Abstract
Ydj1 is a Saccharomyces cerevisiae Hsp40 molecular chaperone that functions with Hsp70 to promote polypeptide folding. We identified Ydj1 as being important for maintaining steady-state levels of protein kinases after screening several chaperones and cochaperones in gene deletion mutant strains. Pulse-chase analyses revealed that a portion of Tpk2 kinase was degraded shortly after synthesis in a ydj1Δ mutant, while the remainder was capable of maturing but with reduced kinetics compared to the wild type. Cdc28 maturation was also delayed in the ydj1Δ mutant strain. Ydj1 protects nascent kinases in different contexts, such as when Hsp90 is inhibited with geldanamycin or when CDC37 is mutated. The protective function of Ydj1 is due partly to its intrinsic chaperone function, but this is minor compared to the protective effect resulting from its interaction with Hsp70. SIS1, a type II Hsp40, was unable to suppress defects in kinase accumulation in the ydj1Δ mutant, suggesting some specificity in Ydj1 chaperone action. However, analysis of chimeric proteins that contained the chaperone modules of Ydj1 or Sis1 indicated that Ydj1 promotes kinase accumulation independently of its client-binding specificity. Our results suggest that Ydj1 can both protect nascent chains against degradation and control the rate of kinase maturation.
Molecular chaperones prevent polypeptides from misfolding and promote acquisition of the folded state. In their absence, nascent polypeptides or proteins that become unfolded aggregate or become targeted for degradation via the ubiquitin proteasome pathway (22). Hsp70 is a molecular chaperone that functions early in protein folding process by interacting with nascent polypeptide chains cotranslationally or immediately after translation. Hsp70 functions in association with cochaperones that facilitate the cycle of nascent chain binding and release. Cochaperones that promote polypeptide loading are members of the Hsp40 family (12, 15). Those that stimulate polypeptide dissociation function in nucleotide exchange and belong to several structurally distinct protein families (1, 25, 28, 46).
Hsp40 chaperones comprise a large and diverse protein family (12, 15). Each member has a conserved “J” domain that functions in stimulating Hsp70's ATPase. Members of type I and type II Hsp40s also interact with unfolded proteins and help to load them onto Hsp70. Once a polypeptide is bound to Hsp70, the Hsp40 J domain stimulates Hsp70's ATPase, which helps stabilize the interaction by leading to closure of a helical lid structure over the polypeptide binding site. Binding of unfolded polypeptides to Hsp70 or Hsp40s is via hydrophobic interactions, and this prevents polypeptide aggregation (18). Polypeptide release is catalyzed by nucleotide exchange factors that facilitate release of ADP from Hsp70, allowing for ATP binding and opening of the lid retaining the polypeptide in the chaperone client-binding site. Cycles of binding and release, catalyzed by Hsp40 and nucleotide exchange factors, can promote folding and prevent aggregation.
In addition to preventing aggregation, chaperones also function in the degradation of misfolded proteins by the proteasome. Inhibition of Hsp90, for example, leads to ubiquitination and degradation of many protein kinases and several nuclear receptors (52). Hsp90 is required in yeast for degradation of heterologously expressed proteins that are not folded by this chaperone, suggesting that it has distinct roles in both folding and degradation (38). In mammalian cells, chaperones promote degradation via interaction with the E3 ubiquitin ligase, Chip, which binds to both Hsp70 and Hsp90 via tetratricopeptide motifs (14, 39). Chip integrates the quality control process by regulating heat shock factor-dependent gene expression and also acting itself as a chaperone (16, 44).
The mechanisms underlying chaperone and cochaperone function in targeting polypeptides toward the proteasome are not well understood, although a common factor is Hsp70. Several genetic and biochemical studies have shown that Hsp70 is important for the degradation of misfolded cytosolic proteins (4, 38, 40). Hsp40s cooperate with Hsp70 in this process, based on the finding that YDJ1 and HLJ1 promote the degradation of cytosolic and membrane bound polypeptides (30, 40, 54). The results from biochemical studies support the view that Hsp40s promote degradation. For example, endoplasmic reticulum-associated degradation of misfolded membrane proteins is promoted by Hdj2, a Ydj1 ortholog (57). A more direct targeting function has been discovered for a neuronal Hsp40, Hsj1, which interacts directly with polyubiquitin chains via a ubiquitin interaction motif, and promotes ubiquitinylation of polypeptides in association with Chip (51).
Protein kinases represent a class of protein whose folding is dependent on the Hsp90 and Cdc37 in addition to the Hsp70/Hsp40 chaperone pair (9). Using in vitro chaperone-kinase assembly reactions, Arlander et al. demonstrated that chaperones bind to a misfolded kinase in a sequential manner, beginning with Hsp70/Hsp40 (2). The interaction of these chaperones with the misfolded kinase was required for subsequent Cdc37 binding, which led to Hsp90 recruitment. This final step is also promoted by the Hsp organizing protein called Hop, which acts as bridge between Hsp70 and Hsp90 (2, 31, 32). Cdc37 interacts primarily with the kinase N-lobe, while Hsp90 interacts with both N-lobe and C-lobe of a kinase catalytic subunit (42, 59). Complementary studies using yeast genetics identified similar chaperone components for kinase folding (17, 29, 31, 32) and showed that Cdc37 has a general role in kinome biogenesis in addition to protecting nascent kinase chains from degradation during or shortly after translation (36). In addition to the folding of nascent kinases, molecular chaperones also function to maintain the mature state of mutant kinases, such as oncogenic forms of Src or Lck (26, 53).
In the present study, we screened for chaperones that affected kinase steady-state levels in a similar manner to Cdc37. Ydj1, a yeast type I Hsp40 chaperone was identified as having such a role. We demonstrate here that Ydj1 protects a cohort of kinases similar to that protected by Cdc37. Our findings support the hypothesis that Ydj1 protects nascent chains during or immediately after synthesis and contrasts with other studies showing that this Hsp40 promotes degradation. In addition, we show that newly synthesized protein kinases that escape triage in ydj1Δ cells are largely protected from further degradation but fold slowly for a period of up to 2 h.
MATERIALS AND METHODS
Strains and plasmids.
In the present study we used yeast strains from the TAP-tagged library (Open Biosystems, Huntsville, AL) based on strain S288C (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) (24). The strain genotype was verified by PCR analysis using primers specific to the tag and kinase gene of interest. The yeast ydj1Δ and erg6Δ knockout strains were constructed by introducing KanMX4 module as described earlier (36). In the ydj1 and erg6 double mutant, YDJ1 was replaced by URA3 and ERG6 replaced by KanMX4.
The H34Q mutant was prepared from pRS315-Ydj1. Amino acid substitution was performed by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The mutation was confirmed by DNA sequencing. pGAL-Ydj1 was constructed by excising a 1.3-kb EcoR1 fragment from pAV2 (8) and ligating into the low-copy-number vector pSEYc58GalP containing the GAL1-10 promoter. Plasmids expressing YDJ1, SIS1, YSY, and SYS in pRS315 were kindly provided by Doug Cyr (University of North Carolina, Chapel Hill). pRS317-ydj1ΔG/F was a gift from E. Craig (University of Wisconsin, Madison). Ydj1ΔG/F was subsequently subcloned into pRS315 by using Not1 and Xma1. The plasmid expressing Escherichia coli dnaJ was described previously (7).
Solubility assay.
Cells expressing TAP-tagged kinases were grown at 30°C, harvested, washed once with sterile water, and resuspended in 500 μl of 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 0.1 mg of pepstatin A/ml. Cells were lysed by bead beating and clarified by centrifugation at 16,000 × g for 10 min at 4°C. A portion (150 μl) of this supernatant was kept aside as total protein, and another 150 μl was spun in a Beckman TLA45 rotor at 100,000 × g for 30 min at 4°C. The supernatant was removed and designated as the soluble fraction. The pellet was washed once with the buffer described above and solubilized with 200 μl of 1× sodium dodecyl sulfate (SDS) sample buffer. Then, 50 μl of 4× SDS sample buffer was added to the total-protein and soluble-fraction samples. Equal amounts of each fraction were analyzed by SDS-polyacrylamide gel electrophoresis, followed by immunoblot analysis with anti-TAP antibody (Open Biosystems).
Western blot analysis.
Western blot analysis was performed on whole-cell extracts prepared in IPP150 (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% NP-40) or in 50 mM Tris (pH 7.5)-0.1 mM EDTA-1% SDS plus Complete protease inhibitor (Boehringer Mannheim) by glass bead lysis. The steady-state kinase level was quantified by Odyssey Li-Cor image processing software. Galactose induction experiments shown in Fig. 5 were conducted by growing cultures in raffinose (2%) overnight. Cultures were diluted to an A600 of 0.2 and supplemented with either galactose or glucose (2% [vol/vol]), followed by incubation for 8 h with shaking at 30°C before extract preparation as described above.
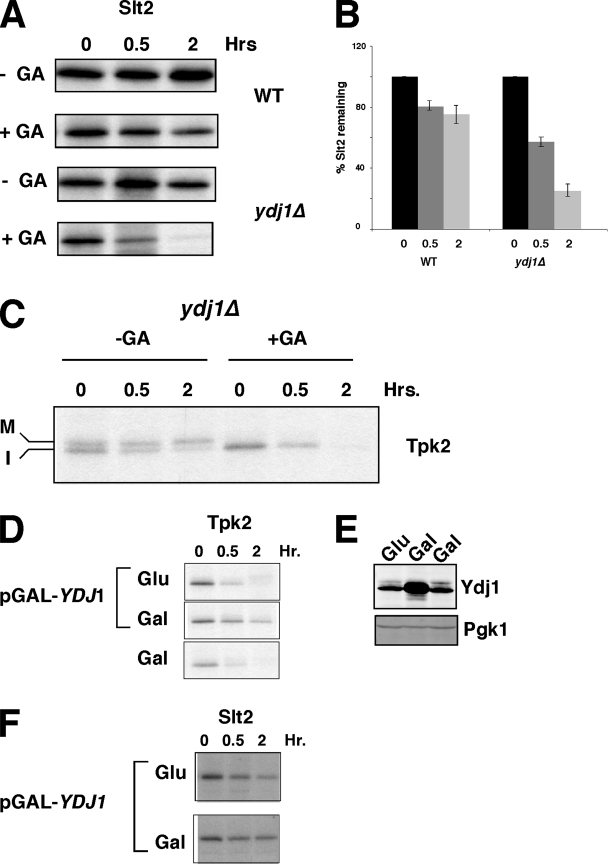
FIG. 5.
Effect of Ydj1 on the stability of nascent protein kinases under conditions that promote their degradation. (A) Effect of GA treatment on the stability of TAP-Slt2 kinase in wild-type and ydj1Δ cells, as indicated. Chase times are given in hours after pulse-labeling in the absence and presence of 50 μM GA. (B) Quantitation of Slt2 based on the data shown in panel A and other data (n = 3 ± the standard error). (C) Pulse-chase analysis of TAP-Tpk2 in the absence and presence of 50 μM GA in ydj1Δ cells. The chase times are indicated in hours. Mature (M) and immature (I) forms of Tpk2 are indicated. (D) Pulse-chase analysis of TAP-Tpk2 from cells containing pGAL-YDJ1 under noninducing condition (glucose; Glu) and inducing conditions (galactose, Gal). The lower panel shows pulse-chase results for Tpk2 in cells containing galactose but not the pGAL-YDJ1 plasmid. (E) Western blot analysis of Ydj1 protein before and after overexpression using the same cultures as in panel D. Pgk1 is shown as a loading control. (F) Effect of YDJ1 overexpression on TAP-Slt2 stability in the presence of GA (50 μM). Inducing and noninduced states are as described in panel D.
Polyclonal antisera to Hsp90, Ydj1, and Sis1 were described previously (8, 35, 43). Hsp70 mouse monoclonal antisera was purchased from Stressgen (spa-822). Antisera to E. coli dnaJ was kindly provided by Ulrich Hartl. Anti-TAP was purchased from Open Biosystems.
Pulse-labeling.
Pulse-labeling and immunoprecipitations were performed as described previously (5).
Kinase assays.
Kinase assay of wild-type and ydj1Δ cells expressing TAP-tagged forms of Tpk2 and Rim11 were performed as described previously (36). Briefly, wild-type and ydj1Δ strains were grown overnight at 30°C in 100 ml of synthetic complete medium (minus histidine) to log phase. Cell extracts were prepared in IPP150 buffer, and the amount of protein was quantified by using Bradford reagent. Tpk2 was immunoprecipitated from 0.1 to 0.5 mg of extract from wild-type cells and from 3 mg of extracts from ydj1Δ mutant cells. The cell extracts were diluted to 0.45 ml in IPP150 and incubated with 50 μl of 50% (vol/vol) immunoglobulin G-Sepharose resin (GE) for 1 h at 4°C. The resin was washed and divided into two aliquots. One aliquot was used for the kinase assay, while the other was used for Western blot analysis with anti-TAP. The Western blots were quantified by using a Li-Cor Odyssey infrared detection system. Kinase assays for Tpk2 were performed by using a Pep-Tag nonradioactive protein kinase assay kit and quantified by spectrophotometry of agarose gel slices as described by the manufacturer (Promega), while Rim11 was assayed by measuring its ability to phosphorylate myelin basic protein (Upstate Biotechnology) and quantified by phosphorimaging.
RESULTS
Ydj1 influences the steady-state levels of protein kinases.
We searched for molecular chaperones and cochaperones besides Cdc37 that could affect the levels of yeast protein kinases, starting with C-terminal TAP tagged Tpk2, a homologue of mammalian cAMP-dependent protein kinase (PKA). The chaperones or cochaperones AHA1, CPR6, CPR7, HCH1, SBA1, SSE1, STI1, and YDJ1 were deleted from strains expressing a TAP-TPK2. These genes encode proteins that modulate Hsp90's ATPase activity (e.g., Aha1, Hch1, Sti1, and Sba1), act as chaperones (Cpr6, Cpr7, Sba1, Sse1 and Ydj1), and/or have peptidyl prolyl isomerase activity (Cpr6 and Cpr7 [37, 41]).
Tpk2 levels were assessed by Western blotting in comparison to a wild-type strain, using Pgk1 as a loading control. The results demonstrated that only upon deletion of the Hsp40 chaperone YDJ1 were Tpk2 levels reproducibly lowered compared to the wild type (Fig. 1A and data not shown). Reduction in kinase levels was associated with increased amounts of Hsp90 and Hsp70, although Pgk1 levels were unaffected by any of the chaperone deletions. Similar reductions in kinase levels and increases in Hsp90/Hsp70 were observed in a ydj1Δ strain expressing TAP-Rim11 (Fig. 1B). These increases suggest that deletion of YDJ1 promotes a stress response in these cells.
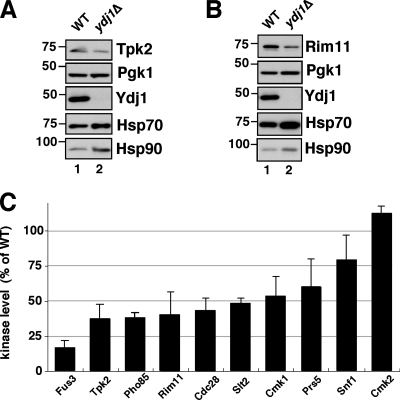
FIG. 1.
Effect of YDJ1 deletion on protein kinase levels. (A) Steady-state level of Tpk2 in wild-type and ydj1Δ yeast cells; (B) Rim11 in wild-type and ydj1Δ cells. Wild-type and ydj1Δ yeast cells were grown overnight at 30°C and kinases identified with anti-TAP. Western blots of the same samples were performed with anti-Pgk1, anti-Ydj1, anti-Hsp70, and anti-Hsp90. Pgk1 levels are shown as a loading control. (C) Steady-state levels of kinases in ydj1Δ strain. The results are expressed as the percentages of levels in wild-type cells in each case based on Western blot analysis. The bars indicate the standard deviation from three to four independent experiments. WT, wild type.
In a previous study we analyzed 65 different kinases for stability in a yeast strain mutated for CDC37, a chaperone essential for protein kinase folding (36). Among the 65 kinases tested, 51 had reduced levels compared to those present in the wild-type strain. We therefore tested a sampling of kinases that were unstable or stable in the cdc37 mutant for a similar phenotype in the ydj1Δ mutant. For these experiments we analyzed Cdc28, Cmk1, Fus3, Pho85, and Slt2 as examples of kinases that were unstable in the cdc37 mutant, and Cmk2 and Snf1 as kinases that were stable. Prs5, a component of ribosylphosphotransferase, was added to this analysis as an example of a kinase-related protein that was unaffected by mutation in CDC37. In accord with results from the cdc37 mutant, we observed that Cdc28, Cmk1, Fus3, Pho85, and Slt2 had reduced levels in the ydj1Δ mutant (Fig. 1C). The mean reduction was ∼2-fold, but Fus3 levels were reduced the most at 6-fold compared to the wild type. Strikingly, kinases that were stable in the cdc37 mutant, Cmk2, and Snf1 displayed the least dependence on Ydj1 for their steady-state levels. Cmk2 steady-state levels were completely unaffected by the loss of Ydj1. Prs5, which was unaffected by mutation in CDC37, was reduced in the ydj1Δ mutant.
The fate of residual Tpk2 kinase that was synthesized and stable in a ydj1Δ mutant was addressed in two ways. First, we investigated whether the kinase accumulated in a soluble form or whether it aggregated. Lysates containing Tpk2 synthesized in the presence or absence of Ydj1 protein were centrifuged at 100,000 × g for 1 h. A small enrichment of Tpk2 was observed in the pellet fraction in the ydj1Δ mutant, although the vast majority of the kinase was soluble. Next, we determined the specific activity of Tpk2 synthesized in the ydj1Δ mutant. Since there was less Tpk2 at steady state in the ydj1Δ mutant compared to the wild type, we first generated a standard curve of Tpk2 activity from the wild-type strain (Fig. 2B). The specific activity of a single concentration of Tpk2 isolated from the ydj1Δ mutant was then determined by comparison with the standard curve. As shown in Fig. 2B, the specific activity of Tpk2 in both strains was similar, although slightly reduced in the ydj1Δ mutant. Therefore, Tpk2 is capable of acquiring the native state in the absence of Ydj1, but in reduced amounts. Similar findings were recorded for Rim11, which also has a very similar specific activity when derived from wild-type or ydj1Δ mutant strains, although its steady-state levels are reduced in the absence of Ydj1 (Fig. 1 and 2C).
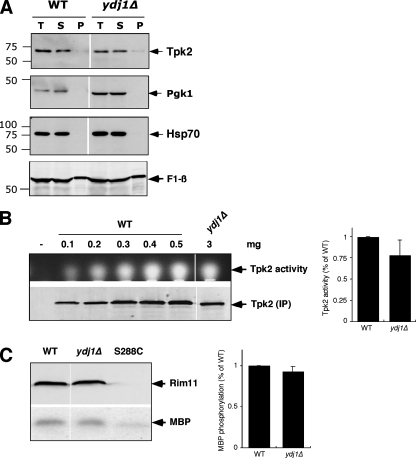
FIG. 2.
Solubility and activity of protein kinases in ydj1Δ mutant strains. (A) Solubility assay of Tpk2 kinase. The solubility of Tpk2 was assayed in wild-type and ydj1Δ cells by comparing equal amounts of total lysate (T), supernatant (S), and pellet (P) fractions by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting. The same blot was analyzed with anti-Hsp70, anti-Pgk1, and anti-Atp2, which encodes the β subunit of the mitochondrial F1 ATPase. (B) Tpk2 activity in wild-type and ydj1Δ mutant strains. The upper panel shows the Tpk2 activity after immunoprecipitation of the kinase from various amounts of wild-type extract (expressed in milligrams) and from a 3-mg portion of extracts from ydj1Δ cells. The panel shows the fluorescence from a labeled peptide whose migration in an agarose gel depends on phosphorylation by immunoprecipitated Tpk2. The lower panel shows the amount of Tpk2 in each reaction as determined by Western blotting. The bar graph at the right shows the quantitation of Tpk2 activity after normalization to the amount of enzyme in the assay. Bars indicate the standard error of the mean values of three independent experiments. (C) Assay of Rim11 activity. The upper panel shows Western blot results of TAP-Rim11 immunoprecipitates and the amount of 32P-labeled myelin basic protein (MBP) used in the assay. The panel on the right shows the MBP data quantified. Bars represent the standard error (n = 3). WT, wild type.
Ydj1 affects the rate of protein kinase maturation.
Previous studies showed that Cdc37 protects newly made protein kinases from degradation either during or shortly after translation (36). Since Ydj1 functions upstream of Cdc37 in in vitro kinase-chaperone assembly reactions (2), it seemed likely that Ydj1 might behave in similar manner, especially given that both chaperones interact with the same cohort of protein kinases.
We addressed the role of Ydj1 in biogenesis of newly translated Tpk2 and Cdc28 by pulse-chase analysis. For Tpk2, pulse-chase experiments revealed the presence of two distinct bands after denaturing gel electrophoresis (Fig. 3A). Based on previous studies with mammalian PKA, these represent immature (inactive, nonphosphorylated; faster migrating) and mature (active, autophosphorylated; slower migrating) conformers (56). This was verified with phosphatase treatment of 35S-labeled wild-type cell extracts, which resulted in partial conversion of Tpk2 from the slow-migrating form to the fast-migrating form (PKA is relatively resistant to phosphatase treatment) (47). Mammalian PKA autophosphorylates in cells at two sites: Ser197 and Ser338 (27). The former is in the activation loop and is necessary for enzyme activity, while the latter facilitates stabilization of the kinase N-lobe (3, 56). It is likely, therefore, that the immature form of the kinase is an unstable conformer.
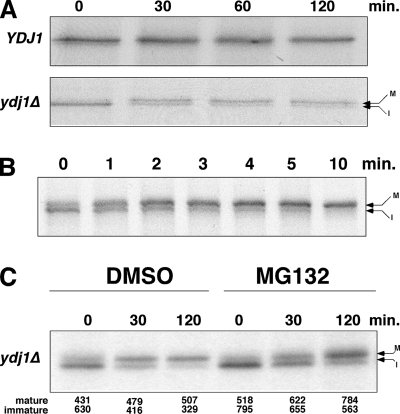
FIG. 3.
Pulse-chase analysis of Tpk2 kinase in the wild type and the ydj1Δ mutant. (A) Pulse-chase analysis of TAP-tagged Tpk2 in wild-type (YDJ1) and ydj1Δ mutant cells. The chase times are indicated in minutes. (B) Pulse-chase analysis of wild-type cells after a 2-min pulse and chase from 0 to 10 min. (C) Pulse-chase analysis of TAP-Tpk2 in ydj1Δ cells. Cells were treated with dimethyl sulfoxide or 100 μM MG132 for 30 min before labeling. The strain background is erg6Δ, which improves MG132 permeability. The chase times are indicated in minutes. Mature (M, phosphorylated) and immature (I, nonphosphorylated) forms of Tpk2 are denoted by arrows. The relative amounts of mature and immature kinase in each lane were quantified, and the numbers appear below the panel.
In the wild-type strain, the faster-migrating form of Tpk2 is a minor component after a 10-min pulse-labeling that disappears after 30 min of chase. In the ydj1Δ mutant, there is a lower level of Tpk2 after the pulse, a finding consistent with decreased levels found at steady state (Fig. 3A). Also, the faster-migrating band is more prominent at zero time of chase and appears to convert into the slower-migrating band over the 2-h chase time. This interpretation is based on the apparent decrease in faster-migrating band intensity and an increase in the amount of the slower-migrating band after 30 min of chase. The faster-migrating band then slowly disappears over the next 90 min (Fig. 3A and C). The relationship between the two forms of Tpk2 was investigated further in wild-type cells using short (2 min) pulse-labeling and chase reactions (Fig. 3B). Under these conditions, most Tpk2 was in the faster-migrating form after the short pulse that converts to the slower-migrating mature form completely within 10 min. In ydj1Δ cells this conversion takes 60 to 120 min (Fig. 3A and C).
When the pulse-chase reaction was performed in the ydj1Δ strain with the proteasome inhibitor MG132 (Fig. 3C), we noted increased Tpk2 levels after the pulse compared to the dimethyl sulfoxide-treated sample. This suggests that some nascent Tpk2 is targeted for degradation rapidly after translation in a manner similar to that of the quality control we observed in a cdc37 mutant strain. In addition, the ratio of immature to mature Tpk2 was clearly altered in the two samples at 30 min of chase. In the MG132- treated sample, there were increased amounts of immature kinase, suggesting that some of the nascent Tpk2 was targeted for degradation in absence of Ydj1. However, it is also clear that a large proportion of the immature kinase managed to adopt the mature state after 120 min of chase, even in the presence of MG132. This shows that, in the absence of Ydj1, the nascent Tpk2 fate is in the balance, with some being susceptible to degradation while most can adopt the mature state.
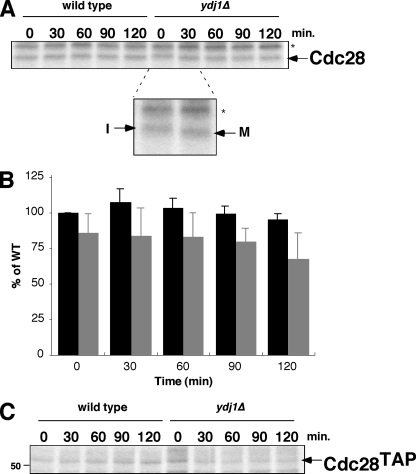
A similar function for Ydj1 in affecting the rate of protein kinase maturation was characterized for Cdc28 (Fig. 4). In this case, there were similar levels of kinase produced after pulse-labeling in both wild-type and ydj1Δ mutant strains. However, Cdc28 in the ydj1Δ mutant had a slower migration at zero time of pulse and quantitatively converted into a faster migrating band after 30 min. In wild-type cells, Cdc28 always appears as the faster-migrating band, even after pulse-labeling. Based on previous studies with Cdc28 (20, 23) and mammalian Cdk2 (6), the slower-migrating band is the immature, nonphosphorylated form, whereas the faster-migrating band results from phosphorylation with the Cdk activating kinase, Cak1. Note that the immature and mature forms of Cdc28 are the inverse of those observed for Tpk2. Importantly, Ydj1 does not appear to affect the levels of nascent Cdc28 after pulse-labeling as dramatically as does mutation in CDC37 (compare Fig. 4A to Fig. 6A), only the rate with which it is converted from the immature form to the mature form. However, the kinase does appear to be degraded to some extent over the 2 h of chase, a finding consistent with the decrease we observed under steady-state conditions. These studies were conducted with nontagged Cdc28, but similar findings were made with TAP-Cdc28 (Fig. 4C). These combined studies show that Ydj1 functions to protect nascent kinases from degradation and also functions to promote efficient folding and/or maturation. The effect of Ydj1 on the rate of maturation is remarkable. In wild-type cells both Cdc28 and Tpk2 are almost quantitatively in the mature state within the 10-min pulse-labeling period. In the absence of Ydj1, its takes up to 2 h for Tpk2 to become fully mature, while for Cdc28 the fully mature state was generated within 30 min.
FIG. 4.
Pulse-chase analysis of Cdc28. (A) Pulse-chase analysis of untagged Cdc28 immunoprecipitated with anti-PSTAIRE (which recognizes a peptide in the α-C helix) in wild-type and ydj1Δ mutant cells. Chase times are indicated in minutes. Cdc28 is denoted by arrow. The band labeled with an asterisk is nonspecific. The inset shows the difference in mobility of Cdc28 at 0 and 30 min of chase time. I, immature form of Cdc28; M, mature form. (B) Quantitation of Cdc28 levels in wild-type (WT) and ydj1Δ cells after phosphorimaging. The results are expressed as the percentages of the levels in wild-type cells at 0-min chase. Bars indicate the standard deviations of three independent experiments. Bars: ▪, wild type;  , ydj1Δ mutant. (C) Pulse-chase analysis of TAP-tagged Cdc28 in wild-type and ydj1Δ cells as described above.
, ydj1Δ mutant. (C) Pulse-chase analysis of TAP-tagged Cdc28 in wild-type and ydj1Δ cells as described above.
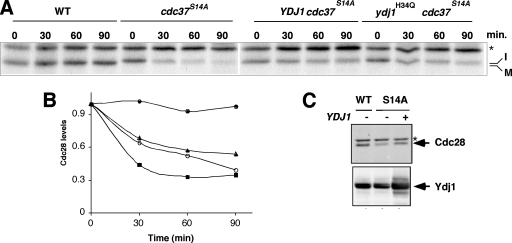
FIG. 6.
Ydj1 protects against degradation of unstable Cdc28 in a cdc37 mutant strain. (A) Pulse-chase analysis of Cdc28 in wild-type and cdc37S14A mutant cells without or with overexpression of YDJ1 or the mutant ydj1H34Q. Chase times are given in minutes. Immature (I) and mature (M) forms of Cdc28 are indicated. The band labeled with an asterisk is nonspecific. (B) Quantitation of the data shown in panel A. Symbols: •, wild type; ▪, cdc37S14A mutant; ▴, cdc37S14A mutant with YDJ1 overexpressed; ○, cdc37S14A mutant with ydj1H34Q overexpressed. (C) Western blot analysis of Cdc28 in wild-type and cdc37S14A mutant cells (S14A) either with (+) or without (−) YDJ1 overexpression. The lower panel shows the Ydj1 levels. The band labeled with an asterisk is nonspecific. WT, wild type.
Ydj1 protects nascent kinases against degradation promoted by Hsp90 inhibition or mutation in CDC37.
Previous analyses of Ydj1 suggested that it promotes polypeptide degradation (30, 40, 54), whereas in our case it appears to protect newly synthesized kinases from this same fate. We therefore addressed the extent of Ydj1's protective function in nascent protein kinase quality control using other models that enhance degradation. The first model involved use of the Hsp90 inhibitor, geldanamycin (GA), which promotes degradation of nascent kinases in yeast (9, 36). For Slt2 kinase, we observed that GA treatment resulted in a modest decline in kinase levels over a 2-h period (Fig. 5A and B). In contrast, Slt2 levels were reduced by ca. 80% in the presence of GA in ydj1Δ cells. These findings show that Ydj1 protects Slt2 from degradation when Hsp90 is inhibited. Tpk2, in contrast, is degraded in the presence of GA in wild-type cells or ydj1Δ mutant cells to a similar extent (compare Fig. 5C with the glucose panel in Fig. 5D).
Previous studies have noted differential degradation of kinases in GA-treated cells derived from human tumors and healthy tissues (11, 48, 58), but the underlying basis for this has never been fully established. One possibility is that chaperones controlling kinase degradation could be modulated. To test this, we extended the findings presented above by analyzing whether YDJ1 overexpression had any further capability to protect nascent kinases from degradation in the presence of GA. YDJ1 was inducibly overexpressed from a galactose-regulated promoter in wild-type cells for these experiments (Fig. 5D and E). In the presence of glucose, which repressed YDJ1 overexpression, GA promotes the degradation of Tpk2 over a 2-h period as described previously (36). However, this effect was suppressed by YDJ1 overexpression (Fig. 5D, middle panel). This effect was not due to galactose replacing glucose in the medium since galactose by itself did not influence the degradation of Tpk2 in the presence of GA (Fig. 5D, lower panel). A similar finding was made for Slt2 kinase (Fig. 5F). Ydj1 is therefore not saturating for its capacity to protect nascent protein kinases from degradation in the presence of GA. For Tpk2, the protective action for Ydj1 in the presence of GA appears to be a gain of function since deletion of the chaperone did not influence GA-dependent kinase degradation (Fig. 5C and D).
The studies described above place Ydj1 in a similar role to Cdc37 as a protector of nascent kinases. We next explored whether these roles were redundant with each other. If this were so, then overexpression of YDJ1 might suppress degradation occurring due to mutation in CDC37. Our model for these studies utilized degradation of Cdc28 in the cdc37S14A mutant. As shown in Fig. 6A, Cdc28 is degraded rapidly in the cdc37S14A mutant, as described previously (36). In contrast, YDJ1 overexpression from a multicopy plasmid results in a reduced rate of degradation of Cdc28 in the cdc37S14A mutant (Fig. 6A and the quantification shown in Fig. 6B). Importantly, a similar finding was observed when the ydj1H34Q mutant was similarly overexpressed in the cdc37S14A mutant (Fig. 6A and B). The ydj1H34Q mutant contains a point mutation in the J domain that inhibits interaction of Ydj1 with Hsp70 (49). The ability of ydj1H34Q to phenocopy wild-type Ydj1 suggests a direct role for Ydj1 in chaperoning nascent kinases that is independent of Hsp70. In contrast, overexpression of either the YDJ1 or the ydj1H34Q mutant in cdc37S14A only delays the degradation of Cdc28, it does not completely prevent it. This is revealed by the failure of Cdc28 to achieve the mature state when YDJ1 is overexpressed in cdc37S14A in the pulse-chase analysis (Fig. 6A and data not shown). Furthermore, Western blot analysis of Cdc28 shows that its levels are only marginally higher in the cdc37S14A mutant overexpressing YDJ1 (Fig. 6C). These findings suggest that Ydj1 has overlapping functions with Cdc37 in stabilizing nascent kinases. The overlapping function is manifest in the way Ydj1 delays Cdc28 degradation in the cdc37S14A mutant, although it clearly fails to promote maturation. This is probably related to Cdc37 chaperone function directly or indirectly in recruiting Hsp90.
Specificity of Ydj1 function in protein kinase maturation.
The experiments described above suggest a model in which Ydj1 protects nascent kinases in association with Cdc37. In addition, Ydj1 contributes to the rate of kinase maturation. The results of experiments with ydj1H34Q suggested a model in which the chaperone action of Ydj1 functioned to protect nascent kinases independently of Ydj1's interaction with Hsp70 (Fig. 6A and B). However, when Tpk2 levels were assayed in the ydj1Δ strain expressing ydj1H34Q from a low-copy-number plasmid (Fig. 7A), they were similar to those found in the ydj1Δ mutant alone. Similar findings were made when Tpk2 maturation was followed by pulse-chase analysis (data not shown). This indicates that under normal conditions, Ydj1 function in Tpk2 maturation involves Hsp70.
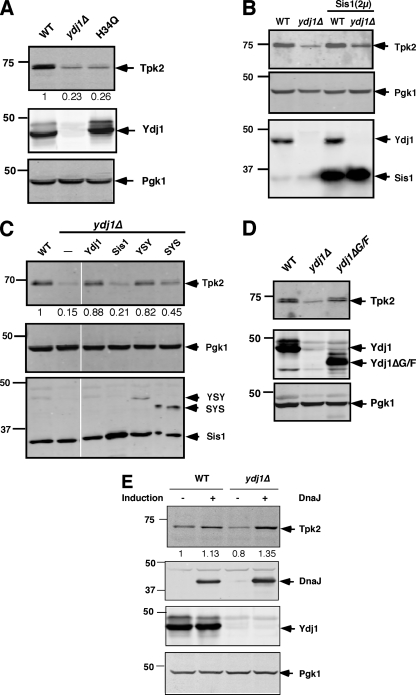
FIG. 7.
Steady-state TAP-Tpk2 levels in ydj1 mutants. (A) Western blot analysis of TAP-Tpk2 (Tpk2) in wild-type and ydj1Δ cells without or with the ydj1H34Q mutant expressed from a low-copy-number plasmid. The middle panel shows Ydj1 levels, and the lower panel shows the levels of Pgk1 as a loading control. (B) Western blot analysis of wild-type and ydj1Δ mutants without or with overexpression of SIS1. The middle panel shows the levels of Pgk1 as a loading control, while the lower panel shows the levels of Ydj1 and Sis1. (C) Western blot of TAP-Tpk2 in wild-type and ydj1Δ cells with or without Sis1, YSY, and SYS chimeric chaperones as indicated. The middle panel shows Pgk1, and the lower panel shows the levels of Sis1 and Ydj1/Sis1 chimeras using anti-Sis1. (D) Effect of Ydj1's G/F region in kinase accumulation. Tpk2 levels were analyzed in wild-type and Ydj1ΔG/F-expressing cells in a ydj1Δ background. (E) Effect of bacterial DnaJ in kinase accumulation. The Tpk2 levels were measured after overexpression of DnaJ in wild-type and ydj1Δ yeast strains. DnaJ expression was carried out after induction with galactose for 8 h. Tpk2 was identified with anti-TAP antibody. The kinase levels were quantified and are shown as a percentage of the wild-type levels. Western blot anlayses with the same samples were performed with anti-Pgk1, anti-DnaJ, and anti-Ydj1. Pgk1 levels served as a loading control. WT, wild type.
To further address the relationship between Ydj1 and Hsp70 in kinase maturation, we overexpressed the type II Hsp40, Sis1, in the ydj1Δ mutant. Previous studies showed that Sis1, a type II Hsp40, partially suppressed the slow-growth phenotype of a ydj1Δ mutant (8); accordingly, Sis1 may compensate for the loss of Ydj1 in kinase biogenesis. However, SIS1 overexpression was largely unable to suppress reduced levels of Tpk2 in the ydj1Δ mutant strain (Fig. 7B). This finding suggested some specificity in the way in which Ydj1 and Hsp70 promote kinase maturation.
Both Ydj1 and Sis1 have distinct chaperone modules that are exchangeable and can specify Sis1 or Ydj1 action in a chimera (19). When we assayed these chimeras, however, those containing the Sis1 chaperone module (YSY; where Y represents Ydj1 domains, and S represents the Sis1 chaperone domain) were more effective at suppressing the loss of Tpk2 than those containing the Ydj1 chaperone module (SYS; Fig. 7C). These findings with Sis1 and Sis1/Ydj1 chimeras suggest that the relationship between Ydj1 and Hsp70 for kinase folding is more complex than expected. They cannot be attributed just to J-domain regulation of Hsp70's ATPase or to specificity in polypeptide-binding by Ydj1. Similarly, deletion of the Ydj1 G/F domain, which separates the J domain from the chaperone-binding module, had little effect on Tpk2 maturation (Fig. 7D).
Finally, we investigated whether the specificity of Ydj1 could be retained by another type I dnaJ protein with the same domain organization. We analyzed E. coli dnaJ for this purpose, which is similar to Ydj1, although it is missing the farnesylation motif. The expression of dnaJ led to complete suppression of the ydj1Δ phenotype for Tpk2 and resulted in even greater levels of kinase in both wild-type and ydj1Δ strains, suggesting it is a more effective chaperone (Fig. 7E). These combined results provide insight into the importance of the C-terminal and perhaps dimerization domains of type I Hsp40s in protecting nascent chains from degradation, while the farnesylation of the Ydj1, which helps to anchor it to membranes (10), is not required for this function.
DISCUSSION
As polypeptide chains emerge from the ribosome, they interact with several chaperone machines. Among these are components of the RAC complex, which includes ribosome-binding Hsp70 and Hsp40 chaperones that promote efficient polypeptide translation (50). Other members of the Hsp70 and Hsp110 families also participate in nascent chain binding (55). More specific chaperones interact with nascent chains from distinct protein families, such as Cdc37, which protects nascent protein kinases from rapid degradation by the proteasome. In the present study, we characterized the type I Hsp40, Ydj1, as also having a protective role in protein kinase biogenesis at the earliest stages posttranslation. Ydj1 also has a role in kinase biogenesis that is distinct from Cdc37, by controlling the rate of kinase maturation as well as the yield of mature kinase.
Ydj1's ability to protect nascent protein kinases results from a direct effect and an indirect one via Hsp70. Of the two, it is the latter that has the greatest contribution to stabilizing protein kinases. In the cdc37S14A mutant, overexpression of wild-type YDJ1 or the mutant ydj1H34Q led to a temporary stabilization of Cdc28. Since the ydj1H34Q mutant cannot interact with Hsp70, these findings suggest that the intrinsic chaperone function of Ydj1 promoted the stabilization, albeit weakly. Furthermore, the overall effects of expressing ydj1H34Q in place of wild-type Ydj1 were negligible at steady state for Tpk2 (Fig. 7A), suggesting that Hsp70 binding is required for complete protection against degradation. As noted previously, Ste11 kinase synthesized in a ydj1Δ mutant had decreased amounts of bound Hsp70 (32), reinforcing the notion that Ydj1 interacts with nascent kinases in order to protect them, but this effect must be integrated with subsequent binding to Hsp70 to avoid degradation. Notably, Hsp90 and Cdc37 could still bind to Ste11 even in a ydj1Δ strain (32). Such binding likely represents a quality control step in kinase maturation that is at least partially distinct from that managed by Ydj1. This is because even in the absence of Ydj1 there is a strong effect of GA in promoting kinase degradation (Fig. 5C). These data therefore point to a role for both Hsp70 and Hsp90 chaperone machines in protecting newly synthesized kinases from degradation, as well as helping them to fold. Interestingly, both chaperones are upregulated in cells with YDJ1 deleted (Fig. 1).
The notion that both Hsp70 and Hsp90 contribute to kinase stability fits with our finding that Tpk2 degradation is biphasic. The first phase is during the 10-min pulse-labeling period, in a manner similar to the way in which Tpk2 is degraded in the cdc37 mutant (36). Like Cdc37, therefore, Ydj1 protects nascent chains and, in its absence, the newly made kinase is largely directed toward the ubiquitin/proteasome system. Tpk2 that escapes initial triage either folds slowly or is subsequently degraded in a slower process. We did not observe the fast degradation of Cdc28 in a ydj1Δ strain, a finding consistent with a similar finding for the cdc37 mutant (Fig. 6) (36). Instead, there is a delay in Cdc28 maturation, followed by a slow decrease in kinase levels. For both Cdc28 and Tpk2, however, the rate of kinase degradation is much more pronounced in the cdc37 mutant than in the ydj1Δ strain. Despite this difference, Ydj1 and Cdc37 both contribute in a similar manner to the folding process; each stabilizes nascent kinases and helps promote binding to another chaperone, Hsp70 in the case of Ydj1 and Hsp90 in the case of Cdc37. Although Ydj1 is an abundant protein, it is still limiting in its capacity to protect newly synthesized kinases. When overexpressed, Ydj1 promotes greater stability of Tpk2 and Slt2 kinases in the presence of GA. Similarly, overexpressed Ydj1 or ydj1H34Q delayed the degradation of unstable Cdc28 in cdc37S14A mutant cells (Fig. 5 and Fig. 6).
Although our results provide strong support for a protective role for Ydj1 in kinase biogenesis, this is not a universal function. There have been several published reports showing that there is reduced degradation of unstable proteins in a mutant of YDJ1 called ydj1-151 (30, 40, 54). However, we found that the expression of ydj1-151 largely suppressed the effect of complete YDJ1 deletion and led to the accumulation of folded Tpk2 as measured by enzyme activity (data not shown). Our model, therefore, is that Ydj1 functions to promote kinase folding but is not involved in targeting misfolded kinases to the degradation machinery, at least under nonstress conditions.
Besides acting to protect nascent kinases, Ydj1 also affects the rate at which maturation takes place. This effect occurred in ydj1Δ and ydj1H34Q mutants (Fig. 3 and data not shown), indicating that the effect depends more on the relationship between Ydj1 and Hsp70 than on the intrinsic chaperone action of Ydj1 itself. The basis for the reduced rate of kinase maturation in the ydj1 mutations, however, remains obscure. One possibility relates to the finding that ydj1 mutants grow slowly and thus have reduced rates of metabolic processes. However, pulse-chase analyses of Tpk2 maturation in other chaperone mutants that grow slowly (cpr7Δ and sse1Δ) suggested that this is not the case (not shown).
The ability of Ydj1 to protect nascent kinases has some specificity since SIS1 overexpression was unable to suppress defects in kinase biogenesis in a ydj1Δ mutant. The lack of suppression occurs despite the finding that SIS1 overexpression can improve growth of ydj1Δ cells (8). Furthermore, the growth of ydj1Δ cells can be improved by just increasing the abundance of the J domain, suggesting that impairment to Hsp70's functional cycle is the main factor that limits growth of ydj1Δ cells (45). Tests of which domains impart the functional specificity were largely negative but demonstrated that the chaperone modules specific to type I and type II Hsp40s were not responsible (Fig. 7). Further support for this view derives from the finding that mammalian Hdj2 (type I) and Hsp40 (type I) are interchangeable for their function in folding of progesterone receptor and Chk1 protein kinase (13, 21). Based on this, the specificity with which we observed Ydj1 function may have more to do with how Sis1 is unable to interact with kinases or nuclear receptors in the presence of Hsp70. Previous studies showed that Ydj1 is capable of refolding luciferase in association with Hsp70 in vitro, whereas Sis1 does not function as efficiently in this capacity (34). The basis for their differential function is due in part to each having a distinct chaperone module, which can specify the function of Sis1 or Ydj1 in chimeric molecules (19). We note, however, that Sis1 also has a more stable interaction with the Hsp70 C terminus than does Ydj1 (33). The C-terminal region of Hsp70 interacts with the peptide-binding groove in the chaperone module of Sis1, suggesting that there could be competitive interactions. This might preclude Sis1 binding to nascent kinases in the presence of Hsp70. We speculate that such binding of Hsp70 to the Sis1 chaperone module is disrupted in the YSY chimera and that this is why it proves so effective compared to Sis1 alone (Fig. 7).
In conclusion, our combined findings suggest a model in which Ydj1 has two specific functions in protein kinase folding. The first involves protecting the newly synthesized kinase chain from degradation, and the second involves promoting efficient folding and maturation. Ydj1 by itself can protect nascent kinases, but not to the same extent that it can in association with Hsp70. The unexpected finding that Ydj1 can affect the rate of kinase maturation presents a new paradigm for its action in cellular quality control.
Acknowledgments
We thank Doug Cyr, Betty Craig, Costa Georgopolis, and Ulrich Hartl for providing plasmids and antisera used in this study. We also thank Maria Theodoraki, Doug Cyr, and Jeff Brodsky for discussions and insight and Paul Lee for initial contributions to the study.
This study was supported by NIH grant GM70596.
Footnotes
Published ahead of print on 28 April 2008.
REFERENCES
- 1.Alberti, S., C. Esser, and J. Hohfeld. 2003. BAG-1: a nucleotide exchange factor of Hsc70 with multiple cellular functions. Cell Stress Chaperones 8225-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arlander, S. J., S. J. Felts, J. M. Wagner, B. Stensgard, D. O. Toft, and L. M. Karnitz. 2006. Chaperoning checkpoint kinase 1 (Chk1), an Hsp90 client, with purified chaperones. J. Biol. Chem. 2812989-2998. [DOI] [PubMed] [Google Scholar]
- 3.Batkin, M., I. Schvartz, and S. Shaltiel. 2000. Snapping of the carboxyl terminal tail of the catalytic subunit of PKA onto its core: characterization of the sites by mutagenesis. Biochemistry 395366-5373. [DOI] [PubMed] [Google Scholar]
- 4.Bercovich, B., I. Stancovski, A. Mayer, N. Blumenfeld, A. Laszlo, A. L. Schwartz, and A. Ciechanover. 1997. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J. Biol. Chem. 2729002-9010. [DOI] [PubMed] [Google Scholar]
- 5.Brodsky, J. L., J. G. Lawrence, and A. J. Caplan. 1998. Mutations in the cytosolic DnaJ homologue, YDJ1, delay and compromise the efficient translation of heterologous proteins in yeast. Biochemistry 3718045-18055. [DOI] [PubMed] [Google Scholar]
- 6.Brown, N. R., M. E. Noble, A. M. Lawrie, M. C. Morris, P. Tunnah, G. Divita, L. N. Johnson, and J. A. Endicott. 1999. Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity. J. Biol. Chem. 2748746-8756. [DOI] [PubMed] [Google Scholar]
- 7.Caplan, A. J., D. M. Cyr, and M. G. Douglas. 1992. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell 711143-1155. [DOI] [PubMed] [Google Scholar]
- 8.Caplan, A. J., and M. G. Douglas. 1991. Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein. J. Cell Biol. 114609-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caplan, A. J., A. K. Mandal, and M. A. Theodoraki. 2007. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 1787-92. [DOI] [PubMed] [Google Scholar]
- 10.Caplan, A. J., J. Tsai, P. J. Casey, and M. G. Douglas. 1992. Farnesylation of YDJ1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J. Biol. Chem. 26718890-18895. [PubMed] [Google Scholar]
- 11.Castro, J. E., C. E. Prada, O. Loria, A. Kamal, L. Chen, F. J. Burrows, and T. J. Kipps. 2005. ZAP-70 is a novel conditional heat shock protein 90 (Hsp90) client: inhibition of Hsp90 leads to ZAP-70 degradation, apoptosis, and impaired signaling in chronic lymphocytic leukemia. Blood 1062506-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheetham, M. E., and A. J. Caplan. 1998. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 328-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cintron, N. S., and D. Toft. 2006. Defining the requirements for Hsp40 and Hsp70 in the Hsp90 chaperone pathway. J. Biol. Chem. 28126235-26244. [DOI] [PubMed] [Google Scholar]
- 14.Connell, P., C. A. Ballinger, J. Jiang, Y. Wu, L. J. Thompson, J. Hohfeld, and C. Patterson. 2001. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 393-96. [DOI] [PubMed] [Google Scholar]
- 15.Craig, E. A., P. Huang, R. Aron, and A. Andrew. 2006. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 1561-21. [DOI] [PubMed] [Google Scholar]
- 16.Dai, Q., C. Zhang, Y. Wu, H. McDonough, R. A. Whaley, V. Godfrey, H. H. Li, N. Madamanchi, W. Xu, L. Neckers, D. Cyr, and C. Patterson. 2003. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 225446-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dey, B., A. J. Caplan, and F. Boschelli. 1996. The Ydj1 molecular chaperone facilitates formation of active p60v-src in yeast. Mol. Biol. Cell 791-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan, C. Y., S. Lee, and D. M. Cyr. 2003. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones 8309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan, C. Y., S. Lee, H. Y. Ren, and D. M. Cyr. 2004. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol. Biol. Cell 15761-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrell, A., and D. O. Morgan. 2000. Cdc37 promotes the stability of protein kinases Cdc28 and Cak1. Mol. Cell. Biol. 20749-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felts, S. J., L. M. Karnitz, and D. O. Toft. 2007. Functioning of the Hsp90 machine in chaperoning checkpoint kinase I (Chk1) and the progesterone receptor (PR). Cell Stress Chaperones 12353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frydman, J. 2001. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70603-647. [DOI] [PubMed] [Google Scholar]
- 23.Gerber, M. R., A. Farrell, R. J. Deshaies, I. Herskowitz, and D. O. Morgan. 1995. Cdc37 is required for association of the protein kinase Cdc28 with G1 and mitotic cyclins. Proc. Natl. Acad. Sci. USA 924651-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425737-741. [DOI] [PubMed] [Google Scholar]
- 25.Harrison, C. 2003. GrpE, a nucleotide exchange factor for DnaK. Cell Stress Chaperones 8218-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartson, S. D., E. A. Ottinger, W. Huang, G. Barany, P. Burn, and R. L. Matts. 1998. Modular folding and evidence for phosphorylation-induced stabilization of an hsp90-dependent kinase. J. Biol. Chem. 2738475-8482. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, D. A., P. Akamine, E. Radzio-Andzelm, M. Madhusudan, and S. S. Taylor. 2001. Dynamics of cAMP-dependent protein kinase. Chem. Rev. 1012243-2270. [DOI] [PubMed] [Google Scholar]
- 28.Kabani, M., J. M. Beckerich, and J. L. Brodsky. 2002. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 224677-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura, Y., I. Yahara, and S. Lindquist. 1995. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science 2681362-1365. [DOI] [PubMed] [Google Scholar]
- 30.Lee, D. H., M. Y. Sherman, and A. L. Goldberg. 1996. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 164773-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, P., J. Rao, A. Fliss, E. Yang, S. Garrett, and A. J. Caplan. 2002. The Cdc37 protein kinase-binding domain is sufficient for protein kinase activity and cell viability. J. Cell Biol. 1591051-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, P., A. Shabbir, C. Cardozo, and A. J. Caplan. 2004. Sti1 and Cdc37 can stabilize Hsp90 in chaperone complexes with a protein kinase. Mol. Biol. Cell. [DOI] [PMC free article] [PubMed]
- 33.Li, J., Y. Wu, X. Qian, and B. Sha. 2006. Crystal structure of yeast Sis1 peptide-binding fragment and Hsp70 Ssa1 C-terminal complex. Biochem. J. 398353-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, Z., and D. M. Cyr. 1998. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J. Biol. Chem. 27327824-27830. [DOI] [PubMed] [Google Scholar]
- 35.Luke, M. M., A. Sutton, and K. T. Arndt. 1991. Characterization of SIS1, a Saccharomyces cerevisiae homologue of bacterial dnaJ proteins. J. Cell Biol. 114623-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandal, A. K., P. Lee, J. A. Chen, N. Nillegoda, A. Heller, S. Distasio, H. Oen, J. Victor, D. M. Nair, J. L. Brodsky, and A. J. Caplan. 2007. Cdc37 has distinct roles in protein kinase quality control that protect nascent chains from degradation and promote posttranslational maturation. J. Cell Biol. 176319-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayr, C., K. Richter, H. Lilie, and J. Buchner. 2000. Cpr6 and Cpr7, two closely related Hsp90-associated immunophilins from Saccharomyces cerevisiae, differ in their functional properties. J. Biol. Chem. 27534140-34146. [DOI] [PubMed] [Google Scholar]
- 38.McClellan, A. J., M. D. Scott, and J. Frydman. 2005. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121739-748. [DOI] [PubMed] [Google Scholar]
- 39.Meacham, G. C., C. Patterson, W. Zhang, J. M. Younger, and D. M. Cyr. 2001. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3100-105. [DOI] [PubMed] [Google Scholar]
- 40.Park, S. H., N. Bolender, F. Eisele, Z. Kostova, J. Takeuchi, P. Coffino, and D. H. Wolf. 2007. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol. Biol. Cell 18153-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearl, L. H., and C. Prodromou. 2006. Structure and mechanism of the hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 75271-294. [DOI] [PubMed] [Google Scholar]
- 42.Prince, T., and R. L. Matts. 2004. Definition of protein kinase sequence motifs that trigger high-affinity binding of Hsp90 and Cdc37. J. Biol. Chem. 27939975-39981. [DOI] [PubMed] [Google Scholar]
- 43.Rao, J., P. Lee, S. Benzeno, C. Cardozo, J. Albertus, D. M. Robins, and A. J. Caplan. 2001. Functional interaction of human Cdc37 with the androgen receptor but not with the glucocorticoid receptor. J. Biol. Chem. 2765814-5820. [DOI] [PubMed] [Google Scholar]
- 44.Rosser, M. F., E. Washburn, P. J. Muchowski, C. Patterson, and D. M. Cyr. 2007. Chaperone functions of the E3 ubiquitin ligase CHIP. J. Biol. Chem. 28222267-22277. [DOI] [PubMed] [Google Scholar]
- 45.Sahi, C., and E. A. Craig. 2007. Network of general and specialty J. protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. USA 1047163-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaner, L., and K. A. Morano. 2007. All in the family: atypical Hsp70 chaperones are conserved modulators of Hsp70 activity. Cell Stress Chaperones 121-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shoji, S., K. Titani, J. G. Demaille, and E. H. Fischer. 1979. Sequence of two phosphorylated sites in the catalytic subunit of bovine cardiac muscle adenosine 3′:5′-monophosphate-dependent protein kinase. J. Biol. Chem. 2546211-6214. [PubMed] [Google Scholar]
- 48.Theodoraki, M. A., M. Kunjappu, D. W. Sternberg, and A. J. Caplan. 2007. Akt shows variable sensitivity to an Hsp90 inhibitor depending on cell context. Exp. Cell Res. 3133851-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai, J., and M. G. Douglas. 1996. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J. Biol. Chem. 2719347-9354. [DOI] [PubMed] [Google Scholar]
- 50.Wegrzyn, R. D., and E. Deuerling. 2005. Molecular guardians for newborn proteins: ribosome-associated chaperones and their role in protein folding. Cell Mol. Life Sci. 622727-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westhoff, B., J. P. Chapple, J. van der Spuy, J. Hohfeld, and M. E. Cheetham. 2005. HSJ1 is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. Curr. Biol. 151058-1064. [DOI] [PubMed] [Google Scholar]
- 52.Whitesell, L., and S. L. Lindquist. 2005. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5761-772. [DOI] [PubMed] [Google Scholar]
- 53.Xu, Y., and S. Lindquist. 1993. Heat-shock protein hsp90 governs the activity of pp60v-src kinase. Proc. Natl. Acad. Sci. USA 907074-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yaglom, J. A., A. L. Goldberg, D. Finley, and M. Y. Sherman. 1996. The molecular chaperone Ydj1 is required for the p34CDC28-dependent phosphorylation of the cyclin Cln3 that signals its degradation. Mol. Cell. Biol. 163679-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yam, A. Y., V. Albanese, H. T. Lin, and J. Frydman. 2005. Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J. Biol. Chem. 28041252-41261. [DOI] [PubMed] [Google Scholar]
- 56.Yonemoto, W., M. L. McGlone, B. Grant, and S. S. Taylor. 1997. Autophosphorylation of the catalytic subunit of cAMP-dependent protein kinase in Escherichia coli. Protein Eng. 10915-925. [DOI] [PubMed] [Google Scholar]
- 57.Younger, J. M., H. Y. Ren, L. Chen, C. Y. Fan, A. Fields, C. Patterson, and D. M. Cyr. 2004. A foldable CFTR{Delta}F508 biogenic intermediate accumulates upon inhibition of the Hsc70-CHIP E3 ubiquitin ligase. J. Cell Biol. 1671075-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yun, B. G., and R. L. Matts. 2005. Differential effects of Hsp90 inhibition on protein kinases regulating signal transduction pathways required for myoblast differentiation. Exp. Cell Res. 307212-223. [DOI] [PubMed] [Google Scholar]
- 59.Zhao, Q., F. Boschelli, A. J. Caplan, and K. T. Arndt. 2004. Identification of a conserved sequence motif that promotes Cdc37 and cyclin D1 binding to Cdk4. J. Biol. Chem. 27912560-12564. [DOI] [PubMed] [Google Scholar]