Abstract
The nucleolar protein nucleostemin (NS) is essential for cell proliferation and early embryogenesis. Both depletion and overexpression of NS reduce cell proliferation. However, the mechanisms underlying this regulation are still unclear. Here, we show that NS regulates p53 activity through the inhibition of MDM2. NS binds to the central acidic domain of MDM2 and inhibits MDM2-mediated p53 ubiquitylation and degradation. Consequently, ectopic overexpression of NS activates p53, induces G1 cell cycle arrest, and inhibits cell proliferation. Interestingly, the knockdown of NS by small interfering RNA also activates p53 and induces G1 arrest. These effects require the ribosomal proteins L5 and L11, since the depletion of NS enhanced their interactions with MDM2 and the knockdown of L5 or L11 abrogated the NS depletion-induced p53 activation and cell cycle arrest. These results suggest that a p53-dependent cell cycle checkpoint monitors changes of cellular NS levels via the impediment of MDM2 function.
The tumor suppressor protein p53 responds to diverse stresses to regulate many target genes whose protein products induce cell cycle arrest, apoptosis, senescence, and DNA repair (28, 48). In unstressed cells, p53 is maintained at low levels by its inhibitor, MDM2, an E3 ubiquitin ligase that ubiquitylates and targets p53 for proteasome-mediated degradation (12, 15, 16, 20) through a feedback mechanism (33, 49). In response to various stresses, p53 is stabilized and activated largely through the inhibition of MDM2. These stresses include DNA damage and oncogenic and nucleolar stresses (22). DNA damage induces phosphorylation of both p53 and MDM2, unties the p53-MDM2 feedback loop, and consequently activates p53 (2, 5, 25, 39, 41). Oncogenic stress induces the expression of the tumor suppressor protein ARF, which in turn prevents MDM2 from targeting p53, leading to p53 activation (29, 40, 55). Recently, a number of external and internal insults were shown to induce nucleolar stress by disrupting the nucleolar structure, activating p53 (37). Also, the perturbation of ribosomal biogenesis by interfering with rRNA synthesis, processing, and ribosome assembly, as exemplified by the treatment of actinomycin D or fluorouracil (5-FU) (1, 13, 43), serum starvation (4), expression of dominant-negative mutant Bop1 (32), or genetic disruption of ribosomal proteins S6 and TIF-IA (31, 51), can cause ribosomal stress and subsequent p53 activation. It is now believed that in response to ribosomal stress caused by some of the aforementioned agents, several nucleolar proteins, including B23 (also called nucleophosmin) (21) and C23 (also called nucleolin) (38), and ribosomal proteins, such as L5, L11, L23, and S7 (6, 8-10, 18, 23, 52), can bind to MDM2 directly and inhibit its activity toward p53, resulting in p53 activation. Hence, these findings demonstrate a critical role for these nucleolar proteins in transmitting nucleolar stress signals to p53 and initiating p53-dependent cell growth arrest or apoptosis.
Interestingly, another recently identified nucleolar protein called nucleostemin (NS) has been shown to regulate cell cycle progression as well. NS possesses two putative GTP-binding motifs and can be readily detected in neural stem cells, embryonic stem cells, hematopoietic primitive cells, and tumor cell lines (46). However, the level of NS decreases drastically prior to cell cycle exit upon differentiation of the stem cells, suggesting that this protein may play a crucial role in stem cell proliferation (46). Indeed, NS is essential for early embryogenesis as deletion of this gene caused embryonic lethality at E3.5 (3, 54). Consistently, the depletion of NS levels by small interfering RNA (siRNA) in cultured cells also blocks cell cycle progression (24, 46). Astonishingly, the overexpression of NS in cultured neural stem cells, cancer cells, and mouse embryonic fibroblasts interferes with cell cycle progression as well. These studies suggest that the physiological level of NS is so finely monitored that its imbalance would be detrimental to cell proliferation (3, 46, 54). Interestingly, the overexpression of NS was shown to arrest cell proliferation possibly by acting on p53 via the N-terminal basic domain of NS and this NS domain was also required for NS-induced cell death (46). However, how NS activates p53 and whether it does so by suppressing MDM2 activity remain unknown. In addition, it is unclear how the reduction of NS levels brings about cell cycle arrest as well.
In an attempt to address the above questions, we found that both the overexpression and the reduction of NS activate p53 through inhibition of MDM2 activity. On one hand, NS, when overexpressed, stabilized p53 and induced p53-dependent cell cycle arrest by directly binding to MDM2 and inhibiting its suppression activity toward p53. On the other hand, the knockdown of NS by siRNA induced p53 activation via enhancement of the binding of the ribosomal proteins L5 and L11 to MDM2. These results not only reveal a novel and critical role for NS in the regulation of MDM2 function but also divulge another role for L11 and L5 in transmitting the stress signal resulting from the loss of NS to the p53-MDM2 pathway.
MATERIALS AND METHODS
Cell lines, plasmids, and antibodies.
Human H1299, U2OS, and SJSA cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin, and 0.1 mg/ml streptomycin at 37°C in a 5% CO2 humidified atmosphere, as described previously (10). An expression plasmid encoding Flag-tagged NS (Flag-NS) was constructed by inserting full-length NS cDNA into a pcDNA3-2Flag vector at BamHI and XbaI sites. The cDNA was amplified by PCR from an NS cDNA clone (Clontech), using the primers 5′-CGCGGATCCATGAAAAGGCCTAAGTTAAAG-3′ (P1) and 5′-CGCTCTAGATTACACATAATCTGTACTGAAG-3′ (P2). All the Flag-tagged deletion mutants of NS were generated using PCR and were cloned into the pcDNA3-2Flag vector. The Flag-NS with double point mutations in the G1 motif (Flag-NSG1dm; Gly 261 to Val and Gly 265 to Val) was generated by a two-step site-specific mutagenesis. The primers used were 5′-CGGGTTGGAGTAATTGTTTTCCCAAATGTGGGG-3′ and 5′-CCCCACATTTGGGAAAACAATTACTCCAACCCG-3′ for the G261V mutation and 5′-GTTTTCCCAAATGTGGTGAAAAGCAGCATTATC-3′ and 5′-GATAATGCTGCTTTTCACCACATTTGGGAAAAC-3′ for the G265V mutation. The green fluorescence protein-NS (GFP-NS) expression plasmid was cloned by inserting full-length NS cDNA amplified by PCR into the pEGFP-C1 (Clontech) vector at the BglII and HindIII sites. The primers used were P1 and 5′-CCCAAGCTTGGGTTACACATAATCTGTACTGAAG-3′ (P3). GFP-NS with double point mutations in the G1 motif (GFP-NSG1dm) and GFP-NSct, containing C-terminal amino acids 268 to 549 of NS, were cloned by PCR using Flag-NSG1dm and Flag-NSct as templates, respectively. The bacterial expression vector pet24a-His-NS was constructed by inserting PCR-amplified full-length NS cDNA into pet24a (Novagen) at the BamHI and XhoI sites. The primers used were P1 and 5′-CGCCTCGAGCACATAATCTGTACTGAAGTC-3′ (P4). The glutathione S-transferase (GST)-NS bacterial expression vector was constructed by subcloning the full-length NS from pet24a-His-NS into the pEGX.4T.1 vector (Pharmacia Biotech). The GST fusion NS fragments pEGX.4T.1-NS47-270 and pEGX.4T.1-NSct were cloned by inserting PCR products into the pEGX.4T.1 vector at the BamHI and XhoI sites. The hemagglutinin-MDM2 (HA-MDM2) expression vector and the pet24a-His-MDM2 plasmid have been described previously (10). The V5-tagged deletion mutants of MDM2 were constructed by inserting PCR products into the pcDNAs-V5 vector. The plasmid encoding V5-tagged p14ARF (designated ARF hereafter) has been described previously (9). Anti-L5 (8), anti-L11 (7), anti-HA, and anti-MDM2 (2A10 and 4B11) (8, 10) antibodies have been previously described. Anti-p21 (NeoMarkers), anti-p53 (DO-1, Santa Cruz Biotechnology, Santa Cruz, CA), anti-MDM2 (SAM14, Santa Cruz Biotechnology), anti-NS (Chemicon), anti-Flag (Sigma), and anti-V5 (Invitrogen) were purchased. Rabbit polyclonal anti-NS antibodies were generated by using purified His-tagged full-length NS protein expressed in Escherichia coli as an antigen.
Cotransfection, IB, and co-IP analyses.
Cells were transfected with plasmids as described in the legends to Fig. 1 to 4, using TransIT-LT1 reagents, following the manufacturer's protocol (Mirus Bio Corp.). Cells were harvested at 48 h posttransfection and lysed in lysis buffer consisting of 50 mM Tris-HCl (pH 8.0), 0.5% Nonidet P-40, 1 mM EDTA, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 0.25 μg/ml pepstatin A, and 1 mM leupeptin. Equal amounts of cleared cell lysates were used for immunoblotting (IB) analysis as described previously (10). Coimmunoprecipitation (co-IP) assays were conducted as described previously (10). Bound proteins were detected by IB using antibodies as described in the legends to Fig. 1, 2, and 6.
FIG. 1.
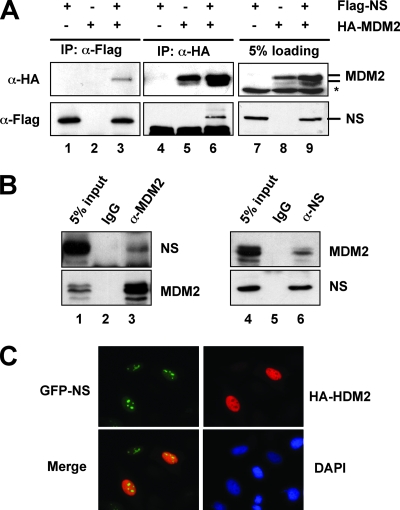
NS interacts with MDM2 in cells. (A) Ectopically expressed NS interacts with ectopically expressed MDM2 in cells. H1299 cells were transfected with Flag-NS and HA-MDM2 individually or together. Cell lysates were immunoprecipitated with anti-Flag (lanes 1 to 3) or anti-HA (lanes 4 to 6) antibodies, followed by IB with anti-Flag or anti-HA antibodies. Asterisk, nonspecific anti-HA antibody-reacting bands. (B) Endogenous NS interacts with endogenous MDM2 in cells. Cell lysates from SJSA cells were immunoprecipitated with monoclonal anti-MDM2 (4B11) antibodies (lane 3) or mouse immunoglobulin G (IgG; lane 2), polyclonal anti-NS antibodies (lane 6), or rabbit IgG (lane 5), followed by anti-MDM2 and anti-NS antibodies. (C) NS colocalizes with MDM2 in the nucleoplasm. H1299 cells were cotransfected with GFP-NS and HA-MDM2. Forty-eight hours posttransfection, the cells were immunostained with anti-MDM2 antibody, followed by staining with goat anti-mouse secondary antibody (red) and DAPI.
FIG. 4.
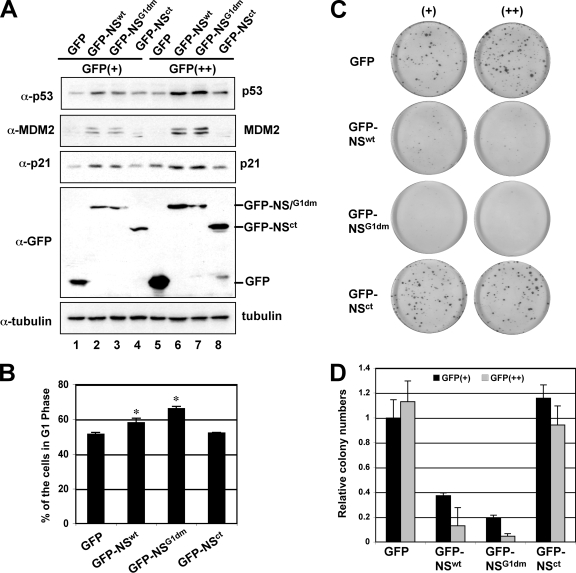
Overexpression of NS activates p53, induces G1 arrest, and inhibits cell proliferation. (A) Overexpression of NS induces and activates p53. U2OS cells were transfected with the GFP, GFP-NS, GFP-NSG1dm, or GFP-NSct plasmid. The GFP-expressing cells were then sorted by FACS into GFP dim-positive (GFP+) and GFP bright-positive (GFP++) populations. Gating scales are shown in Fig. S2B in the supplemental material. The cell lysates were assayed by IB for the expression of p53, MDM2, p21, and GFP-NS. (B) Overexpression of NS induces G1 arrest. U2OS cells were transfected with GFP, GFP-NS, GFP-NSG1dm, or GFP-NSct. Forty-eight hours posttransfection, the GFP-expressing cells were gated for cell cycle analysis. The mean percentage of cells in G1 phase is shown. The histograms of PI staining from one representative experiment are shown in Fig. S2C in the supplemental material. Asterisk, P < 0.01, compared to that of GFP. (C and D). Overexpression of NS inhibits cell proliferation, as determined by colony formation assay. U2OS cells were transfected with the GFP, GFP-NS, GFP-NSG1dm, or GFP-NSct plasmid and sorted into GFP+ and GFP++ populations as described for panel A. Equal numbers of cells were plated and grown in media containing G418 for 3 to 4 weeks. The average numbers of colonies are shown in panel D.
FIG. 2.
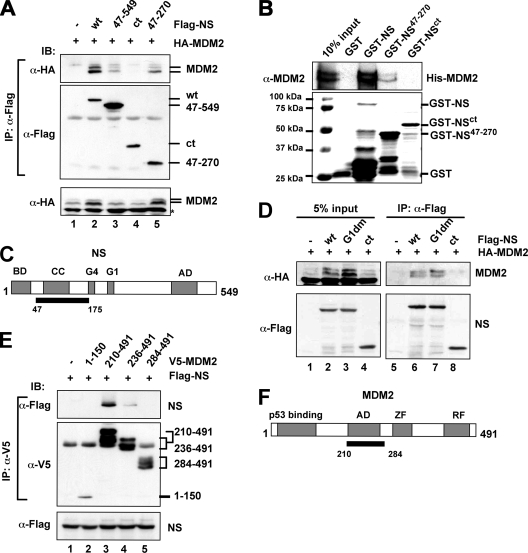
The coiled-coil domain-containing region of NS binds to the central acidic domain of MDM2. (A) MDM2 binds to the coiled-coil domain-containing region of NS in cells. H1299 cells were transfected with Flag-tagged full-length NS (wt) or its deletion mutants, together with the HA-MDM2 plasmid. Cell lysates were immunoprecipitated with the anti-Flag antibody, followed by IB using the indicated antibodies. The lysates were also loaded directly onto a sodium dodecyl sulfate (SDS) polyacrylamide gel for IB, using the anti-HA antibody (bottom panel). (B) MDM2 binds to NS in vitro. About 200 ng of purified GST alone, full-length GST-NS, or GST-NS deletion mutants immobilized on glutathione beads was incubated with 200 ng of His-MDM2 purified from bacteria. Bound MDM2 was detected by IB with anti-MDM2 antibodies. The GST-NS fusion proteins were visualized by Coomassie blue staining. (C) Schematic diagram of NS protein indicates the MDM2-binding domain (black bar). BD, basic domain; AD, acidic domain; CC, coiled-coil domain; G4 and G1, putative GTP binding motifs. (D) GTP binding activity of NS is not required for NS binding to MDM2. H1299 cells were transfected with HA-MDM2, together with Flag-NS, Flag-NSG1dm, Flag-NSct, or the control Flag vectors. Cell lysates were immunoprecipitated with the anti-Flag antibody, followed by IB using the indicated antibodies. The lysates were also loaded directly onto an SDS gel for IB using the indicated antibodies (left panels). (E) NS binds to the central acidic domain of MDM2. H1299 cells were transfected with plasmids encoding V5-tagged MDM2 fragments, together with the indicated Flag-NS plasmid. Cell lysates were immunoprecipitated with an anti-V5 antibody, followed by IB using the indicated antibodies. The lysates were also loaded directly onto an SDS gel for IB using anti-Flag antibody (bottom panel). (F) Schematic diagram of MDM2 protein indicating the NS binding acidic domain (AD) (black bar). ZF, zinc finger domain; RF, ring finger domain.
FIG. 6.
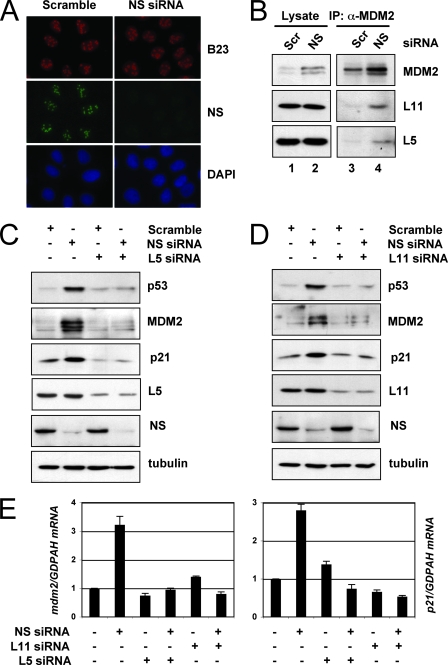
The knockdown of NS does not lead to nucleolar disruption, but its activation of p53 requires the ribosomal proteins L5 and L11. (A) The knockdown of endogenous NS does not disrupt the nucleolus. U2OS cells were transfected with scrambled (Scr) or NS siRNA as indicated. The cells were then stained with anti-B23 (red) and anti-NS (green) antibodies, as well as with DAPI for DNA. (B) The knockdown of endogenous NS enhances the interaction of MDM2 with L5 and L11. U2OS cells were transfected with scrambled or NS siRNA. Cell lysates were subjected to co-IP using anti-MDM2 (4B11) antibodies, followed by IB to detect the level of L11, L5, and MDM2. (C) The knockdown of L5 abolished the induction of p53 by the knockdown of NS. U2OS cells were transfected with scrambled siRNA, with NS siRNA, or with L5 siRNA as indicated. Cell lysates were subjected to IB to detect the expression of p53, MDM2, p21, L5, or NS, as indicated. (D) The knockdown of L11 abolished the induction of p53 by the knockdown of NS. U2OS cells were transfected with scrambled siRNA, with NS siRNA, or with L11 siRNA as indicated. The cell lysates were subjected to IB to detect the expression of p53, MDM2, p21, L11, or NS, as indicated. (E) The knockdown of L5 or L11 abolished the induced expression of the p21 and mdm2 mRNA by the knockdown of NS. U2OS cells were transfected with scrambled siRNA, NS siRNA, L5 siRNA, or L11 siRNA as indicated. Total RNAs were extracted and subjected to RT reactions, followed by real-time PCR assays. Relative expression of the p21 and mdm2 genes was normalized against the expression of GAPDH.
GST fusion protein association assays.
The His-tagged MDM2 protein was expressed in E. coli and purified through a Ni-nitrilotriacetic acid (Qiagen) column and eluted by 0.5 M imidazole. GST-NS, GST-NS47-270, and GST-NSct proteins were purified using glutathione-Sepharose 4B beads. Protein-protein interaction assays were conducted as described previously, using fusion protein-containing glutathione beads (10). Purified GST-NS, GST-NS47-270, GST-NSct, or GST proteins (200 ng) were incubated with His-MDM2 (200 ng) protein for 30 min at room temperature. After bound proteins were washed, they were analyzed by IB using monoclonal anti-MDM2 (2A10 and 4B11) antibodies.
In vivo ubiquitylation assays.
H1299 cells were transfected with plasmids as described in the legend to Fig. 3. The cells were treated with 20 μM of MG132 for 6 h before they were harvested and then harvested at 36 h posttransfection. In vivo ubiquitylation assays were conducted as described previously (10). Eluted proteins were analyzed by IB with anti-p53 (DO-1) antibodies.
FIG. 3.
NS inhibits MDM2-mediated p53 ubiquitylation and degradation. (A) NS inhibits MDM2-mediated p53 degradation. H1299 cells were transfected with the indicated plasmids. Cell lysates were subjected to IB using antibodies as indicated on the left. (B) NS stabilizes MDM2 independently of p53. H1299 cells were transfected with HA-MDM2 in the presence of Flag-NS, Flag-NSG1dm, Flag-NSct, or the control Flag plasmids. Cell lysates were immunoprecipitated with the anti-Flag antibody, followed by IB using the indicated antibodies. Cell lysates were subjected to IB using the antibodies indicated on the left. (C) NS inhibits MDM2-mediated p53 ubiquitylation. H1299 cells were transfected with the indicated plasmids. The transfected cells were treated with MG132 (20 μM) for 6 h before they were harvested. Ubiquitylated p53 species were detected by IB with the anti-p53 (DO-1) antibody (upper panel). Ubiquitylated p53 [p53-(Ub)n] is indicated. The expression of total p53, MDM2, and NS proteins is shown in the lower panels. Asterisk indicates nonspecific anti-HA antibody-reacting bands.
RNA interference (RNAi).
The RNAi-mediated knockdown of endogenous L5, L11, and NS was performed essentially as described previously (10). The target sequences for L5, L11, and the control scrambled II RNA have been described previously (8, 10). The target sequences for NS were 5′-AAGCTGTACTGCCAAGAAC-3′ (NS siRNA-1, used for all experiments except where indicated) and 5′-AAGAACTAAAACAGCAGCAGA-3′ (NS siRNA-2) (46). All the siRNA duplexes with a 3′ dTdT overhang were synthesized by Dharmacon (Lafayette, CO). These siRNA duplexes (0.2 μM) were introduced into cells by using SilentFect (Bio-Rad), following the manufacturer's protocol. Cells were harvested at 48 to 72 h after transfection for IB, immunofluorescence (IF) staining, reverse transcription (RT)-real-time PCR, and cell cycle analyses.
Real-time RT-PCR analyses.
Total RNA was isolated from cells by using Qiagen RNeasy minikits (Qiagen, Valencia, CA). RTs were performed as described previously (8). Quantitative real-time PCR was performed on an ABI 7300 real-time PCR system (Applied Biosystems) using Sybr green mix (Applied Biosystems) as described previously (43). All reactions were carried out in triplicate. Relative gene expression was calculated by using the cycle threshold method, following the manufacturer's instructions. The primers for p21, mdm2, and GAPDH have been described previously (43).
Cell cycle analysis.
U2OS cells were transfected with plasmids or siRNAs as described in legends to Fig. 4 to 7. Cells transfected with GFP, GFP-NS, GFP-NSG1dm, or GFP-NSct were stained with 500 μl of propidium iodide (PI; Sigma) stain buffer (50 μg/ml PI, 30 μg/ml polyethylene glycol 8000, 200 μg/ml RNase A, 0.1% Triton X-100, 0.38 M NaCl [pH 7.2]) at 37°C for 30 min. GFP-positive cells were gated and analyzed for DNA content, using a Becton Dickinson FACScan flow cytometer. Data were analyzed using CellQuest and Modfit software programs.
FIG. 7.
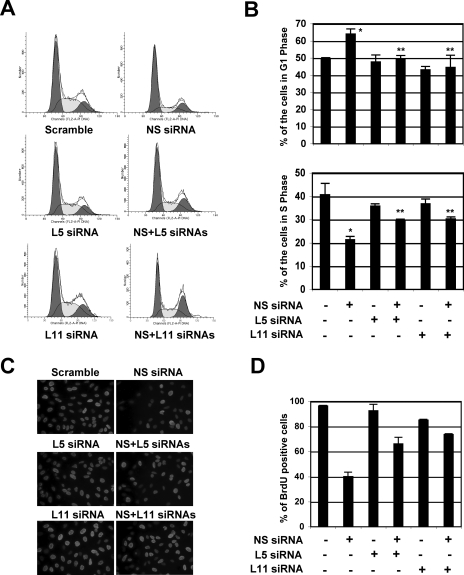
NS knockdown-induced G1 arrest requires the ribosomal proteins L5 and L11. (A and B) U2OS cells were transfected with scrambled, NS, L5, or L11 siRNA as indicated. Seventy-two hours posttransfection, the transfected cells were harvested and stained with PI for cell cycle analysis. The histograms of PI staining from one representative experiment are shown in panel A. The mean percentages of cells in G1 or S phase are shown in panel B. *, P < 0.01 compared to scrambled RNA-transfected cells; **, P < 0.01 compared to NS siRNA-transfected cells. (C and D) Knocking down L5 or L11 partially abolished the NS knockdown-mediated inhibition of cell proliferation. U2OS cells were transfected with scrambled, NS, L5, or L11 siRNA, as indicated. The cells were incubated with BrdU at 48 h posttransfection for another 20 h. The cells were then fixed and stained with anti-BrdU antibodies (red) and DAPI (blue) (E). The average of the BrdU-positive cells is shown in panel F.
BrdU incorporation assays.
Bromodeoxyuridine (BrdU) incorporation assays were conducted as described previously (24). Cells were incubated in the presence of 10 μM BrdU for 20 h. Cells were then fixed with 95% ethanol and 5% acetic acid and treated with 2 M HCl containing 1% Triton X-100. The cells were stained with monoclonal anti-BrdU (Roche) antibody and then stained with Alexa Fluor 546 (red) goat anti-mouse antibodies and 4′,6′-diamidino-2-phenylindole (DAPI). Stained cells were analyzed with a Zeiss Axiovert 25 fluorescence microscope.
Fluorescence-activated cell sorting (FACS) analysis.
To isolate cells expressing GFP, GFP-NS, GFP-NSG1dm, or GFP-NSct proteins, U2OS cells were transfected with the above-described individual plasmids for 48 h and then subjected to cell sorting on a flow cytometer (FacstarPlus; Becton Dickinson) based on GFP fluorescence intensity. GFP-expressing cells were separated into dim-positive (+) and bright-positive (++) cells. Sorting gates are shown in Fig. S2 in the supplemental material.
Cell proliferation assay.
For visual examination of cell proliferation rates, equal amounts of GFP-positive U2OS cells expressing GFP, GFP-NS, GFP-NSG1dm, or GFP-NSct proteins, sorted as described above, were plated in DMEM containing 10% FBS and 0.5 mg/ml G418. Medium was changed every 5 days. After cells were cultured for 3 to 4 weeks, they were stained with 0.5% crystal violet in 50% ethanol for colony analysis.
RESULTS
NS interacts with MDM2 in cells and in vitro.
It has been shown that the overexpression of NS inhibits cell proliferation (46, 54). This effect may be partially attributed to its ability to bind p53 (46). To gain further insight into the mechanisms underlying this inhibition, we sought to test whether NS also interacts with MDM2, because MDM2 is a master regulator of p53 (12, 15, 16, 20). To do so, we cotransfected p53-deficient human lung non-small cell carcinoma H1299 cells with Flag-NS and HA-MDM2, followed by co-IP-IB assays. As shown in Fig. 1A, MDM2 was specifically coimmunoprecipitated with NS by the anti-Flag antibody in cells cotransfected with both HA-MDM2 and Flag-NS (Fig. 1A, lane 3). Conversely, NS was specifically coimmunoprecipitated with MDM2 by the anti-HA antibody in cells cotransfected with HA-MDM2 (Fig. 1A, lane 6). To determine whether endogenous NS and MDM2 also interact with each other, cell lysates from SJSA cells were used because these cells express high levels of MDM2 (8). As shown in Fig. 1B, endogenous NS and MDM2 were specifically coimmunoprecipitated by antibodies against either of these two proteins. These results suggest that NS interacts with MDM2 in cells. To determine whether their binding is direct or not, GST fusion protein-protein association assays were conducted using His-tagged MDM2 and GST-NS proteins purified from bacteria. As shown in Fig. 2B, GST-NS (Fig. 2B, lane 3), but not GST alone (Fig. 2B, lane 2), interacted directly with His-MDM2, verifying that NS does directly bind to MDM2 in vitro.
NS shuttles constantly between the nucleolus and the nucleoplasm in a GTP-regulated manner (45). To determine in which compartment NS interacts with MDM2, H1299 cells were cotransfected with GFP-NS and HA-MDM2, followed by IF staining using anti-MDM2 antibodies. As shown in Fig. 1C, ectopic MDM2 was expressed primarily in the nucleoplasm, while GFP-NS was expressed predominantly in the nucleolus, with a portion of GFP-NS also present in the nucleoplasm. We did not observe significant relocalization of MDM2 into the nucleolus by GFP-NS (data not shown). These results suggest that NS likely interacts with MDM2 in the nucleoplasm.
MDM2 binds to the coiled-coil domain-containing region of NS.
To further characterize the MDM2-NS interaction, we first tried to map the MDM2 binding domain in the NS protein by generating a set of Flag-tagged deletion mutants of NS. Wild-type (wt) Flag-NS and its deletion mutants were cotransfected with HA-MDM2, followed by co-IP assays. As shown in Fig. 2A, HA-MDM2 bound to both of the NS deletion mutants, NS47-549 (Fig. 2A, lane 3) and NS47-270 (Fig. 2A, lane 5), though to a lesser extent than it did to the wt NS (lane 2). In contrast, the deletion mutant NSct containing amino acids 268 to 549 did not bind to MDM2 (Fig. 2A, lane 4). This was also confirmed in an in vitro GST pull-down assay. As shown in Fig. 2B, His-MDM2 bound to the GST-NS47-270 fragment (Fig. 2B, lane 4), although less efficiently than it did to the GST-fused wt NS (Fig. 2B, lane 3), but not to the GST-NSct mutant (Fig. 2B, lane 5) or to GST alone (Fig. 2B, lane 2). Because an N-terminal fragment (NS1-175) containing the basic domain and the coiled-coil domain, but not GTP-binding motifs, still bound to MDM2 (see Fig. S1A in the supplemental material), the minimal domain of NS required for MDM2 binding encompasses the internal N-terminal amino acids 47 to 175. These results suggest that MDM2 binds to the central coiled-coil domain-containing region of NS (Fig. 2C).
NS is a putative GTP-binding protein and contains two GTP-binding motifs (G4 and G1). Its cellular localization and possibly its activity appear to be regulated by intracellular GTP levels (45, 46). A point mutation in the conserved G1 motif, G256V, in rat NS (G261 in human NS) completely abolished the GTP-binding activity of NS (45). To determine whether GTP binding regulates NS interaction with MDM2, we mutated two critical residues, Gly 261 and 265, in the G1 motif to valine (G261V and G265V), producing the NSG1dm mutant. Using similar transfection-co-IP assays, we found that the Flag-NSG1dm mutant, but not the N-terminally deleted mutant (Flag-NSct), coimmunoprecipitated with MDM2 as efficiently as the Flag-tagged wt NS did (Fig. 2D). Consistent with the above result, these results show that the GTP-binding domain is not required for NS to bind to MDM2, thus suggesting that the MDM2-NS interaction is independent of the GTP-binding activity of NS.
NS binds to the central acidic domain of MDM2.
To map the NS binding domain in MDM2, we also generated a panel of V5-tagged MDM2 deletion mutants. Similar assays were performed using anti-V5 antibodies. As shown in Fig. 2E, a mutant deleted of the N-terminal 210 residues (MDM2210-491) was coimmunoprecipitated with Flag-NS (Fig. 2E, lane 3). Further deletion of a portion of the acidic domain (MDM2236-491) markedly reduced the binding of MDM2 to NS (Fig. 2E, lane 4), while further deletion of the entire acidic domain (MDM2284-491) abolished the MDM2-NS binding (Fig. 2E, lane 5). Also, the N-terminal fragment containing the p53-binding domain (MDM21-150) did not bind to NS. These results suggest that the central acidic domain of MDM2 is essential for binding to NS (Fig. 2F). It has been shown that p53 binds to NS at the N-terminal basic domain (45). Thus, p53 and MDM2 bind to two different domains of NS. Consistent with this, we observed that NS formed a ternary complex with both p53 and MDM2 in cells when all the proteins were expressed (data not shown).
The central acidic domain has been shown to be critical for MDM2-mediated p53 degradation (19, 26). Indeed, several MDM2 regulators, including the well-studied tumor suppressor ARF (53), suppress MDM2 activity by binding to this acidic domain. Although both ARF and NS bind to the same central region of MDM2, they did not appear to compete with each other for binding to MDM2 (see Fig. S1B in the supplemental material). Instead, these three proteins could form a complex in cells when they are overexpressed (data not shown). These results suggest that ARF and NS might bind to distinct domains within the central region of MDM2 or to different monomers of the MDM2 oligomers, as it was recently shown that MDM2 could form a homo-oligomer (35, 47).
Overexpression of NS inhibits MDM2-mediated p53 ubiquitylation and degradation.
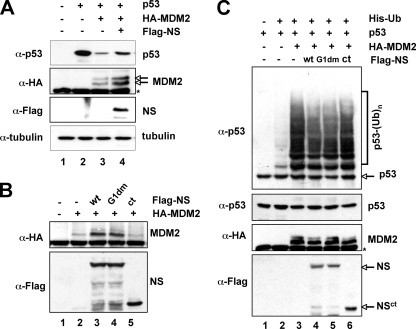
Next, we wanted to determine if NS binding regulates MDM2-mediated p53 ubiquitylation and degradation. We introduced exogenous proteins into H1299 cells as shown in Fig. 3A. As expected (12, 15, 16, 20), overexpression of MDM2 remarkably reduced p53 levels (Fig. 3A, lane 3). In contrast, further expression of NS partially rescued MDM2-mediated p53 degradation (Fig. 3A, lane 4). Interestingly, as for other MDM2 regulators (9, 23, 50), overexpression of NS also stabilized MDM2 independently of p53 (Fig. 3B, lane 3; Fig. 2D, lane 2; and Fig. 1A, lane 9). These results suggest that excess expression of NS blocks MDM2-mediated degradation of p53 and MDM2 itself. To determine if this stabilization of p53 is due to the inhibition of MDM2-mediated p53 ubiquitylation, H1299 cells were transfected with exogenous MDM2, p53, and NS and treated with the proteasome inhibitor MG132 for 6 h for in vivo ubiquitylation assays. As shown in Fig. 3C and by the findings of others (12, 15, 16), MDM2 ubiquitylated p53 (Fig. 3C, lane 2). In contrast, the expression of NS (Fig. 3C, lane 3) inhibited MDM2-mediated p53 ubiquitylation. These results indicate that NS can stabilize p53 by alleviating MDM2-mediated p53 ubiquitylation and degradation.
To determine whether the inhibition of MDM2-mediated p53 ubiquitylation by NS requires its MDM2- or GTP-binding activity, Flag-NSG1dm (G261V and G265V), which is defective in GTP binding, but not in MDM2 binding, and the MDM2-binding-defective Flag-NSct mutant (Fig. 2D) were used as controls for the in vivo ubiquitylation assays. As shown in Fig. 3C, the Flag-NSG1dm, but not Flag-NSct, mutant inhibited MDM2-mediated ubiquitylation, suggesting that the MDM2-binding, but not GTP-binding, activity of NS is crucial for NS inhibition of MDM2 activity toward p53. Consistently, the Flag-NSG1dm, but not Flag-NSct, mutant stabilized MDM2 independently of p53 in cells (Fig. 3B, lanes 4 and 5). This effect is similar to that of ARF and L11 (9, 50).
Overexpression of NS activates endogenous p53 and induces G1 cell cycle arrest.
To determine whether overexpression of NS induces and activates endogenous p53, we first introduced Flag-NS into human osteosarcoma U2OS cells that contain wt p53. Interestingly, overexpression of Flag-NS induced p53 in a dose-dependent fashion (see Fig. S2A in the supplemental material). Correspondingly, the levels of the p53 targets, p21cip1 (designated p21 hereafter) and MDM2, were also induced (see Fig. S2A in the supplemental material). Consistently, overexpression of NS prolonged the half-life of endogenous p53 in U2OS cells (data not shown). To further confirm the effect of NS overexpression on p53-dependent suppression of cell proliferation, we performed GFP-based cell sorting assays. U2OS cells were transfected with GFP, GFP-NS, GFP-NSG1dm, or GFP-NSct. Forty-eight hours posttransfection, GFP-expressing cells were sorted by a flow cytometer based on GFP fluorescence intensity and separated into two populations: GFP dim-positive (GFP+) and GFP bright-positive (GFP++) cells. The gating scale is shown in Fig. S2B in the supplemental material. Cell lysates were used for IB. As shown in Fig. 4A, overexpression of GFP-NS (Fig. 4A, lanes 2 and 6) and GFP-NSG1dm (Fig. 4A, lanes 3 and 7), but not GFP-NSct (Fig. 4A, lanes 4 and 8), induced p53 levels in a dose-dependent manner, compared to those of GFP-expressing cells (Fig. 4A, lanes 1 and 5). Higher expression levels of GFP-NS and GFP-NSG1dm induced higher levels of p53 (Fig. 4A, compare lanes 6 and 7 to 2 and 3). The induced p53 was transcriptionally active as p21 and MDM2 levels were also induced in GFP-NS- and GFP-NSG1dm- expressing cells, but not GFP-NSct-expressing cells, in a dose-dependent manner (Fig. 4A). These results, together with the results from Fig. 3, suggest that overexpression of NS induces and activates p53 by blocking MDM2-mediated p53 degradation and that this effect requires the binding of NS to MDM2 but does not require the GTP-binding activity of NS.
Next, to test whether the p53 and p21 induced by NS mediates cell cycle arrest, we transfected U2OS cells with GFP-NS, GFP-NSG1dm, GFP-NSct, and GFP as a control and then gated GFP-positive cells for cell cycle analysis. As shown in a representative histogram in Fig. S2C in the supplemental material and summarized in Fig. 4B, a significant portion of cells expressing GFP-NS (58.1% ± 2.5%, P < 0.01) and GFP-NSG1dm (66.2% ± 1.3%, P < 0.01), but not GFP-NSct (52.2% ± 0.3%), were arrested in the G1 phase compared with that of GFP-expressing cells (51.4% ± 1.1%). Therefore, ectopic overexpression of NS induces G1 cell cycle arrest.
To determine whether overexpression of NS inhibits cell proliferation, we conducted colony formation assays. Equal amounts of the above-described sorted GFP, GFP-NS, GFP-NSG1dm, and GFP-NSct cells (both the GFP+ and the GFP++ cells) were plated in medium containing G418. As shown in Fig. 4C and D, expression of GFP-NS and GFP-NSG1dm, but not GFP-NSct, inhibited the colony formation in a dose-dependent manner, compared to that of cells expressing GFP alone. Of note, the induction of G1 arrest (Fig. 4B) and the inhibition of cell proliferation (Fig. 4C and D) by GFP-NSG1dm were much more efficient than that of the wt GFP-NS-expressing cells. These results are consistent with the observation by Tsai and McKay, who showed that the GTP-binding-defective mutants of NS are those that are actually the most efficient at inducing cell cycle exit and p53-dependent apoptosis (46). Taken together, our results suggest that an aberrantly high level of NS by overexpression significantly reduced cell proliferation rate and thus is detrimental to cells.
Knockdown of NS by siRNA also induces p53 and G1 cell cycle arrest.
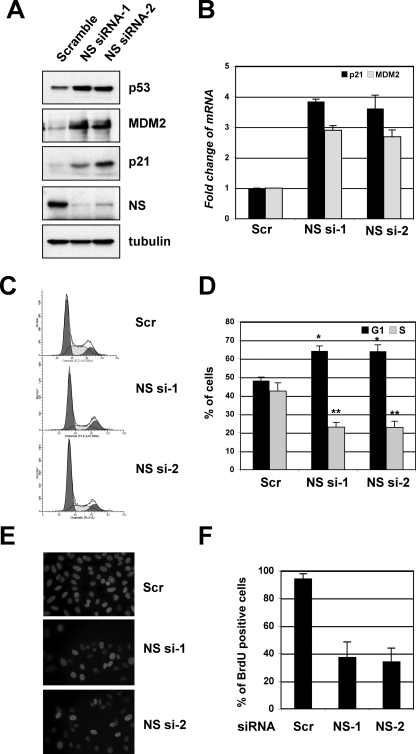
To determine the physiological relevance of NS in the regulation of the MDM2-p53 pathway, we performed siRNA-mediated knockdown experiments. Surprisingly, the knockdown of endogenous NS also induced p53 levels, as well as the levels of the p53 targets p21 and MDM2 (Fig. 5A). p53 activation by the knockdown of NS was confirmed by two siRNAs against two different sequences in the NS gene; thus, this effect was less likely an off-target effect of siRNA transfection. Consistently, the knockdown of NS induced the mRNA expression of p21 and MDM2, as determined by RT-real-time PCR assays (Fig. 5B). Furthermore, the knockdown of NS by two individual siRNAs drastically induced cell cycle arrest in the G1 phase (Fig. 5C and D) and inhibited cell proliferation, as determined by BrdU incorporation assays (Fig. 5E and F). These results show that the depletion of NS also induces p53 and G1 arrest.
FIG. 5.
Knockdown of endogenous NS activates p53 and induces G1 cell cycle arrest. (A) The knockdown of NS induces p53 levels. U2OS cells were transfected with scrambled siRNA or with one of the two NS siRNAs against different sequences. Cell lysates were assayed for the expression of p53, MDM2, p21, and NS, as indicated, by using IB assays. (B) The knockdown of endogenous NS increases the mRNA expression of the p53 target genes mdm2 and p21. Total RNAs were extracted from U2OS cells transfected with siRNAs as in panel A and subjected to RT reactions, followed by real-time PCR assays. The relative expression of the p21 and mdm2 genes was normalized against the expression of GAPDH. Scr, scrambled RNA-transfected cells. (C and D) The knockdown of endogenous NS induces G1 arrest. U2OS cells were transfected with siRNA, as described in panel A. Seventy-two hours posttransfection, the cells were harvested and stained with PI for cell cycle analysis. The histograms of PI staining from one representative experiment are shown in panel C. The mean percentages of cells in G1 or S phase are shown in panel D. *, P < 0.05; **, P < 0.01, compared to the scrambled RNA-transfected cells. (E and F) The knockdown of endogenous NS reduces cell proliferation. U2OS cells were transfected with scrambled siRNA or with one of the two above-described NS siRNAs. The cells were incubated with BrdU at 48 h posttransfection for another 20 h. The cells were then fixed and stained with anti-BrdU antibodies (red) and DAPI (blue) (E). The average of BrdU-positive cells is shown in panel F.
Knockdown of NS by siRNA does not disrupt the nucleolus but enhances the interaction of MDM2 with ribosomal proteins L5 and L11.
NS is an essential nucleolar protein that shuttles constantly between the nucleolus and the nucleoplasm (45) and is expressed in most proliferating cells (46). Treatment of cells with either actinomycin D or MPA redistributes NS from the nucleolus to the nucleoplasm (45). Thus, it is possible that NS is an integrated component of the nucleolus and that depletion of NS could trigger a nucleolar stress that, in turn, activates p53. Because nucleolar stresses usually lead to the disruption of the nucleolus (37), we first examined the nucleolus integrity upon the NS knockdown by using IF staining. To our surprise, the knockdown of NS did not significantly alter the nucleolar structure, as determined by the nucleolar marker B23 (Fig. 6A). This result is consistent with a previous observation showing that NS deficiency does not cause nucleolar disruption in embryonic blastocysts (3).
Next, we determined whether the knockdown of NS by siRNA could still activate the ribosomal protein-MDM2-p53 pathway without disrupting the nucleolus. Here, we investigated only the ribosomal proteins L5 and L11, but not L23, since the knockdown of L23 itself also induces p53 (10, 18), by performing transfection of U2OS cells with scrambled or NS siRNAs, followed by co-IP assays with anti-MDM2 antibodies. As shown in Fig. 6B, the NS knockdown indeed enhanced the interaction between MDM2 and L5 or L11. These results suggest that the interaction between MDM2 and L5 or L11 is also responsive to the depletion of NS by siRNA.
Knockdown of L5 or L11 by siRNA alleviates NS depletion-induced p53 activation, G1 arrest, and cell proliferation.
The enhanced interaction of MDM2 with L5 and L11 suggests that these ribosomal proteins may play a role in NS knockdown-induced p53 activation. Therefore, we determined if the reduction of the L5 or L11 level by siRNAs would influence the NS knockdown-induced p53 level and activity in U2OS cells. As shown in Fig. 6, reduction of either the L5 (Fig. 6C) or the L11 (Fig. 6D) level by siRNA markedly inhibited the NS knockdown-induced levels of p53 compared with that of the scrambled RNA-transfected cells. Consistently, the induction of p21 and MDM2 protein levels by the NS knockdown was drastically impaired by siRNA against L5 or L11 but not by the scrambled sequence (Fig. 6C and D). This impairment was also true for the mRNA levels of p21 and MDM2, as measured by real-time RT-PCR assays (Fig. 6E). In line with this, the ablation of either L5 or L11 by siRNA partially, but significantly, reduced the NS knockdown-induced G1 arrest (Fig. 7A and B). Also, the knockdown of either L5 or L11 significantly, though partially, relieved the NS knockdown-mediated inhibition of cell proliferation, as determined by BrdU incorporation assays (Fig, 7C and D). These results reveal that L5 and L11 also play a role in NS knockdown-caused p53 activation and inhibition of cell proliferation.
DISCUSSION
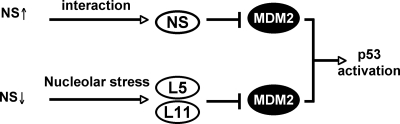
It has been shown that a physiologically optimal level of NS is essential for normal cell homeostasis. Both aberrant overexpression and depletion of NS inhibit cell proliferation (46, 54). In this study, we have further illustrated the mechanisms underlying the role of NS in cell cycle regulation. We show that both aberrantly high and low levels of NS induce G1 arrest and reduce cell proliferation rates through the activation of p53 as a result of the inhibition of MDM2. Upon ectopic overexpression, excess NS molecules bind directly to MDM2 and inhibit MDM2-mediated p53 ubiquitylation and degradation. When the level of NS is significantly reduced by siRNA, a stress occurs, leading to the association of ribosomal proteins L5 and L11 with MDM2 and subsequently the inhibition of MDM2 function. As a result, p53 is activated in both cases (Fig. 8). Therefore, p53 acts as a key cell cycle checkpoint that senses the aberrant cellular levels of NS via distinct mechanisms that funnel into the inhibition of MDM2.
FIG. 8.
Schematic model illustrating potential mechanisms underlying p53 activation by aberrantly high levels of NS upon overexpression or by siRNA-caused low levels of NS (see text for further discussion).
Our domain mapping studies show that NS binds to the central acidic region of MDM2. This region has been shown to be critical for mediating p53 degradation (19, 26). Indeed, many MDM2 regulatory proteins, such as L5, L23, and ARF, target this region and regulate MDM2-mediated p53 turnover (10, 11, 18, 27, 42). Similar to the effect of these proteins on MDM2 function, the binding of NS to MDM2 also leads to the inhibition of E3 ligase activity of MDM2 toward p53, and thus, consequent p53 activation. Also, overexpression of NS markedly inhibits MDM2 degradation. Hence, these effects are reminiscent of the role of ARF in regulating the p53-MDM2 pathway.
Since MDM2 locates mostly in the nucleoplasm (36, 44), while NS resides predominantly in the nucleolus (45, 46), how do these proteins interact in cells? It is possible that under normal growth conditions, the steady-state nucleoplasmic NS is extremely low and would not significantly interact with MDM2. That is why we can detect only a weak binding for the endogenous proteins in normally cultured cells. However, when NS is overexpressed, a significant portion of the protein is accumulated in the nucleoplasm and thus is able to interact with MDM2. Consistent with this, the GTP-binding-defective mutant GFP-NSG1dm, which was expressed mostly in nucleoplasm while retaining partial nucleolus localization (data not shown), interacted with MDM2 somewhat more efficiently than did wt NS (Fig. 2D). The dynamic nature of both proteins in nuclear shuttling would allow their interaction to respond to cell growth and stress signals. It is conceivable that in response to certain cellular stresses, NS may be redistributed from the nucleolus to the nucleoplasm, where it can target MDM2.
It has been shown that p53 can bind to NS at the N-terminal basic domain (46), whereas here we show that MDM2 interacts with the coiled-coil domain of NS. Together, these studies suggest that NS could bind to p53 and MDM2 simultaneously. Indeed, we detected a ternary complex that contains all three proteins (data not shown). Thus, MDM2 does not compete with p53 for NS binding. This result is similar to the regulation of the MDM2-p53 pathway by other nucleolar proteins, such as ARF and ribosomal proteins, which do not disrupt the p53-MDM2 binding. Likewise, they associate with both p53 and MDM2, and inhibit MDM2-mediated p53 ubiquitylation and degradation (8-10, 17, 18, 27, 52).
Since the role of NS in regulating the p53-MDM2 circuit is similar to that of ARF, one question would be whether NS possesses intrinsic tumor suppressor activity. NS is highly expressed in proliferative cells, and it is conceivably responsive to high proliferative signals. Our data suggest that abnormally high levels of NS are detrimental to cells due to p53 activation by NS via inhibition of MDM2. Thus, it is possible that NS may provide cells with a surveillance mechanism to check uncontrolled cell growth and tumorigenesis. However, to our knowledge, no genetic alterations such as mutations or deletions in the NS gene have been documented so far. This might be partially because that loss of NS function also activates p53, and mice deleted of NS are embryonic lethal (see discussion below). Chromosome translocation, mutation, and deletion in the B23 locus have been reported in leukemias and lymphomas (14). Thus, it would be interesting and worthwhile to screen genetic alterations of the NS gene in primary cancers in the future.
Similar to the overexpression of NS, depletion of NS by siRNA also induces p53. While the manuscript for this report was in preparation, Ma and Pederson also showed that knocking down NS by siRNA induced p53-dependent G1 cell cycle arrest (24). Our further analysis reveals that ribosomal proteins L5 and L11 are required for the NS depletion-induced p53 levels, activation, and cell cycle arrest, suggesting that depletion of NS triggers a stress that activates p53. Unlike other nucleolar stresses such as those induced by treatment of 5-FU, actinomycin D, or the genetic knockdown of TIA-IA (23, 43, 51), but like the case of S6 knockdown (30), the knockdown of NS by siRNA did not apparently disrupt the nucleolus, which is consistent with the observation that NS deficiency does not cause nucleolar disruption in embryonic blastocysts (3). This raises a question whether the nucleolar disruption is absolutely required for a nucleolar stress to occur. Because NS is localized at the distinct region of the nucleolus that is not actively involved in the ribosomal biogenesis (34), how NS depletion causes nucleolar stress remains to be determined. Alternatively, it is possible that the reduction of NS by siRNA may cause a nucleolus-independent stress that also activates the L5/L11-MDM2-p53 pathway, as our results clearly show that the ribosomal proteins L5 and L11 are required for NS depletion-induced p53 activation. In addition, NS depletion by siRNA enhances the interaction of MDM2 with L5 and L11. Another speculation would be that NS may regulate the nucleolar import of ribosomal proteins for ribosome assembly and that the depletion of NS would lead to the accumulation of free L5 and L11 in the nucleoplasm, where they find MDM2 to interact with. These speculations remain to be investigated in the future.
Intriguingly, heterozygous deletion of NS in mice did not trigger significant p53 activation (54). This result suggests that the loss of one copy of the NS gene might not reduce NS to a level that would sufficiently trigger a stress. Also, a p53 knockout did not rescue the lethality by the NS knockout in mice (3), suggesting that NS may have p53-independent function essential for early embryogenesis. Consistently, the knockdown of L5 and L11 only partially rescued the NS knockdown-induced cell cycle arrest and inhibition of cell proliferation (Fig. 7), although the induction of p53 by NS siRNA treatment was completely abolished by knockdown of L5 or L11 (Fig. 6). Uncovering this p53-independent function would provide a complete picture for better understanding the role of NS in cell proliferation.
Supplementary Material
Acknowledgments
We thank Jayme Gallegos for proofreading the manuscript and other members of the laboratory for active discussion and Lizz Scaletta for assistance with FACS.
M.-S.D. is supported by NIH/NCI grant K99-CA127134 and a Biomedical Research Grant (BRG) from Indiana University. This work is supported by grants to H.L. from NIH/NCI (CA095441, CA93614, and CA079721).
Footnotes
Published ahead of print on 21 April 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ashcroft, M., Y. Taya, and K. H. Vousden. 2000. Stress signals utilize multiple pathways to stabilize p53. Mol. Cell. Biol. 203224-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 2811674-1677. [DOI] [PubMed] [Google Scholar]
- 3.Beekman, C., M. Nichane, S. De Clercq, M. Maetens, T. Floss, W. Wurst, E. Bellefroid, and J. C. Marine. 2006. Evolutionarily conserved role of nucleostemin: controlling proliferation of stem/progenitor cells during early vertebrate development. Mol. Cell. Biol. 269291-9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhat, K. P., K. Itahana, A. Jin, and Y. Zhang. 2004. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 232402-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 2811677-1679. [DOI] [PubMed] [Google Scholar]
- 6.Chen, D., Z. Zhang, M. Li, W. Wang, Y. Li, E. R. Rayburn, D. L. Hill, H. Wang, and R. Zhang. 2007. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 265029-5037. [DOI] [PubMed] [Google Scholar]
- 7.Dai, M. S., H. Arnold, X. X. Sun, R. Sears, and H. Lu. 2007. Inhibition of c-Myc activity by ribosomal protein L11. EMBO J. 263332-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai, M. S., and H. Lu. 2004. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 27944475-44482. [DOI] [PubMed] [Google Scholar]
- 9.Dai, M. S., D. Shi, Y. Jin, X. X. Sun, Y. Zhang, S. R. Grossman, and H. Lu. 2006. Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. J. Biol. Chem. 28124304-24313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai, M. S., S. X. Zeng, Y. Jin, X. X. Sun, L. David, and H. Lu. 2004. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 247654-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elenbaas, B., M. Dobbelstein, J. Roth, T. Shenk, and A. J. Levine. 1996. The MDM2 oncoprotein binds specifically to RNA through its RING finger domain. Mol. Med. 2439-451. [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, S., J. P. Jensen, R. L. Ludwig, K. H. Vousden, and A. M. Weissman. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 2758945-8951. [DOI] [PubMed] [Google Scholar]
- 13.Gilkes, D. M., L. Chen, and J. Chen. 2006. MDMX regulation of p53 response to ribosomal stress. EMBO J. 255614-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grisendi, S., C. Mecucci, B. Falini, and P. P. Pandolfi. 2006. Nucleophosmin and cancer. Nat. Rev. Cancer 6493-505. [DOI] [PubMed] [Google Scholar]
- 15.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387296-299. [DOI] [PubMed] [Google Scholar]
- 16.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 42025-27. [DOI] [PubMed] [Google Scholar]
- 17.Honda, R., and H. Yasuda. 1999. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1822-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin, A., K. Itahana, K. O'Keefe, and Y. Zhang. 2004. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 247669-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai, H., D. Wiederschain, and Z. M. Yuan. 2003. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol. Cell. Biol. 234939-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387299-303. [DOI] [PubMed] [Google Scholar]
- 21.Kurki, S., K. Peltonen, L. Latonen, T. M. Kiviharju, P. M. Ojala, D. Meek, and M. Laiho. 2004. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 5465-475. [DOI] [PubMed] [Google Scholar]
- 22.Lindstrom, M. S., C. Deisenroth, and Y. Zhang. 2007. Putting a finger on growth surveillance: insight into MDM2 zinc finger-ribosomal protein interactions. Cell Cycle 6434-437. [DOI] [PubMed] [Google Scholar]
- 23.Lohrum, M. A., R. L. Ludwig, M. H. Kubbutat, M. Hanlon, and K. H. Vousden. 2003. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3577-587. [DOI] [PubMed] [Google Scholar]
- 24.Ma, H., and T. Pederson. 2007. Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol. Biol. Cell 182630-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maya, R., M. Balass, S. T. Kim, D. Shkedy, J. F. Leal, O. Shifman, M. Moas, T. Buschmann, Z. Ronai, Y. Shiloh, M. B. Kastan, E. Katzir, and M. Oren. 2001. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 151067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meulmeester, E., R. Frenk, R. Stad, P. de Graaf, J. C. Marine, K. H. Vousden, and A. G. Jochemsen. 2003. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol. Cell. Biol. 234929-4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Midgley, C. A., J. M. Desterro, M. K. Saville, S. Howard, A. Sparks, R. T. Hay, and D. P. Lane. 2000. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 192312-2323. [DOI] [PubMed] [Google Scholar]
- 28.Oren, M. 2003. Decision making by p53: life, death and cancer. Cell Death Differ. 10431-442. [DOI] [PubMed] [Google Scholar]
- 29.Palmero, I., C. Pantoja, and M. Serrano. 1998. p19ARF links the tumour suppressor p53 to Ras. Nature 395125-126. [DOI] [PubMed] [Google Scholar]
- 30.Panic, L., J. Montagne, M. Cokaric, and S. Volarevic. 2007. S6-haploinsufficiency activates the p53 tumor suppressor. Cell Cycle 620-24. [DOI] [PubMed] [Google Scholar]
- 31.Panić, L., S. Tamarut, M. Sticker-Jantscheff, M. Barkić, D. Solter, M. Uzelac, K. Grabus¡ić, and S. Volarević. 2006. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol. Cell. Biol. 268880-8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pestov, D. G., Z. Strezoska, and L. F. Lau. 2001. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G1/S transition. Mol. Cell. Biol. 214246-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picksley, S. M., and D. P. Lane. 1993. The p53-mdm2 autoregulatory feedback loop: a paradigm for the regulation of growth control by p53? Bioessays 15689-690. [DOI] [PubMed] [Google Scholar]
- 34.Politz, J. C., I. Polena, I. Trask, D. P. Bazett-Jones, and T. Pederson. 2005. A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol. Biol. Cell 163401-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poyurovsky, M. V., C. Priest, A. Kentsis, K. L. Borden, Z. Q. Pan, N. Pavletich, and C. Prives. 2007. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2690-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth, J., M. Dobbelstein, D. A. Freedman, T. Shenk, and A. J. Levine. 1998. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 17554-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubbi, C. P., and J. Milner. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 226068-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saxena, A., C. J. Rorie, D. Dimitrova, Y. Daniely, and J. A. Borowiec. 2006. Nucleolin inhibits Hdm2 by multiple pathways leading to p53 stabilization. Oncogene 257274-7288. [DOI] [PubMed] [Google Scholar]
- 39.Schon, O., A. Friedler, M. Bycroft, S. M. Freund, and A. R. Fersht. 2002. Molecular mechanism of the interaction between MDM2 and p53. J. Mol. Biol. 323491-501. [DOI] [PubMed] [Google Scholar]
- 40.Sherr, C. J., and J. D. Weber. 2000. The ARF/p53 pathway. Curr. Opin. Genet. Dev. 1094-99. [DOI] [PubMed] [Google Scholar]
- 41.Siliciano, J. D., C. E. Canman, Y. Taya, K. Sakaguchi, E. Appella, and M. B. Kastan. 1997. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 113471-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stott, F. J., S. Bates, M. C. James, B. B. McConnell, M. Starborg, S. Brookes, I. Palmero, K. Ryan, E. Hara, K. H. Vousden, and G. Peters. 1998. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 175001-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun, X. X., M. S. Dai, and H. Lu. 2007. 5-Fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J. Biol. Chem. 2828052-8059. [DOI] [PubMed] [Google Scholar]
- 44.Tao, W., and A. J. Levine. 1999. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc. Natl. Acad. Sci. USA 963077-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai, R. Y., and R. D. McKay. 2005. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J. Cell Biol. 168179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai, R. Y., and R. D. McKay. 2002. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 162991-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uldrijan, S., W. J. Pannekoek, and K. H. Vousden. 2007. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 26102-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408307-310. [DOI] [PubMed] [Google Scholar]
- 49.Wu, X., J. H. Bayle, D. Olson, and A. J. Levine. 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 71126-1132. [DOI] [PubMed] [Google Scholar]
- 50.Xirodimas, D., M. K. Saville, C. Edling, D. P. Lane, and S. Lain. 2001. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene 204972-4983. [DOI] [PubMed] [Google Scholar]
- 51.Yuan, X., Y. Zhou, E. Casanova, M. Chai, E. Kiss, H. J. Grone, G. Schutz, and I. Grummt. 2005. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol. Cell 1977-87. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Y., G. W. Wolf, K. Bhat, A. Jin, T. Allio, W. A. Burkhart, and Y. Xiong. 2003. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 238902-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, Y., and Y. Xiong. 2001. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 12175-186. [PubMed] [Google Scholar]
- 54.Zhu, Q., H. Yasumoto, and R. Y. Tsai. 2006. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol. Cell. Biol. 269279-9290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 122424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.