (Amersham Pharmacia Biotech) according to the manufacturer's instructions, using polyclonal antibodies specific for Tim17, Tim23, Tim50, Tim44, Pam16, and Pam18 (9). All results presented were obtained in a minimum of two independent experiments.
RESULTS
The N-terminal segment of Tim44 interacts with Ssc1.
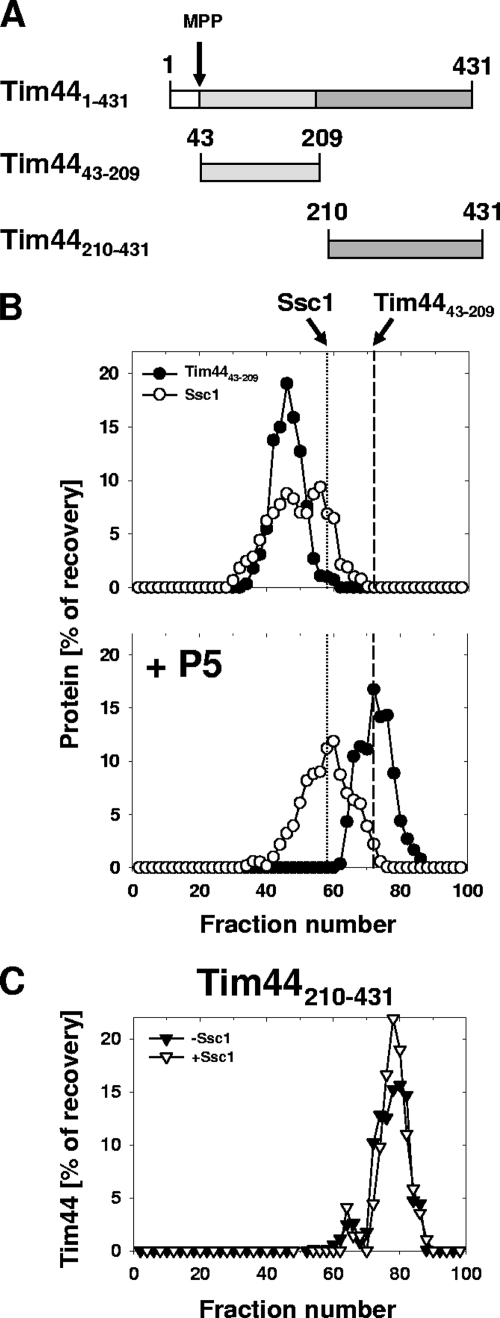
As a first step in our analysis, we set out to find a fragment of Tim44 that retained the ability to bind Ssc1. During purification of Tim44, we observed a stable degradation product that encompassed amino acids 210 to 431 (Fig. 1A). Thus, as an initial test, we proceeded to purify Tim4443-209, providing us with two fragments that encompassed the entirety of the mature form of Tim44. Since the N-terminal 42 amino acids of the Tim44 preprotein are removed upon translocation into the matrix, Tim4443-209 represents the N-terminal segment of functional Tim44. The interaction of the purified truncated Tim44 proteins with Ssc1 was assessed by size-exclusion chromatography. Ssc1 with ATP bound [Ssc1(ATP)] was incubated with the Tim44 fragments prior to being loaded on the column. Similar to what had been observed with full-length proteins (7, 23), a portion of Tim4443-209 coeluted with Ssc1 (Fig. 1B, top panel). However, no interaction between Ssc1 and Tim44210-431 was observed, as there was no significant change in migration of Tim44210-431 in the presence of Ssc1 (Fig. 1C and data not shown).
FIG. 1.
Interaction of Tim44 fragments with Ssc1. (A) Diagram of Tim44 and two fragments used in this study. Tim4443-209, N terminus of mature Tim44; Tim44210-431, C terminus. The arrow indicates the site of cleavage by the matrix processing protease (MPP) to generate mature Tim44. (B) (Top) Tim4443-209 and full-length Ssc1 were incubated together at a molar ratio of 2:1 (Tim44 to Ssc1) for 60 min in the presence of ATP. (Bottom) Same as top panel, except that the Tim4443-209-Ssc1 mixture was incubated for an additional 5 min with substrate peptide P5 prior to analysis. The mixtures were subjected to chromatography, and eluted fractions were analyzed by SDS-PAGE and immunoblotting using Tim44- or Ssc1-specific antibodies. Migration of Ssc1 alone (short dashes) and Tim4443-209 alone (long dashes) is indicated by vertical lines. (C) Tim44210-431 and full-length Ssc1 were incubated together in the presence of ATP (+Ssc1). As a control, Tim44210-431 alone was subjected to chromatography (−Ssc1). Analysis of samples was done as described for panel B.
As previously shown (7, 23), the Tim44-Ssc1(ATP) complex is destabilized upon addition of a peptide substrate, such as a portion of the matrix-targeting sequence of chicken aspartate aminotransferase (CALLLSAPRR), called P5, in vitro. To test whether this peptide substrate-induced destabilization could also be observed with the N-terminal fragment of Tim44, we added peptide P5 to a preformed Tim4443-209-Ssc1(ATP) complex prior to loading it on the size-exclusion column. The elution profiles of both proteins were shifted to positions corresponding to those of the individual proteins alone (Fig. 1B; data not shown). Thus, the peptide substrate efficiently facilitated the destabilization of the Tim4443-209-Ssc1(ATP) complex, indicating that the N-terminal segment of mature Tim44 (residues 43 to 209) is sufficient for physical interaction with Ssc1 and for responding to peptide binding by Ssc1.
Mutational analysis of the N-terminal segment of Tim44.
We first asked if either the N- or C-terminal fragment of Tim44 was capable of substituting for the full-length protein in vivo. TIM44 mutant plasmids were transformed into a tim44Δ strain harboring a plasmid carrying a wild-type copy of TIM44 as well as the URA3 gene. Plasmids harboring the mutant genes designated tim44Δ210-431 and tim44Δ43-209, thus producing Tim4443-209 and Tim44210-431, respectively, upon removal of the presequence (amino acids 1 to 42) by the matrix processing protease, were used for this analysis. The presence of the URA3 gene allowed for selection of cells that had lost the plasmid carrying the wild-type TIM44 gene, as 5-FOA is toxic to cells having a functional uracil biosynthetic pathway but not to cells lacking a functional URA3 gene (see Materials and Methods). No colonies were obtained after incubating the transformants on 5-FOA-containing plates at 23, 30, or 37°C. We concluded that neither Tim4443-209 nor Tim44210-431 is capable of substituting for Tim44 in vivo, even though they are expressed at levels similar to that of wild-type protein and imported into the mitochondrial matrix (data not shown). We concluded that both segments of Tim44 play important roles in vivo. As described below, we focused our analysis on the N-terminal fragment because of its ability to interact with Ssc1.
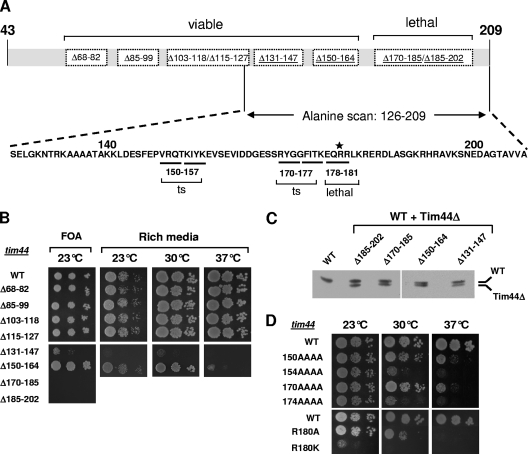
As a first step, we constructed eight mutant TIM44 genes, each with a deletion of 12 to 17 codons predicted to encode an entire alpha helix (Fig. 2A and data not shown). We also included in our analysis tim44Δ185-202, a previously reported null mutant (27). All TIM44 genes were under the control of the native TIM44 promoter. To test the ability of the genes to function in vivo, we utilized the tester strain as described above (Fig. 2B). tim44Δ170-185 and tim44Δ185-202 cells were not recovered on 5-FOA plates. The other six strains expressing only the Tim44 variant were viable. The four strains with Tim44 deletions within the interval of amino acids 68 to 127 grew as well as the wild type at a variety of temperatures on several different carbon sources (Fig. 2B and data not shown). However, all strains with deletions within the region of amino acids 131 to 202 had severe consequences. The tim44Δ131-147 mutant grew very slowly at all temperatures tested; the tim44Δ150-164 mutant grew robustly at 23 and 30°C but poorly at 37°C. To determine whether any phenotypic effects were due to expression levels, extracts from tester strains harboring both plasmids, the one carrying wild-type TIM44 and one carrying a mutant TIM44 gene under the control of the native TIM44 promoter, were subjected to immunoblotting using Tim44-specific antibodies. Wild-type Tim44 migrated slightly more slowly than the mutant proteins did, allowing comparison of protein levels. Similar levels of mutant and wild-type protein were expressed in each case (Fig. 2C), indicating that amino acids within residues 131 to 202 of full-length Tim44 are critical for normal cell growth.
FIG. 2.
Mutational analysis of TIM44. (A) Overview of the phenotypes of tim44 mutants. (Top) Eight deletion mutants lacking 12 to 17 amino acids within the interval of residues 68 to 202 of Tim44 (dotted rectangles). Deletion mutations causing phenotypic effects are underlined (see panel B). (Bottom) Alanine scanning of the segment containing residues 126 to 209. Twenty-one quadruple mutants substituting four alanines for four consecutive native residues were constructed. Amino acid substitutions leading to growth defects are underlined. Residue R180, which is critical for Tim44's in vivo function, is labeled with an asterisk. ts: temperature sensitive. (B) Growth phenotypes of tim44Δ strains carrying deletions within the coding sequence for the N-terminal segment of Tim44. Tenfold serial dilutions of cells were plated on the indicated medium. (Left) Medium containing 5-FOA. tim44Δ strains expressing the indicated plasmid-encoded mutant and wild-type (WT) TIM44 genes were expressed at 23°C for 4 days. (Right) Strains recovered from plating on 5-FOA were spotted onto rich glucose-based medium (rich) and incubated for 3 days (23°C) or 2 days (30°C and 37°C). (C) Protein expression. Cell extracts of tim44Δ strains expressing both the indicated mutant and WT TIM44 genes were analyzed by SDS-PAGE and immunoblotting using Tim44-specific antibodies. Cells harboring both WT and mutant TIM44 genes on different plasmids were grown at 23°C in minimal medium to select for the presence of both plasmids. See Materials and Methods for details. (D) Growth phenotypes of TIM44 amino acid substitution mutants. Cells were plated onto rich medium and incubated at the indicated temperatures for 3 days.
Deletion mutations can cause phenotypic effects due to drastic effects on overall protein conformation. Therefore, alanine scanning mutagenesis was performed with the segment of TIM44 encoding amino acids 126 to 210, creating 21 mutants, with substitutions of four consecutive native residues in each case. Sixteen mutants displayed no obvious growth defect on either fermentable or nonfermentable carbon sources at our three test temperatures (data not shown). These included the mutants carrying substitutions within the segments deleted in the tim44Δ131-147 and tim44Δ185-202 mutants, suggesting that the effects caused by the deletions were indirect, likely affecting the overall protein conformation. On the other hand, the mutant encoding alanines at positions 178 to 181, referred to as tim44178AAAA, was inviable. tim44150AAAA, tim44154AAAA, tim44170AAAA, and tim44174AAAA mutants displayed differing severities of temperature-sensitive growth defects (Fig. 2D). All grew similarly to a wild-type strain at 23°C, with the tim44154AAAA and tim44174AAAA mutants displaying strong defects at 30°C as well as 37°C.
Single alanine substitution mutations were made in the regions encoding segments that displayed the most severe phenotypes when four amino acids were altered, i.e., amino acids 154 to 157, 174 to 177, and 178 to 181. Only very mild or no growth defects were observed for the single alanine substitution mutants in these intervals, with the sole exception being the R180 substitution. The tim44R180A mutant grew robustly at 23°C but not at 30 and 37°C; the conservative substitution R180K had a more severe effect, as the tim44R180K mutant was unable to form colonies at 30°C and grew only poorly at 23°C (Fig. 2D). Thus, R180 is a critical residue, with additional changes in adjacent residues leading to the inviability of the tim44178AAAA mutant. In the case of the tim44174AAAA and tim44154AAAA mutants, the phenotypic effect was probably due to cumulative effects of the individual alterations. For further analysis, we focused on amino acid substitution mutants that showed significant in vivo phenotypes, choosing the tim44R180A, tim44R180K, tim44174AAAA, and tim44154AAAA mutants.
Substitutions in Tim44's N-terminal region cause protein import defects.
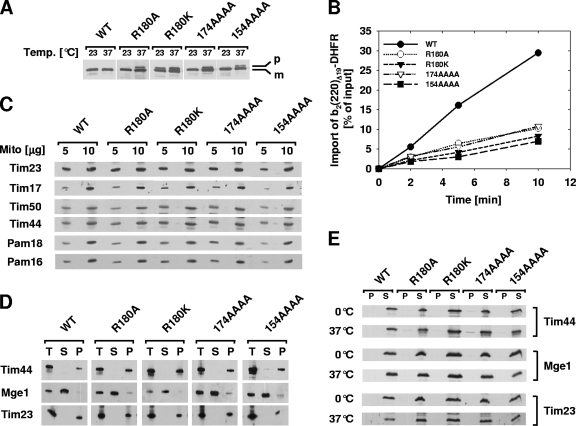
As a first step in the analysis of the four TIM44 mutants, we asked whether the observed growth phenotypes correlated with defects in the import of mitochondrial preproteins in vivo. Import of the chaperonin Hsp60 into mitochondria was assessed by monitoring the removal of the N-terminal targeting sequence, which is cleaved off by the matrix processing peptidase. Wild-type and mutant cells were grown at 23°C and then shifted to 37°C, the temperature at which all the mutant strains showed a growth defect (Fig. 2D). In wild-type cells, cleavage of the presequence occurs rapidly (12, 18); thus, the Hsp60 preprotein was undetectable (Fig. 3A). However, the tim44 mutant strains accumulated the precursor form of Hsp60 at the nonpermissive temperature of 37°C, indicating an import defect in vivo. The tim44R180K mutant also accumulated precursor at 23°C, consistent with its more severe growth defect (Fig. 2D).
FIG. 3.
Translocation defect of TIM44 mutants. (A) Accumulation of a preprotein in vivo. Cell extracts from tim44 mutant strains grown at 23°C and subsequently shifted to 37°C for 6 h were analyzed by SDS-PAGE and immunoblotting using Hsp60-specific antibodies. p, premature Hsp60; m, mature Hsp60. (B) Import of the recombinant precursor cytochrome b2(220)Δ19-DHFR into tim44 mitochondria. Radiolabeled cytochrome b2(220)Δ19-DHFR was incubated with energized mitochondria for the indicated times. Following protease treatment to remove unimported precursor, cytochrome b2(220)Δ19-DHFR was detected by SDS-PAGE and autoradiography. (C) Steady-state protein levels in tim44 mitochondria. Five and 10 μg of total mitochondrial protein was subjected to SDS-PAGE and immunoblotting using antibodies against the indicated components of the TIM23 translocase and the PAM. (D) Fractionation of tim44 mitochondria. Purified mitochondria were fractionated by sonication followed by ultracentrifugation. The distributions of the respective Tim44 variants in the total (T) and soluble (S) fractions and the pellet containing the membranes (P) were monitored using Tim44-specific antibodies. As controls, the presence of the matrix-soluble Mge1 and the inner membrane protein Tim23 was also assessed. (E) Solubility of mutant Tim44 proteins after temperature upshift. Energized mitochondria were incubated for 30 min at the indicated temperatures. After solubilization with detergent, mitochondrial lysates were subjected to ultracentrifugation to separate them into supernatant (S) and pellet (P) fractions that were subsequently subjected to SDS-PAGE and immunoblotting using antibodies against Tim44, Mge1, and Tim23.
Import of a recombinant precursor protein into mitochondria isolated from the respective tim44 mutant strains was also tested. We utilized cytochrome b2(220)Δ19-DHFR, a precursor protein composed of mouse DHFR fused to the N-terminal 220 amino acids of cytochrome b2 with a deletion of 19 amino acids within the inner membrane sorting signal to ensure sorting to the matrix rather than to the inner membrane (42). Radiolabeled cytochrome b2(220)Δ19-DHFR was incubated with isolated wild-type and tim44 mitochondria. After 10 min, approximately 30% of the precursor offered had been imported into wild-type mitochondria (Fig. 3B). In contrast, only 5 to 10% was imported into tim44 mitochondria in the same period. To test whether the observed import defects were due to altered expression of either the mutant Tim44 proteins or components of the import motor and the translocon, we monitored the steady-state levels of components of the PAM import motor and the TIM23 translocase. No apparent differences in the protein levels in the mutants were detected compared to the wild type (Fig. 3C). We therefore concluded that the four tim44 mutants are defective in translocation of preproteins into the mitochondrial matrix.
Next, we performed a series of tests to generally assess proper membrane association of the mutant Tim44 proteins. To test the possibility that the protein import defect might be due to a mislocalization of the Tim44 variants within submitochondrial compartments, we carried out fractionation experiments by sonication of isolated mitochondria and immunoblot analyses of soluble and membrane fractions (Fig. 3D). Like wild-type Tim44, the majority of the mutant Tim44 proteins were localized to the insoluble membrane fraction. Furthermore, the Tim44 mutant proteins could be extracted from the membrane by alkaline carbonate extraction, as is characteristic of membrane-associated proteins (data not shown). Also, since the temperature sensitivity of a TIM44 mutant, tim44-8, was reported to be due to aggregation of the mutant protein at elevated temperatures (5), we tested the solubility of the mutant Tim44 proteins as in that study. Energized mitochondria from wild-type and mutant strains were incubated at 0°C or 37°C, lysed with detergent, and separated into soluble and insoluble fractions by ultracentrifugation. Tim23 and Tim44 were found exclusively in the soluble fraction, as was the matrix-soluble nucleotide exchange factor Mge1 (Fig. 3E), indicating that the observed phenotypes of the tim44 strains are not caused by depletion of functional Tim44 caused by protein aggregation. In sum, our results support the idea that the alterations present in Tim44R180A, Tim44R180K, Tim44174AAAA, and Tim44154AAAA directly affect the ability of Tim44 to carry out its in vivo functions.
N-terminal region of Tim44 functionally interacts with Ssc1.
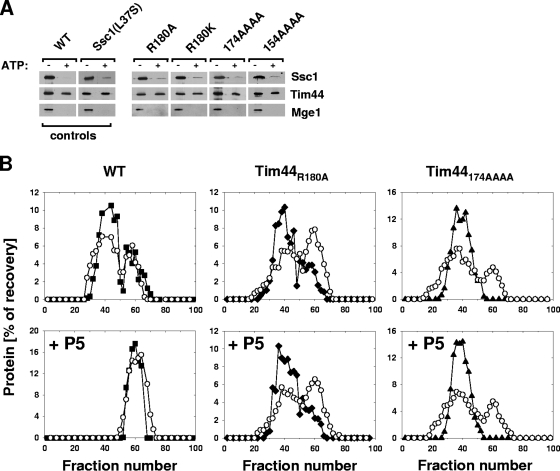
Since Tim44 tethers Ssc1 to the translocon, we next assessed the Ssc1-Tim44 interaction by coprecipitation from mitochondrial extracts, using Tim44-specific antibodies. Two control extracts, from the wild type and an ssc1L37S mutant, were tested. As expected, Mge1 and Ssc1 efficiently coprecipitated with Tim44 from wild-type extracts in the absence of ATP, but no detectable coprecipitation was observed in the presence of ATP (Fig. 4A). This lack of precipitation in the presence of ATP, even though purified Ssc1 and Tim44 interact under these conditions (23), is likely due to the presence in the extracts of Hsp70 client proteins, such as partially unfolded proteins, that destabilize the Tim44-Hsp70 interaction. In the case of Ssc1L37S (7), the interaction between the two proteins is not destabilized as efficiently upon peptide substrate binding by the mutant Ssc1 protein. In coprecipitation experiments with ssc1L37S extracts, coimmunoprecipitation is observed in the presence of ATP, albeit at much lower levels than in its absence. Ssc1 was efficiently coprecipitated from extracts of tim44R180A, tim44R180K, tim44174AAAA, and tim44154AAAA mutants in the absence of ATP. However, in addition, Ssc1 coimmunoprecipitated with all of the Tim44 variants at a significant level when ATP was present, similar to what was observed with ssc1L37S extracts (Fig. 4A).
FIG. 4.
Interaction of Tim44 variants with Ssc1. (A) In organellar. Mitochondrial lysates prepared in the presence (+) or absence (−) of ATP were subjected to immunoprecipitation with Tim44-specific antibodies, followed by immunoblotting with the indicated antibodies. (B) In vitro. (Top) Indicated Tim44 proteins (closed symbols) and Ssc1 (open circles) were incubated in the presence of ATP and then subjected to size-exclusion chromatography. Eluted fractions were analyzed by SDS-PAGE and immunoblotting using Tim44- or Ssc1-specific antibodies. (Bottom) Same as top panels, except that peptide substrate (P5) was added after formation of the Tim44-Ssc1 complex, prior to chromatography.
The results from experiments with mutant extracts suggested that although the mutant proteins interact with Ssc1, destabilization by substrate peptide binding might be affected. To test this idea more directly, we purified Tim44R180A, Tim44174AAAA, and as a control, wild-type Tim44. Their interaction with Ssc1 in the presence of ATP was monitored by size-exclusion chromatography. When they were incubated together, portions of Ssc1 and Tim44 migrated as a complex, as expected (7, 23). Wild-type Tim44-Ssc1 complexes eluted at about fraction number 40, whereas the noncomplexed proteins eluted at fractions around fraction 60. Both Tim44 variants formed a complex with Ssc1 in the presence of ATP, showing profiles similar to that of the wild-type control (Fig. 4B, top panels; data not shown). To address the effect of peptide on mutant Tim44-Ssc1 interactions, we added peptide to preformed Ssc1(ATP)-Tim44 complexes and subsequently monitored the effect on complex stability by chromatography. Consistent with the observations in mitochondrial lysates (Fig. 4A), the wild-type Tim44-Ssc1 complex was efficiently dissociated upon addition of peptide substrate, as no complex was observed (Fig. 4B, bottom left panel). In contrast, preformed complexes of Tim44R180A or Tim44174AAAA with Ssc1(ATP) remained intact during the course of the experiment. Like wild-type protein, the mutant proteins formed a complex with Ssc1(ADP) which was not destabilized upon peptide addition (data not shown). Together, these results indicate that the amino acid substitutions within Tim44 alter the structure of the protein such that a Tim44-Ssc1 complex is not rapidly destabilized upon addition of peptide substrate.
Reduced association of Tim44 variants with the TIM23 translocase.
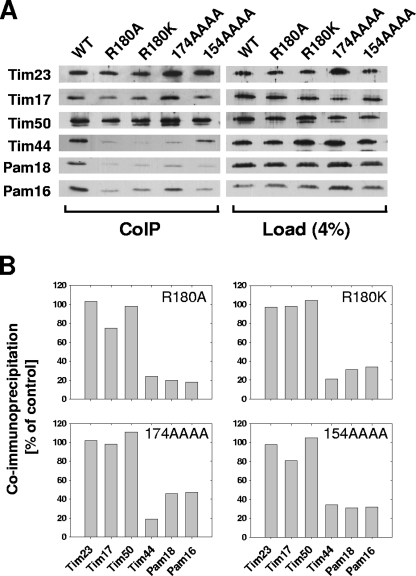
Since Tim44 acts as a tether for the components of the import motor of the translocon, we tested whether alterations in Tim44 affected its own association with the translocon. To do so, we carried out coimmunoprecipitation experiments with lysates generated by treatment of mitochondria with digitonin, which disrupts the membrane but preserves the interaction between the PAM components and the translocon (9). Using affinity-purified antibodies against Tim23, efficient coprecipitation of two other components of the translocon, Tim17 and Tim50, as well as Tim44 and components of the import motor, Pam16 and Pam18, was observed (Fig. 5A). However, only between 15 and 30% as much of the variant Tim44 proteins was coprecipitated, even though the steady-state protein levels of Tim23, Tim17, and Tim50 in the solubilized fraction (loading control) were similar in all mitochondria tested (Fig. 5B; also see Fig. 3C). As expected, the relative amounts of coprecipitated Pam16 and Pam18 were also significantly reduced. These results indicate that the mutations in the N-terminal region of Tim44 compromise the association of Tim44 with the translocon.
FIG. 5.
Association of the import motor with the TIM23 translocase. (A) Mitochondria were solubilized by treatment with digitonin. Solubilized material was subjected to immunoprecipitation using Tim23-specific antibodies cross-linked to protein A beads. Precipitates were analyzed by SDS-PAGE and immunoblotting using antibodies specific for the indicated proteins (IP). A total of 4% solubilized material was used as a loading control (load). (B) Signals were quantified by densitometry and plotted as percentages of the wild-type control reaction value.
DISCUSSION
The results presented here indicate that a small segment of Tim44, a central component of the presequence translocase-associated import motor, is critical for at least the following two events that are important for effective protein import into the mitochondrial matrix in vivo: (i) the polypeptide substrate-induced destabilization of the Ssc1-Tim44 complex and (ii) the association of Tim44, and thus the entire import motor, with the translocon itself.
In addition to these general conclusions, our analysis revealed an unexpected functional resiliency of Tim44. Despite comprehensive alanine scanning mutagenesis, only one residue of the N-terminal segment, R180, was found to result in significant defects in vivo when altered individually. The functional resiliency of the N-terminal segment of Tim44, coupled with the demonstration of its sufficiency for interaction with Ssc1, suggests that the two proteins may interact over a broad interface. More structural information about a Tim44-Ssc1 complex is required to test this hypothesis. But the fact that the identified mutations caused defects in destabilization of the Tim44-Ssc1 complex upon binding of peptide substrate suggests that this aspect of the interaction may be particularly sensitive to small conformational changes in Tim44. The observation that the phenotypic effect of a conservative substitution in the highly conserved R180 residue (1), that is, R180K, was even more pronounced than that of the alanine substitution, is consistent with this idea, as it suggests that it is not simply the charge of this region of Tim44 that is critical.
The mutant proteins analyzed here associated less stably with the TIM23 translocase, in addition to being defective in destabilization of the Ssc1-Tim44 interaction. Thus, it is possible that these sequences directly interact with the translocon. It is tempting to speculate that this region of Tim44 may serve as a “sensor,” being responsive to the presence of a polypeptide in the channel and transmitting a signal to the import motor. Such a signal could serve several roles to make import a more efficient process. (i) A signal could activate Pam18 such that its stimulation of Ssc1's ATPase activity is coordinated with binding of the incoming polypeptide. Such coordination would serve to minimize futile cycles of ATP hydrolysis by Ssc1. Consistent with that idea, not only is Tim44 required for the association of the Pam16-Pam18 heterodimer with the translocon (20, 31), but amino acid substitutions within the interval of amino acids 76 to 99 in Tim44 are able to suppress the effects of amino acid substitutions in the N terminus of Pam16, which is important for translocon association (9). (ii) A signal could also facilitate destabilization of Tim44's interaction with Ssc1, which is triggered by the interaction of Ssc1 with the polypeptide chain. Such facilitation not only would coordinate the release of Ssc1 from the translocon with stabilization of its interaction with the translocating polypeptide but would also permit binding of another Ssc1(ATP) at the translocon channel.
Our results also demonstrate the importance of the C-terminal region of Tim44 in vivo, as the N-terminal segment alone cannot support cell viability. Previous work has led to the suggestion that this domain may be responsible for directly interacting with the inner membrane. Structural analysis of Tim44's C-terminal domain revealed a large hydrophobic pocket that was proposed to mediate Tim44's association with the inner mitochondrial membrane (17). Further evidence for the C-terminal domain being involved in membrane association comes from the observation that the C-terminal domain of Tim44 penetrates into phospholipid vesicles (44). In support of this idea, N-terminally truncated Tim44 is detected exclusively in the membrane fraction of mitochondria isolated from strains expressing both the truncated Tim44 version and wild-type Tim44 (data not shown). It seems likely that both the N- and C-terminal regions play a role in Tim44's association with the translocon, with the N terminus of Tim44 interacting directly with the translocon, while the C-terminal domain serves to stabilize the interaction by its direct interaction with phospholipids of the inner membrane.
In conclusion, our finding that small alterations within Tim44 result in multiple functional changes suggests that a finely tuned system has evolved in which interdependent and concerted protein-protein interactions ensure efficient protein translocation across the mitochondrial inner membrane. This is consistent with the fact that, overall, the mitochondrial protein import machinery is one of the most complex protein translocation systems known. A mechanistic understanding of the function of this system at the molecular level is just beginning to emerge.
Acknowledgments
We thank Robert Jensen for Tim17 antibody and Justin Hines, Thomas Lee, and Amy Prunuske for helpful comments on the manuscript.
This work was supported by National Institutes of Health grant GM278709 (to E.A.C.) and by a research fellowship of the Deutsche Forschungsgemeinschaft (to D.S.).
Footnotes
Published ahead of print on 21 April 2008.
REFERENCES
- 1.Bauer, M. F., K. Gempel, A. S. Reichert, G. A. Rappold, P. Lichtner, K. D. Gerbitz, W. Neupert, M. Brunner, and S. Hofmann. 1999. Genetic and structural characterization of the human mitochondrial inner membrane translocase. J. Mol. Biol. 28969-82. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, M. F., C. Sirrenberg, W. Neupert, and M. Brunner. 1996. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell 8733-41. [DOI] [PubMed] [Google Scholar]
- 3.Becker, L., M. Bannwarth, C. Meisinger, K. Hill, K. Model, T. Krimmer, R. Casadio, K. N. Truscott, G. E. Schulz, N. Pfanner, and R. Wagner. 2005. Preprotein translocase of the outer mitochondrial membrane: reconstituted Tom40 forms a characteristic TOM pore. J. Mol. Biol. 3531011-1020. [DOI] [PubMed] [Google Scholar]
- 4.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197345-346. [DOI] [PubMed] [Google Scholar]
- 5.Bomer, U., A. C. Maarse, F. Martin, A. Geissler, A. Merlin, B. Schonfisch, M. Meijer, N. Pfanner, and J. Rassow. 1998. Separation of structural and dynamic functions of the mitochondrial translocase: Tim44 is crucial for the inner membrane import sites in translocation of tightly folded domains, but not of loosely folded preproteins. EMBO J. 174226-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donzeau, M., K. Kaldi, A. Adam, S. Paschen, G. Wanner, B. Guiard, M. F. Bauer, W. Neupert, and M. Brunner. 2000. Tim23 links the inner and outer mitochondrial membranes. Cell 101401-412. [DOI] [PubMed] [Google Scholar]
- 7.D'Silva, P., Q. Liu, W. Walter, and E. A. Craig. 2004. Regulated interactions of mtHsp70 with Tim44 at the translocon in the mitochondrial inner membrane. Nat. Struct. Mol. Biol. 111084-1091. [DOI] [PubMed] [Google Scholar]
- 8.D'Silva, P. D., B. Schilke, W. Walter, A. Andrew, and E. A. Craig. 2003. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl. Acad. Sci. USA 10013839-13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Silva, P. R., B. Schilke, M. Hayashi, and E. A. Craig. 2008. Interaction of the J-protein heterodimer, Pam18/Pam16, of the mitochondrial import motor with the translocon of the inner membrane. Mol. Biol. Cell 19424-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Silva, P. R., B. Schilke, W. Walter, and E. A. Craig. 2005. Role of Pam16's degenerate J domain in protein import across the mitochondrial inner membrane. Proc. Natl. Acad. Sci. USA 10212419-12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frazier, A. E., J. Dudek, B. Guiard, W. Voos, Y. Li, M. Lind, C. Meisinger, A. Geissler, A. Sickmann, H. E. Meyer, V. Bilanchone, M. G. Cumsky, K. N. Truscott, N. Pfanner, and P. Rehling. 2004. Pam16 has an essential role in the mitochondrial protein import motor. Nat. Struct. Mol. Biol. 11226-233. [DOI] [PubMed] [Google Scholar]
- 12.Gambill, B. D., W. Voos, P. J. Kang, B. Miao, T. Langer, E. A. Craig, and N. Pfanner. 1993. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J. Cell Biol. 123109-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geissler, A., A. Chacinska, K. N. Truscott, N. Wiedemann, K. Brandner, A. Sickmann, H. E. Meyer, C. Meisinger, N. Pfanner, and P. Rehling. 2002. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111507-518. [DOI] [PubMed] [Google Scholar]
- 14.Hill, K., K. Model, M. T. Ryan, K. Dietmeier, F. Martin, R. Wagner, and N. Pfanner. 1998. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature 395516-521. [DOI] [PubMed] [Google Scholar]
- 15.Horst, M., P. Jeno, N. G. Kronidou, L. Bolliger, W. Oppliger, P. Scherer, U. Manning-Krieg, T. Jascur, and G. Schatz. 1993. Protein import into yeast mitochondria: the inner membrane import site protein ISP45 is the MPI1 gene product. EMBO J. 123035-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, E. W. 1991. Three proteolytic systems in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2667963-7966. [PubMed] [Google Scholar]
- 17.Josyula, R., Z. Jin, Z. Fu, and B. Sha. 2006. Crystal structure of yeast mitochondrial peripheral membrane protein Tim44p C-terminal domain. J. Mol. Biol. 359798-804. [DOI] [PubMed] [Google Scholar]
- 18.Kang, P. J., J. Ostermann, J. Shilling, W. Neupert, E. A. Craig, and N. Pfanner. 1990. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348137-143. [DOI] [PubMed] [Google Scholar]
- 19.Koehler, C. M. 2004. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 20309-335. [DOI] [PubMed] [Google Scholar]
- 20.Kozany, C., D. Mokranjac, M. Sichting, W. Neupert, and K. Hell. 2004. The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat. Struct. Mol. Biol. 11234-241. [DOI] [PubMed] [Google Scholar]
- 21.Krayl, M., J. H. Lim, F. Martin, B. Guiard, and W. Voos. 2007. A cooperative action of the ATP-dependent import motor complex and the inner membrane potential drives mitochondrial preprotein import. Mol. Cell. Biol. 27411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kronidou, N. G., W. Oppliger, L. Bolliger, K. Hannavy, B. S. Glick, G. Schatz, and M. Horst. 1994. Dynamic interaction between Isp45 and mitochondrial hsp70 in the protein import system of the yeast mitochondrial inner membrane. Proc. Natl. Acad. Sci. USA 9112818-12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, Q., P. D'Silva, W. Walter, J. Marszalek, and E. A. Craig. 2003. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science 300139-141. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Q., J. Krzewska, K. Liberek, and E. A. Craig. 2001. Mitochondrial Hsp70 Ssc1: role in protein folding. J. Biol. Chem. 2766112-6118. [DOI] [PubMed] [Google Scholar]
- 25.Maarse, A. C., J. Blom, L. A. Grivell, and M. Meijer. 1992. MPI1, an essential gene encoding a mitochondrial membrane protein, is possibly involved in protein import into yeast mitochondria. EMBO J. 113619-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, J., K. Mahlke, and N. Pfanner. 1991. Role of an energized inner membrane in mitochondrial protein import. Delta psi drives the movement of presequences. J. Biol. Chem. 26618051-18057. [PubMed] [Google Scholar]
- 27.Merlin, A., W. Voos, A. C. Maarse, M. Meijer, N. Pfanner, and J. Rassow. 1999. The J-related segment of Tim44 is essential for cell viability: a mutant Tim44 remains in the mitochondrial import site, but inefficiently recruits mtHsp70 and impairs protein translocation. J. Cell Biol. 145961-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milisav, I., F. Moro, W. Neupert, and M. Brunner. 2001. Modular structure of the TIM23 preprotein translocase of mitochondria. J. Biol. Chem. 27625856-25861. [DOI] [PubMed] [Google Scholar]
- 29.Mokranjac, D., A. Berg, A. Adam, W. Neupert, and K. Hell. 2007. Association of the Tim14.Tim16 subcomplex with the TIM23 translocase is crucial for function of the mitochondrial protein import motor. J. Biol. Chem. 28218037-18045. [DOI] [PubMed] [Google Scholar]
- 30.Mokranjac, D., S. A. Paschen, C. Kozany, H. Prokisch, S. C. Hoppins, F. E. Nargang, W. Neupert, and K. Hell. 2003. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 22816-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mokranjac, D., M. Sichting, W. Neupert, and K. Hell. 2003. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 224945-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neupert, W., and M. Brunner. 2002. The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol. 3555-565. [DOI] [PubMed] [Google Scholar]
- 33.Neupert, W., and J. M. Herrmann. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76723-749. [DOI] [PubMed] [Google Scholar]
- 34.Pfanner, N., and K. N. Truscott. 2002. Powering mitochondrial protein import. Nat. Struct. Biol. 9234-236. [DOI] [PubMed] [Google Scholar]
- 35.Rassow, J., A. C. Maarse, E. Krainer, M. Kubrich, H. Muller, M. Meijer, E. A. Craig, and N. Pfanner. 1994. Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J. Cell Biol. 1271547-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rehling, P., N. Wiedemann, N. Pfanner, and K. N. Truscott. 2001. The mitochondrial import machinery for preproteins. Crit. Rev. Biochem. Mol. Biol. 36291-336. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, H. C., J. Berthold, M. F. Bauer, K. Dietmeier, B. Guiard, M. Brunner, and W. Neupert. 1994. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature 371768-774. [DOI] [PubMed] [Google Scholar]
- 38.Sickmann, A., J. Reinders, Y. Wagner, C. Joppich, R. Zahedi, H. E. Meyer, B. Schonfisch, I. Perschil, A. Chacinska, B. Guiard, P. Rehling, N. Pfanner, and C. Meisinger. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA 10013207-13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slutsky-Leiderman, O., M. Marom, O. Iosefson, R. Levy, S. Maoz, and A. Azem. 2007. The interplay between components of the mitochondrial protein translocation motor studied using purified components. J. Biol. Chem. 28233935-33942. [DOI] [PubMed] [Google Scholar]
- 41.Truscott, K. N., W. Voos, A. E. Frazier, M. Lind, Y. Li, A. Geissler, J. Dudek, H. Muller, A. Sickmann, H. E. Meyer, C. Meisinger, B. Guiard, P. Rehling, and N. Pfanner. 2003. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 163707-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voisine, C., E. A. Craig, N. Zufall, O. von Ahsen, N. Pfanner, and W. Voos. 1999. The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell 97565-574. [DOI] [PubMed] [Google Scholar]
- 43.Voos, W., O. von Ahsen, H. Muller, B. Guiard, J. Rassow, and N. Pfanner. 1996. Differential requirement for the mitochondrial Hsp70-Tim44 complex in unfolding and translocation of preproteins. EMBO J. 152668-2677. [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss, C., W. Oppliger, G. Vergeres, R. Demel, P. Jeno, M. Horst, B. de Kruijff, G. Schatz, and A. Azem. 1999. Domain structure and lipid interaction of recombinant yeast Tim44. Proc. Natl. Acad. Sci. USA 968890-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]