Abstract
Acetylation of Saccharomyces cerevisiae histone H3 on K56 by the histone acetyltransferase (HAT) Rtt109 is important for repairing replication-associated lesions. Rtt109 purifies from yeast in complex with the histone chaperone Vps75, which stabilizes the HAT in vivo. A whole-genome screen to identify genes whose deletions have synthetic genetic interactions with rtt109Δ suggests Rtt109 has functions in addition to DNA repair. We show that in addition to its known H3-K56 acetylation activity, Rtt109 is also an H3-K9 HAT, and we show that Rtt109 and Gcn5 are the only H3-K9 HATs in vivo. Rtt109's H3-K9 acetylation activity in vitro is enhanced strongly by Vps75. Another histone chaperone, Asf1, and Vps75 are both required for acetylation of lysine 9 on H3 (H3-K9ac) in vivo by Rtt109, whereas H3-K56ac in vivo requires only Asf1. Asf1 also physically interacts with the nuclear Hat1/Hat2/Hif1 complex that acetylates H4-K5 and H4-K12. We suggest Asf1 is capable of assembling into chromatin H3-H4 dimers diacetylated on both H4-K5/12 and H3-K9/56.
Changes in chromatin structure greatly influence all aspects of DNA metabolism, including DNA replication, repair, and transcription. Remodeling of the chromatin structure at the nucleosomal level occurs by a variety of methods. One such remodeling is the direct covalent modification of histones, examples of which include acetylation, methylation, ubiquitination, and phosphorylation (6). Histone acetylation is catalyzed by histone acetyltransferases (HATs), proteins that mediate the transfer of an acetyl moiety from acetyl (acetyl-coenzyme A CoA) to the ɛ-amino group of target lysine residues. HATs that acetylate nucleosomes are usually components of large multiprotein complexes, including NuA4, SAS-C, and SAGA. The enzymatic removal of these acetyl groups from lysines is performed by histone deacetylases (HDACs).
Histones can also be posttranslationally modified prior to their deposition into chromatin. On histone H4, lysines 5 and 12 (H4-K5 and H4-K12) on the N-terminal tail are acetylated in S phase prior to deposition during DNA replication in most if not all eukaryotes (8, 46). The deposition-associated H4-K5/12 HAT is the evolutionarily conserved Hat1 protein, part of a protein complex that in Saccharomyces cerevisiae may include Hat2 and Hif1 (3, 32, 38). Unlike the strong evolutionary conservation of deposition-associated acetylation sites in H4, there is some variability in H3 sites among organisms. In Drosophila and Tetrahymena, H3-K14/23 and H3-K9/14, respectively, are associated with deposition-related acetylation (46), while in S. cerevisiae, H3-K9 is the major acetylated lysine in the H3 N-terminal tail associated with newly synthesized H3 (26). The identity of the S. cerevisiae HAT responsible for H3-K9 acetylation (H3-K9ac) on newly synthesized H3 is unclear, although Gcn5 has been suggested as a candidate (1, 45). Nucleosomal H3-K9ac is catalyzed in vivo by Gcn5 acting, in this case, as the catalytic subunit of the transcriptional coactivator complex SAGA. H3-K9ac levels peak at the 5′ ends of genes and likely function to promote transcription (16, 28) by helping TFIID bind to chromatin (52).
Recent mass spectrometry approaches reveal that certain lysine residues are also acetylated in the globular domains of newly synthesized histones (11). The acetylation of lysine 56 on H3 (H3-K56ac) is one such abundant modification in S. cerevisiae. It is associated with newly synthesized H3 that is subsequently incorporated into chromatin and persists until G2/M, when it is deacetylated by the Hst3/Hst4 HDACs (7, 27, 36, 54). One identified function of H3-K56ac is to promote the efficient repair of replication-associated lesions through S phase (27), perhaps by altering nucleosome mobility (11).
During DNA replication, newly synthesized, acetylated histones are associated with the histone H3/H4 histone chaperones, CAF-I and Asf1 (12). CAF-I assembles H3-H4 dimers onto replicating DNA through a physical interaction with PCNA during DNA replication (44), whereas Asf1 is required for S-phase progression in the presence of DNA damage and physically interacts with the PCNA-loading complex RFC (43). CAF-I has been shown to bind H3-K56ac (27), while Asf1 is absolutely required for acetylation of the residue in vivo (36). The Drosophila RCAF (replication-coupling assembly factor) protein complex was identified based on its ability to facilitate the assembly of nucleosomes onto newly replicated DNA in vitro. RCAF consists of Drosophila Asf1 bound to H3-H4 dimers that are specifically acetylated at H3-K14 (functionally equivalent to S. cerevisiae H3-K9ac with respect to chromatin assembly) and H4-K5/12 (50). In addition to being required for the generation of H3-K56ac, Asf1 is necessary to maintain wild-type levels of H3-K9ac in S. cerevisiae (1, 2), presumably on newly synthesized H3. Although it has been suggested that Gcn5 might be responsible for Asf1-associated H3-K9ac (1), conclusive evidence is lacking.
We and others have recently shown that S. cerevisiae Rtt109 is a HAT with strong substrate and site specificity for H3-K56, an activity that in vitro is enhanced in the presence of Asf1 (10, 13, 18, 39). We have also previously shown that Rtt109 copurifies with another protein, Vps75 (25), an H3-H4 histone chaperone of the Nap1 family (41). The function of this physical interaction and the potential role(s) of Vps75 in histone acetylation and histone deposition are currently unknown. In this study we show that one function of Vps75 is to stabilize Rtt109. A genome-wide screen for genes that genetically interact with rtt109Δ reveals that it has important functions in maintaining genome stability but functions in other cellular processes. We demonstrate that Vps75 can stimulate Rtt109 to acetylate H3-K9 in vitro and that Rtt109 and Gcn5 are the only H3-K9 HATs in vivo in S. cerevisiae. In addition, we show that Rtt109, Vps75, and Asf1 function together in vivo in a Gcn5-independent H3-K9 acetylation pathway. Finally we demonstrate that Asf1 physically interacts with the deposition-associated nuclear Hat1 protein complex. We suggest that Asf1 assembles into chromatin via CAF-1 or other histone chaperones a form of the H3-H4 dimer which is potentially acetylated on H4-K5/12 and H3-K9/56.
MATERIALS AND METHODS
Strains used in this study.
See Table S1 in the supplemental material for strains used in this study. Construction of histone point mutants in the MSY421 background was as described previously (36).
Rad53 in situ kinase assays.
Rad53 kinase assays were performed as described previously (31).
Protein expression.
Full-length Rtt109, Asf1, or Asf1N was cloned into pET14b or pET28a. A plasmid encoding GST-Vps75 (41) was kindly provided by Luke Selth (Cancer Research United Kingdom). Recombinant proteins were expressed and purified using standard methods and subsequently dialyzed into tandem affinity purification (TAP) dialysis buffer (25).
Cell cycle synchronization.
Strains used for α-factor synchronization were deleted for BAR1 and blocked in G1 as described previously (24).
HAT assays.
HAT assays were performed using either chicken core histones (Upstate) or Xenopus recombinant histone H3 (Upstate) as a substrate. HAT assays were incubated for 45 min (except for those in Fig. 2B and C, which were incubated for 30 min) at 30° in a 30-μl volume containing either 3 μg (core histones) or 0.75 μg (rH3) total substrate, 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 3.3 nCi [14C]acetyl-CoA (60 mCi/mmol), and 1 mM phenylmethylsulfonyl fluoride. Five-microliter aliquots of tandem affinity-purified protein complexes from yeast (25) or recombinant proteins (adjusted to 0.5 μg/μl) were added to each reaction. For enzyme titration experiments, the respective recombinant proteins were diluted in TAP elution buffer either 1:1, 1:2, 1:5, or 1:10 and used as described above. Reactions were stopped by addition of 30 μl 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading dye and boiled. Aliquots (20% of total volume) were run on several 15% SDS-PAGE gels. One gel was fixed and stained with Coomassie blue, saturated with Enlightning (Perkin Elmer), dried under vacuum, and either exposed to film or imaged using a Typhoon phosphorimager. Other gels were treated with a silver stain or transferred to nitrocellulose for Western analysis, as required.
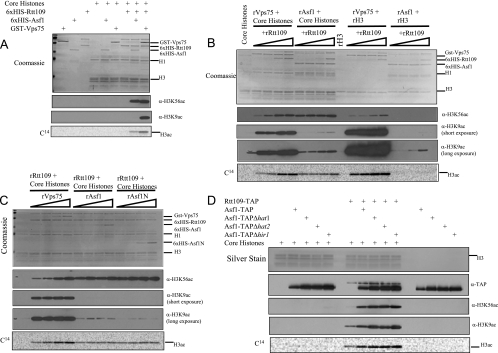
FIG. 2.
Rtt109 acetylates both H3-K56 and H3-K9 in vitro. (A) Rtt109 is an H3-K9 HAT in vitro. HAT assays contained the indicated recombinant factors in the presence of [14C]acetyl-coA using chicken core histones as a substrate. Reactions were separated by 15% SDS-PAGE and either Coomassie or silver stained, treated for fluorography, and then imaged or were immunoblotted with the indicated antibodies (α-H3K56ac and -H3K9ac [anti-H3-K56ac and -H3-K9ac antibodies, respectively]). (B) HAT assays performed as described for panel A, with the exception that core histones (chicken) or recombinant H3 (Xenopus) was used as a substrate and that several dilutions of rRtt109 were used, as indicated. (C) HAT assays performed as described for panel A, with the exception that several dilutions of the respective histone chaperones were used, as indicated. (D) Asf1 stimulates the HAT activity of Rtt109-Vps75 toward both H3-K9 and H3-K56. Factors were affinity purified from the indicated yeast strains, and HAT assays were performed as for panel A. α-TAP, anti-TAP antibody.
SGA.
Synthetic genetic arrays (SGA) were performed as described previously (40) using KLY21 (see Table S1 in the supplemental material) as a query strain against the entire nonessential deletion collection. SGA analysis was performed at 30°C and 37°C. All genetic interactions observed at 30°C were also observed at 37°C.
Generation of WCE and Western blotting.
Whole-cell extracts (WCE) were generated using trichloroacetic acid as described previously (22). WCE were separated on 15% SDS-PAGE, transferred to nitrocellulose, and blotted with the indicated antibodies. Antibodies and dilutions used were anti-H3 (1:2,000; Abcam), anti-H3-K9ac (1:10,000; Abcam), anti-H3-K56ac (1:5,000; Upstate), anti-γH2A (1:3,000; Abcam), anti-H3-K18ac (1:15,000; Lake Placid Biologicals), anti-H3-K27ac (1:5,000; Lake Placid Biologicals), anti-H3-K14ac (1:5,000; Lake Placid Biologicals), anti-H3-K23ac (1:5,000; Lake Placid Biologicals), anti-H4-K5ac (1:10,000; Upstate), anti-H4-K8ac (1:20,000; Upstate), anti-H4-K12 (1:5,000; Abcam), anti-CBP (1:3,000; Open Biosystems), anti-Clb2 (1:5,000; Santa Cruz), and anti-Med2 (1:3,000; Santa Cruz).
Protein stability assay.
The analysis of Rtt109 stability was performed as described previously (5). Briefly, exponentially growing TAP-tagged strains were grown to an optical density (OD) of ∼0.5, and cycloheximide (Sigma) was added to 35 μg/ml. Twenty-five-milliliter samples were taken at the indicated time points after addition of the drug, and whole-cell extracts were generated by the trichloroacetic acid method described above. Six-hundred-nanometer-OD samples were read for each time point to confirm growth arrest characteristic of cycloheximide treatment.
RESULTS
Roles of Rtt109 chaperones in S-phase DNA repair and Rtt109 stability.
In the course of our studies on the H3-K56 HAT Rtt109, we determined that rtt109Δ cycling cells exhibit elevated levels of DNA damage-associated γH2A (Fig. 1A), a phosphorylated form of histone H2A associated with DNA double-strand breaks (DSBs) (15). This phenotype is also seen when PPH3, the gene encoding the γH2A phosphatase, is deleted (23) (see Fig. S1 in the supplemental material). Supporting the idea that H3-K56ac is important for the repair of S-phase-specific DNA damage (27), deletion of either ASF1 or RTT109 in cycling cells ablates H3-K56ac (Fig. 1A) (10, 13, 19, 36, 39, 49) and leads to elevated levels of γH2A (Fig. 1A) (demonstrated previously for asf1Δ [33]) and Rad53 phosphorylation (Fig. 1B) (13, 35), the latter a proxy for checkpoint activation. In addition, the H3-K56R, rtt109Δ, and asf1Δ strains are highly sensitive to DSB-causing chemicals, such as methyl methanesulfonate (MMS) (Fig. 1C) (13, 18, 39). Western blot analysis of WCE taken during a synchronous cell cycle after release from a G1 block shows that γH2A levels in wild-type and rtt109Δ cells are very low in G1 and rise as cells progress through S phase (see Fig. S2A in the supplemental material). In a wild-type strain, where replication-induced DSBs are quickly repaired, γH2A levels decrease before rising again in the next S phase. However, in rtt109Δ cells, γH2A levels increase to a point within S and then remain relatively stable well into G2/M, as assessed by the presence of the G2/M-specific cyclin Clb2. It is likely that the lack of acetylated H3-K56 in rtt109Δ cells causes a cell cycle delay as a result of difficulty repairing S-phase-specific DNA damage. The fact that we observe a similar pattern of γH2A and Clb2 accumulation when a cell cycle is similarly analyzed for a K56R strain (see Fig. S2B in the supplemental material) supports this idea.
FIG. 1.
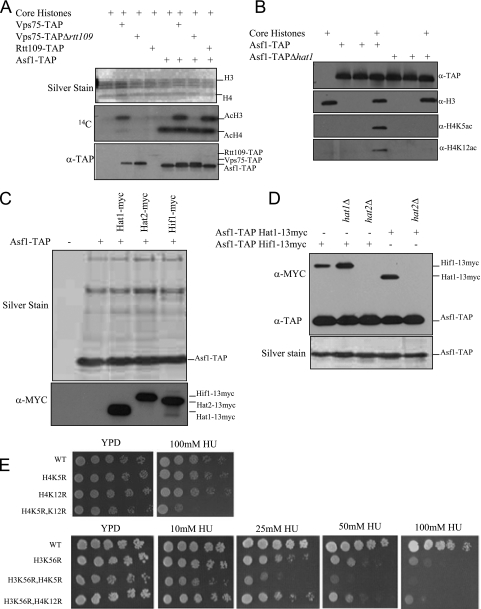
Involvement of Rtt109 chaperones Asf1 and Vps75 in DNA damage response and Rtt109 stability. (A) Lack of H3-K56ac is correlated with elevated γH2A. WCE from the indicated strains were analyzed by immunoblotting after 15% SDS-PAGE with the antibodies shown to the right of each panel (α-γH2A, -H3-K56ac, and -H3, anti-γH2A, -H3-K56ac, and -H3 antibodies, respectively). (B) Activation of the Rad53 checkpoint kinase (*), as monitored by in situ autophosphorylation, observed in rtt109Δ and asf1Δ cells but not in vps75Δ cells. By comparison, the level of Rad53 activation in wild-type (WT) cells treated with MMS (0.1%, 2 h) is shown. The upper autophosphorylating species (†) is independent of DNA damage and serves as a loading control. (C) Inability to acetylate H3-K56 correlates with genotoxin sensitivity. Indicated strains were grown to an OD at 600 nm (OD600) of ≅0.5 before being plated at fivefold serial dilutions on YPD medium with or without MMS. All strains are isogenic with MSY421 (36). (D) Members of the Rtt109 epistasis group (10) all show checkpoint activation in cycling cells, but only the rtt109Δ strain shows elevated γH2A. (E) The Vps75 histone chaperone stabilizes Rtt109. TAP was performed on WCE from the indicated strains and purified complexes visualized by silver staining. Immunoblotting of the WCE used for TAP shows that Rtt109 is expressed and soluble in the vps75Δ background (see panel F). (F) Rtt109-TAP is unstable in the absence of Vps75. WCE were collected at the indicated times after addition of the translation inhibitor cycloheximide (5). Growth curve analysis (OD600) indicates that each strain arrested with similar kinetics (not shown). WCE were analyzed by immunoblotting with the antibodies to the right of the panel.
We have previously shown that in addition to Asf1, Rtt109 is also functionally linked to Rtt101, Mms1, and Mms22 (10). Although the rtt101Δ, mms1Δ, and mms22Δ strains share with the rtt109Δ strain the property of persistent checkpoint activation (Fig. 1D) (37), we observe elevated γH2A only in the rtt109Δ strain (Fig. 1D). As mentioned above, Rtt109 copurifies with the histone chaperone Vps75 (25). Deletion of VPS75, however, does not result in elevation of γH2A (Fig. 1A), checkpoint activation (Fig. 1B), the abolition of H3-K56ac (Fig. 1A) (41, 49), or sensitivity to DSB-inducing chemicals (Fig. 1C) (41, 49).
In the course of analyzing the composition of protein complexes in yeast, we noticed that Rtt109 is difficult to affinity purify in the absence of Vps75 (Fig. 1E). In the reciprocal case, Vps75 can easily be purified in the absence of Rtt109. To test whether Vps75 affects the stability of Rtt109, we treated asynchronous Rtt109-TAP strains that contained either wild-type VPS75 or its deletion with cycloheximide to arrest protein synthesis. We then made WCE for each strain at various time points and showed that in the absence of Vps75, Rtt109 is unstable relative to Rtt109 in the wild-type strain (Fig. 1F), a property not observed for two other proteins. Thus, Rtt109 appears to be degraded quickly in the absence of Vps75. Therefore, one function of Vps75 is to stabilize Rtt109.
Whole-genome SGA reveals that RTT109 has functions in addition to maintaining genome stability.
To further evaluate the function of Rtt109, we used SGA analysis (48), a method for systematic construction of double mutants. Inviable or slow-growing double-mutant meiotic progeny identify functional relationships between genes. When the rtt109Δ strain was crossed to an array of ∼4,700 nonessential deletion mutants and double-mutant haploid strains grown at 30°C, there were 113 double-deletion combinations that resulted in a synthetic sick/synthetic lethal (SS/SL) phenotype (see Table S2A in the supplemental material). We observed an additional 45 SS/SL interactions when the double-mutant haploid cells were placed under stress by growth at 37°C (see Table S2B in the supplemental material).
When the genes identified by their SS/SL genetic interactions with rtt109Δ were clustered by their Gene Ontology (GO) terms, we identified three main enriched functional categories (Table 1). The top category, chromosome organization and biogenesis, is consistent with a role for H3-K56ac and Rtt109 in chromatin assembly. In addition, the telomere organization and biogenesis functional grouping is consistent with the demonstrated role of H3-K56ac in telomeric silencing (55). Although not detected by GO term analysis, we observed a number of SS/SL interactions with genes encoding proteins involved in maintaining genome stability (see Fig. S3 in the supplemental material), which is consistent with Rtt109's demonstrated role in this process. SS/SL genetic interactions were also obtained with a number of other genes unrelated to genome stability (see Fig. S3 in the supplemental material). GO-term analysis also links Rtt109 functionally to the ribosome and translation (Table 1). The SGA data suggest that Rtt109 has other functions in the cell and therefore potentially could have non-H3-K56 targets for its HAT activity.
TABLE 1.
Numbers of genes from nonessential S. cerevisiae collection associated with SS/SL phenotype in combination with rtt109Δ::NAT querya
| Temp (°C) | No. of genes
|
GO term; category, P value | |
|---|---|---|---|
| With SS/SL interaction | With SS/SL BioGrid/Krogan annotation | ||
| 30 | 113 | 34 | Chromosome organization and biogenesis; GO:0007001, 7.23e−5 |
| Telomere organization and biogenesis; GO:0032200, 9.93e−5 | |||
| Small ribosomal subunit; GO:0015935, 9.93e−5 | |||
| 37 | 62 | ||
| 30 + 37 | 159 | Translation; GO:0006412, 4.24e−4 | |
SS/SL phenotype was scored at 30 or 37°C. Biogrid/Krogan refers to the number of genes previously annotated as SS/SL with rtt109Δ by us and other workers. A selection of the top GO terms statistically overrepresented in these SS/SL groups was scored by GOstat (http://gostat.wehi.edu.au).
Rtt109 with Vps75 is an H3-K9 HAT in vitro.
We looked for potential non-H3-K56 substrates for Rtt109 in vitro using HAT assays with Rtt109 in combination with various histone chaperones. Using this assay, we have previously shown that recombinant Rtt109 has H3-K56-specific HAT activity on recombinant H3 and that the H3-K56ac activity of Rtt109 purified from yeast (i.e., Rtt109-TAP-Vps75) is stimulated by Asf1 when core histones are used as a substrate (10). Several other groups have shown similar effects of Asf1 and Vps75 on the H3-K56ac activity of Rtt109 using recombinant proteins (13, 19, 49). To separate Rtt109 from Vps75, we expressed it as a six-His-tagged recombinant protein (rRtt109) and performed HAT assays either alone or in combination with recombinant full-length six-His-Asf1 (rAsf1) or recombinant glutathione S-transferase (GST)-Vps75. In addition, we tested rRtt109 in combination with the H3-H4 histone chaperone Rtt106 (20) purified from yeast (Rtt106-TAP), Vps75 purified from yeast in the absence of RTT109 (Vps75-TAPΔrtt109), and recombinant six-His-Hat2 (rHat2, a protein that has similarity to histone chaperones and that binds yeast Hat1 [53]).
Under the conditions of our HAT assay, we did not observe substantial HAT activity on core histones by rRtt109 itself (Fig. 2A) or when rRtt109 was incubated with either Rtt106-TAP or rHat2 (see Fig. S4A in the supplemental material). However, when rRtt109 was incubated with rAsf1, we observed strong H3 acetylation by autoradiography, and as expected, we confirmed using Western blotting that some of the acetylation is H3-K56 specific (see Fig. 4A; also see Fig. S4A in the supplemental material). We similarly observed H3-K56-specific acetylation when rRtt109 was incubated in combination with GST-Vps75 or Vps75-TAP (rtt109Δ) (Fig. 2A) (see Fig. S4A in the supplemental material), in agreement with our previous results and those of others.
FIG. 4.
Chaperone involvement in H3-K9 acetylation by the Gcn5 and Rtt109 HATs in vivo. (A) Deletion of either GCN5 or RTT109 reduces cellular levels of H3-K9ac. The gcn5Δ (JF81) and rtt109Δ (SC218) strains are isogenic with the wild-type (WT) strain BY4741. The second set of rtt109Δ (AT398) and sas3Δ (AT411) strains are isogenic with WT-Y3656. α-H3, -H3-K18ac, -H3-K9ac, and -H3-K56ac, anti-H3, -H3-K18ac, -H3-K9ac, and -H3-K56ac antibodies, respectively. (B) H3-K9ac is absent in the gcn5Δ rtt109Δ double mutant. (C) Double deletion of the GCN5 and RTT109 genes leads to synthetic sickness. Growth analysis of strains in the BY4741 background was performed as described for Fig. 1C. α-H4-K12ac, anti-H4-K12ac antibody. (D) Absence of H3-K9ac in gcn5Δ asf1Δ but not rtt109Δ asf1Δ double mutants. (E) Absence of H3-K9ac in gcn5Δ vps75Δ but not asf1Δ vps75Δ double mutants.
To determine if Rtt109 could acetylate other lysines on H3 in the presence of core histones, we carried out Western blotting of the HAT assays with several commercially available acetyl-H3 antibodies and observed that rRtt109 in combination with GST-Vps75 or Vps75-TAP (rtt109Δ) but not with rAsf1 or other histone chaperones has substantial activity on H3-K9 (Fig. 2A; also see Fig. S4A in the supplemental material). We did not observe a significant signal by rRtt109 (either alone or in combination with a histone chaperone) with our other H3 acetyl antibodies, although we did detect very weak activity on H3-K18—a modification that in our hands is strongly catalyzed on core histones by rGcn5 (see Fig. S4B in the supplemental material).
To better characterize the H3-K9ac activity of Rtt109, we varied the concentration of rRtt109 in combination with either rAsf1 or Vps75 using both core histones or recombinant H3 as a substrate (Fig. 2B). We observed that Rtt109-Vps75 strongly acetylated H3-K56 either on H3 alone or on H3 within core histones (Fig. 2B), in agreement with previous results. In addition, we observed Rtt109-Vps75 to have strong activity on H3-K9 on H3 alone or on H3 within core histones (Fig. 2B). Also in agreement with previous results, Rtt109-Asf1 was able to strongly acetylate H3-K56 within core histones but not on H3 alone (Fig. 2B). We observed only very weak activity on H3-K9 by Rtt109-Asf1 when H3 was alone within core histones (Fig. 2B). We also performed HAT assays with core histone substrates, varying the concentration of chaperone (rVps75, rAsf1, or rAsf1N, which contains the first 155 amino acids of Asf1, a region sufficient for normal H3-K56ac levels in vivo [36]). As expected from our previous data, Rtt109-Vps75 displayed strong activity on H3-K56 and H3-K9, while Rtt109-Asf1 showed strong activity on H3-K56 only (Fig. 2C). Similarly, Rtt109-Asf1N showed strong activity on H3-K56 but not H3-K9 (Fig. 2C).
Keeping in mind the demonstrated importance of Asf1 in maintaining cellular H3-K9ac levels (1, 2), we were interested to see whether the presence of Vps75 could alter the ability of Asf1 to stimulate the H3-K9ac activity of Rtt109 on H3. We therefore performed HAT assays using purified Rtt109 protein complexes (Rtt109-Vps75) from yeast in combination with Asf1 purified from either a wild-type strain or those carrying deletions of HAT1, HAT2 (either deletion prevents the copurification of an H4-specific HAT activity from yeast; see below), or HIR1 (which prevents physical association of the HIR histone chaperone complex with Asf1 [17]). Under our in vitro conditions, all Asf1-containing protein complexes tested enhanced the H3-K9- and H3-K56-specific HAT activity of the Rtt109-TAP-Vps75 complex on core histones (Fig. 2D).
Rtt109 is required in vivo for full acetylation of H3-K9.
We used Western blotting on WCE made from deletion mutants to determine whether Rtt109 is an H3-K9 HAT in vivo. It has been shown previously that H3-K9 levels are reduced in asf1Δ or gcn5Δ mutant strains (1, 2). We first determined that rtt109Δ cells have reduced levels of H3-K9ac, similar to those of asf1Δ cells (Fig. 3A) and that vps75Δ cells also show reduced H3-K9ac levels (Fig. 3A). The role of Asf1 in H3-K9 acetylation is likely related to its role as a histone chaperone, since it is not required for proper expression of Rtt109 or Vps75 (Fig. 3B). In addition, Vps75 is not required for expression of Asf1, suggesting that its effect on H3-K9ac is also likely to be direct (Fig. 3B).
FIG. 3.
Rtt109 acetylates H3-K9 in vivo. In panels A through E, immunoblotting on WCE from the indicated strains was performed with the indicated antibodies after 15% SDS-PAGE. (A) Individual deletion of RTT109, VPS75, or ASF1 results in reduced levels of H3-K9ac. WT, wild type; α-H3-K9ac, -H3-K18ac, and -H3-K4me2, anti-H3-K9ac, -H3-K18ac, and -H3-K4me2 antibodies. (B) VPS75 and ASF1 do not control each other's expression. α-TAP, -H3-K56ac, and -H3, anti-TAP, -H3-K56ac, and -H3 antibodies, respectively. (C) Deletion of RTT109 reduces the peak of S-phase-specific accumulation of H3-K9ac. Strains were synchronized in G1/S with α-factor before release into the cell cycle. Passage through a single cycle takes ∼90 min under optimal conditions. Synchronized progress was monitored by expression of the G2/M cyclin Clb2. Immunostaining of H3 was used as a loading control. α-CLB2, anti-CLB2 antibody. (D) The Hst3/Hst4 HDACs deacetylate H3-K56ac but not H3-K9ac. The hst3Δ hst4Δ double mutant is isogenic with the WT (7). (E) H3-K9ac is independent of H3-K56ac. H3-K9ac levels are indistinguishable from WT levels in an unacetylatable H3-K56R mutant, as are H3-K56ac levels in an H3-K9R mutant. Strains are isogenic with MSY421 (36). (F) Inability to acetylate H3-K9 has no effect on DNA repair/genome stability. Genotoxin sensitivity assays are performed as described for Fig. 1C. Strains are isogenic with MSY421 (36).
It was previously demonstrated that expression of ASF1 is required for S-phase accumulation of H3-K9ac (1). To determine if Rtt109 is also required, at least in part, for the S-phase-specific accumulation of H3-K9ac, we analyzed protein extracts generated from synchronized wild-type and rtt109Δ cells by Western blotting. In a wild-type cell cycle, H3-K9ac is low in G1, accumulates through S, and then decreases again in G2/M (Fig. 3C). In an rtt109Δ cell cycle, H3-K9ac also starts very low in G1 and then accumulates through S phase but to a lesser extent than in the wild-type cell cycle. At the time scale used in our analysis, we do not see the subsequent decrease in H3-K9ac levels observed in G2/M as a consequence of the delay in G2/M suffered by rtt109Δ cells (also seen in Fig. S2A in the supplemental material), consistent with a previous analysis that observed a significant G2/M delay in rtt109Δ cells by fluorescence-activated cell sorting (13). Our results indicate that rtt109Δ eliminates most but not all of the S-phase accumulation of H3-K9ac. Deacetylation of H3-K56 occurs in G2/M because of the HDAC pair Hst3/Hst4 (7). We determined that while levels of H3-K56ac are considerably elevated, H3-K9ac levels are not changed in an hst3Δ/hst4Δ background, indicating that the H3-K56ac HDACs do not deacetylate H3-K9 (Fig. 3D).
To further understand the function of H3-K9 acetylation in S. cerevisiae, we generated an H3-K9R strain. Western blotting of WCE made from this strain demonstrates that, as expected, no H3-K9ac signal is observed for H3-K9R (Fig. 3E), showing the specificity of the antibody. In addition, H3-K9ac levels are reduced in the rtt109Δ strain but not in the H3-K56R strain, indicating that Rtt109-catalyzed H3-K9ac does not depend on H3-K56ac (and also confirming the specificity of the antibody). The reciprocal is also true, since H3-K56ac levels are wild type in the H3-K9R mutant (Fig. 3E). To determine if Rtt109's H3-K9ac activity is important for DNA repair, we assessed the ability of H3-K9R point mutants to grow in the presence of DNA-damaging chemicals (Fig. 3F). The H3-K9R mutant grows as well as the wild type in the presence of DNA-damaging agents (as has been previously shown [34]) and does not enhance the drug hypersensitivity of H3-K56R when the two lysines are mutated (H3-K9R/K56R, Fig. 3F), suggesting that H3-K9ac does not have a role in DNA damage repair, further supporting our SGA results for a genome stability-independent role for the HAT.
Rtt109 functions with Vps75 and Asf1 to generate H3-K9ac.
It has been reported previously that Gcn5 is partially responsible for H3-K9ac in vivo (1). We verified this result by Western blot analysis (Fig. 4A). In addition, we found that gcn5Δ lacks a different acetylation, H3-K18ac (Fig. 4A), a result consistent with previously published in vivo data (29) and our in vitro result using rGcn5, which shows HAT activity on this residue when core histones are used as a substrate (see Fig. S4B in the supplemental material).
The fact that rtt109Δ and gcn5Δ cells both have reduced H3-K9ac levels raises the possibility that Gcn5 and Rtt109 HATs may together control all H3-K9ac in budding yeast. We therefore made a gcn5Δ rtt109Δ double mutant and analyzed both its growth (Fig. 4B) and the levels of H3-K9ac by Western blotting (Fig. 4C). Under our conditions, deletion of GCN5 does not cause sensitivity to DNA-damaging agents as observed with the rtt109Δ strain (Fig. 4B). Deletion of GCN5 has been previously shown to cause sensitivity to DNA-damaging chemicals (9) but at higher concentrations than used in the present study. In contrast, gcn5Δ rtt109Δ double mutants show growth defects both on rich medium (yeast extract-peptone-dextrose [YPD]) and on YPD supplemented with chemicals that cause DNA damage (Fig. 4B). These results indicate that Rtt109 and Gcn5 might function in parallel pathways for DNA damage repair. Since the H3-K9R/K56R mutant phenotype is not synthetic for DNA damage sensitivity, as is gcn5Δ rtt109Δ, the Gcn5 contribution likely reflects the diverse targets of this particular HAT besides H3-K9, including other H3 or H4 targets (45) or even nonhistone substrates (51). By Western blotting, the rtt109Δ and gcn5Δ strains separately showed reduced levels of H3-K9ac (Fig. 4C), with the gcn5Δ strain lacking H3-K18ac and the rtt109Δ strain showing no H3-K56ac, both as expected (Fig. 4C). In contrast, the gcn5Δ rtt109Δ double mutant showed a complete loss of H3-K9ac, indicating that Rtt109 and Gcn5 are the only H3-K9 HATs in vivo in S. cerevisiae.
To determine which of these two HATs functions in conjunction with Asf1, since it is likely that the HAT responsible for most deposition-related H3-K9ac functions with this histone chaperone, we generated gcn5Δ asf1Δ and rtt109Δ asf1Δ double mutants and analyzed their cellular levels of H3-K9ac. The rtt109Δ asf1Δ mutant had low levels of H3-K9ac, similar to results for each respective single mutant (Fig. 4D). By contrast, the gcn5Δ asf1Δ double mutant, like the gcn5Δ rtt109Δ double mutant, lacked H3-K9ac. Thus, our in vivo results strongly suggest that Rtt109 and Asf1 function together in a Gcn5-independent pathway to generate H3-K9ac.
To determine if Rtt109's protein binding partner, Vps75, functions in the H3-K9ac pathway along with Asf1 and Rtt109, we also generated gcn5Δ vps75Δ double mutants and analyzed them by Western blotting. We found that, like the gcn5Δ rtt109Δ and gcn5Δ asf1Δ mutants, the gcn5Δvps75Δ double mutants are also null for H3-K9ac (Fig. 4E). These results imply that two histone chaperones, Asf1 and Vps75, are both required in vivo for the generation of Rtt109-catalyzed H3-K9ac.
Asf1 interacts with the Hat1 complex that acetylates H4-K5/12.
The complex relationship of Rtt109 with Asf1 and Vps75 raised the interesting possibility that other HATs or histone-modifying enzymes could associate with those histone chaperones. In our previous analysis of Rtt109, we used purified complexes from yeast to show that Asf1 stimulates the H3-K56ac activity of Rtt109 (10) (Fig. 5A). We also noticed that Asf1 copurified from yeast with a robust H4-specific HAT activity (Fig. 5A). We then analyzed Asf1 copurifying proteins by using SDS-PAGE, silver stain analysis, and matrix-assisted laser desorption ionization-time-of-flight mass spectrometry and identified Hat1 as a novel Asf1 copurifying protein. We subsequently deleted HAT1 from Asf1-TAP and eliminated the copurifying H4-specific HAT activity (Fig. 5B). Hat1 and Hat2 are components of Hat-B, the cytoplasmic HAT that acetylates H4 at K5 and K12 (30). In addition, a nuclear Hat1 complex exists, containing Hat1, Hat2, and Hif1 (3, 32). We tagged these three components separately in the Asf1-TAP background and determined that Asf1-TAP copurifies with the nuclear Hat1 complex containing Hif1 (Fig. 5C). To determine which component of the nuclear Hat1 complex interacts with Asf1, we deleted either HAT1 or HAT2 from Asf1-TAP carrying either Hat1-Myc or Hif1-Myc (which does not affect their respective expression; see Fig. S5A in the supplemental material) and analyzed copurifying proteins by tandem affinity purification and Western blotting (Fig. 5D). Deletion of HAT1 does not affect the interaction of Hif1 and Hat2 (see Fig. S5B in the supplemental material) or that of Hif1 and Asf1 (Fig. 5D). However, deletion of HAT2 from either strain prevented the association of Asf1-TAP with either tagged Hif1 or Hat1 (Fig. 5D). It has been previously shown that Hat2 mediates the interaction of Hif1 and Hat1 (32) (see Fig. S5B in the supplemental material). If Hif1 itself mediated the interaction of Asf1 and Hat1, then deletion of HAT2 should prevent the Asf1-Hat1 interaction but not that of Asf1-Hif1. Since both interactions are prevented, we conclude one function of Hat2 is to mediate a physical association between Asf1 and the nuclear HAT complex.
FIG. 5.
Asf1 interacts with the nuclear Hat1 complex. (A) Asf1 copurifies with H4-specific HAT activity that is independent of its ability to promote H3-K56ac. Core histone acetylation assays and their analyses are as described in the legend for Fig. 3. α-TAP, anti-TAP antibody. (B) Asf1 copurifies with Hat1-dependent H4-K5- and -K12-specific HAT activity. In vitro HAT assays are as described for Fig. 3. α-H3, -H4K5ac, and -H4K12ac, anti-H3, -H4-K5ac, and -H4-K12ac antibodies, respectively. (C) Asf1 interacts with the nuclear Hat1 complex. Immunoblotting was carried out with anti-TAP or anti-Myc (α-MYC) after tandem affinity purification of Asf1-TAP and 10% SDS-PAGE. (D) Association of the Asf1 histone chaperone with the nuclear Hat1 complex (Hat1-Hat2-Hif1) is dependent on the Hat2 subunit. Silver staining or immunoblotting was carried out after tandem affinity purification of Asf1-TAP followed by 10% SDS-PAGE. (E) Acetylation of H4-K5 and H3-K56 may perform parallel roles in DNA repair. The unacetylatable H4-K5R/H3-K56R double mutant is synthetically sensitive to the S-phase inhibitor HU relative to the corresponding single mutants. Growth analysis of strains in an MSY421 background (36) was performed as described for Fig. 1C. WT, wild type.
To determine if deposition-related acetylation sites in H4 and H3 are related, we made a collection of single or multiple point mutations in H4-K5, H4-K12, and H3-K56 and analyzed them for their sensitivity to hydroxyurea (HU). Individual H4-K5R or H4-K12R point mutants (carrying mutations which prevent acetylation of these lysines) are resistant to HU, but an H4-K5R/K12R double mutant does show hypersensitivity (Fig. 5E). In addition, when H4-K5R is combined with H3-K56R, we observed that the double mutant is sicker when grown on HU than H3-K56R (Fig. 5E), suggesting that H4-K5ac might also have a role in the repair of S-phase DNA damage.
DISCUSSION
Control of Rtt109 specificity by the chaperones Asf1 and Vps75.
We have shown that in addition to its function as an H3-K56 HAT, Rtt109 is an H3-K9 HAT in S. cerevisiae. The generation of H3-K9ac by Rtt109 requires both Asf1 and Vps75 in vivo. Our in vitro data are consistent with the in vivo data: when using recombinant proteins, Rtt109-based in vitro H3-K9ac HAT activity is strongly enhanced by Vps75 when core histones or H3 alone is used as a substrate (Fig. 2A). In addition, we have shown when using protein complexes from yeast that Rtt109-Vps75 can promote H3-K9 acetylation in vitro on core histones in a manner that is stimulated by Asf1 (Fig. 2D).
The in vitro role of Asf1 in promoting Rtt109-based H3-K9ac appears to be mechanistically quite different from its role in stimulating Rtt109-based H3-K56ac. Rtt109 specificity is determined in vivo by the histone chaperones it is using: Vps75 and/or Asf1. A model in which the substrate (H3-H4) or the enzyme itself (Rtt109) is altered by the chaperone in a manner that leads to a specific acetylation can explain the Asf1 stimulation of H3-K56ac by Rtt109. Indeed, this is consistent with structural and modeling analyses of yeast Asf1 that suggest the K56 residue on H3 is solvent exposed when bound to Asf1 as an H3-H4 dimer (4). Similarly, with respect to Rtt109's H3-K9ac activity, a structural change could occur upon H3-H4 binding by Vps75 that makes H3-K9 more accessible to Rtt109. Alternately, a structural change could occur on H3-H4 upon binding by Asf1, rendering H3-K9 inaccessible to Rtt109.
In addition, specificity is likely determined at least in part by Rtt109 itself, with the substrate made accessible when it is brought to Rtt109 by the histone chaperone. Supporting this idea is the fact that the Rtt109 target sites H3-K9 and H3-K56 are the only lysine residues in H3 to be followed by ST residues. It is formally possible that KST represents a minimal target sequence (“motif”) for Rtt109-based acetylation and that histone chaperones are required to properly position a particular substrate for acetylation.
One possible explanation of the fact that Rtt109-Asf1 does not strongly catalyze H3-K9ac in vitro is that Asf1 could have an inhibitory effect on modification of sites in the N termini of histones. An inhibitory effect on N-terminal sites of acetylation by rAsf1 on H3-H4 dimers has been reported previously. The SAS complex (Sas2, Sas4, and Sas5) acetylates H3 (K14) and H4 (K16) in vitro and has been shown to physically interact with Asf1. However, when rAsf1-H3-H4 was incubated with the recombinant SAS complex, acetylation on both histones was significantly reduced (47). In addition, rAsf1 strongly inhibits the H4-K5ac activity of rHat1 in an in vitro HAT assay using core histones from chicken as a substrate (J. Fillingham and J. F. Greenblatt, unpublished data). In this respect, Asf1 may be acting like the human protein INHAT, which blocks the action of PCAF and p300/CBP, presumably through histone masking (42).
Why would Asf1 block N-terminal modification of the H3-H4 dimer? One proposed function for histone chaperones is to protect acetylated histones from the action of nuclear HDACs before their assembly into chromatin (36). It is possible that Asf1 is able to block the N-terminal tails of bound H3-H4 dimers to prevent their spurious modification by inappropriate enzymatic activities. Asf1 could then recruit appropriate enzymes for specific modification depending on the particular situation (e.g., Rtt109-Vps75 and/or the nuclear Hat1 complex). The ability to overcome this hypothetical Asf1-mediated block could be a property of HAT-associated proteins (Hat2 and/or Hif1 in the case of Hat1 and Vps75 in the case of Rtt109) and/or Asf1-associated proteins, such as HIR-C (17) or Rad53 (14). In this respect, it is worth noting that Asf1, when purified from yeast, enhanced the H3-K9ac activity of Rtt109-Vps75 purified from yeast (Fig. 2D). The exact mechanism behind this observation remains unclear but is at least consistent with this model. In addition, the genes encoding Hat2 and components of HIR-C have a similar set of genetic interactions, indicating they could function in the same pathway (N. J. Krogan, unpublished observation).
Why does Vps75 stabilize Rtt109?
We have shown here that a function of Vps75 is to stabilize Rtt109. The biological role of this stabilization is currently unclear. Rtt109 and Vps75 appear to be approximately equimolar when copurified via Rtt109-TAP (25) (Fig. 1E). However, when copurified via Vps75-TAP, it appears that Vps75 is in excess to Rtt109, suggesting Rtt109 may not exist in the cell without Vps75. It seems likely, therefore, that Asf1 recruits Rtt109 in conjunction with Vps75 even when, as for H3-K56ac, Vps75 is not strictly needed for the acetylation event.
Function of Rtt109-based H3-K9ac.
Because H4-K5 appears to be partially redundant with H3-K56 with respect to sensitivity to HU (Fig. 5E), we suggest that a shared function of these deposition-related acetylation marks is to promote genome stability through S phase. In contrast, H3-K9R is not synthetic with H3-K56R when cells are grown in the presence of HU or MMS (Fig. 3F), indicating that H3-K9ac may have a function other than promotion of genome stability/DNA repair during chromatin assembly in S phase. Since Rtt109 has been suggested to be an S-phase-specific HAT due to its role in H3-K56ac generation and because H3-K9ac has a well-documented role in transcription, we suggest the possibility that one function of the Rtt109-based H3-K9ac associated with newly synthesized H3 is to promote transcription of S-phase-specific genes. Such a hypothesis would be consistent with the synthetic genetic interactions of rtt109Δ observed with deletions of transcription-related genes (see Fig. S3 in the supplemental material). We are currently testing this hypothesis using chromatin immunoprecipitation-chip and microarray technologies.
Model for histone modifications associated with S-phase chromatin assembly.
We suggest that Rtt109 is an H3-K9ac HAT associated with S-phase chromatin assembly. Asf1 recruits Rtt109 (or Rtt109-Vps75) to bound H3-H4 dimers to acetylate H3 at K56 via a physical interaction (which has been demonstrated in vitro and in vivo using a cross-linking agent [18, 49]). In the case of H3-K9ac, there are two possible models consistent with our in vitro and in vivo data. In the first, Asf1 recruits Rtt109-Vps75 to its H3-H4 substrate to generate H3-K9ac. In the second, Rtt109-Vps75 acetylates H3 associated with Vps75; this acetylated H3 is then “passed” to Asf1, which prevents its deacetylation before assembly into chromatin. In addition, Asf1 can recruit the nuclear Hat1 complex to acetylate H4 at K5 and K12. We suggest that Asf1 has the potential to assemble into chromatin itself or via other histone chaperones, such as CAF-1, H3-H4 dimers that are acetylated on H4 at K5 and K12, as well as on H3 at K9 and K56 (Fig. 6). These predictions match very closely the actual acetylation pattern found on H3 and H4, which copurify with the Drosophila RCAF complex that contains Drosophila Asf1 (50). The H3 HAT activity of Gcn5 during S-phase chromatin assembly remains unclear.
FIG. 6.
Model illustrating the roles of histone chaperones and HATs during chromatin assembly.
The S. cerevisiae genome encodes a variety of histone chaperones, including one that appears to function in transcriptional silencing, Rtt106 (20, 21). Our results suggest that Rtt106 does not modify Rtt109 specificity. Identification of protein-protein interactions for histone chaperones like Rtt106 might help determine whether they interact physically with histone-modifying enzymes; given their requirement for repression, one possibility is that they might be recruiting HDACs. If this is the case, as we suggest for chromatin deposition HATs, it would be interesting to know if these chaperones are involved in depositing hypoacetylated, rather than hyperacetylated, histones into chromatin. Thus, a more general model contemplates various chaperones acting in concert with HATs and/or HDACs (or even other histone-modifying enzymes) prior to chromatin assembly to provoke either a specific histone acetylation pattern (see Fig. 6) or a specific hypoacetylation pattern. This is in contrast to HATs (e.g., Gcn5 and Esa1) and HDACs (e.g., Sir2 and Rpd3) that act on nucleosomes.
Supplementary Material
Acknowledgments
This work was supported by grants to J.F.G. from the Canadian Institutes of Health Research (CIHR) and the National Cancer Institute of Canada with funds from the Canadian Cancer Society. J.F. was supported by a CIHR Fellowship. M.-C.K. was supported by a grant from the Speaker's Fund for Biomedical Research: Toward the Science of Patient Care, awarded by the City of New York.
We thank Guoquing Zhong and Joyce Li for excellent technical assistance, William Kuo for help with data presentation, Tania Roberts and Eduard Nedea for yeast strains and for helpful discussions during the course of this work, Wolfgang Fischle for recombinant Gcn5, and Luke Selth for recombinant GST-Vps75.
Footnotes
Published ahead of print on 5 May 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adkins, M. W., J. J. Carson, C. M. English, C. J. Ramey, and J. K. Tyler. 2007. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J. Biol. Chem. 2821334-1340. [DOI] [PubMed] [Google Scholar]
- 2.Adkins, M. W., and J. K. Tyler. 2004. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem. 27952069-52074. [DOI] [PubMed] [Google Scholar]
- 3.Ai, X., and M. R. Parthun. 2004. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol. Cell 14195-205. [DOI] [PubMed] [Google Scholar]
- 4.Antczak, A. J., T. Tsubota, P. D. Kaufman, and J. M. Berger. 2006. Structure of the yeast histone H3-ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics. BMC Struct. Biol. 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belle, A., A. Tanay, L. Bitincka, R. Shamir, and E. K. O'Shea. 2006. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. USA 10313004-13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. [DOI] [PubMed] [Google Scholar]
- 7.Celic, I., H. Masumoto, W. P. Griffith, P. Meluh, R. J. Cotter, J. D. Boeke, and A. Verreault. 2006. The sirtuins Hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr. Biol. 161280-1289. [DOI] [PubMed] [Google Scholar]
- 8.Chicoine, L. G., I. G. Schulman, R. Richman, R. G. Cook, and C. D. Allis. 1986. Nonrandom utilization of acetylation sites in histones isolated from Tetrahymena. Evidence for functionally distinct H4 acetylation sites. J. Biol. Chem. 2611071-1076. [PubMed] [Google Scholar]
- 9.Choy, J. S., and S. J. Kron. 2002. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell. Biol. 228215-8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, S. R., K. M. Miller, N. L. Maas, A. Roguev, J. Fillingham, C. S. Chu, M. Schuldiner, M. Gebbia, J. Recht, M. Shales, H. Ding, H. Xu, J. Han, K. Ingvarsdottir, B. Cheng, B. Andrews, C. Boone, S. L. Berger, P. Hieter, Z. Zhang, G. W. Brown, C. J. Ingles, A. Emili, C. D. Allis, D. P. Toczyski, J. S. Weissman, J. F. Greenblatt, and N. J. Krogan. 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446806-810. [DOI] [PubMed] [Google Scholar]
- 11.Cosgrove, M. S. 2007. Histone proteomics and the epigenetic regulation of nucleosome mobility. Expert Rev. Proteomics 4465-478. [DOI] [PubMed] [Google Scholar]
- 12.De Koning, L., A. Corpet, J. E. Haber, and G. Almouzni. 2007. Histone chaperones: an escort network regulating histone traffic. Nat. Struct. Mol. Biol. 14997-1007. [DOI] [PubMed] [Google Scholar]
- 13.Driscoll, R., A. Hudson, and S. P. Jackson. 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315649-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emili, A., D. M. Schieltz, J. R. Yates III, and L. H. Hartwell. 2001. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell 713-20. [DOI] [PubMed] [Google Scholar]
- 15.Fillingham, J., M. C. Keogh, and N. J. Krogan. 2006. GammaH2AX and its role in DNA double-strand break repair. Biochem. Cell Biol. 84568-577. [DOI] [PubMed] [Google Scholar]
- 16.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 111640-1650. [DOI] [PubMed] [Google Scholar]
- 17.Green, E. M., A. J. Antczak, A. O. Bailey, A. A. Franco, K. J. Wu, J. R. Yates III, and P. D. Kaufman. 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 152044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han, J., H. Zhou, B. Horazdovsky, K. Zhang, R. M. Xu, and Z. Zhang. 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315653-655. [DOI] [PubMed] [Google Scholar]
- 19.Han, J., H. Zhou, Z. Li, R. M. Xu, and Z. Zhang. 2007. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by Asf1 is required for replisome integrity. J. Biol. Chem. 28228587-28596. [DOI] [PubMed] [Google Scholar]
- 20.Huang, S., H. Zhou, D. Katzmann, M. Hochstrasser, E. Atanasova, and Z. Zhang. 2005. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc. Natl. Acad. Sci. USA 10213410-13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, S., H. Zhou, J. Tarara, and Z. Zhang. 2007. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 262274-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao, C. F., and M. A. Osley. 2003. In vivo assays to study histone ubiquitylation. Methods 3159-66. [DOI] [PubMed] [Google Scholar]
- 23.Keogh, M. C., J. A. Kim, M. Downey, J. Fillingham, D. Chowdhury, J. C. Harrison, M. Onishi, N. Datta, S. Galicia, A. Emili, J. Lieberman, X. Shen, S. Buratowski, J. E. Haber, D. Durocher, J. F. Greenblatt, and N. J. Krogan. 2006. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature 439497-501. [DOI] [PubMed] [Google Scholar]
- 24.Keogh, M. C., T. A. Mennella, C. Sawa, S. Berthelet, N. J. Krogan, A. Wolek, V. Podolny, L. R. Carpenter, J. F. Greenblatt, K. Baetz, and S. Buratowski. 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20660-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A. P. Tikuisis, T. Punna, J. M. Peregrin-Alvarez, M. Shales, X. Zhang, M. Davey, M. D. Robinson, A. Paccanaro, J. E. Bray, A. Sheung, B. Beattie, D. P. Richards, V. Canadien, A. Lalev, F. Mena, P. Wong, A. Starostine, M. M. Canete, J. Vlasblom, S. Wu, C. Orsi, S. R. Collins, S. Chandran, R. Haw, J. J. Rilstone, K. Gandi, N. J. Thompson, G. Musso, P. St Onge, S. Ghanny, M. H. Lam, G. Butland, A. M. Altaf-Ul, S. Kanaya, A. Shilatifard, E. O'Shea, J. S. Weissman, C. J. Ingles, T. R. Hughes, J. Parkinson, M. Gerstein, S. J. Wodak, A. Emili, and J. F. Greenblatt. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440637-643. [DOI] [PubMed] [Google Scholar]
- 26.Kuo, M. H., J. E. Brownell, R. E. Sobel, T. A. Ranalli, R. G. Cook, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383269-272. [DOI] [PubMed] [Google Scholar]
- 27.Masumoto, H., D. Hawke, R. Kobayashi, and A. Verreault. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436294-298. [DOI] [PubMed] [Google Scholar]
- 28.Millar, C. B., and M. Grunstein. 2006. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 7657-666. [DOI] [PubMed] [Google Scholar]
- 29.Morris, S. A., B. Rao, B. A. Garcia, S. B. Hake, R. L. Diaz, J. Shabanowitz, D. F. Hunt, C. D. Allis, J. D. Lieb, and B. D. Strahl. 2007. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J. Biol. Chem. 2827632-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parthun, M. R., J. Widom, and D. E. Gottschling. 1996. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell 8785-94. [DOI] [PubMed] [Google Scholar]
- 31.Pellicioli, A., C. Lucca, G. Liberi, F. Marini, M. Lopes, P. Plevani, A. Romano, P. P. Di Fiore, and M. Foiani. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 186561-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poveda, A., M. Pamblanco, S. Tafrov, V. Tordera, R. Sternglanz, and R. Sendra. 2004. Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J. Biol. Chem. 27916033-16043. [DOI] [PubMed] [Google Scholar]
- 33.Prado, F., F. Cortes-Ledesma, and A. Aguilera. 2004. The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO Rep. 5497-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin, S., and M. R. Parthun. 2002. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 228353-8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramey, C. J., S. Howar, M. Adkins, J. Linger, J. Spicer, and J. K. Tyler. 2004. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol. Cell. Biol. 2410313-10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Recht, J., T. Tsubota, J. C. Tanny, R. L. Diaz, J. M. Berger, X. Zhang, B. A. Garcia, J. Shabanowitz, A. L. Burlingame, D. F. Hunt, P. D. Kaufman, and C. D. Allis. 2006. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 1036988-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts, T. M., I. W. Zaidi, J. A. Vaisica, M. Peter, and G. W. Brown. 2008. Regulation of rtt107 recruitment to stalled DNA replication forks by the cullin rtt101 and the rtt109 acetyltransferase. Mol. Biol. Cell 19171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz-Garcia, A. B., R. Sendra, M. Galiana, M. Pamblanco, J. E. Perez-Ortin, and V. Tordera. 1998. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J. Biol. Chem. 27312599-12605. [DOI] [PubMed] [Google Scholar]
- 39.Schneider, J., P. Bajwa, F. C. Johnson, S. R. Bhaumik, and A. Shilatifard. 2006. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J. Biol. Chem. 28137270-37274. [DOI] [PubMed] [Google Scholar]
- 40.Schuldiner, M., S. R. Collins, J. S. Weissman, and N. J. Krogan. 2006. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods 40344-352. [DOI] [PubMed] [Google Scholar]
- 41.Selth, L., and J. Q. Svejstrup. 2007. Vps75, a new yeast member of the NAP histone chaperone family. J. Biol. Chem. 28212358-12362. [DOI] [PubMed] [Google Scholar]
- 42.Seo, S. B., P. McNamara, S. Heo, A. Turner, W. S. Lane, and D. Chakravarti. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104119-130. [DOI] [PubMed] [Google Scholar]
- 43.Sharp, J. A., E. T. Fouts, D. C. Krawitz, and P. D. Kaufman. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11463-473. [DOI] [PubMed] [Google Scholar]
- 44.Shibahara, K., and B. Stillman. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96575-585. [DOI] [PubMed] [Google Scholar]
- 45.Sklenar, A. R., and M. R. Parthun. 2004. Characterization of yeast histone H3-specific type B histone acetyltransferases identifies an ADA2-independent Gcn5p activity. BMC Biochem. 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sobel, R. E., R. G. Cook, C. A. Perry, A. T. Annunziato, and C. D. Allis. 1995. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. USA 921237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutton, A., W. J. Shia, D. Band, P. D. Kaufman, S. Osada, J. L. Workman, and R. Sternglanz. 2003. Sas4 and Sas5 are required for the histone acetyltransferase activity of Sas2 in the SAS complex. J. Biol. Chem. 27816887-16892. [DOI] [PubMed] [Google Scholar]
- 48.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 2942364-2368. [DOI] [PubMed] [Google Scholar]
- 49.Tsubota, T., C. E. Berndsen, J. A. Erkmann, C. L. Smith, L. Yang, M. A. Freitas, J. M. Denu, and P. D. Kaufman. 2007. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell 25703-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka, and J. T. Kadonaga. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402555-560. [DOI] [PubMed] [Google Scholar]
- 51.VanDemark, A. P., M. M. Kasten, E. Ferris, A. Heroux, C. P. Hill, and B. R. Cairns. 2007. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol. Cell 27817-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vermeulen, M., K. W. Mulder, S. Denissov, W. W. Pijnappel, F. M. van Schaik, R. A. Varier, M. P. Baltissen, H. G. Stunnenberg, M. Mann, and H. T. Timmers. 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 13158-69. [DOI] [PubMed] [Google Scholar]
- 53.Verreault, A., P. D. Kaufman, R. Kobayashi, and B. Stillman. 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 8795-104. [DOI] [PubMed] [Google Scholar]
- 54.Xu, F., K. Zhang, and M. Grunstein. 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121375-385. [DOI] [PubMed] [Google Scholar]
- 55.Xu, F., Q. Zhang, K. Zhang, W. Xie, and M. Grunstein. 2007. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell 27890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.