Abstract
Interleukin-1 (IL-1)-induced mRNA expression of ccl2 (also called MCP-1), a prototypic highly regulated inflammatory gene, is severely suppressed in cells lacking c-Jun or Jun N-terminal protein kinase 1 (JNK1)/JNK2 genes and is only partially restored in cells expressing a c-Jun(SS63/73AA) mutant protein. We used chromatin immunoprecipitation to identify three c-Jun-binding sites located in the far 5′ region close to the transcriptional start site and in the far 3′ region of murine and human ccl2 genes. Mutational analysis revealed that the latter two sites contribute to ccl2 transcription in response to the presence of IL-1 or of ectopically expressed c-Jun-ATF-2 dimers. Further experiments comparing wild-type and c-Jun-deficient cells revealed that c-Jun regulates Ser10 phosphorylation of histone H3, acetylation of histones H3 and H4, and recruitment of histone deacetylase 3 (HDAC3), NF-κB subunits, and RNA polymerase II across the ccl2 locus. c-Jun also coimmunoprecipitated with p65 NF-κB and HDAC3. Based on DNA microarray analysis, c-Jun was required for full expression of 133 out of 162 IL-1-induced genes. For inflammatory genes, these data support the idea of an activator function of c-Jun that is executed by multiple mechanisms, including phosphorylation-dependent interaction with p65 NF-κB and HDAC3 at the level of chromatin.
The inflammatory response is characterized by the coordinated induction or repression of a large number of inflammatory genes. These genes control leukocyte infiltration, innate or adaptive immune responses, and tissue remodeling. Prototypic proinflammatory cytokines such as interleukin-1 (IL-1) or tumor necrosis factor (TNF) can activate or repress several hundred genes within the same cell (53).
Many of the gene-regulatory properties of IL-1 or TNF are transduced through the NF-κB and mitogen-activated protein (MAP) kinase signaling pathways. After binding to cell surface receptors, both cytokines activate MAP kinase kinase kinases such as TAK1 that subsequently activate the IκB kinase-NF-κB signaling pathway as well as the MAP kinases Jun N-terminal protein kinase (JNK) and p38. After signal-mediated destruction of inhibitory (IκB) proteins, NF-κB proteins translocate to the nucleus and bind to their cognate DNA-elements. Nuclear NF-κB is a dimer composed of two of the five subunits RelA (p65), RelB, c-Rel, p100/p52, and p105/p50. Depending on their cellular abundance, different homo- or heterotypic combinations of NF-κB subunits generate transcriptionally active or inactive NF-κB complexes (14, 25, 29, 36, 37).
The JNK group of protein kinases consists of 10 highly homologous enzymes that are derived from three genes by alternative splicing: the short JNKs (45 to 48 kDa) JNK1α1, JNK1β1, JNK2α1, JNK2β1, and JNK3α1 and the long JNKs (54 to 57 kDa) JNK1α2, JNK1β2, JNK2α2, JNK2β2, and JNK3α2 (13).
One of the most important functions of JNK is the regulation of the transcription factor activator protein 1 (AP-1) (9, 24, 44, 45, 54, 61). AP-1 is a homo- or heterodimer composed of members of the Jun (i.e., c-Jun, JunB, and JunD) and Fos (i.e., c-Fos, FosB, and Fos-related antigen [Fra1, Fra2]) protein families. Fos and Jun proteins can also dimerize to many other basic leucine zipper proteins such as ATF, C/EBP, Maf, and NF-E2, increasing the number of the potential AP-1 factors that bind to a given AP-1 site (4, 9, 49).
We previously isolated JNK as IL-1-stimulated protein kinase from rabbit liver (28). Compelling evidence from many laboratories indicates that JNK is an important effector in the regulation of inflammatory gene expression. Our own work showed that JNK cooperates with NF-κB signals to increase the extent of chemokine expression, such as that by interleukin-8 (18-20). However, the JNK substrates that mediate this effect have been elusive. c-Jun is the best-characterized JNK substrate and forms a high-affinity interaction with JNK; in fact, JNK has been isolated using c-Jun as bait in affinity purification (8, 16). JNK can phosphorylate c-Jun at at least four sites (Ser63, Ser73, Thr91, and Thr93) within the N-terminal transactivation domain (Fig. 1A). A c-Jun(SS63/73AA) mutant can rescue the otherwise lethal phenotype of c-Jun ablation in mice (2). c-Jun is also phosphorylated within the DNA-binding domain at Thr239 by glycogen synthase kinase 3β and at Ser243 by an unknown protein kinase (34) (Fig. 1A). Despite these findings, the physiological role of individual c-Jun phosphorylations in the regulation of c-Jun or JNK target genes is still unclear.
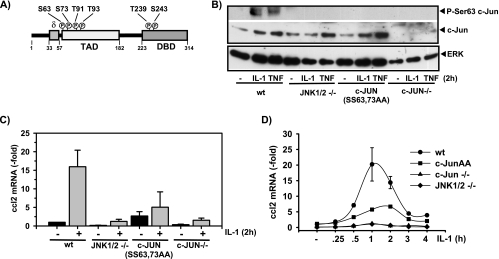
FIG. 1.
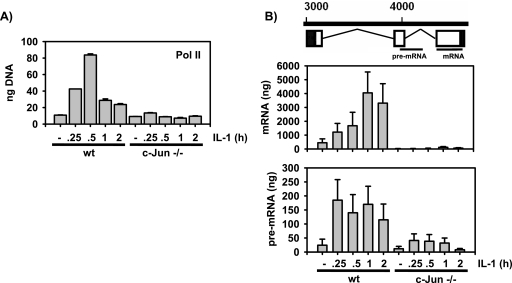
JNK- and c-Jun-dependent ccl2 mRNA expression in genetically altered mouse fibroblasts. (A) Schematic representation of the c-Jun structure, indicating the N-terminal transactivation domain (TAD), the C-terminal DNA-binding domain (DBD), and the JNK-binding domain (δ). Each circled “P” indicates a phosphorylation site. (B) Wild-type (wt) embryonic fibroblast cell lines, cells lacking JNK1 and JNK2 genes (JNK1/2 −/−) or c-Jun (c-Jun−/−), or cells that were isolated from mice carrying a c-Jun allele mutated in two of the four JNK phosphoacceptor sites (c-Jun SS63/73AA) were kept in low-concentration (0.1%) serum for 48 h. Thereafter, cells were treated for 2 h with 10 ng/ml IL-1α or were left untreated. Phosphorylation and expression of c-Jun were analyzed by Western blotting of whole-cell extracts. ERK antibodies were used to control for protein loading. (C) The cells described for panel B were kept in low-concentration (0.1%) serum for 48 h. Thereafter, they were treated for 2 h with IL-1α (10 ng/ml). ccl2 mRNA expression was determined by TaqMan real-time PCR using total RNA. The values shown represent mean ccl2 expression ± the standard errors of the means of the results from three independent experiments relative to the unstimulated wild-type control cell results. (D) ccl2 mRNA expression was determined as described for panel C for cells kept in low-concentration serum that were treated for the indicated times with IL-1α (10 ng/ml). Expression of ccl2 was analyzed as described for panel C. The values shown represent mean ccl2 expression ± the standard errors of the means of the results from two independent experiments relative to the unstimulated wild-type control cell results.
A major mechanistic advance was the observation that phosphorylation of c-Jun by JNK dissociates a repressor complex which contains histone deacetylase 3 (HDAC3) and activates c-Jun transcriptional activity by derepression in reporter gene assays (58). This model suggests that c-Jun acts largely as a repressor whose negative modulatory role is switched off by JNK-dependent phosphorylation and is supported by RNA interference-mediated suppression of Drosophila c-Jun or JNK (26). In the latter study c-Jun or JNK was found to counteract NF-κB-mediated gene expression (26).
Despite the abundant occurrence of putative AP-1 sites in numerous mammalian genes, relatively little is known about the role of c-Jun in regulation of individual inflammatory genes. Recently, compound deletion of c-Jun and JunB in keratinocytes revealed a role for this AP-1 complex in negative regulation of genes that are upregulated in psoriasis and arthritis (62).
Previously, by means of small interfering RNA directed against c-Jun and a cell-permeable peptide which disrupts the c-Jun-JNK interaction, we demonstrated that IL-1-induced expression of ccl2 in human primary fibroblasts requires c-Jun (21). The chemokine ccl2 (also called MCP-1) is an important mediator of leukocyte invasion during acute and chronic inflammation (7, 12, 31). ccl2 is highly inducible in mice and humans. A contribution of NF-κB subunits and of SP-1 to transcriptional regulation of murine ccl2 has been demonstrated by chromatin immunoprecipitation experiments and by analysis of knockout fibroblasts (3, 40, 41). In contrast, a direct role of c-Jun in transcriptional regulation of ccl2 has not been demonstrated.
In the present study we investigated the contribution of c-Jun to ccl2 transcriptional regulation in genetically modified mouse fibroblasts and in human cells. We provide compelling evidence that c-Jun binds to multiple sites within the ccl2 gene, including the 3′ region of the ccl2 locus, and acts as an activator for inflammatory gene expression by multiple mechanisms, including direct interaction with p65 NF-κB and HDAC3.
MATERIALS AND METHODS
Cells and materials.
MRC5 embryonic lung fibroblasts were from the American Type Culture Collection (no. CCL-171), Manassas, VA. HEK293IL-1R cells have been described previously (52). The following immortalized murine embryonic fibroblast (Mef) lines were used throughout this study and have been described previously (2, 42): wild-type Mefs, Mefs deficient for JNK1/JNK2 or for c-Jun, and Mefs isolated from c-Jun(SS63/73AA) mutant mice. For the experiments shown in Fig. 6D, wild-type Mef lines derived from MK2/MK5 wild-type embryos according to reference 46 or derived from glycogen synthase kinase 3β wild-type control embryos (17) (data not shown) were used.
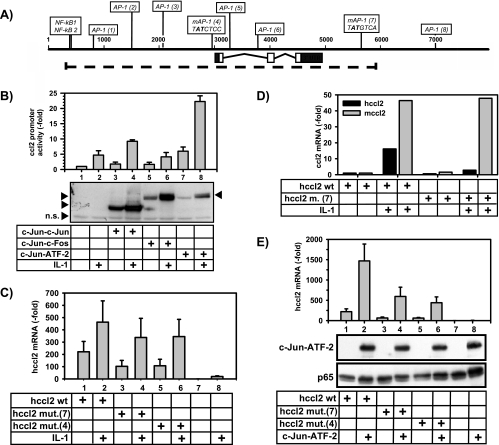
FIG. 6.
The 5′ and 3′ c-Jun binding sites mediate ccl2 mRNA synthesis in response to single-chain c-Jun dimers or IL-1. (A) Schematic representation of the region of the human ccl2 gene (dotted line) that was transfected in HEK293IL-1R cells or murine fibroblasts. The positions and nucleotide sequences of the mutated AP-1-binding sites (mAP-1) adjacent to the TSS and downstream of the 3′ untranslated region are indicated. Boldface characters represent mutated nucleotides. (B) HEK293IL-1R cells were transfected with 1 μg of the indicated c-Jun-containing dimer constructs, 1 μg of a luciferase reporter gene driven by 2.8 kb of the murine ccl2 promoter (pluc2 2.8/2 [53]), and 1 μg of simian virus 40 β-galactosidase. At 24 h later, cells were stimulated for 4 h with IL-1α (10 ng/ml) or were left untreated. Luciferase activity in cell extracts was normalized for β-galactosidase activity. Data shown represent the mean normalized luciferase activity ± standard errors of the means of the results of two experiments. Lysates from one experiment were analyzed for expression of the single-chain AP-1 dimers by use of c-Jun antibodies, as indicated by arrows. Note that IL-1 induces phosphorylation and stabilization of the AP-1 dimers. n.s., a nonspecifically detected antigen that served as a loading control. (C) HEK293IL-1R cells were transfected with empty vector (lanes 7 and 8) or with 5 μg of wild-type human ccl2 expression vectors (lanes 1 and 2) or with versions carrying single-site mutations in the 5′ [hcc12 mut.(4)] (lanes 5 and 6) or 3′ [hcc12 mut.(7)] (lanes 3 and 4) AP-1 sites, as illustrated in panel A. One microgram of pGL2 Luc was cotransfected. At 24 h later, cells were stimulated with IL-1α (10 ng/ml) for 2 h or were left untreated. Thereafter, relative human ccl2 mRNA expression levels were analyzed by TaqMan real-time PCR and normalized for luciferase activity. Data shown represent mean hccl2 mRNA expression ± standard errors of the means of the results of five independent experiments relative to the unstimulated vector control results. (D) Human and murine ccl2 mRNA expression was analyzed using murine fibroblasts stably transfected with wild-type human ccl2 expression vectors or the version carrying a single-site mutation in the 3′ AP-1 site [hcc12 m. (7)] as indicated in panel A. Cells were stimulated for 2 h with IL-1α (10 ng/ml) or were left untreated. The results represent normalized hccl2 mRNA expression corrected for integrated hccl2 gene copies (black bars) and normalized endogenous murine cc12 (mccl2) mRNA expression (gray bars). Data shown represent comparisons to the unstimulated wild-type control value (set as 1). Data represent the results of one of two experiments. See Materials and Methods for details. (E) HEK293IL-1R cells were transfected with empty vector (lanes 7 and 8) or were transfected with 5 μg of wild-type human ccl2 expression vectors (lanes 1 and 2) or with versions carrying single-site mutations in the 5′ [hcc12 mut.(4)] (lanes 5 and 6) or 3′ [hcc12 mut.(7)] (lanes 3 and 4) AP-1 sites as illustrated in panel A. One microgram of the c-Jun-ATF-2 encoding construct and pGL2 Luc was cotransfected. At 24 h later human ccl2 mRNA expression was analyzed by TaqMan real-time PCR and normalized for Luc activity. Results represent mean hccl2 mRNA expression ± standard errors of the means of the results of six independent experiments relative to the vector control results. Lysates from one experiment were analyzed for expression of the c-Jun-ATF-2 dimers by use of c-Jun antibodies as indicated. Antibodies against p65 NF-κB were used to control for protein loading.
Unless stated otherwise, cells were cultured in Dulbecco's modified Eagle's medium complemented with 10% fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. The medium for the MRC-5 fibroblasts was completed with 1 mM sodium pyruvate and 0.1 mM nonessential amino acids.
Antibodies against the following proteins or peptides were used in this study: c-Jun (catalog no. sc-1694), ATF2 (sc-6233), extracellular signal-regulated kinase 2 (ERK2) (sc-154), polymerase II (sc-899), p65 NF-κB (sc-372), p50 NF-κB (sc-114), CBP (sc-583), HDAC1 (sc-6298), HDAC3 (sc-11417), Med 1 (TRAPP220; sc-8998), and Med 17 (CRSP7; sc-12453), all from Santa Cruz; acetyl(K5, 8, 12, 16)-histone H4 (catalog no. 06-866), acetyl(K9, 14)-histone H3 (06-599), and phospho-Ser10-histone H3 (06-570) from Upstate Biotechnology/Biomol; histone H3 (catalog no. 9712), histone H4 (2592), phospho-Ser73, and c-Jun phospho-Ser63 from Cell Signaling; and phospho-Ser73 c-Jun (Biosource), rabbit immunoglobulin G (IgG) (Rockland), and rabbit IgG (Santa Cruz).
Horseradish peroxidase-coupled secondary antibodies and protein A/protein G-Sepharose were from GE Healthcare. Human recombinant IL-1α was produced as described previously (27). TNF-α was purchased from Strathmann, and SP600125 was from Tocris. Trichostatin A (TSA) was from Sigma (catalog no. T8552) and was dissolved in ethyl alcohol. Other reagents were from Sigma-Aldrich or Fisher and were of analytical grade or better.
Cloning of wild-type and mutant human ccl2 genes and transfections.
The human ccl2 gene (hccl2) (5.6 kb) was amplified from MRC5 genomic DNA by use of the primers se (5′-CAGTATCTGGAATGCAGGCTC-3′) and as (5′-GCAGCATTTCACCCTGCATGGCTCATT]) and of Accuzyme DNA polymerase (Bioline), 1% dimethyl sulfoxide, and the following PCR conditions: 2 min at 96°C and 40 cycles of 95°C (30 s), 60°C (30 s), and 72°C (12 min), followed by a final extension reaction at 72°C (30 min). The PCR fragment was cloned into pCR-Blunt II-TOPO (Invitrogen). AP-1 sites 4 and 7 (as shown in Fig. 2; also see Fig. 6) were mutated using Stratagene QuikChange protocols, and mutations were verified by DNA sequencing.
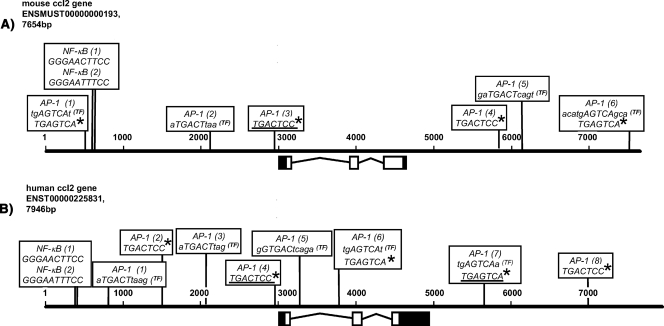
FIG. 2.
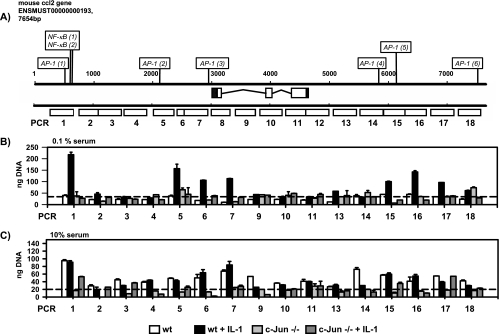
Schematic representation of AP-1 binding sites in human and mouse ccl2 loci. The 5′ and 3′ untranslated regions (black boxes), the three exons (white boxes), and 3,000 bp of upstream and downstream flanking sequences of the murine (A) and human (B) ccl2 genomic sequences are shown. Sequences were taken from the ENSEMBL database using the indicated accession numbers. Nucleotides of both genomic loci are aligned according to scale and are indicated by numbers. The AP-1-binding sites adjacent to the TSS (TGACTCC) that were previously verified by mutational analysis (30, 40, 41, 47, 60) and a downstream AP-1 site (TGAGTCA) that was suggested by DNase hypersensitive site mapping in the human gene locus (10) are underlined. Asterisks indicate AP-1 sites with identical sequences in other regions of the mouse and human genes. Additional AP-1-binding sites predicted by TRANSFAC Professional, version 10.3, are indicated (TF). Note that TRANSFAC uses uppercase lettering for the core sequence of a predicted binding site. Only sites located on the positive strand are shown. NF-κB (1) and NF-κB (2), experimentally verified binding sites in the distal regulatory region of the ccl2 enhancer (40, 51).
MK2/MK5 wild-type Mefs were transfected in 12-well plates with 1.25 μg of pCR-Blunt II-hCCL2 plasmids and 0.5 μg of pBSPACdeltaP by use of jetPEI (Biomol). Stably transfected pools of cells were selected in 0.75 μg/ml puromycin. Numbers of integrated hccl2 copies were determined from genomic DNA by use of TaqMan probe A (see Fig. 5) and quantitated by real-time PCR using serially diluted MRC5 DNA (256 to 1.28 ng). A relative copy number value was obtained by comparing nanogram values of integrated DNA of hccl2 mutants to those obtained with integrated wild-type hccl2 DNA (set at a value of 1). mRNA transcribed from the integrated human ccl2 genes was detected by use of TaqMan reverse transcription PCR (RT-PCR) (see below).
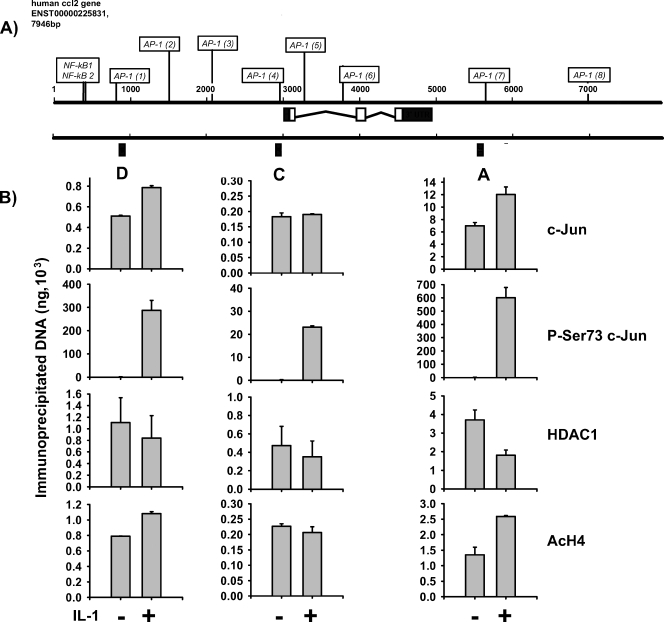
FIG. 5.
c-Jun recruitment to the human ccl2 locus. (A) Schematic representation of the human ccl2 locus. Black boxes and capital letters (D, C, and A) indicate the genomic regions that were analyzed by TaqMan PCR. (B) Human MRC5 fibroblasts cultured in 10% serum were treated for 2 h with 10 ng/ml IL-1α or were left untreated. (A) After in vivo cross-linking, chromatin was isolated from all cultures and immunoprecipitated with the indicated antibodies. Genomic DNA fragments from the indicated A, C, or D region of human ccl2 were amplified and quantitated by real-time PCR using TaqMan probes and serially diluted chromatin to generate standard curves. See Materials and Methods for details. Data represent the results from two independent experiments ± standard errors of the means.
Cycle threshold (CT) values obtained for hccl2 were normalized by subtracting the CT values obtained for murine β-actin to yield ΔCT values. Relative changes of mRNA expression determined as the ratio (R) of mRNA expression of stimulated cells to that of unstimulated cells were calculated according to the following equation: R = 2−[ΔCT(stimulated) − ΔCT(unstimulated)]. These values were divided by the relative copy number to correct for integration events. Similar results were obtained when hccl2 mRNA amounts of the transfected cells were quantitated against a standard curve derived from serially diluted MRC5 cDNA (2.5 μg to 2.5 ng, corresponding to approximately 7.5 × 105 to 7.5 × 102 mRNA copies of a single-copy gene).
Transient transfections of HEK293IL-1R cells were performed by the calcium phosphate method as described previously (18) or by use of jetPEI (Biomol) with expression vectors pCG-c-Jun-c-Jun, pCG-c-Jun-ATF-2, and pCG-c-Jun-c-Fos (1) and simian virus 40 β-galactosidase and pluc2.8/2-ccl2 promoter Luc-(53). pGL2 Luc was obtained from Promega.
DNA oligonucleotides.
All primer pairs and TaqMan probes used in this study for PCR with chromatin immunoprecipitation (ChIP) DNA or cDNA are indicated in Table S1 in the supplemental material.
RT-PCR.
A total of 1 μg of total RNA was prepared as previously described (18) and transcribed into cDNA by use of Moloney murine leukemia virus RT (BioLabs) in a total volume of 40 μl. A 2-μl volume of this reaction mixture was used to amplify cDNAs by use of assays on demand (Applied Biosystems) for murine (Mm00441242_m1) or human (Hs00234140_m1) ccl2 and murine (Mm00607939_s1) or human (Hs00174103_m1) β-actin on an ABI 7500 real-time PCR instrument. The threshold CT value for each individual PCR product was calculated by use of the instrument's software, and CT values obtained for ccl2 were normalized by subtracting the CT values obtained for β-actin. The resulting ΔCT values were then used to calculate relative changes of mRNA expression as the ratio (R) of mRNA expression of stimulated to unstimulated cell results according to the following equation: R = 2−[ΔCT(stimulated) − ΔCT(unstimulated].
Measurement of ccl2 mRNA and pre-mRNA derived from cDNA was performed by real-time PCR using a SensiMix Sybr green kit (Quantace) with specific primers for ccl2 mRNA and pre-mRNA as depicted here (see Fig. 7B). PCR products were quantitated against a standard curve derived from serially diluted genomic DNAs of c-Jun wild-type Mefs (200 ng to 0.1 ng).
FIG. 7.
c-Jun is required for RNA polymerase II recruitment and activity during ccl2 mRNA synthesis. (A) Cells were kept in low-concentration serum as described for Fig. 1. IL-1-induced recruitment of polymerase II was analyzed by ChIP using untreated cells or IL-1-stimulated cells at the indicated time points. wt, wild type. (B) cDNAs derived from cells kept in low-concentration serum as described for Fig. 1 were analyzed for expression of ccl2 pre-mRNA and mature mRNA. The regions of murine ccl2 transcripts that were examined are indicated in the upper panel. Amounts of PCR products were quantitated against a standard curve derived from serially diluted genomic DNA by use of Sybr green.
ChIP.
Three to five 175-cm2 flasks of confluent fibroblasts, treated as described in the figure legends, were used for each set of conditions. Proteins bound to DNA were cross-linked in vivo with 1% formaldehyde in warm phosphate-buffered saline (PBS). After 5 min of incubation at room temperature, this solution was replaced by cold PBS containing 0.125 M glycine to stop the cross-linking. Then the supernatant was removed, and the cells were washed with and scraped into ice-cold PBS. Cells were collected at 500 × g at 4°C, washed again in ice-cold PBS, and then lysed in ChIP-RIPA buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM EDTA, and freshly added 1% aprotinin). Lysates were cleared by sonication (four times on ice at 1 min per time) and centrifuged at 15,000 × g at 4°C for 20 min. Supernatants were collected and stored in aliquots at −80°C for subsequent ChIP experiments.
A total of 50 μl of the initial lysates (input samples) was diluted to 200 μl with Tris-EDTA (TE) buffer (10 mM Tris [pH 7.5], 1 mM EDTA) containing 1% SDS and 50 μg/ml RNase A. After 30 min at 37°C, proteinase K (0.5 mg/ml) was added and samples were incubated for at least 6 h at 37°C followed by at least 6 h at 65°C. Samples were diluted to 500 μl in PB loading buffer (Qiagen), and DNA was purified using QIAquick spin columns (Qiagen) according to the manufacturer's instructions. DNA was eluted with TE buffer, and the DNA concentrations were measured by use of NanoDrop. Equal amounts of chromatin (ranging from 20 to 40 μg) were used for the immunoprecipitations, and volumes were adjusted to 500 μl with ChIP-RIPA buffer.
A total of 10 μl (2 μg) of Santa Cruz antibodies or 4 μl of other antibodies was added to 500 μl of lysates, and the mixture was rotated at 4°C overnight. Then 40 μl of a protein A/protein G mixture, preequilibrated in ChIP-RIPA buffer, was added to the lysates, and incubation continued for 2 h at 4°C. Beads were collected by centrifugation, washed two times in 1.4 ml of ChIP-RIPA buffer, once in high-salt buffer (10 mM Tris [pH 7.5], 2 M NaCl, 1% NP-40, 0.5% deoxycholate, 1 mM EDTA), once more in ChIP-RIPA buffer (cold), and once in TE buffer at room temperature and were finally resuspended in 55 μl of elution buffer (10 mM Tris [pH 7.5], 1 mM EDTA, 1% SDS). Samples were vigorously mixed for 15 min at 25°C and then centrifuged. The supernatant (50 μl) was diluted to 200 μl with TE buffer and was treated as described above for the input samples. The DNA was purified using QIAquick spin columns, eluted with TE buffer, and stored at −20°C until further use.
For Re-ChIP experiments, 50 μl of supernatant eluted from the first immunoprecipitation was diluted to 300 μl with ChIP-RIPA buffer and was incubated with the second antibody at 4°C overnight. A 40-μl volume of a protein A/protein G mixture preequilibrated in ChIP-RIPA buffer was added to the lysates, and immunoprecipitation of protein-DNA complexes was continued as described above.
Quantification of ChIP DNA by real-time PCR.
PCR products derived from ChIP were quantitated by real-time PCR using an ABI 7500 instrument (Applied Biosystems). The reaction mixture contained 3 μl of ChIP or input DNA, 0.5 μM of primers, and 12.5 μl of Sybr green Mastermix (Applied Biosystems) in a total volume of 25 μl. PCR cycles were as follows: 95°C for 10 min followed by 40 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 1 min. Melting curve analysis revealed a single PCR product. Serial dilutions of input DNA revealed that PCR results were linear from 100 ng to 0.1 ng and were used to calculate absolute amounts of PCR products by use of the instrument's software. For the experiments shown (see Fig. 5), TaqMan probes and corresponding primer pairs were designed and used to quantitate genomic fragments of human ccl2 captured by ChIP.
Amplification of ChIP DNA by LM PCR.
Aliquots of ChIP DNA as shown in Fig. 3 were amplified by two rounds of ligation-mediated (LM) PCR using unidirectional primer pairs and following published protocols (38). For quantification, 3 μl of amplicon DNA (8.3 ng/μl) or 3 μl of a serial dilution from amplicon input DNA (ranging from 100 to 0.1 ng) was used as a PCR template. PCR was performed using a SensiMix Sybr green kit (Quantace) and the following PCR conditions: 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 30 s, and 72°C for 1 min.
FIG. 3.
Analysis of recruitment of c-Jun to the mouse ccl2 locus. (A) Schematic representation of 18 genomic regions of the murine ccl2 gene (indicated by white boxes) analyzed by PCR. (B and C) Wild-type or c-Jun-deficient (−/−) fibroblasts were kept in either low-concentration (0.1%) (B) or high-concentration (10%) (C) serum for 2 days. Then the cells were treated for 2 h with 10 ng/ml IL-1α or were left untreated, as indicated. After in vivo cross-linking, chromatin was isolated from all cultures and immunoprecipitated with c-Jun antibodies. Immunopurified DNA was amplified by LM PCR, and the amounts of genomic fragments of mouse ccl2 associated with c-Jun were determined by real-time PCR. Standard curves for each individual PCR were generated using LM PCR products derived from total chromatin. Error bars indicate standard deviations from two replicate measurements. See Materials and Methods for details. Dashed lines indicate the mean signal levels obtained from all ChIP experiments as obtained using cells lacking c-Jun and therefore indicate the mean levels of unspecific binding of genomic ccl2 fragments.
Western blotting.
Cells were lysed in Triton cell lysis buffer (10 mM Tris [pH 7.05], 30 mM NaPPi, 50 mM NaCl, 1% Triton X-100, 2 mM Na3VO4, 50 mM NaF, 20 mM β-glycerophosphate and freshly added 0.5 mM phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, 0.5 μg/ml pepstatin, 1 μg/ml microcystin), or else nuclear and cytosolic extracts were prepared as previously described (20). Cell extract proteins were separated using 7.5 to 12.5% SDS-polyacrylamide gel electrophoresis and electrophoretically transferred to polyvinylidene difluoride membranes (Immobilon; Millipore). After being blocked with 5% dried milk in Tris-buffered saline (TBS)-0.05% Tween (TBST) overnight, membranes were incubated for 4 to 24 h with primary antibodies, washed in TBST, and incubated for 2 to 4 h with the peroxidase-coupled secondary antibody. Proteins were detected by using a Pierce or Millipore enhanced chemiluminescence system.
Coimmunoprecipitation experiments.
For HEK293IL-1R cells, 1 to 1.5 mg of nuclear extract was incubated with 2 μg of primary antibodies in 500 μl of IP buffer A (TBS [pH 7.4], 50 mM NaF, 1 mM Na3VO4, 2mM dithiothreitol, 1% Triton X-100) overnight. Antibodies were captured with Trueblot anti-rabbit IgG beads (catalog no. 00-800; eBioscience), beads were washed three times in IP buffer A, and proteins were eluted for 10 min at 95°C in SDS sample buffer containing 50 mM dithiothreitol. For Mef cell lines, 1 to 1.5 mg of nuclear extract was diluted in 500 μl of IP buffer B (50 mM Tris-HCl [pH 7.8], 150 mM NaCl, 1% NP-40) and was precleared with 25 μl of Trueblot IgG beads for 30 min on ice. Immunoprecipitation of supernatant proteins was continued in IP buffer B as described above. Proteins were separated on 10% SDS-polyacrylamide gel electrophoresis and subjected to Western blotting. Blots were hybridized with the indicated primary antibodies (at 1:1,000 in 5% milk-TBST at 4°C overnight) followed by TrueBlot horseradish-coupled anti-rabbit IgG (catalog no. 18-8816; eBioscience) diluted 1:1,000 in 5% milk-TBST for 1 h at room temperature.
DNA microarray experiments.
Approximately 300 ng of total RNA was used to prepare fluorophore-labeled cRNA by use of an Amino-Allyl MessageAmp II aRNA kit (catalog no. AM1753) from Ambion. Samples from unstimulated cells were labeled with Cy3, and samples from IL-1-stimulated cells were labeled with Cy5. Approximately 2.6 μg of Cy3- or Cy5-labeled cRNAs was mixed and cohybridized onto a mouse oligonucleotide microarray (V2) (catalog no. G4121B [design identification no. 013326]; Agilent) according to published protocols from Agilent. Hybridized arrays were scanned at maximal resolution on an Affymetrix 428 scanner at variable PMT voltage settings. Fluorescence intensity values from Cy3 or Cy5 channels were processed using Imagene 4.2 software (Biodiscovery) and normalized by an intensity-dependent nonlinear strategy. Spots of poor signal-to-noise ratios or bad signal quality were flagged and removed prior to further analysis using Excel. Values obtained using probes that occurred multiple times or that measured the same gene were averaged, and heat map visualization was performed by importing log 2-transformed ratio data into the Mayday program (http://www.zbit.uni-tuebingen.de/pas/mayday.html).
RESULTS AND DISCUSSION
Regulation of mouse ccl2 mRNA expression by c-Jun and JNK.
To analyze the role of the JNK-c-Jun pathway in ccl2 gene expression in a genetic model, we made use of previously established fibroblast cell lines that lack the JNK1 and JNK2 genes, cells isolated from mice with a c-Jun allele mutated in two of the four JNK phosphoacceptor sites (SS63/73AA) (Fig. 1A), or cells that lack c-Jun. Expression and phosphorylation of c-Jun were assessed using whole-cell extracts (Fig. 1B). When cultured for 2 days in low-concentration serum (0.1%), wild-type fibroblasts expressed low amounts of c-Jun protein; levels were increased by about two- to threefold upon IL-1 or TNF treatment (Fig. 1B). JNK1/JNK2-deficient and c-JunAA cells expressed basal amounts of c-Jun that were comparable to wild-type cell amounts (Fig. 1B). In JNK1/JNK2-deficient cells, TNF, but not IL-1, induced c-Jun protein (Fig. 1B). c-Jun protein induction in response to both cytokines was restored in c-JunAA cells (Fig. 1B). Both JNK1/JNK2-deficient and c-JunAA cells lacked c-Jun phosphorylation at Ser63, as expected (Fig. 1B).
Under these conditions IL-1 induced a rapid and transient 15- to 20-fold increase in ccl2 mRNA levels in wild-type cells (Fig. 1C and D). In cells in which c-Jun or JNK genes were ablated, constitutive as well as IL-1-induced ccl2 mRNA expression was severely impaired and could be rescued only partially by the mutant c-JunAA (Fig. 1C and D). It should also be noted that in unstimulated cells expressing the JunAA mutant, basal ccl2 levels were modestly increased, indicating that these phosphorylation sites mediate a negative regulatory role of c-Jun for basal ccl2 expression (Fig. 1C and D). Similarly, expression of ccl2 in response to TNF was impaired in the absence of c-Jun or JNK and was only partially restored by c-JunAA (data not shown).
When cultured in 10% serum (i.e., in the presence of higher growth factor concentrations), wild-type fibroblasts expressed increased amounts of c-Jun protein (data not shown). We noticed that under these conditions, IL-1 induced only a moderate 1.5- to 4-fold increase in ccl2 mRNA expression in wild-type cells, but as seen with cells cultured in 0.1% serum, ccl2 mRNA levels were profoundly suppressed in the absence of c-Jun (data not shown). Thus, inducible expression of ccl2 is sensitive to both cytokine induction and changes in growth factor concentrations, but in each case such expression requires c-Jun.
Collectively, the results of the experiments represented by Fig. 1 clearly indicate that the presence of c-Jun and its inducible phosphorylation by JNK at Ser63/Ser73 are required for physiological expression of ccl2 in response to proinflammatory cytokines. Apparently, in IL-1-stimulated cells, c-Jun acts as an activator for ccl2 expression, in contrast to a previous report on other inflammatory genes in which c-Jun was suggested to act predominantly as a repressor (39).
Identification of several c-Jun-binding sites within the murine and human ccl2 loci.
Experimental evidence for a role of c-Jun in ccl2 mRNA regulation is scarce. Overexpression of wild-type c-Jun or mutant c-Jun proteins, or of antisense RNA for c-Jun, resulted in upregulation or inhibition of ccl2 expression in some studies (35, 50, 57, 60). None of these studies proved that the effects were direct.
Only two candidate c-Jun-binding sites have been previously characterized. Mutation of an AP-1-binding site (TGACTCC) in close proximity to the transcriptional start site (TSS) was found to reduce basal and TNF-inducible activity of a 0.5-kb human ccl2 or of a 2.6-kb murine ccl2 promoter fragment fused to a luciferase reporter gene (40, 47, 60).
Furthermore, by means of DNase I hypersensitive site mapping, a putative TNF-regulated AP-1 site (TGAGTCA) was found in the human ccl2 gene located approximately 600 bp downstream of the polyadenylation site. This site was occupied by c-Jun and Fra1 in vitro, as assessed by electrophoretic mobility shift assay, but its functional relevance has not been validated (10). Moreover, as indicated in Fig. 2, identical sequences are also found at several other locations of the human and mouse ccl2 loci. So far, in vivo binding of c-Jun to these two sites has not been demonstrated.
AP-1 cis elements are highly degenerated (9). Accordingly, bioinformatic analysis of the human and murine ccl2 genes, performed using an array of published DNA-binding matrices for AP-1 deposited in the TRANSFAC Professional database (version 10.3) (33), revealed several additional potential AP-1 binding sites that were scattered across more than 7 kb of genomic sequence. Data for these sites, as well as for the experimentally suggested ones, are summarized in Fig. 2. In total, we identified six AP-1 sites for the murine ccl2 gene and eight for the human ccl2 gene. However, it should be noted that use of less-stringent search criteria or of other programs resulted in even greater numbers of predicted AP-1-binding sites (results not shown).
Alignment of the murine and human ccl2 loci suggested that although the exact positions of these AP-1 sites differ, the overall patterns displayed considerable similarity. In particular, the locations of AP-1-binding elements in the far upstream region, around the TSSs, and within the far downstream region of the genes mirrored each other (Fig. 2).
As this analysis suggested a putatively very complex pattern of c-Jun binding to the ccl2 gene, we sought to use ChIP systematically to map the binding of c-Jun to the murine ccl2 locus. Genomic DNA was immunoprecipitated from wild-type or c-Jun-deficient cells by use of c-Jun antibodies and was amplified by LM PCR. Tiled PCR fragments of 400 bp were designed to cover around 7 kb of genomic sequence of the ccl2 locus (Fig. 3A). Amounts of corresponding PCR products that were amplified from amplicons were quantitated by real-time PCR using standard curves, derived from serially diluted input chromatin, for each individual PCR. As depicted in Fig. 3B and C, enriched c-Jun binding was detected in several regions across the ccl2 locus. In cells cultured in low-concentration serum, specific c-Jun binding was detectable only after IL-1 stimulation (Fig. 3B), whereas in cells cultured in high-concentration serum, specific c-Jun binding was constitutive and appeared to occur more broadly, suggesting that under these conditions c-Jun occupied more binding sites (Fig. 3C). Essentially, three regions of c-Jun binding were identified: (i) approximately 2.5 kb upstream of the TSS (PCR fragment 1); (ii) immediately adjacent to the TSS (PCR fragments 5 to 7) and (iii) approximately 2 kb downstream of the polyadenylation signal in the 3′ region of the locus (PCR fragments 15 to 17).
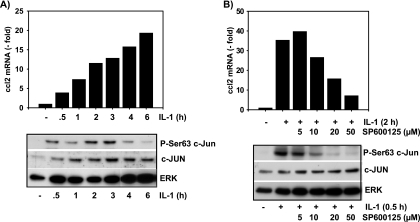
To further extend these results to human cells, we also investigated nontransformed MRC5 fibroblasts. In similarity to murine fibroblast results, IL-1 induces a rapid increase in ccl2 mRNA levels (Fig. 4A). IL-1 also increases the constitutive amount of c-Jun protein as well as its phosphorylation at Ser63 (Fig. 4A). The JNK inhibitor SP600125 suppressed IL-1-induced c-Jun phosphorylation as well as ccl2 mRNA expression in a dose-dependent manner (Fig. 4B). SP600125 did not inhibit constitutive or inducible expression of c-Jun. Of note, half-maximal suppression of ccl2 required 10 to 20 μM SP600125, while half-maximal suppression of c-Jun phosphorylation occurred at around 5 to 10 μM (Fig. 4B). At a concentration of 50 μM SP600125, c-Jun phosphorylation was completely inhibited, whereas ccl2 mRNA expression was suppressed by around 80% (Fig. 4B). Hence, strong suppression of JNK activity (as assessed by measuring c-Jun phosphorylation) substantially decreased IL-1-inducible expression of ccl2, suggesting that, as seen in murine cell experiments, ccl2 is under the control of the JNK-Jun pathway.
FIG. 4.
JNK-and c-Jun-dependent ccl2 mRNA expression in human fibroblasts. (A) Human MRC5 fibroblasts were cultivated in 10% fetal calf serum and were thereafter stimulated for the indicated times with IL-1α (10 ng/ml) or were left untreated. Relative ccl2 mRNA expression levels were analyzed by TaqMan real-time PCR. In parallel, cells were lysed and expression and phosphorylation of c-Jun were analyzed by Western blotting of whole-cell extracts. (B) Cells were treated for 30 min with the indicated concentrations of SP600125. Thereafter, cells were stimulated for the indicated times with 10 ng/ml IL-1α or were left untreated. ccl2 mRNA and expression and phosphorylation of c-Jun were analyzed as described for panel A. ERK antibodies were used to control for protein loading.
Based on the analysis shown in Fig. 2 and on the results obtained with the murine ccl2 gene (Fig. 3), three genomic regions in the human ccl2 gene were analyzed by ChIP (Fig. 5A). To quantitate binding of c-Jun and its modified forms to the ccl2 gene, we established TaqMan real-time PCR probes for the three genomic fragments. As shown in Fig. 5B, we found constitutive binding of c-Jun to AP-1 sites in the far upstream region, adjacent to the TSS, and at the previously characterized downstream AP-1 site. IL-1 induced a moderate increase in c-Jun binding to the far upstream and the far downstream sites but not to the site adjacent to the TSS. There was a strong increase in binding of c-Jun phosphorylated at Ser73 in all three regions, suggesting that the major IL-1-mediated signal is the JNK-dependent phosphorylation of prebound c-Jun (Fig. 5B). As it was previously suggested that c-Jun phosphorylation disrupts the protein-protein interaction of c-Jun with HDAC3 (58), we analyzed the recruitment of two HDACs, HDAC3 and HDAC1, to the human ccl2 locus. For HDAC1 alone (Fig. 5B), but not for HDAC3 (data not shown), there was an IL-1-induced decrease in binding to the ccl2 gene which was paralleled by an increase in acetylation of histone H4 at the far upstream site and at the far downstream region (Fig. 5B). Hence, the analysis of the human ccl2 gene revealed a pattern resembling that of c-Jun-binding obtained for the murine gene. It further suggested that phosphorylation of c-Jun is involved in modulation of HDAC1 binding and histone acetylation at distinct regions of the gene.
Mutational analysis of c-Jun-binding sites that were identified by ChIP.
To assess the functional significance of the c-Jun binding sites that were identified in the ChIP experiments shown in Fig. 3 and 5 we activated the ccl2 gene by use of specific c-Jun-containing AP-1 dimers and also mutated the potential c-Jun binding sites within the context of the human ccl2 locus. Ectopically expressed AP-1 single-chain proteins have been previously used as specific tools to analyze the contribution of AP-1 homo- or heterodimers to promoter activation (1). In line with the recruitment of endogenous c-Jun to the ccl2 gene, AP-1 dimers composed of c-Jun-c-Jun, or c-Jun-ATF2, but not c-Jun-c-Fos activated by about two- to fivefold a luciferase reporter gene driven by a previously described 2.8-kb fragment of the murine ccl2 promoter and enhancer (53, 55) (Fig. 6B). This effect was further enhanced by IL-1 (Fig. 6B).
To our knowledge, inducible recruitment of c-Jun to the 3′ region of an AP-1 target gene has not been described before. To assess the relevance of this site for ccl2 mRNA expression, 5.6 kb of the human ccl2 locus was amplified by PCR, subcloned, and transiently (Fig. 6C and E) or stably (Fig. 6D6D) transfected into human HEK293IL-1R cells or murine fibroblasts, respectively. Likewise, as shown in Fig. 6A, point mutations of the AP-1-binding site adjacent to the TSS (AP-1 site 4) or within the far downstream region (AP-1 site 7) were generated.
Transfected cells were then analyzed for expression of human ccl2 mRNA transcribed from the wild-type or mutated genes.
Both types of experiments confirmed that both AP-1-binding sites contribute to basal and inducible ccl2 mRNA synthesis in response to IL-1 (Fig. 6C and D) or c-Jun-ATF-2 (Fig. 6E).
ChIP analysis confirmed that c-Jun and HDAC3 were recruited to both regions of the wild-type hccl2 gene but not to mutant versions in which both AP-1 sites were mutated (data not shown).
The mutational experiments shown in Fig. 6C to E confirm the contribution of the AP-1-binding site, adjacent to the TSS, to ccl2 transcription, as previously suggested using reporter gene assays (40, 47, 60). But they also demonstrate a comparable contribution of the 3′ AP-1 site to ccl2 induction.
c-Jun is required for RNA polymerase II recruitment and activation at the ccl2 gene.
To further characterize the gene-regulatory events that might be under the control of c-Jun, we measured the recruitment of RNA polymerase II to the ccl2 promoter (Fig. 7A). In cells kept in low-concentration serum—the condition which results in the strongest ccl2 mRNA regulation in response to IL-1—binding of RNA polymerase II was transiently increased up to eightfold upon IL-1 treatment (Fig. 7A). As assessed by absolute quantification, expression of mature mRNA was induced about 8- to 10-fold by IL-1 (Fig. 7B, upper graph). Under these conditions, no inducible RNA polymerase II recruitment or mRNA expression in the absence of c-Jun was found (Fig. 7A and B). We also analyzed the synthesis of ccl2 pre-mRNA as a measure of polymerase II activity. In wild-type cells, expression of pre-mRNA was low but was rapidly and strongly induced by IL-1 in parallel to the generation of mature mRNA (Fig. 7B, lower graph). The mature ccl2 mRNA was about 15 to 20 times more abundant than pre-mRNA (Fig. 7B), indicating that the initial long transcript is rapidly converted to the mature mRNA species. In c-Jun-deficient cells, expression of basal and IL-1-inducible ccl2 pre-mRNA was strongly suppressed (Fig. 7B). Taken together, the data shown in Fig. 7 indicate that under low-concentration-serum conditions, IL-1 mainly upregulates recruitment of RNA polymerase to induce ccl2 transcription. This effect strictly requires c-Jun.
c-Jun mediates histone modifications and recruitment of NF-κB.
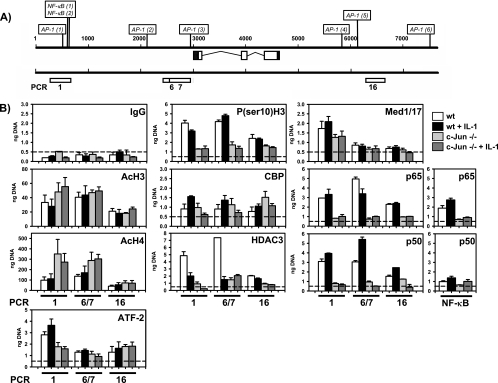
To further analyze putative c-Jun-dependent coregulators of ccl2 transcription, we chose the three major c-Jun-binding regions identified by ChIP analysis (Fig. 3 and 5), namely, the 5′ upstream region, the region around the TSS, and the 3′ downstream region represented by PCR fragments 1, 6 and 7, and 16, as shown in Fig. 3 and 8A. One additional primer pair exactly covering the two known NF-κB-binding sites within the murine locus was also used (Fig. 8B). As the AP-1 sites in serum-starved cells are sparsely occupied by c-Jun prior to IL-1 stimulation (Fig. 3B), we performed this analysis with cells kept in 10% serum to examine the role of c-Jun under constitutive as well as IL-1-stimulated conditions.
FIG. 8.
c-Jun mediates histone modifications, histone deacetylase binding, and NF-κB recruitment at the murine ccl2 gene locus. (A) Schematic representation of the three selected genomic regions of the murine ccl2 gene (indicated by white boxes) that were analyzed by PCR. (B) Wild-type (wt) or c-Jun-deficient (−/−) fibroblasts were treated for 2 h with 10 ng/ml IL-1α or were left untreated. After in vivo cross-linking, chromatin was isolated from all cultures and immunoprecipitated with the indicated antibodies. Genomic DNA fragments from the indicated regions of mouse ccl2 were amplified by real-time PCR and amounts of DNA quantitated using a standard curve. Data represent mean amounts of DNA (in nanograms) ± standard errors of the means of the results of at least two independent experiments. The dashed lines indicate nonspecifically bound DNA fragments as determined by the IgG control ChIP. “NF-κB” indicates a primer pair that covers the region around the two NF-κB-binding sites as shown in panel A.
HDAC3 was constitutively associated with ccl2 genomic DNA at all three regions examined (Fig. 8B). IL-1 induced a significant decrease of HDAC3 levels. In c-Jun-deficient cells, HDAC3 binding was low in unstimulated as well as in IL-1-stimulated cells. These data demonstrate that c-Jun is essential for recruitment of HDAC3 to the endogenous ccl2 locus. Reduced HDAC3 binding in cells lacking c-Jun was associated with increased levels of histone H4 acetylation at the 5′ and TSS regions, but not at the 3′ region, whereas levels of histone H3 acetylation were only weakly increased at the 5′ region and were not affected at the TSS and 3′ regions.
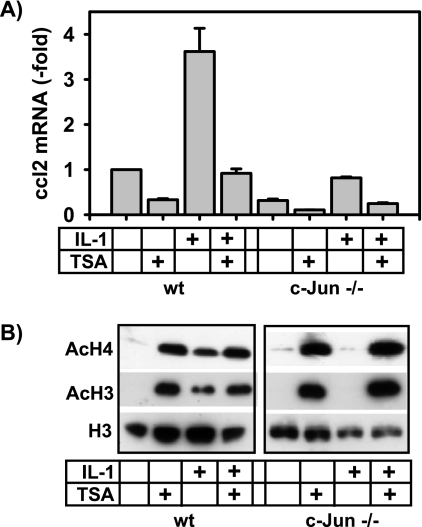
Hence, in the case of ccl2, hyperacetylation of histones correlates with inhibition of transcription in c-Jun-deficient cells, whereas, in general, increased acetylation of histones often correlates with increased mRNA synthesis (23). The data derived from ChIP analysis were supported by the results of experiments that used the general histone deacetylase inhibitor TSA, which strongly suppressed constitutive as well as IL-1-inducible ccl2 mRNA expression (Fig. 9A). Moreover, TSA further suppressed the low level of mRNA expression of ccl2 in c-Jun-deficient cells (Fig. 9A). Treatment of wild-type cells with IL-1 caused a modest increase in total cellular histone H3 and H4 acetylation (Fig. 9B) but did not affect local histone H3 or H4 acetylation at the three regions of the ccl2 gene (Fig. 8B). In inverse correlation to this observation, IL-1-inducible total cellular histone acetylation was absent in c-Jun-deficient cells (Fig. 9B) but was increased locally at the ccl2 gene at 5′ and TSS regions (Fig. 8B). In contrast to the results seen with IL-1, pharmacological blockade of HDAC activity by TSA increased total acetylation of histones H3 and H4 in wild-type and c-Jun−/− cells at comparable levels (Fig. 9B). These data suggest that in the absence of c-Jun, turnover of histone acetylation at specific regions of the ccl2 gene is disturbed in an HDAC3-dependent, stimulus- and gene-specific manner. In addition to the clear requirement of c-Jun for HDAC3 recruitment, we also noted that in c-Jun-deficient cells, IL-1-inducible recruitment of CBP was decreased at the 5′ and TSS regions but not at the 3′ region (Fig. 8B), suggesting that c-Jun also regulates access of histone acetylases such as CBP to local histones across the ccl2 gene.
FIG. 9.
Inhibition of histone deacetylase activity suppresses ccl2 mRNA synthesis. (A) Basal and IL-1-inducible ccl2 mRNA expression in wild-type (wt) or c-Jun-deficient fibroblasts was analyzed using cells that were pretreated for 2 h with 500 ng/ml of the HDAC inhibitor TSA. Then, cells were treated for a further 2 h with IL-1α (10 ng/ml) or were left untreated as indicated. ccl2 mRNA expression was determined by TaqMan real-time PCR. Data represent mean ccl2 expression ± standard errors of the means of the results of two independent experiments relative to the unstimulated wild-type control cell results. (B) The inhibitory effect of TSA on HDAC activity was validated in nuclear extracts from wild-type or c-Jun-deficient fibroblasts by Western blotting using antibodies directed against acetylated histone H3 and histone H4.
p38 MAP kinase-dependent phosphorylation of Ser10 of histone H3 has been associated with increased mRNA synthesis of certain human inflammatory genes, including ccl2 (43), and is executed by the protein kinases MSK1/MSK2 (6, 48). Saccani et al. (43) have already suggested that H3 phosphorylation promotes recruitment of p65 NF-κB. For ccl2, we found that in all three regions, phosphorylation of Ser10 of histone H3 was significantly reduced in c-Jun-deficient cells (Fig. 8B). Interestingly, a H3 mutant in which Ser10 was mutated to alanine suppressed epidermal growth factor-induced AP-1 activity, providing independent evidence for a link between Ser10 phosphorylation and AP-1-mediated gene expression (5). Hence, our data reinforce the conclusions of both aforementioned studies but provide novel evidence for a crucial role of c-Jun in H3 phosphorylation under the conditions used in this study.
We observed no significant effect of c-Jun on the recruitment of selected components of the basal transcriptional machinery, i.e., TATA-binding protein, a component of TFIID (data not shown). Recruitment of Mediator 1 and Mediator 17, subunits of the mediator complex (56), was mainly restricted to the 5′ and TSS regions and was reduced in c-Jun-deficient cells at the 5′ region but not at the TSS region (Fig. 8B).
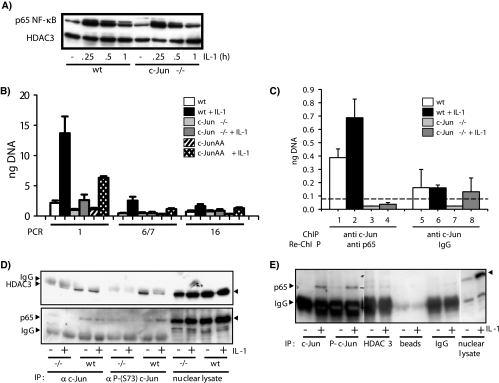
We further investigated the recruitment of ATF-2 and NF-κB subunits, transcription factors whose relevance for ccl2 transcription was evident from the experiments shown in Fig. 6 and from previous studies (41, 51). ATF-2 was constitutively bound at all three regions of the ccl2 gene (Fig. 8B). IL-1 induced recruitment of ATF-2 modestly at the 5′ site, and levels of ATF-2 bound to the ccl2 gene dropped in c-Jun-deficient cells only at this site (Fig. 8B). Binding of p65 and p50 NF-κB subunits was found at the 5′, the TSS, and the downstream regions. Under conditions of high serum concentrations, there was no induction by IL-1, with the exception of p50 recruitment to the TSS site (Fig. 8B). Binding of NF-κB subunits to the 5′ region most likely occurs through the two well-defined conserved NF-κB cis elements (40, 51). Binding to the other regions may occur through unknown or less-conserved NF-κB sites or indirectly via protein-protein interactions. In line with this result, ChIP analysis of regions across chromosome 22 also revealed recruitment of p65 NF-κB to coding and noncoding regions of many genes by use of the same antibody as used in our study (32). Importantly, NF-κB binding was suppressed at all three regions in c-Jun-deficient cells (Fig. 8B).
This effect was not due to a general defect in proximal NF-κB signaling, as constitutive and IL-1-inducible nuclear translocation of p65 NF-κB was unaffected (Fig. 10A).
FIG. 10.
c-Jun forms chromatin-associated protein-protein complexes with p65 NF-κB at the ccl2 locus in a phosphorylation-dependent manner. Cells were kept in low-concentration serum as described for Fig. 1. wt, wild type. (A) Western blot analysis of constitutive and IL-1-induced translocation of p65 NF-κB in nuclear extracts from wild-type and c-Jun-deficient cells. Antibodies against HDAC3 were used to control for protein loading. (B) Wild-type, c-Jun-deficient (−/−), or c-JunAA fibroblasts were treated for 1 h with 10 ng/ml IL-1α or were left untreated. After in vivo cross-linking, chromatin was isolated from all cultures and immunoprecipitated with antibodies against p65 NF-κB. Genomic DNA fragments from the indicated regions of mouse ccl2 represented in Fig. 8A were amplified by real-time PCR, and amounts of DNA were quantitated using a standard curve. Data represent the mean amounts of DNA (in nanograms) ± standard errors of the means of the results of at least two independent experiments. (C) Chromatin from wild-type or c-Jun-deficient (−/−) cells was immunoprecipitated with anti-c-Jun antibodies (ChIP) followed by a second round of immunoprecipitation (Re-ChIP) with anti-p65 antibodies or control IgG. ccl2 genomic DNA fragments associated with c-Jun and with p65 were detected using primer pair 1 as shown in Fig. 8A and quantified by real-time PCR as described for panel B. Data represent the mean amounts of DNA (in nanograms) ± standard errors of the means of the results of at least two independent experiments. The dashed line indicates the mean signal obtained from all ChIP experiments that served as controls (lanes 3 to 8) and therefore indicates the mean of unspecific binding results for genomic ccl2 fragments. (D) Fibroblasts were stimulated for 1 h with 10 ng/ml IL-1α or were left untreated. HDAC3 (upper panel) or p65 NF-κB (lower panel) was coimmunoprecipitated from nuclear extracts of wild-type (wt) or c-Jun-deficient (−/−) cells by use of c-Jun or phospho-c-Jun (P-c-Jun)-specific antibodies. Proteins in immune complexes or in nuclear lysates were detected by Western blotting. (E) HEK293IL-1R cells were stimulated for 30 min with 10 ng/ml IL-1α or were left untreated. p65 NF-κB was coimmunoprecipitated from nuclear extracts by use of the indicated antibodies or IgG beads and was detected in immune complexes or nuclear lysates by Western blotting. The blot membrane was cut into three pieces that were processed and exposed identically.
This strict requirement of c-Jun for NF-κB binding was unexpected and was therefore investigated in more detail. Serum-starved cells, which have little c-Jun bound to the ccl2 gene, show much lower p65 binding to all three regions but considerable stronger induction by IL-1 (Fig. 10B). As seen with serum-treated cells, NF-κB recruitment was severely suppressed in c-Jun-deficient cells and could be restored only partially in c-JunAA cells, suggesting that c-Jun phosphorylation is required not only for HDAC3 recruitment (as shown in Fig. 8) but also for p65 NF-κB recruitment (Fig. 10B). Normally, ChIP does not reveal whether immunoprecipitated proteins are bound to the same genetic elements or to different genetic elements contained in the pool of all cross-linked complexes. However, Re-ChIP experiments clearly indicated that c-Jun and p65 NF-κB form chromatin-associated complexes at the same ccl2 fragment. These complexes were not enriched with material from c-Jun-deficient cells by specific antibodies or IgG control antibodies, demonstrating the specificity of the Re-ChIP assays (Fig. 10C). Coimmunoprecipitation experiments using c-Jun or phospho-c-Jun-specific antibodies confirmed an interaction of endogenous c-Jun with p65 NF-κB or HDAC3, respectively (Fig. 10D and E). An interaction of c-Jun and p65 in transient overexpression systems or cell-free assays has been reported previously (49). Hence, our results corroborate the results of those earlier experiments but also suggest that c-Jun, p65, and HDAC3 form chromatin-associated protein-protein complexes at specific regions of a gene in an IL-1-dependent manner. Apparently, phosphorylation of c-Jun favors interaction with p65 NF-κB over interaction with HDAC3 (Fig. 10D). Moreover, throughout our study we found that it was much easier to detect these interactions after in vivo cross-linking compared to coimmunoprecipitation. This may reflect different levels of efficiency of ChIP/Re-ChIP compared to coimmunoprecipitation but may also result from local enrichment or stabilization of c-Jun-p65 or c-Jun-HDAC3 complexes, respectively, when they are bound to chromatin.
These data can be reconciled by a model in which c-Jun is required for histone modifications, recruitment of p65/p50 NF-κB, and regulation of RNA polymerase II binding and activity. In IL-1-stimulated cells, phosphorylation of c-Jun promotes dissociation of HDACs, which results in a modest increase in CBP binding and histone acetylation across the ccl2 locus. We propose that this rearranges chromatin structure to facilitate recruitment of the strong transcriptional activator p65 NF-κB. p65 not only binds to its NF-κB-binding site but also interacts directly with c-Jun. As assessed by ChIP (Fig. 3, 5, 8, and 10), c-Jun, HDAC1/HDAC3, CBP, and p65/p50 can be found at the 5′ region and at the TSS region as well as at the 3′ region of the ccl2 locus. This suggests that the far 5′ and 3′ ends of the ccl2 gene may actually adopt a flexible structure whereby multiple protein-DNA contacts are established to all four proteins. This higher-order complex does not form in the absence of c-Jun. In line with this interpretation, Teferedegne et al. (51) recently suggested that the far 5′ upstream region of ccl2 contacts the transcription factor Sp1 at the proximal promoter region, an interaction that requires p65 NF-κB and CBP. Hence, the entire ccl2 locus may actually adopt a quite flexible structure that facilitates rapid recruitment and activation of the RNA polymerase II holoenzyme. In the absence of c-Jun, the balance of HDAC/CBP binding is disturbed. As a result, histones are “frozen” in a hyperacetylated state that prohibits correct folding of the 5′ and 3′ region, IL-1-mediated recruitment of p65 NF-κB, and subsequent polymerase II activation. This situation allows only very little ccl2 mRNA synthesis and resembles that seen after treatment of cells with TSA.
A similar phenomenon has been recently described for the c-Jun and c-Fos genes, which, like ccl2, are very rapidly induced by numerous stimuli (15). Hyperacetylation of histones within these genes by treatment with TSA also resulted in suppression of mRNA synthesis. In that important study it was therefore suggested that increased mRNA synthesis requires dynamic turnover of histone acetylation and phosphorylation at the same histone tails rather than arrest of histones in the modified state (15). As discussed above, our data support this model. Presumably, IL-1 promotes acetylation turnover, and our ChIP experiments detected only the small fraction of histones acetylated at a given time point. In contrast, pharmacological inhibition of HDACs by TSA or, as shown in our experiments, by removal of HDACs from the ccl2 gene in the absence of c-Jun arrests a larger fraction of histones in the acetylated state and hence suppresses mRNA expression.
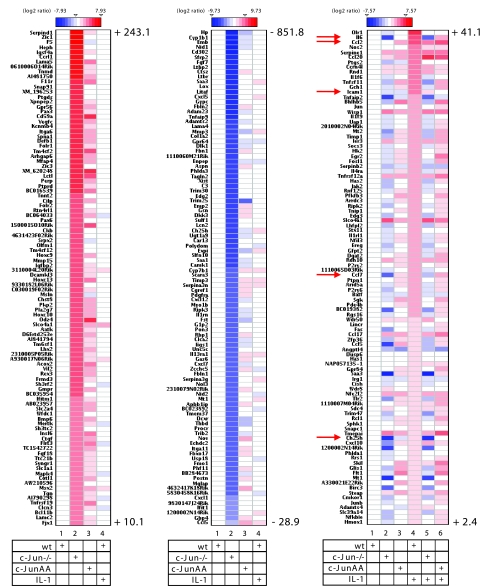
To explore whether the activator function of c-Jun represents a more general phenomenon, the RNAs from the experiment shown in Fig. 1C were analyzed by high-density DNA microarray experiments. Figure 11 summarizes the results for the 100 most strongly differentially expressed genes as described below. We identified 162 genes whose expression was upregulated more than twofold by IL-1 in wild-type cells. mRNA levels of 133 of these genes were reduced by more than 50% in IL-1-stimulated cells lacking c-Jun (Fig. 11, right panel; compare lane 4 with lane 5). Expression of a total of 69 of these genes was already downregulated by more than twofold in unstimulated cells (Fig. 11, right panel; compare lane 1 with lane 2). For 35 out of the 162 IL-1-regulated genes, mRNA expression levels reached at least 80% of the wild-type levels in c-JunAA cells (Fig. 11, right panel; compare lane 4 with lane 6), indicating that for these genes c-Jun phosphorylation is not required. Furthermore, in c-JunAA cells expression of 47 of the 162 induced genes was already upregulated more than twofold in the absence of IL-1 (Fig. 11, right panel; compare lane 1 with lane 3).
FIG. 11.
Identification of c-Jun-dependent genes by high-density DNA microarray experiments. RNAs from three independent preparations as shown in Fig. 1C were pooled, and expression of 18,194 genes was analyzed by oligonucleotide DNA microarray experiments. Ratios of expression values relative to unstimulated wild-type cell values were log2 transformed and are visualized as heatmaps, as indicated by color scales. The three panels shown indicate the top-ranking 100 genes that are upregulated in c-Jun-deficient cells (left panel), downregulated in c-Jun-deficient cells (middle panel), or induced by IL-1 (right panel). Numbers indicate highest and lowest severalfold regulation results for upregulated genes (left panel, lane 2), downregulated genes (middle panel, lane 2), and IL-1-regulated genes (right panel, lane 4). The full data set is available upon request. Details are described in Materials and Methods. Arrows indicate visualized mRNA expression values for genes (ccl2, il6, icam1, ccl7, and ch25h) as validated using real-time PCR (Fig. 1D; also see Fig. S1 in the supplemental material).
Real-time PCR validation experiments for il6, ccl7, icam1, and ch25h confirmed that mRNA expression of these genes is strictly dependent on c-Jun and JNK-mediated c-Jun phosphorylation, suggesting that they are regulated by mechanisms similar to those seen in ccl2 experiments (see Fig. S1 in the supplemental material).
We identified 674 genes that were upregulated from 3.0- to 243-fold and 819 genes that were downregulated by 3.0- to 852-fold in the absence of c-Jun. The c-JunAA mutant rescued most of the downregulated genes and about a third of the upregulated genes (Fig. 11, left and middle panels; compare lanes 2 to lanes 3).
While the results obtained with the latter two groups of genes underscore the dual functions of c-Jun as a repressor or activator of gene expression, they largely represent genes that are not regulated by IL-1 (Fig. 11, left and middle panels; compare lanes 2 to lanes 4). Hence, compared to the IL-1-stimulated situation, for these genes c-Jun might be integrated into different corepressor or coactivator complexes rather than those described in our study. Using a cDNA microarray carrying probes for 15,000 mainly unknown genes, Florin et al. (11) found a group of more than 1,000 c-Jun- or JunB-dependent genes, of which 101 were regulated by IL-1. As that group used a different DNA microarray and did not analyze the role of c-Jun phosphorylation in gene expression, their results cannot be directly compared to ours on a gene-to-gene basis. However, both DNA microarray studies together clearly identified several large classes of c-Jun response genes.
In summary, our investigation of ccl2 shows that c-Jun (and AP-1) binding is scattered across a large region of an AP-1 response gene and that “classical” analyses of 5′ promoter regions fused to reporter genes fail to detect these additional control mechanisms. Our functional analysis using gain- or loss-of-function approaches combined with mutational analysis of the ccl2 locus suggests that c-Jun homo- or c-Jun-ATF-2 heterodimers activate ccl2 whereas c-Fos is less important. In support of the latter observation, ccl2 mRNA expression was also not affected in cells lacking c-Fos (22). We found that the pattern of c-Jun binding to the ccl2 locus was less distinct in the presence of serum (Fig. 3C), leaving the possibility that the additional candidate AP-1 cis elements predicted by the TransFAC analysis (Fig. 2) may actually be occupied by c-Jun in other situations. The combination of all mechanisms described in this report multiplies the options for c-Jun-dependent-specific gene control.
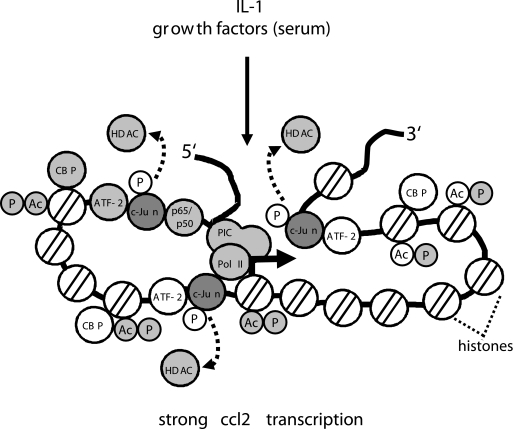
Collectively, the results presented here confirm but also significantly extend previous results with respect to the mechanisms of c-Jun-mediated transcriptional regulation obtained by using a reporter gene system (58) or c-Jun-deficient macrophages (39). Those studies suggested that c-Jun mainly represses genes (39) and that gene induction by c-Jun involves activation by derepression (58). However, c-Jun was also shown by the latter group to interact with RNA helicases (59). Our report now suggests that the role of c-Jun in regulation of inflammatory genes is more complex. As exemplified for the ccl2 gene, lack of c-Jun has several profound effects on inducible inflammatory mRNAs, affecting nucleosomal, histone-dependent mechanisms of gene control, recruitment of coactivators/corepressors, and polymerase II activity (summarized in Fig. 12).
FIG. 12.
Schematic representation of the roles of c-Jun in regulation of ccl2 transcription. c-Jun binds to several regions within the ccl2 gene. As deduced from ChIP experiments, it is involved in multiple protein-protein and protein-DNA contacts that are established across the entire locus. Proteins whose recruitment, activity, or modifications are altered in c-Jun-deficient cells are shaded in gray. For details, see text. PIC, preinitiation complex.
We also demonstrate that JNK/c-Jun and NF-κB pathways converge at the level of chromatin, indicating a novel level of cross-talk between these two important signaling systems.
Thus, our systematic analysis of c-Jun-mediated ccl2 mRNA expression establishes an example of the enormous combinatorial potential of c-Jun-mediated specific gene control during inflammation.
Supplementary Material
Acknowledgments
We thank Judith Ashouri and Oliver Dittrich-Breiholz for critically reading the manuscript. We thank Jeremy Saklatvala, Matthias Gaestel, and James Woodgett for the gift of IL-1 or murine fibroblasts.
This work was supported by grants KR-1143/5-1, Kr1143/4-3, Kr1143/6-1, and SFB566/B06 (to M.K.) from the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 28 April 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bakiri, L., K. Matsuo, M. Wisniewska, E. F. Wagner, and M. Yaniv. 2002. Promoter specificity and biological activity of tethered AP-1 dimers. Mol. Cell. Biol. 224952-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behrens, A., M. Sibilia, and E. F. Wagner. 1999. Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nat. Genet. 21326-329. [DOI] [PubMed] [Google Scholar]
- 3.Boekhoudt, G. H., Z. Guo, G. W. Beresford, and J. M. Boss. 2003. Communication between NF-κB and Sp1 controls histone acetylation within the proximal promoter of the monocyte chemoattractant protein 1 gene. J. Immunol. 1704139-4147. [DOI] [PubMed] [Google Scholar]
- 4.Chinenov, Y., and T. K. Kerppola. 2001. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 202438-2452. [DOI] [PubMed] [Google Scholar]
- 5.Choi, H. S., B. Y. Choi, Y. Y. Cho, H. Mizuno, B. S. Kang, A. M. Bode, and Z. Dong. 2005. Phosphorylation of histone H3 at serine 10 is indispensable for neoplastic cell transformation. Cancer Res. 655818-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clayton, A. L., and L. C. Mahadevan. 2003. MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett. 54651-58. [DOI] [PubMed] [Google Scholar]
- 7.Daly, C., and B. J. Rollins. 2003. Monocyte chemoattractant protein-1 (CCL2) in inflammatory disease and adaptive immunity: therapeutic opportunities and controversies. Microcirculation 10247-257. [DOI] [PubMed] [Google Scholar]
- 8.Dérijard, B., M. Hibi, I. H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R. J. Davis. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 761025-1037. [DOI] [PubMed] [Google Scholar]
- 9.Eferl, R., and E. F. Wagner. 2003. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3859-868. [DOI] [PubMed] [Google Scholar]
- 10.Finzer, P., U. Soto, H. Delius, A. Patzelt, J. F. Coy, A. Poustka, H. zur Hansen, and F. Rosl. 2000. Differential transcriptional regulation of the monocyte-chemoattractant protein-1 (MCP-1) gene in tumorigenic and non-tumorigenic HPV 18 positive cells: the role of the chromatin structure and AP-1 composition. Oncogene 193235-3244. [DOI] [PubMed] [Google Scholar]
- 11.Florin, L., L. Hummerich, B. T. Dittrich, F. Kokocinski, G. Wrobel, S. Gack, M. Schorpp-Kistner, S. Werner, M. Hahn, P. Lichter, A. Szabowski, and P. Angel. 2004. Identification of novel AP-1 target genes in fibroblasts regulated during cutaneous wound healing. Oncogene 237005-7017. [DOI] [PubMed] [Google Scholar]
- 12.Gu, L., S. Tseng, R. M. Horner, C. Tam, M. Loda, and B. J. Rollins. 2000. Control of TH2 polarization by the chemokine monocyte chemoattractant protein-1. Nature 404407-411. [DOI] [PubMed] [Google Scholar]
- 13.Gupta, S., T. Barrett, A. J. Whitmarsh, J. Cavanagh, H. K. Sluss, B. Dérijard, and R. J. Davis. 1996. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 152760-2770. [PMC free article] [PubMed] [Google Scholar]
- 14.Hayden, M. S., and S. Ghosh. 2004. Signaling to NF-κB. Genes Dev. 182195-2224. [DOI] [PubMed] [Google Scholar]
- 15.Hazzalin, C. A., and L. C. Mahadevan. 2005. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS Biol. 3e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hibi, M., A. Lin, T. Smeal, A. Minden, and M. Karin. 1993. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 72135-2148. [DOI] [PubMed] [Google Scholar]
- 17.Hoeflich, K. P., J. Luo, E. A. Rubie, M. S. Tsao, O. Jin, and J. R. Woodgett. 2000. Requirement for glycogen synthase kinase-3beta in cell survival and NF-κB activation. Nature 40686-90. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann, E., A. Thiefes, D. Buhrow, O. Dittrich-Breiholz, H. Schneider, K. Resch, and M. Kracht. 2005. MEK1-dependent delayed expression of Fos-related antigen-1 counteracts c-Fos and p65 NF-κB-mediated interleukin-8 transcription in response to cytokines or growth factors. J. Biol. Chem. 2809706-9718. [DOI] [PubMed] [Google Scholar]
- 19.Holtmann, H., J. Enninga, S. Kalble, A. Thiefes, A. Dorrie, M. Broemer, R. Winzen, A. Wilhelm, J. Ninomiya-Tsuji, K. Matsumoto, K. Resch, and M. Kracht. 2001. The MAPK kinase kinase TAK1 plays a central role in coupling the interleukin-1 receptor to both transcriptional and RNA-targeted mechanisms of gene regulation. J. Biol. Chem. 2763508-3516. [DOI] [PubMed] [Google Scholar]
- 20.Holtmann, H., R. Winzen, P. Holland, S. Eickemeier, E. Hoffmann, D. Wallach, N. L. Malinin, J. A. Cooper, K. Resch, and M. Kracht. 1999. Induction of interleukin-8 synthesis integrates effects on transcription and mRNA degradation from at least three different cytokine- or stress-activated signal transduction pathways 2. Mol. Cell. Biol. 196742-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holzberg, D., C. G. Knight, O. Dittrich-Breiholz, H. Schneider, A. Dorrie, E. Hoffmann, K. Resch, and M. Kracht. 2003. Disruption of the c-JUN-JNK complex by a cell-permeable peptide containing the c-JUN delta domain induces apoptosis and affects a distinct set of interleukin-1-induced inflammatory genes. J. Biol. Chem. 27840213-40223. [DOI] [PubMed] [Google Scholar]
- 22.Hu, E., E. Mueller, S. Oliviero, V. E. Papaioannou, R. Johnson, and B. M. Spiegelman. 1994. Targeted disruption of the c-fos gene demonstrates c-fos-dependent and -independent pathways for gene expression stimulated by growth factors or oncogenes. EMBO J. 133094-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 24.Karin, M., Z. Liu, and E. Zandi. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9240-246. [DOI] [PubMed] [Google Scholar]
- 25.Karin, M., Y. Yamamoto, and Q. M. Wang. 2004. The IKK NF-κB system: a treasure trove for drug development. Nat. Rev. Drug Discov. 317-26. [DOI] [PubMed] [Google Scholar]
- 26.Kim, T., J. Yoon, H. Cho, W. B. Lee, J. Kim, Y. H. Song, S. N. Kim, J. H. Yoon, J. Kim-Ha, and Y. J. Kim. 2005. Downregulation of lipopolysaccharide response in Drosophila by negative crosstalk between the AP1 and NF-κB signaling modules. Nat. Immunol. 6211-218. [DOI] [PubMed] [Google Scholar]
- 27.Kracht, M., and J. Saklatvala. 2002. Transcriptional and post-transcriptional control of gene expression in inflammation. Cytokine 2091-106. [DOI] [PubMed] [Google Scholar]
- 28.Kracht, M., O. Truong, N. F. Totty, M. Shiroo, and J. Saklatvala. 1994. Interleukin 1 alpha activates two forms of p54 alpha mitogen-activated protein kinase in rabbit liver. J. Exp. Med. 1802017-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Q., and I. M. Verma. 2002. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2725-734. [DOI] [PubMed] [Google Scholar]
- 30.Lim, S. P., and A. Garzino-Demo. 2000. The human immunodeficiency virus type 1 Tat protein up-regulates the promoter activity of the beta-chemokine monocyte chemoattractant protein 1 in the human astrocytoma cell line U-87 MG: role of SP-1, AP-1, and NF-κB consensus sites. J. Virol. 741632-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, B., B. J. Rutledge, L. Gu, J. Fiorillo, N. W. Lukacs, S. L. Kunkel, R. North, C. Gerard, and B. J. Rollins. 1998. Abnormalities in monocyte recruitment and cytokine expression in monocyte chemoattractant protein 1-deficient mice. J. Exp. Med. 187601-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martone, R., G. Euskirchen, P. Bertone, S. Hartman, T. E. Royce, N. M. Luscombe, J. L. Rinn, F. K. Nelson, P. Miller, M. Gerstein, S. Weissman, and M. Snyder. 2003. Distribution of NF-κB-binding sites across human chromosome 22. Proc. Natl. Acad. Sci. USA 10012247-12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matys, V., O. V. Kel-Margoulis, E. Fricke, I. Liebich, S. Land, A. Barre-Dirrie, I. Reuter, D. Chekmenev, M. Krull, K. Hornischer, N. Voss, P. Stegmaier, B. Lewicki-Potapov, H. Saxel, A. E. Kel, and E. Wingender. 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34D108-D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morton, S., R. J. Davis, A. McLaren, and P. Cohen. 2003. A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 223876-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakayama, K., A. Furusu, Q. Xu, T. Konta, and M. Kitamura. 2001. Unexpected transcriptional induction of monocyte chemoattractant protein 1 by proteasome inhibition: involvement of the c-Jun N-terminal kinase-activator protein 1 pathway. J. Immunol. 1671145-1150. [DOI] [PubMed] [Google Scholar]
- 36.Natoli, G. 2006. Tuning up inflammation: how DNA sequence and chromatin organization control the induction of inflammatory genes by NF-κB. FEBS Lett. 5802843-2849. [DOI] [PubMed] [Google Scholar]
- 37.Natoli, G., S. Saccani, D. Bosisio, and I. Marazzi. 2005. Interactions of NF-κB with chromatin: the art of being at the right place at the right time. Nat. Immunol. 6439-445. [DOI] [PubMed] [Google Scholar]
- 38.Oberley, M. J., J. Tsao, P. Yau, and P. J. Farnham. 2004. High-throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 376315-334. [DOI] [PubMed] [Google Scholar]
- 39.Ogawa, S., J. Lozach, K. Jepsen, D. Sawka-Verhelle, V. Perissi, R. Sasik, D. W. Rose, R. S. Johnson, M. G. Rosenfeld, and C. K. Glass. 2004. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc. Natl. Acad. Sci. USA 10114461-14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ping, D., G. H. Boekhoudt, E. M. Rogers, and J. M. Boss. 1999. Nuclear factor-κB p65 mediates the assembly and activation of the TNF-responsive element of the murine monocyte chemoattractant-1 gene. J. Immunol. 162727-734. [PubMed] [Google Scholar]
- 41.Ping, D., P. L. Jones, and J. M. Boss. 1996. TNF regulates the in vivo occupancy of both distal and proximal regulatory regions of the MCP-1/JE gene. Immunity 4455-469. [DOI] [PubMed] [Google Scholar]
- 42.Sabapathy, K., K. Hochedlinger, S. Y. Nam, A. Bauer, M. Karin, and E. F. Wagner. 2004. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol. Cell 15713-725. [DOI] [PubMed] [Google Scholar]
- 43.Saccani, S., S. Pantano, and G. Natoli. 2002. p38-dependent marking of inflammatory genes for increased NF-κB recruitment. Nat. Immunol. 369-75. [DOI] [PubMed] [Google Scholar]
- 44.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 202390-2400. [DOI] [PubMed] [Google Scholar]
- 45.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4E131-E136. [DOI] [PubMed] [Google Scholar]
- 46.Shi, Y., A. Kotlyarov, K. Laabeta, A. D. Gruber, E. Butt, K. Marcus, H. E. Meyer, A. Friedrich, H. D. Volk, and M. Gaestel. 2003. Elimination of protein kinase MK5/PRAK activity by targeted homologous recombination. Mol. Cell. Biol. 237732-7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shyy, J. Y., M. C. Lin, J. Han, Y. Lu, M. Petrime, and S. Chien. 1995. The cis-acting phorbol ester “12-O-tetradecanoylphorbol 13-acetate”-responsive element is involved in shear stress-induced monocyte chemotactic protein 1 gene expression. Proc. Natl. Acad. Sci. USA 928069-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soloaga, A., S. Thomson, G. R. Wiggin, N. Rampersaud, M. H. Dyson, C. A. Hazzalin, L. C. Mahadevan, and J. S. Arthur. 2003. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 222788-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stein, B., A. S. Baldwin, Jr., D. W. Ballard, W. C. Greene, P. Angel, and P. Herrlich. 1993. Cross-coupling of the NF-κB p65 and Fos/Jun transcription factors produces potentiated biological function. EMBO J. 123879-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeshita, A., Y. Chen, A. Watanabe, S. Kitano, and S. Hanazawa. 1995. TGF-beta induces expression of monocyte chemoattractant JE/monocyte chemoattractant protein 1 via transcriptional factor AP-1 induced by protein kinase in osteoblastic cells. J. Immunol. 155419-426. [PubMed] [Google Scholar]
- 51.Teferedegne, B., M. R. Green, Z. Guo, and J. M. Boss. 2006. Mechanism of action of a distal NF-κB-dependent enhancer. Mol. Cell. Biol. 265759-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thiefes, A., A. Wolf, A. Doerrie, G. A. Grassl, K. Matsumoto, I. Autenrieth, E. Bohn, H. Sakurai, R. Niedenthal, K. Resch, and M. Kracht. 2006. The Yersinia enterocolitic effector YopP inhibits host cell signalling by inactivating the protein kinase TAK1 in the IL-1 signalling pathway. EMBO Rep. 7838-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thiefes, A., S. Wolter, J. F. Mushinski, E. Hoffmann, O. Dittrich-Breiholz, N. Graue, A. Dorrie, H. Schneider, D. Wirth, B. Luckow, K. Resch, and M. Kracht. 2005. Simultaneous blockade of NF-κB, JNK, and p38 MAPK by a kinase-inactive mutant of the protein kinase TAK1 sensitizes cells to apoptosis and affects a distinct spectrum of tumor necrosis target genes. J. Biol. Chem. 28027728-27741. (Erratum, 280:32564.) [DOI] [PubMed] [Google Scholar]
- 54.Ventura, J.-J., N. J. Kennedy, J. A. Lamb, R. A. Flavell, and R. J. Davis. 2003. c-Jun NH2-terminal kinase is essential for the regulation of AP-1 by tumor necrosis factor. Mol. Cell. Biol. 232871-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner, K., U. Dendorfer, S. Chilla, D. Schlondorff, and B. Luckow. 2001. Identification of new regulatory sequences far upstream of the mouse monocyte chemoattractant protein-1 gene. Genomics 78113-123. [DOI] [PubMed] [Google Scholar]
- 56.Wang, G., M. A. Balamotis, J. L. Stevens, Y. Yamaguchi, H. Handa, and A. J. Berk. 2005. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell 17683-694. [DOI] [PubMed] [Google Scholar]
- 57.Wang, N., L. Verna, S. Hardy, J. Forsayeth, Y. Zhu, and M. B. Stemerman. 1999. Adenovirus-mediated overexpression of c-Jun and c-Fos induces intercellular adhesion molecule-1 and monocyte chemoattractant protein-1 in human endothelial cells. Arterioscler. Thromb. Vasc. Biol. 192078-2084. [DOI] [PubMed] [Google Scholar]
- 58.Weiss, C., S. Schneider, E. F. Wagner, X. Zhang, E. Seto, and D. Bohmann. 2003. JNK phosphorylation relieves HDAC3-dependent suppression of the transcriptional activity of c-Jun. EMBO J. 223686-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Westermarck, J., C. Weiss, R. Saffrich, J. Kast, A. M. Musti, M. Wessely, W. Ansorge, B. Seraphin, M. Wilm, B. C. Valdez, and D. Bohmann. 2002. The DEXD/H-box RNA helicase RHII/Gu is a co-factor for c-Jun-activated transcription. EMBO J. 21451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamada, M., S. Kim, K. Egashira, M. Takeya, T. Ikeda, O. Mimura, and H. Iwao. 2003. Molecular mechanism and role of endothelial monocyte chemoattractant protein-1 induction by vascular endothelial growth factor. Arterioscler. Thromb. Vasc. Biol. 231996-2001. [DOI] [PubMed] [Google Scholar]
- 61.Yang, D. D., C. Y. Kuan, A. J. Whitmarsh, M. Rincon, T. S. Zheng, R. J. Davis, P. Rakic, and R. A. Flavell. 1997. Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the Jnk3 gene. Nature 389865-870. [DOI] [PubMed] [Google Scholar]
- 62.Zenz, R., R. Eferl, L. Kenner, L. Florin, L. Hummerich, D. Mehic, H. Scheuch, P. Angel, E. Tschachler, and E. F. Wagner. 2005. Psoriasis-like skin disease and arthritis caused by inducible epidermal deletion of Jun proteins. Nature 437369-375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.