Abstract
The vertebrate Ets transcriptional repressor Tel (ETV6) and its invertebrate orthologue, Yan, are both indispensable for development, and they orchestrate cell growth and differentiation by binding to DNA, thus inhibiting gene expression. To trigger cell differentiation, these barriers to transcriptional activation must be relieved, and it is established that posttranslational modifications, such as phosphorylation and sumoylation, can specifically impair the repressive functions of Tel and Yan and are crucial for modulating their transcriptional activity. To date, however, relatively little is known about the control of Tel and Yan protein degradation. In recent years, there has been a concentrated effort to assign functions to the large number of F-box proteins encoded by both vertebrate and invertebrate genomes. Here, we report the identification and characterization of a previously unreported, evolutionarily conserved F-box protein named Fbl6. We isolated both human and Drosophila melanogaster fbl6 cDNA and show that the encoded Fbl6 protein binds to both Tel and Yan via their SAM domains. We demonstrate that both Tel and Yan are ubiquitinated, a process which is stimulated by Fbl6 and leads to proteasomal degradation. We recently established that the sumoylation of Tel on lysine 11 negatively regulates its repressive function and that the sumoylation of Tel monomers, but not that of Tel oligomers, may sensitize Tel for proteasomal degradation. Here, we found that Fbl6 regulates Tel/Yan protein stability and allows appropriate spatiotemporal control of gene expression by these repressors.
Cell fate is determined by programs of gene expression, which are strictly regulated spatiotemporally by a complex network of interacting molecular mechanisms that control the balance between transcription, translation, and degradation. The Drosophila melanogaster transcription factor Yan and its vertebrate orthologue, Tel (ETV6), are well placed to interrogate these processes. An abundance of molecular and genetic evidence shows that these unique Ets transcription factor repressors are indispensable for normal cell differentiation (15, 19, 22, 27, 35, 36, 45, 46). Importantly, the disruption of normal Tel function leads to neoplasia (10, 11). A finely controlled interplay between posttranscriptional and posttranslational modifications regulates their functions (42). In the case of Tel, the observed heterogeneity of Tel proteins in cells (37, 29, 43) can be accounted for by at least two posttranscriptional mechanisms—the use of an alternative initiation codon (37, 29) and alternative splicing (38; M. G. Roukens and D. A. Baker, unpublished data). Together, these processes can produce Tel isoforms that differentially control Tel function. Yan, on the other hand, appears to be regulated posttranscriptionally by a microRNA, miR7, that might act in a tissue-specific fashion and that limits Yan protein translation by binding to the 3′ untranslated region of yan mRNA (21). Posttranslationally, phosphorylation and sumoylation play pivotal roles in modulating the activities of Tel and Yan, particularly by impairing the repression of transcription by these factors. Specifically, phosphorylation of Yan is a trigger for its downregulation (2, 33, 35, 40, 41), and sumoylation of Tel is PIAS dependent (37) and inhibits the repression of gene expression (5, 37, 49). To date, however, relatively little is known about the control of Tel/Yan protein degradation.
There are vital mechanisms of protein degradation for controlling the timing of the action of proteins and, ultimately, the timing of cellular processes in general. One such crucial mechanism of degradation is mediated by the process of ubiquitination, which impinges on virtually all known eukaryotic cellular processes (13). Ubiquitin is a 76-amino-acid polypeptide that can be covalently bonded to target proteins in a number of ways (28). Monoubiquitination has been shown to play an essential role in endocytosis (16) and in the subcellular targeting of proteins (20). Polyubiquitination, in contrast, is almost exclusively associated with protein degradation and turnover, either via the proteasome or through endocytosis and lysosomal sorting (47). Mechanistically, ubiquitination is well defined and involves the concerted action of at least three different catalytic components, namely the E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin ligase) enzymes (48). A crucial aspect of ubiquitination-driven degradation is how substrate specificity is achieved. One way is through the recruitment of F-box proteins. F-box proteins are characterized by the presence of an N-terminal F-box domain that is approximately 50 amino acids long (1), and they are broadly classified into three families: the FBW family, which contains WD repeats; the FBL family, which contains leucine-rich repeats; and the FBX family, which consists of F-box proteins with other protein interaction domains. These domains are usually located in the C terminus of the protein, and they associate with substrates and link them to the ubiquitinating machinery via the F-box domain (4, 14).
We have performed yeast two-hybrid screens to identify proteins that associate with both Tel and Yan, and by this means, we have uncovered an F-box protein named Fbl6. Our biochemical and genetic analyses of human and Drosophila tissue culture cells, as well as of Drosophila embryos, suggest that F-box-mediated ubiquitination of Tel and Yan promotes their downregulation.
MATERIALS AND METHODS
Cell-based ubiquitination assays.
Cells were transfected with the appropriate plasmids and then incubated for 6 h with or without the proteasome inhibitor MG132 24 to 36 h posttransfection. His-ubiquitin pulldowns were performed with 50 μl of Ni-nitrilotriacetic acid beads (Qiagen) for 3 h at room temperature in 6 ml of 6 M guanidinium-HCl, 0.1 M Na2HPO4, 0.1 M NaH2PO4, 0.01 M Tris-HCl (pH 8.0), 20 mM imidazole, and 10 mM β-mercaptoethanol (buffer A). The beads were successively washed twice with 1 ml of each of the following buffers: (i) buffer A plus 0.2% Triton X-100; (ii) 8 M urea, 0.1 M Na2HPO4, 0.1 M NaH2PO4, 0.01 M Tris-HCl (pH 8.0), 20 mM imidazole, 10 mM β-mercaptoethanol, and 0.2% Triton X-100 (buffer B); and (iii) 8 M urea, 0.1 M Na2HPO4, 0.1 M NaH2PO4, 0.01 M Tris-HCl (pH 6.3), 20 mM imidazole, 10 mM β-mercaptoethanol, and 0.2% Triton X-100 (buffer C). Ubiquitinated proteins were eluted in 60 μl of urea sample buffer, which consisted of 37.5% buffer C, 39.3% 3× Laemmli buffer, 20 mM imidazole, and 3.2% β-mercaptoethanol. The samples were boiled and analyzed by Western blotting.
Immunofluorescence.
Cells were grown on glass coverslips and transfected using Fugene 6 (Roche). Cells were fixed after 24 h with 4% paraformaldehyde for 15 min at room temperature (all of the following steps were done at room temperature) and permeabilized in 0.2% Triton X-100-phosphate-buffered saline for 5 min. Cells were washed with phosphate-buffered saline and blocked with 5% goat serum for 1 h, incubated with primary antibodies for 1 h, washed, and incubated with secondary antibodies for 30 min. Following extensive washing, the cells were mounted and immunostaining was visualized with a Leica DM5500 B microscope.
Cell culture, biochemistry, and molecular biology.
A schematic representation of all of the constructs used in this study can be found in Fig. S3 in the supplemental material and also in the relevant figures. Mammalian cell lines were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and antibiotics. Drosophila Schneider cells were cultured at 25°C in Schneider cell medium (Sigma) supplemented with 10% fetal bovine serum. Mammalian cells were transfected with Fugene 6 (Roche) and insect cells with Effectene reagent (Qiagen), according to the manufacturers' protocol. In general, 1 μg of each construct was transfected into cells seeded at 40 to 60% confluence in 6-cm tissue culture dishes, and cells were lysed 24 to 48 h posttransfection. TelHA,TelFlag, YanHA, and YanFlag constructs were fused in frame with either a hemagglutinin (HA) or a Flag epitope tag and cloned into either the pCS2 expression vector for expression in mammalian cells or the pMT vector (Invitrogen) for expression in insect cells. Glutathione S-transferase (GST)-Tel was cloned in frame with GST in the pGEX-2TK vector. Mutants were generated by PCR. For immunoprecipitations, cells were lysed in 1 ml of either radioimmunoprecipitation assay-sodium dodecyl sulfate buffer (1 mM EDTA, 50 mM Tris [pH 8.0], 150 mM NaCl, 10% glycerol, 1% Triton X-100) or protein lysis buffer (50 mM Tris [pH 7.5], 1% NP-40, 0.1% sodium dodecyl sulfate, 0.5% Na deoxycholate, 150 mM NaCl) with protease inhibitors (phenylmethylsulfonyl fluoride, trypsin, pepstatin A, leucine, aprotinin) and NaF. Cell lysates were syringed and centrifuged. Immunoprecipitations were carried out with 0.75 μl of either anti-HA polyclonal antibody (Abcam) or anti-V5 monoclonal antibody (Invitrogen). Lysates were preincubated with the antibody for 1 h, after which suitable beads, protein A-Sepharose 4 fast flow (Amersham Pharmacia Biotech) for HA immunoprecipitations or protein G-Sepharose (Sigma-Aldrich) for V5 immunoprecipitations, were added for a further 2 h of incubation. Associated proteins were analyzed by Western blotting. For pulldown assays, Fbl6 proteins were labeled with [35S]methionine by using the TNT coupled reticulocyte in vitro translation system (Promega) and then incubated with GST-Tel fusions that were immobilized on glutathione-Sepharose beads as previously described (2).
Production and infection with shRNA-expressing lentiviruses.
Short hairpin RNA (shRNA) lentiviral constructs for the knockdown of fbl6 were cloned by insertion of the following oligonucleotides into the BglII/HindIII sites of the pSuper vector: fbl6i#1 (5′-GATCGCAAGTTGTGGCTGACCTATTCAAGAGATAGGTCAGCCACAACTTGCTTTTT-3′ and 5′ AGCTAAAAAGCAAGTTGTGGCTGACCTATCTCTTGAATAGGTCAGCCACAACTTGC-3′) and fbl6i#2 (5′-GATCGCATCAACCGTAATAGCATTTCAAGAGAATGCTATTACGGTTGATGCTTTTT-3′ and 5′-AGCTAAAAAGCATCAACCGTAATAGCATTCTCTTGAAATGCTATTACGGTTGATGC-3′). The pSuper constructs were subsequently digested with PstI and XhoI, and the fragment was placed in the lentiviral PLV-cytomegalovirus-green fluorescent protein vector.
shRNA lentiviral constructs for the knockdown of S-phase kinase-associated protein 1 (Skp1) were obtained from the Mission shRNA library (Sigma-Aldrich clones TRCN 006 496 and TRCN 011 029).
Recombinant lentiviruses were produced by the transfection of 293T cells. Transduction of U2OS cells was performed at a multiplicity of infection of 1, with the addition of Polybrene in a final concentration of 8 μg/ml. One day postinfection, the medium was replaced with fresh medium (supplemented with puromycin for the skp1 lines), and cells were lysed after 48 h.
In vivo 35S labeling.
Cells were washed free of medium and seeded into 6-cm tissue culture dishes (Gibco) in methionine- and serum-free Dulbecco modified Eagle medium (Gibco). Cells were routinely incubated for 3 h, and then the medium was supplemented with 50 μCi 35S-labeled methionine for an additional 3 h of labeling. Labeled HA epitope-tagged Tel proteins were immunoprecipitated from the cell lysates as described above.
Baculovirus production and expression in cells.
Recombinant baculoviruses for expression in SF9 cells were generated using the Bac-to-Bac baculovirus expression system (Invitrogen) according to the manufacturer's manual.
For the ubiquitination assay, SF9 cells were seeded in Grace's medium (Invitrogen) at 40 to 60% confluence in 6-cm culture dishes and infected with recombinant viruses.
Analysis of gene expression by quantitative PCR.
All methods were performed essentially as described previously (37). The following primer sets were deployed for real-time PCR analysis: Telq15 (CCCTGCGCCACTACTACAA) and Telq13 (TGATTTCATCTGGGGTTTTCA), Telq25 (CTTTCGCTATCGATCTCCTCA) and Telq23 (AGGGTGGAAGAATGGTGAAA), hFbl615 (ATGCCCAATCGGTTTTCA) and hFbl613 (AGGACAGCACTCACCTACCAG), hFbl625 (GGATCTTCGTGGCTGTGC) and hFbl623 (CATACAGGCCCAGATGAAGC), hSKP1A15 (GGAGATTCGCAAGACCTTCA) and hSKP1A13 (CACTGGTTCTCTTTGCGTACC), and Gapdh5 (TGCCATGTAGACCCCTTGAAG) and Gapdh3 (ATGGTACATGACAAGGTGCGG).
Drosophila genetics and embryo analysis.
P{GawB}NP0326, Df(2R)stan1, and Df(2R)Exel6060 fly stocks were obtained from the Bloomington Stock Center. Stocks were maintained using balancers expressing the green fluorescent protein. Reverted flies were generated by transposase-mediated precise excision of the P element in P{GawB}NP0326 flies. Precise excision was confirmed by the sequencing of fly genomic DNA. Stocks were maintained under standard conditions, and crosses were performed by following standard procedures. For expression analyses, embryos were carefully staged, and total RNA was prepared using the SV total RNA isolation kit in accordance with the manufacturer's protocol (Promega). Following cDNA synthesis using a TaqMan kit (Roche), expression levels of relevant transcripts were determined by PCR using the following primer sets: the fbl6 sense primer (AGGTCCTGCGTGTGACAAACTCTC) and the fbl6 antisense primer (GAGGCAATGAGTTGGATGTGAAC); the argos sense primer (GGATTTTCTGTTTGATCAGTTC) and the argos antisense primer (ATTATTGGATATTTCATTCACT); the CG18335 sense primer, (ATAACGCCGGAACCGCACTTGGTG) and the CG18335 antisense primer (GTAAACGGAGTCCACCAAACCCTC); the CG13220 sense primer (ACCTGACGAACGTGTTCCTCATCTC) and the CG13220 antisense primer (GGTTCGAATACCTCAGTCCTATATC); the tubulin sense primer (CGTTCACATCCAAGCTGGTCAG) and the tubulin antisense primer (GGGTGCGGAAGCAGATATCG).
Yeast two-hybrid assay and cDNA cloning.
Full-length yan or tel cDNA was cloned into pAS2-1 (Clontech) in frame with the GAL4 DNA binding domain. Independent transformants (approximately 5 × 105) from a GAL4 activation domain Drosophila embryo library (for Yan) or a human bone marrow library (for Tel) were screened in Saccharomyces cerevisiae Hf7c cells according to the manufacturer's protocol (Clontech). We confirmed the specificities of interactions between Yan and Drosophila Fbl6 and between Tel and human Fbl6 by transforming yeast with the cDNA of fbl6, in either the presence or the absence of the yan bait (for Drosophila fbl6), the tel bait (for human fbl6), or an unrelated bait. Full-length cDNAs of the human fbl6 gene and of the Drosophila fbl6 gene were isolated by reverse transcriptase (RT)-PCR using total RNA derived from either insect Schneider cells (for Drosophila fbl6) or U2OS human osteosarcoma cells (for human fbl6).
Antibodies/drugs.
The following antibodies (and dilutions, if applicable) were used: anti-V5 antibody (Invitrogen); anti-Flag mouse M2 monoclonal antibody (Sigma-Aldrich); anti-HA.11 mouse monoclonal antibody (Covance); anti-GST rabbit antibody (My Probe), 1:5,000; anti-HA rabbit polyclonal antibody (Abcam), 1:1,000; human anti-Tel rabbit polyclonal antibody, 1:1,000; anti-Yan monoclonal antibody, 1:500; Drosophila anti-SKPA rabbit polyclonal antibody, 1:1,000; and anti-His polyclonal antibody, 1:1,000. For MG132, experiment cells were incubated with 3 μM MG132 (Calbiochem) for 6 h prior to lysis.
RESULTS
Fbl6, a conserved F-box-containing protein, associates biochemically with both human Tel and Drosophila Yan.
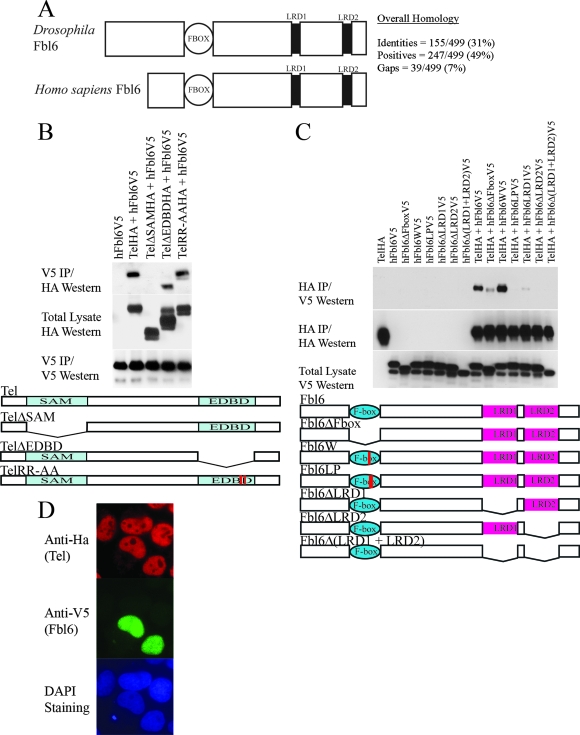
To uncover evolutionarily conserved pathways that might regulate Yan/Tel function, we performed yeast two-hybrid screens using either Drosophila Yan or its human orthologue, Tel, as bait to identify common interacting proteins. Proteins that were recovered from both of the independent screens are highlighted in Fig. S1 in the supplemental material. Since posttranslational modifications are essential modulators of protein function, we centered our subsequent analyses on Fbl6, a putative mediator of protein degradation. Figure 1A (see also Fig. S2 in the supplemental material) schematically shows that Fbl6 harbors an F-box domain (1) and two other conserved sequence blocks, LRD1 and LRD2, which have been previously classified as leucine-rich domains (LRDs) (4). Our subsequent analyses are presented in the five figures included in this article. Figures 1 to 3 describe the regulation of human Tel by human Fbl6, and Fig. 4 and 5 explore the role of Fbl6 in Drosophila Yan function and together suggest an evolutionarily conserved function.
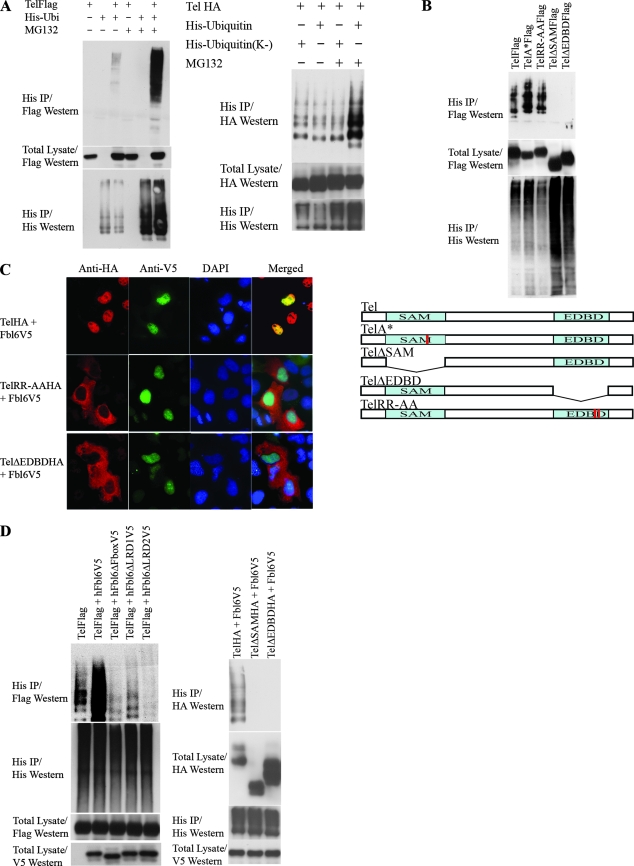
FIG. 1.
(A) Schematic representation of human and Drosophila Fbl6 proteins. Fbl6 is characterized by a conserved, N-terminal F-box domain (1) that is 40 to 50 amino acids long. Two other blocks of conserved sequences, which have previously been defined as leucine-rich regions (LRD1 and LRD2), are represented by black rectangles (4). The alignments of the primary amino acid sequences can be found in Fig. S1 in the supplemental material. (B) The SAM domain of Tel is needed for association with Fbl6 in cells. HA epitope-tagged wild-type Tel, TelΔSAM, TelΔEDBD, and Tel harboring substitutions of alanine for arginine in the Ets DNA-binding motif (KMSRALRHYYK) were each coexpressed with V5 epitope-tagged human Fbl6 in 293T cells. Fbl6 complexes were immunopurified from cell lysates using an antibody directed against the V5 epitope, and associated Tel proteins were detected using an antibody directed against the HA epitope. (C) The LRDs of Fbl6 and an intact F-box domain are essential for interacting with Tel. V5 epitope-tagged full-length Fbl6 or the indicated Fbl6 mutants were coexpressed with Tel in 293T cells. Tel complexes were immunopurified from cell lysates by using an antibody directed against the HA epitope, and associated Fbl6 proteins were detected using an antibody directed against the V5 epitope. Fbl6ΔFbox contains a deletion of the F-box domain. Fbl6W and Fbl6LP have point mutations that have been shown to disrupt the F-box domain (37). Fbl6ΔLRD1 and Fbl6ΔLRD2 harbor deletions of the LRD1 and LRD2 sequences, respectively. Fbl6Δ(LRD1 + LRD2) has a deletion of both LRD1 and LRD2. h, human. (D) Fbl6 and Tel are colocalized in the nucleus. Cells were transfected with the indicated constructs and proteins detected with the indicated antibodies. DAPI, 4′,6′-diamidino-2-phenylindole.
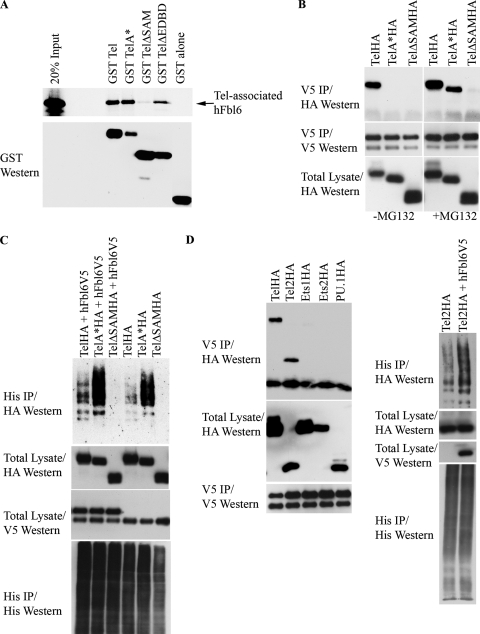
FIG. 3.
(A) Fbl6 binds efficiently with Tel monomers in vitro. Fusions were made between GST and either full-length wild-type Tel, full-length Tel harboring mutations that disrupt the SAM domain (TelΔSAM and TelA*), or TelΔEDBD (see text for details). 35S-labeled Fbl6 was produced by in vitro translation in reticulocyte lysates and assessed for binding to GST-Tel. (B) Inhibition of proteasome function stabilizes the interaction between Tel monomers and Fbl6 in cells. Cells were cotransfected with the indicated Tel constructs along with V5 epitope-tagged Fbl6. Following incubation with (+) or without (−) MG132, Fbl6 complexes were immunopurified from cell lysates by using an antibody directed against the V5 epitope, and the associated Tel protein was detected using an antibody directed against the HA epitope. (C) Tel monomers can bind with Fbl6 and are efficiently ubiquitinated; their ubiquitination is augmented by Fbl6. U2OS cells were cotransfected with the indicated constructs together with His epitope-tagged ubiquitin, and a ubiquitination assay was performed. (D) Fbl6 interacts with Tel2 and stimulates its ubiquitination. HA epitope-tagged versions of the indicated Ets transcription factors were coexpressed with or without Fbl6. Fbl6 complexes were immunopurified from cell lysates using an antibody directed against the V5 epitope, and associated Ets proteins were detected using an antibody directed against the HA epitope. For the ubiquitination assay, cells were transfected with the indicated constructs and processed as described in the legend to Fig. 2A. Western, Western blotting; IP, immunoprecipitation; h, human.
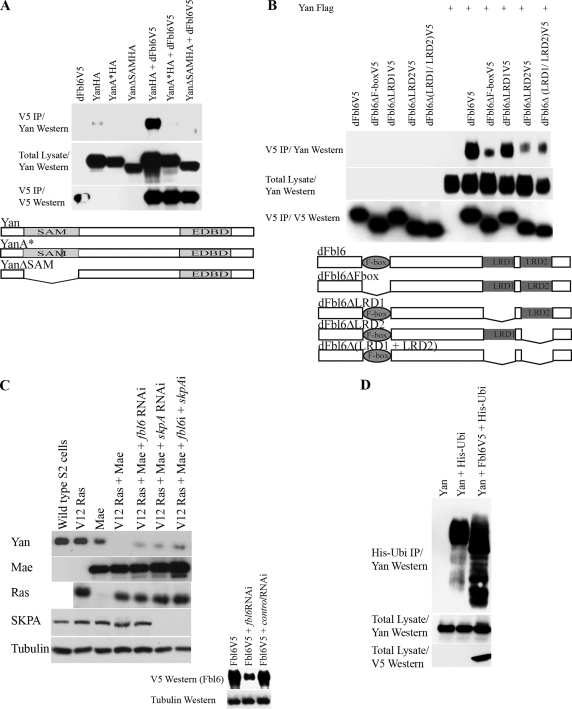
FIG. 4.
(A) The SAM domain of Yan is needed for Yan's association with Fbl6 in cells. HA epitope-tagged wild-type Yan, Yan expressing a deletion of the SAM domain (YanΔSAM), and Yan harboring a substitution of alanine at position 86 for arginine (YanA*) were each coexpressed with V5 epitope-tagged Drosophila Fbl6 in 293T cells. Fbl6 complexes were immunopurified from cell lysates by using an antibody directed against the V5 epitope, and associated Yan proteins were detected using an anti-Yan monoclonal antibody. (B) The LRD2 of Drosophila Fbl6 is essential for interacting with Yan. V5 epitope-tagged full-length Fbl6 or the indicated Fbl6 mutants were coexpressed with Yan in Drosophila S2 cells. Fbl6 complexes were immunopurified from cell lysates by using an antibody directed against the V5 epitope, and associated Yan proteins were detected using an anti-Yan monoclonal antibody. Fbl6ΔF-box contains a deletion of the F-box domain. Fbl6ΔLRD1 and Fbl6ΔLRD2 harbor deletions of the LRD1 and LRD2 sequences, respectively. Fbl6Δ(LRD1 + LRD2) has a deletion of both the LRD1 and LRD2 sequences. (C) Fbl6 regulates Yan downregulation in Drosophila Schneider (S2) cells. Stable S2 cell lines in which expression of the indicated constructs was placed under the control of the metallothionine promoter were established. Cells were transfected with or without the indicated double-stranded RNA (termed fbl6 RNAi [fbl6i] and skpA RNAi [skpAi]), and 2 days later, expression of the indicated constructs was induced with a 600 μM copper sulfate solution. Following a further 24 h of incubation, Western blot analysis was performed with cell lysates by using the indicated antibodies. In the absence of an antibody specific for Drosophila Fbl6, the efficiency of Fbl6 knockdown was assessed by separately targeting V5 epitope-tagged Fbl6 proteins expressed in S2 cells. A nonspecific double-stranded RNA was used as a control (see lower panel). (D) Yan is ubiquitinated, a process which is augmented by Fbl6. The indicated constructs were expressed in insect Sf9 cells using baculoviruses, and a ubiquitination assay was performed (see the legend to Fig. 2). d, Drosophila.
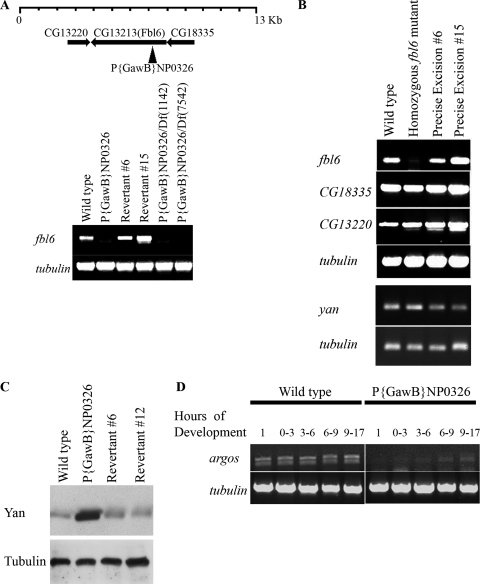
FIG. 5.
Fbl6 regulates Yan downregulation in vivo. (A) P{GawB}NP0326 flies have strongly diminished levels of fbl6 expression. RT-PCR was performed on cDNA prepared from stage-15 embryos derived from the indicated fly lines. Levels of tubulin transcript were determined as a control for the procedure. (B) The P element of P{GawB}NP0326 flies specifically disrupts fbl6 and not adjacent transcripts. RT-PCR was performed as described for panel A. (C) Yan protein levels are elevated in P{GawB}NP0326 embryos. Embryos were carefully staged to stage 12 and lysed in Laemmli sample buffer. Yan protein levels were determined by Western blotting with a Yan monoclonal antibody, and tubulin protein levels were determined as a loading control. (D) Expression of argos is attenuated in P{GawB}NP0326 embryos. cDNA was prepared from total RNA isolated from differently staged embryos. Levels of argos expression and control tubulin expression were determined by RT-PCR.
The Tel SAM domain and Fbl6 LRDs are required for their binding.
We engineered a V5 epitope-tagged version of wild-type human Fbl6 and confirmed the biochemical interaction between Tel and Fbl6 in tissue culture cells (Fig. 1B and C). Furthermore, we found that this interaction requires the SAM domain of Tel (Fig. 1B) and the putative LRDs (LRD1 and LRD2) of Fbl6 (Fig. 1C). Interestingly, disruption of the F-box domain of Fbl6 also impairs binding to Tel (see Fig. 1C). In cells, both Tel and Fbl6 colocalize in the nucleoplasm, and Fbl6 is also found in the nucleolus (Fig. 1D and see Fig. 2C). Recent reports have highlighted differential regulation of protein function by different isoforms of the same F-box protein that are localized in either the nucleolus or the nucleus (31). It would be interesting as a future study to determine whether nuclear and nucleolar Fbl6 proteins play different roles in protein regulation.
FIG. 2.
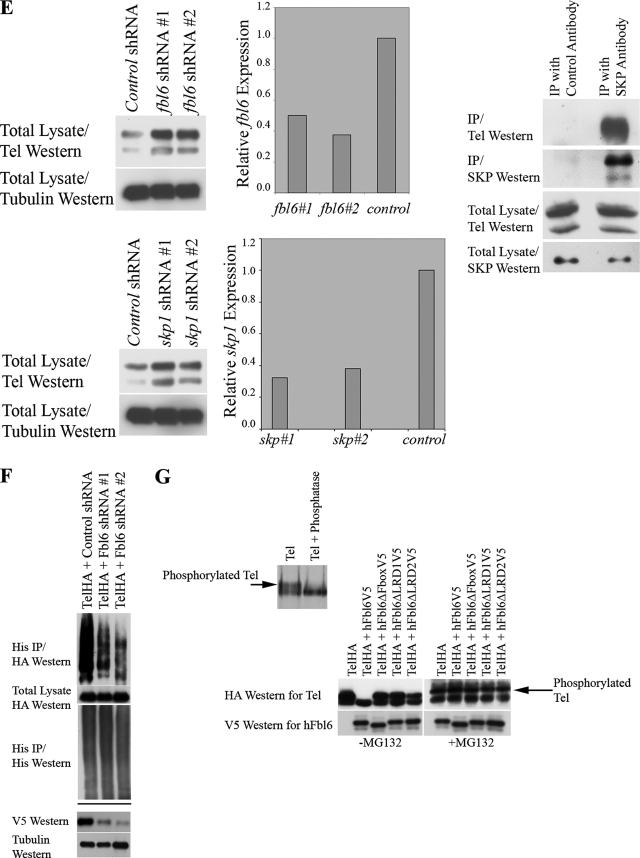
(A) Tel ubiquitination. The left panel shows the results of an experiment in which Flag epitope-tagged wild-type Tel was cotransfected into U2OS cells with (+) or without (−) His epitope-tagged ubiquitin and cells were subsequently incubated in the presence (+) or absence (−) of MG132. Ubiquitinated Tel was recovered from cells lysed in guanidinium, by nickel bead purification (hereafter termed a ubiquitination assay). The right panel shows a ubiquitination assay using HA epitope-tagged Tel and either a wild-type His epitope-tagged ubiquitin or a His epitope-tagged version of ubiquitin in which all of the lysine residues have been mutated (K−). (B) The SAM domain and EDBD of Tel are required for its ubiquitination. U2OS cells were transfected with the indicated constructs (described in the legend to Fig. 1B), and a ubiquitination assay was performed as described for panel A. (C) Mutations that disrupt the EDBD of Tel lead to the mislocalization of Tel to the cytoplasm. Cells were transfected with the indicated constructs, and immunohistochemistry was performed with the antibodies shown. (D to G) Fbl6 promotes the ubiquitination of Tel. (D) U2OS cells were transfected with the indicated constructs (see the legend to Fig. 1C), and a ubiquitination assay was performed as described for panel A. (E) shRNA-mediated knockdown of fbl6 and skp1 leads to stabilization of endogenous steady-state Tel protein levels. U2OS cells were infected with lentiviruses delivering the indicated shRNAs, and following 2 days of puromycin selection, stably infected cells were lysed and proteins levels monitored with the indicated antibodies. Tubulin was used as a loading control for proteins. Quantitative PCR was used to demonstrate effective knockdown of fbl6 and skp1 transcripts. In both cases, quantitative PCR was also used to show that tel transcript levels were unaffected by any of the shRNA treatments (data not shown). The rightmost panel shows that endogenous Tel and endogenous SKP1 form a complex in tissue culture cells. U2OS cells were lysed in protein lysis buffer (see Materials and Methods), and SKP1 was purified from the lysate with a SKP1-specific antibody. Associated endogenous Tel was detected with a Tel-specific antibody. (F) Short-hairpin RNA (shRNA)-mediated knockdown of Fbl6 inhibits Tel ubiquitination. Cells were transiently transfected with the indicated constructs, and a ubiquitination assay was performed 2 days later. In the absence of an effective antibody specific for human Fbl6, the efficiency of Fbl6 knockdown was assessed separately by targeting V5 epitope-tagged Fbl6 expressed in U2OS cells. A nonspecific small interfering RNA was used as a control (see lower panel). (G) Fbl6 strongly promotes the degradation of phosphorylated Tel. The upper panel shows [35S]methionine-labeled Tel that was immunopurified from U2OS cells, following 3 h of labeling, and then treated with 10 U of calf intestinal phosphatase or left untreated for 30 min at 37°C. In the lower panel, cells were transfected with the indicated constructs, incubated in the presence (+) or absence (−) of MG132, and then lysed in Laemmli sample buffer. Proteins were detected with the indicated antibodies. Western, Western blotting; IP, immunoprecipitation; h, human.
Tel is ubiquitinated.
It is firmly established that F-box-containing proteins couple their substrates to the conserved ubiquitinating machinery that targets the protein for proteasome-dependent degradation (14, 18, 48). Thus, we investigated whether Tel is ubiquitinated and whether the ubiquitination of Tel might be regulated by Fbl6. Figure 2A shows that Tel is ubiquitinated in tissue culture cells and that ubiquitinated Tel is targeted for degradation by the proteasome because the incubation of cells with inhibitors of the proteasome stabilizes the fraction of ubiquitin-conjugated Tel. In support of these data, Fig. 2A also shows that Tel that is covalently conjugated by a version of ubiquitin that lacks all lysines (and therefore fails to form chains of ubiquitin that are recognized by the proteasome) is not stabilized following inhibition of the proteasome with MG132. Figure 2B shows that both the SAM domain and the Ets DNA binding domain (EDBD) are indispensable for Tel ubiquitination. Tel with EDBD deleted (TelΔEDBD) can interact with Fbl6 both in cells (Fig. 1B) and in vitro (see Fig. 3A); however, we have shown previously (37) (Fig. 2C) that the deletion of the EDBD leads to the mislocalization of Tel from the nucleus to the cytoplasm. Therefore, to determine that the failure of TelΔEDBD to be ubiquitinated in cells does not result simply from subcellular mislocalization, we engineered a Tel mutant, TelRR-AA. TelRR-AA contains two single point mutations of arginine residues in the EDBD that are known to make contact with DNA (25) and are sufficient to promote Tel mislocalization from the nucleus to the cytoplasm (Fig. 2C). However, unlike TelΔEDBD, TelRR-AA is efficiently ubiquitinated (Fig. 2B), suggesting that the failure of TelΔEDBD to be ubiquitinated does not result simply from its mislocalization from the nucleus. Some possibilities are that lysine residues in the EDBD are targeted for ubiquitin conjugation and/or that the EDBD mediates a protein interaction that is essential for Tel ubiquitination. Indeed, the EDBDs of all known Ets proteins harbor four highly conserved lysine residues (three of which make contact with DNA) (25) that may be substrates for ubiquitin conjugation. Precise definition, by mutagenesis, of the exact site of ubiquitination is hampered by redundancy (M. Alloul-Ramdhani, M. Op Den Brouw, and D. A. Baker, unpublished data); however, improved antibodies and mass spectrometry techniques should, in time, resolve this issue. Collectively, these results show that Tel is ubiquitinated and that ubiquitination requires both the Tel SAM domain and the EDBD.
Fbl6 stimulates Tel downregulation and ubiquitination.
We next explored the role of Fbl6 in Tel ubiquitination. Several lines of evidence support a role for Fbl6 in promoting Tel downregulation and ubiquitination. First, deletion of the SAM domain, which is essential for Fbl6 binding to Tel (Fig. 1B), abolished Tel ubiquitination (Fig. 2B). Second, ectopic expression of wild-type Fbl6 stimulated Tel ubiquitination, whereas Fbl6 harboring mutations that disrupt the interaction with Tel (Fig. 1C) failed to augment Tel ubiquitination (Fig. 2D). Furthermore, in contrast to wild-type Tel, Fbl6 failed to stimulate ubiquitination of either Tel with the SAM domain deleted (TelΔSAM), which is unable to bind Fbl6, or TelΔEDBD. Third, shRNA-mediated knockdown of Fbl6 both stabilized steady-state endogenous Tel protein levels (Fig. 2E) and inhibited ubiquitin conjugation of Tel (Fig. 2F). If ubiquitination of Tel is indeed F-box dependent, one should expect Tel to associate with SKP1, which is a core component of SCF ubiquitin ligases, and one should also expect SKP1 to play a role in regulating steady-state Tel protein levels. Figure 2E shows that endogenous Tel and endogenous SKP1 were copurified as a complex from tissue culture cells, and it also shows that, similarly to lowering fbl6 levels, the shRNA-mediated knockdown of SKP1 stabilized steady-state endogenous Tel protein levels. In cells, antibodies specific for endogenous Tel detect two differently migrating protein species (29, 43), and the higher-molecular-weight form is likely to represent phosphorylated Tel (29). Ectopic expression of Tel also produces two differently migrating proteins, and the higher-molecular-weight Tel form is likely to be phosphorylated Tel because incubation with phosphatase leads to a loss of this band and a concomitant increase in the amount of the faster-migrating, unmodified Tel species (Fig. 2G, upper panel). Figure 2G (lower panel) shows that wild-type Fbl6, but not Fbl6 harboring mutations in either the F-box domain or the leucine-rich regions, strongly promotes the degradation of phosphorylated Tel. This suggests that in cells, phosphorylation of Tel might further sensitize it to Fbl6-mediated downregulation, perhaps by enhancing the affinity of Fbl6 for binding to Tel. Together, these data show that Tel is ubiquitinated and that Fbl6 stimulates Tel downregulation and ubiquitination.
Tel monomers are efficiently ubiquitinated in cells.
Numerous studies have established that the SAM domain is essential for the monomers of Tel to form homotypic oligomers (17, 33, 37). Moreover, we showed earlier that the SAM domain is also indispensable both for the binding of Fbl6 and for Tel ubiquitination (Fig. 1B and 2B). In light of these findings, we compared the ubiquitination of Tel oligomers and that of Tel monomers. First, we established that Fbl6 can associate with both Tel oligomers and Tel monomers. Figure 3A shows that both wild-type Tel and a Tel mutant that is unable to self-associate (TelA*) (37) efficiently interacted with Fbl6 in vitro, whereas TelΔSAM, which is also monomeric, failed to bind Fbl6. In contrast, in cells, wild-type Tel efficiently coimmunoprecipitated with Fbl6, but TelA*, like TelΔSAM, did not (Fig. 3B). However, the interaction between TelA* and Fbl6, but not that between TelΔSAM and Fbl6, was stabilized in the presence of proteasome inhibitors (Fig. 3B), and furthermore, TelA* was far more readily ubiquitinated than the wild-type Tel (Fig. 3C), suggesting that Tel monomers are more labile than Tel oligomers. We recently reported a similar phenomenon with regard to the sumoylation of Tel (37). In cells, Tel is efficiently sumoylated on lysine 11, whereas TelA* is detectably sumoylated on lysine 11 in cells only in the presence of the proteasome inhibitor MG132. Figure 3C shows that the ubiquitination of TelA*, like wild-type Tel ubiquitination, was also stimulated by Fbl6, in contrast to the ubiquitination of TelΔSAM, which is unable to bind Fbl6. Collectively, these findings revealed that Tel monomers can associate with Fbl6 and are efficiently ubiquitinated.
Fbl6 associates with Tel2 and stimulates its ubiquitination.
The SAM domain defines a subfamily of Ets transcription factors, including Tel, Tel2, Ets1, and Ets2 from vertebrates as well as Yan and Pointed P2 from Drosophila (23, 34). Since Fbl6 associates with Tel via its SAM domain, we tested whether Fbl6 also interacts with other SAM domain-containing Ets family members. Figure 3F shows that Fbl6 efficiently associated with both Tel and Tel2 but did not copurify detectably with Ets1 or Ets2, both of which express SAM domains, or PU.1, which does not contain a SAM domain. Tel2 strongly resembles Tel, both structurally and functionally (12, 30, 32), and may have resulted from an earlier gene duplication of the ancestral Tel gene. We found that, like Tel, Tel2 is ubiquitinated and that Fbl6 augments Tel2 ubiquitination (Fig. 3F), suggesting that Tel and Tel2 may share a common F-box-dependent degradation pathway. In the future, it will be of interest to determine whether Fbl6 plays a role in regulating the activities of other factors in vivo.
Drosophila Fbl6 interacts with the Tel orthologue, Yan.
Having established a biochemical and functional interaction between human Tel and Fbl6, we next explored whether this mechanism might be evolutionarily conserved. To that end, we isolated a full-length Drosophila fbl6 (also referred to as CG13213) cDNA from insect Schneider cells. Figure 4A and B show that, like the binding of human Fbl6 to Tel (Fig. 1B and C), Drosophila Fbl6 efficiently associated with Yan, and this interaction depended on the Yan SAM domain (Fig. 4A) and the LRD2 domain of Fbl6 (Fig. 4B). In contrast to Tel-Fbl6 binding, Yan-Fbl6 binding appeared to not require the LRD1 sequence (Fig. 4B).
Fbl6 promotes Yan downregulation and ubiquitination.
Next, we determined whether Fbl6 influences Yan protein stability. To that end, we established a number of stable Schneider S2 cell lines and monitored their endogenous Yan protein levels. We and others have established that in Drosophila, a protein named Mae (modulator of the activity of Ets) orchestrates Yan derepression by binding to Yan, thereby disrupting Yan self-association and binding to DNA and sensitizing it to downregulation that is dependent on mitogen-activated protein kinase (2, 33, 40, 41). Figure 4C shows that ectopic expression of a constitutively active form of Ras alone in Drosophila S2 cells had little detectable effect on steady-state endogenous Yan protein levels. In contrast, ectopic Mae expression alone strongly promoted endogenous Yan downregulation, and this effect was enhanced by activated Ras. Under these conditions, incubation of the cells with MG132 failed to fully rescue Yan protein levels (Roukens and Baker, unpublished), suggesting that Ras/Mae might destabilize Yan by proteasome-independent and proteasome-dependent pathways. However, we found that double-stranded-RNA-mediated knockdown of Fbl6 and SKPA (suppressor of kinetochore protein A), which is a core component of the ubiquitinating machinery (1, 26, 39, 47, 48), inhibited Mae/Ras-dependent Yan downregulation (Fig. 4C). These results suggest that in cells, SKP and Fbl6 play a role in facilitating Yan degradation. To ascertain whether Yan can be ubiquitinated and whether Fbl6 augments Yan ubiquitination, we employed the baculovirus system for Yan expression in insect Sf9 cells (6). Figure 4D shows that ubiquitin was efficiently and covalently conjugated to Yan and that the coexpression of Fbl6 strongly stimulated this ubiquitination. Together, these data suggest that Fbl6 promotes Yan downregulation and stimulates Yan ubiquitination.
Fbl6 regulates Yan protein levels in vivo.
In order to investigate the in vivo role of Fbl6, we monitored Yan protein levels in Drosophila embryos that are lacking expression of fbl6. To that end, we analyzed the enhancer trap line, P{GawB}NP0326, which is viable as a homozygote. Figure 5A schematically shows that P{GawB}NP0326 contains a P element that is inserted into the 5′ untranslated region of fbl6, 60 base pairs upstream of the initiation codon. By precise, transposase-mediated excision of the P element, we recovered several reverted lines that were confirmed by the sequencing of their genomic DNA. Figure 5A shows that while the revertant lines have wild-type levels of the fbl6 transcript, fbl6 expression is strongly reduced in P{GawB}NP0326 embryos. Moreover, P{GawB}NP0326 embryos that were hemizygous for two different chromosomal deletions covering this region also lacked fbl6 expression (Fig. 5B). Band 47F1 on chromosome 2R is a relatively gene-rich region, and we noted two other genes (CG13220 and CG18335) in close proximity to the P-element insertion site in P{GawB}NP0326. Figure 5B shows that whereas expression of fbl6 is disrupted in P{GawB}NP0326 embryos, expression of both CG13220 and CG18335 appears to be unaffected. Together, these analyses suggest that the P element specifically disrupts fbl6. Next, we assessed endogenous Yan protein levels in embryos carefully staged to stage 12. Figure 5C shows that the reverted lines express wild-type Yan protein levels, whereas P{GawB}NP0326 embryos have enhanced Yan protein levels. Coupled to our prior biochemical analysis, this suggests that in vivo, Fbl6 contributes to Yan downregulation. To further validate these findings, we analyzed the expression of argos, an epidermal growth factor receptor signaling antagonist, whose expression is regulated by Yan (7, 8, 9). Consistent with our finding of elevated Yan protein levels in fbl6-deficient embryos, Fig. 5D shows a wave of argos expression during embryo development that is significantly inhibited in P{GawB}NP0326 embryos. Collectively, our analyses suggest that in vivo, Fbl6 participates in the process of Yan downregulation.
Overall, our work suggests that an evolutionarily conserved F-box-protein-dependent degradation pathway participates in the regulation of the activity of both vertebrate Tel and its invertebrate orthologue, Yan. Presumably, this serves to further refine spatiotemporal control of gene expression by these transcriptional repressors.
DISCUSSION
The ancestral Ets repressor Yan and its vertebrate orthologue, Tel (ETV6), play pivotal roles in the control of cell differentiation (15, 19, 22, 27, 35, 36, 45, 46). Because these factors directly and negatively regulate gene expression (19, 22, 27, 35), deciphering the mechanisms that regulate their activity is crucial for understanding the spatiotemporal control of cell differentiation. We recently found that sumoylation of an N-terminal lysine residue encoded by Tel (TelK11) serves to inhibit repression of gene expression by Tel (37). We have further explored Tel/Yan posttranslational regulation, and here we report that both Tel and Yan protein downregulation is promoted by an evolutionarily conserved F-box-protein-dependent mechanism. Specifically, we found that both Tel and Yan can be ubiquitinated and that ubiquitination is facilitated by Fbl6, which sensitizes these proteins for degradation. It is, of course, possible that other F-box proteins may also play a role in regulating Tel/Yan activity, perhaps in a tissue-specific fashion, and future studies should clarify this.
The Tel/Yan SAM domain is required for association with the F-box protein Fbl6.
We found that the SAM domain of both Tel (Fig. 1B) and Yan (Fig. 4A) is required for the binding of Fbl6. The SAM domain defines a subfamily of Ets transcription factors, including Tel, Tel2, Ets1, and Ets2 from vertebrates as well as Yan and Pointed P2 from Drosophila (23, 33), raising the possibility that at least some of these factors are also targeted by Fbl6 or a related F-box protein. Indeed, we found that human Fbl6 also associates with the Tel-related Tel2 protein and promotes its ubiquitination (Fig. 3F). Furthermore, we found biochemical interactions between Drosophila Fbl6 and the SAM domain-containing proteins Pointed P2 (3, 27) (but not P1, which lacks the SAM domain) and Mae (2, 33, 41) in tissue culture cells (Roukens and Baker, unpublished). However, we were unable to detect associations between Fbl6 and either Ets1 or Ets2, each of which harbors a SAM domain, suggesting that there is specificity, perhaps structural, that determines these interactions. Many other families of proteins express SAM domains, such as the polycomb group proteins and components of the mitogen-activated protein kinase cascade (34). It should be interesting to investigate whether Fbl6 (or related F-box proteins) plays a role in regulating their activity.
Tel monomers are especially labile and prone to ubiquitination.
Our data support the idea that Tel/Yan monomers are more susceptible to ubiquitination and degradation than the oligomeric forms of these proteins are. Apparently, and paradoxically, the biochemical interactions between Tel/Yan monomers and Fbl6 were found to be far weaker than the interactions between Tel/Yan oligomers and Fbl6 (Fig. 3B and 4A). However, these interactions were strongly stabilized in the presence of proteasome inhibitors (Fig. 3B), suggesting that the failure to find strong associations between these proteins resulted from the instability of the Tel/Yan monomer and of the Fbl6 complex. Consistent with this, we found that Tel monomers were more readily ubiquitinated than wild-type Tel (Fig. 3C). It is worth noting that, in general, wild-type Tel is detected in cells as a phosphorylated or nonphosphorylated protein species and that the phosphorylation of Tel appears to further sensitize it to Fbl6-mediated degradation (Fig. 2G). In contrast, phosphorylated forms of monomeric Tel are evidently especially unstable and are detected only in the presence of proteasome inhibitors (M. G. Roukens, M. Alloul-Ramdhani, and D. A. Baker, unpublished data). It has previously been described that Tel is negatively regulated by extracellular signal-regulated kinase-induced phosphorylation of serine residues 213 and 257 (24). Phosphorylation of these sites could be a trigger for promoting Tel ubiquitination, although in our ubiquitination assays, we were unable to detect an obvious effect of mutating these residues or an additional putative mitogen-activated protein kinase target, serine residue 22 in the N terminus of Tel (M. Alloul-Ramdhani and D. A. Baker, data not shown). Our previous work also supports the contention that Tel/Yan monomers are relatively labile (37). Monomeric Tel was detectably and very efficiently sumoylated in cells only in the presence of proteasome inhibitors, suggesting that this species of Tel is normally very unstable. The current model of Tel function holds that Tel monomers directly associate via their conserved SAM domains and that the resulting DNA-bound oligomers oppose the transcription-activating apparatus (reviewed in references 34, 42, and 44). Since Fbl6 also interacts with the Tel SAM domain (Fig. 1 and 4), perhaps self-associated Tel oligomers are relatively more resistant to F-box-mediated degradation than are Tel monomers because of reduced accessibility to exposed SAM domains. In the future, it should be of considerable interest to elucidate how Tel sumoylation and ubiquitination are precisely integrated to regulate Tel activity.
Fbl6 regulates Yan protein stability.
Our work suggests that Fbl6 regulates Yan protein levels in vivo. Drosophila embryos that lack fbl6 expression contain significantly elevated levels of Yan protein. Consistent with this, the levels of expression of a downstream target of Yan named argos was sharply reduced. Surprisingly, we were unable to find obvious morphological phenotypes, including during photoreceptor development (such as clear alterations in normal ommatidium development) or wing development, either in embryos, in larvae, or in adult flies that lack fbl6 expression (Alloul-Ramdhani and Baker, unpublished). Moreover, we could not observe any detectable phenotypic consequences for embryos that were deficient in fbl6 expression and also in expressing loss-of-function mutations of mae or the recently identified miR7 microRNA, both of which are also negative regulators of Yan activity in vivo (2, 21, 41, 44; Alloul-Ramdhani and Baker, unpublished). This may reflect genetic redundancy with other F-box-containing proteins or perhaps a broader role for Fbl6 as a regulator of other proteins that might counteract the function of Yan and therefore prevent any obvious mutant phenotypes resulting from elevated Yan protein levels. Indeed, in tissue culture cells, we found biochemical interactions between Fbl6 and known antagonists of Yan function, including the SAM domain-containing proteins Pointed P2 (3, 27) (but not P1, which lacks the SAM domain) and Mae (2, 33, 41) (Roukens and Baker, unpublished). Also, the nature of Tel/Yan degradation may be tissue specific and mediated by distinct or overlapping protein-degrading processes. For example, it has been suggested previously that the PEST sequences of Yan may be determinants of Yan stability (35), which may attract, or act independently of, the F-box-dependent ubiquitination system. In addition, altering the concentration of ubiquitination effectors can have a substantial impact on the function of the targeted proteins, which may influence whether or not a morphological phenotype is discernible. For example, different levels of Mdm2 have profoundly different impacts on p53 function; low levels promote p53 monoubiquitination and subsequent nuclear export, whereas high Mdm2 levels lead to polyubiquitination and proteasomal degradation (20).
Normal cell development and differentiation rely upon an orchestrated program of actions that must be controlled precisely, both spatially and temporally. We describe here an evolutionarily conserved pathway of Tel/Yan downregulation. This complements our recent work on Tel sumoylation and previous studies that delineated the function of these unique repressors (2, 15, 19, 22, 27, 35, 36, 45, 46). Our future work aims to uncover further regulatory features of these crucial cellular mechanisms and to provide a unified framework for how these different processes impinge on Tel/Yan and collaboratively elicit an appropriate cellular response.
Supplementary Material
Acknowledgments
We are indebted to the members of the Department of Molecular and Cellular Biology. We are grateful to Peter ten Dijke and A. G. Jochemsen for their invaluable advice during the course of the work. We are most grateful to those who very kindly provided us with materials, namely Terence Murphy and Bob Duronio for the SKPA antibody, Zhi-Chun Lai (Penn State University) for the Yan antibody, and Richard Carthew for the miR7 fly stocks.
Footnotes
Published ahead of print on 21 April 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bai, C., P. Sen, K. Hofman, L. Ma, M. Goebel, W. Harper, and S. Elledge. 1996. Skp1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86263-274. [DOI] [PubMed] [Google Scholar]
- 2.Baker, D. A., B. Mille-Baker, S. M. Wainwright, D. Ish-Horowicz, and N. J. Dibb. 2001. Mae mediates MAP kinase phosphorylation of Ets transcription factors in Drosophila. Nature 411330-334. [DOI] [PubMed] [Google Scholar]
- 3.Brunner, D., D. K. Ducker, N. Oellers, E. Hafen, H. Scholz, and C. Klambt. 1994. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature 370386-389. [DOI] [PubMed] [Google Scholar]
- 4.Cenciarelli, C., D. S. Chiaur, D. Guardavaccaro, W. Parks, M. Vidal, and M. Pagano. 1999. Identification of a family of human F-box proteins. Curr. Biol. 91177-1179. [DOI] [PubMed] [Google Scholar]
- 5.Chakrabarti, S. R., R. Sood, S. Nandi, and G. Nucifora. 2000. Posttranslational modification of TEL and TELyAML1 by SUMO-1 and cell-cycle-dependent assembly into nuclear bodies. Proc. Natl. Acad. Sci. USA 9713281-13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciccarone, V. C., D. Polayes, and V. A. Luckow. 1997. Generation of recombinant baculovirus DNA in E. coli using baculovirus shuttle vector, p. 213-235, In U. Reischt (ed.), Molecular diagnosis of infectious diseases, vol. 13. Humana Press Inc., Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 7.Freeman, M., C. Klambt, C. S. Goodman, and G. M. Rubin. 1992. The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell 69963-975. [DOI] [PubMed] [Google Scholar]
- 8.Gabay, L., H. Scholz, M. Golembo, A. Klaes, B. Z. Shilo, and C. Klambt. 1996. EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development 1223355-3362. [DOI] [PubMed] [Google Scholar]
- 9.Golembo, M., R. Schweitzer, M. Freeman, and B.-Z. Shilo. 1996. Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development 122223-230. [DOI] [PubMed] [Google Scholar]
- 10.Golub, T. R. 1997. TEL gene rearrangements in myeloid malignancy. Hematol. Oncol. Clin. North Am. 111207-1220. [DOI] [PubMed] [Google Scholar]
- 11.Golub, T. R., G. F. Barker, K. Stegmaier, and G. D. Gilliland. 1997. The TEL gene contributes to the pathogenesis of myeloid and lymphoid leukemias by diverse molecular genetic mechanisms. Curr. Top. Microbiol. Immunol. 22067-79. [DOI] [PubMed] [Google Scholar]
- 12.Gu, X., B.-H. Shin, Y. Akbarali, A. Weiss, J. Boltax, P. Oettgen, and T. A. Libermann. 2001. Tel-2 is a novel transcriptional repressor related to the Ets factor Tel/ETV-6. J. Biol. Chem. 2769421-9436. [DOI] [PubMed] [Google Scholar]
- 13.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67425-479. [DOI] [PubMed] [Google Scholar]
- 14.Ho, M. S., P.-I. Tsai, and C.-T. Chien. 2006. F-box proteins: the key to protein degradation. J. Biomed. Sci. 13181-191. [DOI] [PubMed] [Google Scholar]
- 15.Hock, H., E. Meade, S. Medeiros, J. W. Schindler, P. J. M. Valk, Y. Fujiwara, and S. H. Orkin. 2004. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 182336-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoeller, D., N. Crosetto, B. Blagoev, C. Raiborg, R. Tikkanen, S. Wagner, K. Kowanetz, R. Breitling, M. Mann, H. Stenmark, and I. Dikic. 2006. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat. Cell Biol. 8163-169. [DOI] [PubMed] [Google Scholar]
- 17.Kim, C. A., M. L. Phillips, W. Kim, M. Gingery, H. H. Tran, M. A. Robinson, S. Faham, and J. U. Bowie. 2001. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 204173-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kipreos, E. T., and M. Pagano. 2000. The F-box protein family. Genome Biol. 1reviews3002.1-reviews3002.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai, Z. C., and G. M. Rubin. 1992. Negative control of photoreceptor development in Drosophila by the product of the yan gene, an ETS domain protein. Cell 70609-620. [DOI] [PubMed] [Google Scholar]
- 20.Li, M., C. L. Brooks, F. Wu-Baer, D. Chen, R. Baer, and W. Gu. 2003. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 3021972-1975. [DOI] [PubMed] [Google Scholar]
- 21.Li, X., and R. W. Carthew. 2005. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 1231267-1277. [DOI] [PubMed] [Google Scholar]
- 22.Lopez, R. G., C. Carron, C. Oury, P. Gardellin, O. Bernard, and J. Ghysdael. 1999. TEL is a sequence-specific transcriptional repressor. J. Biol. Chem. 27430132-30138. [DOI] [PubMed] [Google Scholar]
- 23.Mackereth, C. D., M. Schaarpf, L. N. Gentile, S. E. MacIntosh, C. M. Slupsky, and L. P. McIntosh. 2004. Diversity in structure and function of the Ets family PNT domains. J. Mol. Biol. 3421249-1264. [DOI] [PubMed] [Google Scholar]
- 24.Maki, K., H. Arai, K. Waga, K. Sasaki, F. Nakamura, Y. Imai, M. Kurokawa, H. Hirai, and K. Mitani. 2004. Leukemia-related transcription factor TEL is negatively regulated through extracellular signal-regulated kinase-induced phosphorylation. Mol. Cell. Biol. 243227-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mo, Y., B. Vaessen, K. Johnston, and R. Marmorstein. 1998. Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins. Mol. Cell 2201-212. [DOI] [PubMed] [Google Scholar]
- 26.Murphy, T. A. 2003. Drosophila skpA, a component of SCF ubiquitin ligases, regulates centrosome duplication independently of cyclin E accumulation. J. Cell Sci. 1162321-2332. [DOI] [PubMed] [Google Scholar]
- 27.O'Neill, E. M., I. Rebay, R. Tjian, and G. M. Rubin. 1994. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78137-147. [DOI] [PubMed] [Google Scholar]
- 28.Pickart, C. M., and M. J. Eddins. 2004. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 169555-72. [DOI] [PubMed] [Google Scholar]
- 29.Poirel, H., C. Oury, C. Carron, E. Duprez, Y. Laabi, A. Tsapis, S. P. Romana, M. Mauchauffe, M. Le Coniat, R. Berger, J. Ghysdael, and O. A. Bernard. 1997. TEL gene products: nuclear phosphoproteins with DNA binding properties. Oncogene 14349-357. [DOI] [PubMed] [Google Scholar]
- 30.Poirel, H., R. G. Lopez, V. Lacronique, V. Della Valle, M. Mauchauffé, R. Berger, J. Ghysdael, and O. A. Bernard. 2000. Characterization of a novel ETS gene, TELB, encoding a protein structurally and functionally related to TEL. Oncogene 194802-4806. [DOI] [PubMed] [Google Scholar]
- 31.Popov, N., M. Wanzel, M. Madiredjo, D. Zhang, R. Beijersbergen, R. Bernards, R. Moll, S. J. Elledge, and M. Eilers. 2007. The ubiquitin-specific protease USP28 is required for MYC stability. Nat. Cell Biol. 9765-774. [DOI] [PubMed] [Google Scholar]
- 32.Potter, M. D., A. Buijs, B. Kreider, L. van Rompaey, and G. C. Grosveld. 2000. Identification and characterization of a new human ETS-family transcription factor, TEL2, that is expressed in hematopoietic tissues and can associate with TEL1/ETV6. Blood 953341-3348. [PubMed] [Google Scholar]
- 33.Qiao, F., H. Song, C. A. Kim, M. R. Sawaya, J. B. Hunter, M. Gingery, I. Rebay, A. J. Courey, and J. U. Bowie. 2004. Derepression by depolymerization: structural insights into the regulation of Yan by Mae. Cell 118163-173. [DOI] [PubMed] [Google Scholar]
- 34.Qiao, F., and J. U. Bowie. 2005. The many faces of SAM. Sci. STKE 2005re7. [DOI] [PubMed] [Google Scholar]
- 35.Rebay, I., and G. M. Rubin. 1995. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell 81857-866. [DOI] [PubMed] [Google Scholar]
- 36.Rogge, R., P. J. Green, J. Urano, S. Horn-Saban, M. Mlodzik, B.-Z. Shilo, V. Hartenstein, and U. Banerjee. 1995. The role of yan in mediating the choice between cell division and differentiation. Development 1213947-3958. [DOI] [PubMed] [Google Scholar]
- 37.Roukens, M. G., M. Alloul-Ramdhani, A. C. Vertegaal, Z. Anvarian, C. I. Balog, A. M. Deelder, P. J. Hensbergen, and D. A. Baker. 2008. Identification of a new site of sumoylation on Tel (ETV6) uncovers a PIAS-dependent mode of regulating Tel function. Mol. Cell. Biol. 282342-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki, K., Y. Nakamura, K. Maki, K. Waga, F. Nakamura, H. Arai, Y. Imai, H. Hirai, and K. Mitani. 2004. Functional analysis of a dominant-negative DETS TEL/ETV6 isoform. Biochem. Biophys. Res. Commun. 3171128-1137. [DOI] [PubMed] [Google Scholar]
- 39.Schulman, B. A., A. C. Carrano, P. D. Jeffrey, Z. Bowen, E. R. Kinnucan, M. S. Finnin, S. J. Elledge, J. W. Harper, M. Pagano, and N. P. Pavletich. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408381-386. [DOI] [PubMed] [Google Scholar]
- 40.Song, H., M. Nie, F. Qiao, J. U. Bowie, and A. J. Courey. 2005. Antagonistic regulation of Yan nuclear export by Mae and Crm1 may increase the stringency of the Ras response. Genes Dev. 191767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tootle, T. L., P. S. Lee, and I. Rebay. 2003. CRM1-mediated nuclear export and regulated activity of the receptor tyrosine kinase antagonist YAN require specific interactions with MAE. Development 130845-857. [DOI] [PubMed] [Google Scholar]
- 42.Tootle, T. L., and I. Rebay. 2005. Post-translational modifications influence transcription factor activity: a view from the ETS superfamily. Bioessays 27285-298. [DOI] [PubMed] [Google Scholar]
- 43.van Waalwijk van Doorn-Khosrovani, S. B., D. Spensberger, Y. de Knegt, M. Tang, B. Löwenberg, and R. Delwel. 2005. Somatic heterozygous mutations in ETV6 (TEL) and frequent absence of ETV6 protein in acute myeloid leukemia. Oncogene 244129-4137. [DOI] [PubMed] [Google Scholar]
- 44.Vivekanand, P., and I. Rebay. 2006. Intersection of signal transduction pathways and development. Annu. Rev. Genet. 40139-157. [DOI] [PubMed] [Google Scholar]
- 45.Wang, L. C., F. Kuo, Y. Fujiwara, D. G. Gilliland, T. R. Golub, and S. H. Orkin. 1997. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J. 164374-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, L. C., W. Swat, Y. Fujiwara, L. Davidson, J. Visvader, F. Kuo, F. W. Alt, D. G. Gilliland, T. R. Golub, and S. H. Orkin. 1998. The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes Dev. 122392-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2169-178. [DOI] [PubMed] [Google Scholar]
- 48.Willems, A. R., M. Schwab, and M. Tyers. 2004. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695133-170. [DOI] [PubMed] [Google Scholar]
- 49.Wood, L. D., B. J. Irvin, G. Nucifora, K. S. Luce, and S. W. Hiebert. 2003. Small ubiquitin-like modifier conjugation regulates nuclear export of TEL, a putative tumor suppressor. Proc. Natl. Acad. Sci. USA 1003257-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.