Abstract
Human papillomaviruses (HPVs) are involved in the pathogenesis of cancer of the cervix (CaCx). MicroRNA (miRNA) expression analysis using Ambion arrays showed that three miRNAs were overexpressed and 24 underexpressed in cervical cell lines containing integrated HPV-16 DNA compared to the normal cervix. Furthermore, nine miRNAs were overexpressed and one underexpressed in integrated HPV-16 cell lines compared to the HPV-negative CaCx cell line C-33A. Based on microarray and/or quantitative real-time PCR and Northern blot analyses, microRNA-218 (miR-218) was specifically underexpressed in HPV-positive cell lines, cervical lesions and cancer tissues containing HPV-16 DNA compared to both C-33A and the normal cervix. Expression of the E6 oncogene of high-risk HPV-16, but not that of low-risk HPV-6, reduced miR-218 expression, and conversely, RNA interference of E6/E7 oncogenes in an HPV-16 positive cell line increased miR-218 expression. We also demonstrate that the epithelial-cell specific marker LAMB3 is a target of miR-218. We also show that LAMB3 expression is increased in the presence of the HPV-16 E6 oncogene and this effect is mediated through miR-218. These findings may contribute to a better understanding of the molecular mechanisms involved in cervical carcinogenesis.
Keywords: HPVs, microRNA, cervical cancer, oncogene
Introduction
High-risk human papillomaviruses (HPVs) such as types 16 and 18 are causally involved in cervical cancer (zur Hausen, 2002). HPVs are small double-stranded DNA viruses that contain two oncogenes E6 and E7 that are involved in cellular transformation. Most low-grade cervical lesions contain HPV DNA in an episomal state, but in most cases of cervical carcinomas the HPV DNA is found integrated into the host chromosomes, increasing expression of E6 and E7 (Munger and Howley, 2002; Hebner and Laimins, 2006). The E6 protein promotes ubiquitination and proteasomal degradation of the tumor suppressor protein p53 (Huibregtse et al., 1991; Lechner et al., 1992; Band et al., 1993; Thomas et al., 1999) and PDZ domain-containing disc large protein (DLG) (Gardiol et al., 1999). The E7 protein binds to and inactivates the function of the pRB and related tumor suppressor proteins p107 and p130 (Munger and Howley, 2002). E7 also interacts with additional cellular proteins such as TBP, histone H1 kinase and cyclin E (Massimi et al., 1996; Hebner and Laimins, 2006). In addition, E6/E7 expression promotes chromosomal instability, foreign DNA integration and other mutagenic events in the cell (Duensing et al., 2000; Hebner and Laimins, 2006).
MicroRNAs (miRNAs) are small non-coding RNAs that may regulate thousands of mRNA targets (for reviews, see Lewis et al., 2005; Calin and Croce, 2006). MiRNAs are transcribed in the nucleus and after processing they associate with the RISC complex and act as negative regulators of gene expression by binding to their complementary mRNA targets and either repressing translation or promoting mRNA degradation (Kim, 2005). Recently, changes in the expression of miRNAs have been shown to be associated with a variety of human cancers (Calin and Croce, 2006).
In this study, we demonstrate differential expression of several miRNAs in HPV-16 positive cervical cell lines and tissues, as well as in the HPV-18 positive cell line HeLa, compared to the normal cervical tissue and an HPV-negative cervical carcinoma cell line. We also demonstrate that miR-218 and the tumor suppressor gene SLIT2 are specifically downregulated in several HPV-16 positive cervical cell lines and tissues, and this effect is mediated by the E6 oncogene of high-risk HPV-16. Finally, our studies show that LAMB3 is a possible target of miR-218 at the transcriptional level.
Results
Differential expression of microRNAs in cervical cell lines compared to the normal cervix and the HPV-negative cell line C-33A
MiRNA microarray analysis showed that approximately 220 known human miRNAs out of 328 represented on the array were expressed in the normal cervix (Supplementary Table 1). The miRNAs that were most highly expressed in the cervix were miR-145, miR-26a, miR-99a, let-7a, miR-143, let-7b, let-7c, miR-125b, miR-126, and miR-195 in that order. We investigated the miRNA expression profile in normal cervical tissue and cervical carcinoma cell lines SiHa and CaSki containing integrated HPV-16 DNA. We also used two clonal derivatives, 20861 and 201402, of the W12 cell line derived from a low-grade CIN I lesion (Stanley et al., 1989) which contain integrated HPV-16 DNA (Alazawi et al., 2002). SAM analysis of the array data showed that 24 miRNAs, including miR-126, miR-143, miR-145 and miR-195 (four of the ten most highly expressed miRNAs in the normal cervix) were underexpressed in all the integrated HPV-16 cervical cell lines compared to the normal cervix (Table 1). Only three miRNAs, miR-182, miR-183 and miR-210, were found to be overexpressed in the integrated HPV-16 cell lines (Table 1). The array data for individual cell lines compared to the normal cervix is presented in Supplementary Table 2. In the 20863 cell line (a clonal derivative of the W12 cell line) containing episomal HPV-16 DNA, six miRNAs were underexpressed compared to the normal cervix (Supplementary Table 2). A direct comparison of the miRNA expression profiles of integrated vs. episomal HPV-16 cell lines did not reveal any significant differences (data not shown). This, at least in part, may result from the use of only one cell line containing episomal HPV-16 DNA, as well as the use of a 2-fold cut-off value and a q value of zero in our statistical analysis.
Table 1.
MiRNAs differentially expressed in HPV-16 positive cell lines compared to the normal cervix
| MiRNA | HPV-16 integrated Folda |
|---|---|
| Overexpressed | |
| hsa_miR_210 | 7.3 |
| hsa_miR_182 | 6.4 |
| hsa_miR_183 | 5.1 |
| Underexpressed | |
| hsa_miR_126 | −14.5 |
| hsa_miR_145 | −14.1 |
| hsa_miR_451 | −11.3 |
| ambi_miR_7029 | −9.5 |
| hsa_miR_195 | −8.3 |
| hsa_miR_143 | −8 |
| hsa_miR_199b | −7.9 |
| hsa_miR_133a | −7.6 |
| hsa_miR_368 | −7.6 |
| hsa_miR_1 | −7.2 |
| hsa_miR_495 | −6.3 |
| hsa_miR_497 | −6.2 |
| hsa_miR_133b | −6 |
| hsa_miR_223 | −5.4 |
| hsa_miR_146a | −5.1 |
| hsa_miR_218 | −4.8 |
| hsa_miR_126_AS | −4.7 |
| hsa_miR_150 | −4.4 |
| hsa_miR_376a | −4.2 |
| hsa_miR_214 | −4.1 |
| hsa_miR_487b | −4.1 |
| hsa_miR_10b | −3.9 |
| ambi_miR_5021 | −3.6 |
| ambi_miR_7070 | −3.5 |
Mean fold-changes in HPV-16 integrated cell lines CaSki, SiHa, 20861 and 201402.
The q value of all miRNAs was 0.
MicroRNA expression profile of the HPV-18 containing HeLa cell line showed that 14 miRNAs were underexpressed in HeLa cells compared to the normal cervix (Supplementary Table 3). Eight of these miRNAs (miR-1, miR-133b, miR-143, miR-145, miR-214, miR-368, miR-451 and miR-7029) were also found to be underexpressed in cell lines containing integrated HPV-16 DNA (Table 1). Thirteen miRNAs were found to be overexpressed in the HeLa cell line compared to the normal cervix (Supplementary Table 3). Of these, two miRNAs (miR-182 and miR-183) were also overexpressed in the HPV-16 integrated cell lines (Table 1).
Nine miRNAs in cell lines containing integrated HPV-16 DNA were found to be expressed at much higher levels compared to the HPV-negative cervical carcinoma cell line C-33A cells (Table 2). Interestingly, miR-218 was the only miRNA that was underexpressed in the cell lines containing integrated HPV-16 DNA as compared to both the normal cervix and C-33A (Tables 1 and 2). This suggested that miR-218 may be specifically affected in the presence of HPV-16. The array data for individual cell lines compared to C-33A is presented in Supplementary Table 4.
Table 2.
MiRNAs differentially expressed in HPV-16 positive cell lines compared to the HPV-negative cell line C-33A
| MiRNA | HPV-16integrated Folda |
|---|---|
| Overexpressed | |
| hsa_miR_200c | 27.9 |
| hsa_miR_203 | 23.4 |
| hsa_miR_193b | 21.2 |
| hsa_miR_34a | 10.4 |
| hsa_miR_31 | 8.4 |
| hsa_miR_210 | 5.7 |
| hsa_miR_27a | 5.4 |
| hsa_miR_503 | 5.4 |
| hsa_miR_27b | 4.9 |
| Underexpressed | |
| hsa_miR_218 | −7.4 |
Mean fold-changes in HPV-16 integrated cell lines CaSki, SiHa, 20861 and 201402.
The q value of all miRNAs was 0.
The HPV-18 positive HeLa cell line showed overexpression of six miRNAs compared to the HPV-negative cell line C-33A (Supplementary Table 5). Of these, three miRNAs (miR-31, miR-34a and miR-193b) were also overexpressed in integrated HPV-16 cell lines compared to C-33A (Table 2). Finally, the array data for the HPV-negative cell line C-33A showed that 4 miRNAs (miR-143, miR-145, miR-200c and miR-203) were underexpressed as compared to the normal cervix (Supplementary Table 6). In addition, qRT-PCR and Northern blot analyses showed that miR-193b, miR-205 and miR-497 were also downregulated in the C-33A cell line compared to the normal cervix (see below).
Validation of the miRNA microarray expression data by quantitative real-time RT-PCR and Northern blot analyses
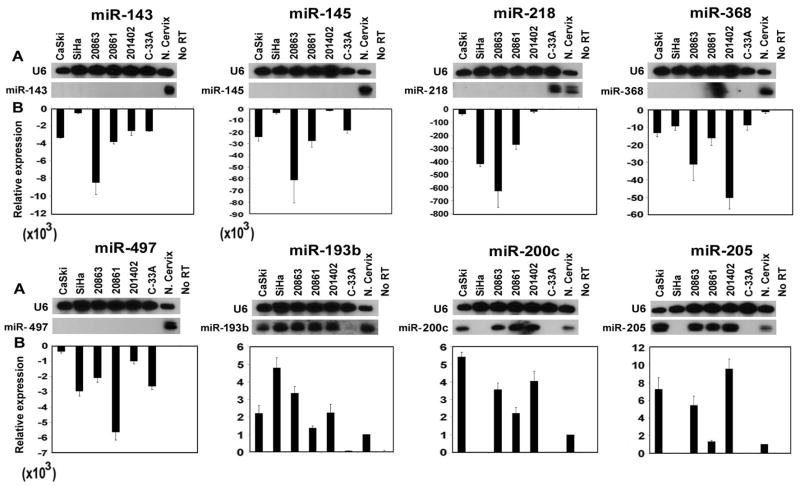
The miRNA microarray results were validated by qRT-PCR and Northern blot analyses of 8 representative miRNAs representing those whose expression was either most affected, was known to be affected in other types of cancers, or one that appeared to be HPV-specific (miR-218) (Figure 1A, B). MicroRNAs 143, 145, 218, 368 and 497 which were found to be downregulated in HPV-16 positive cell lines in array data were also found to be underexpressed based on qRT-PCR and Northern blot analyses (Figure 1A, B), although there were differences in the relative levels of individual miRNAs in various cell lines. The fold-changes observed by qRT-PCR and Northern blot analyses were much greater than those obtained with the microarrays. This was observed consistently and suggests that the results of qRT-PCR and Northern blot analyses are more robust for quantifying differential expression of miRNAs. The qRT-PCR and Northern blot analyses revealed that miR-200c and 205 were upregulated in all the HPV-positive cell lines except SiHa compared to the normal cervix (Figure 1A, B). These two miRNAs were not detectable in SiHa and the HPV-negative cell line C-33A (Figure 1A, B). The “band” seen in the Northern blot for miR-368 in the 20861 sample is a gel artifact and does not represent a miRNA signal.
Figure 1.
Confirmation of miRNA microarray expression data in various cervical cell lines and normal cervical tissue. (A) Northern blot analysis. The housekeeping splicing-related small U6 RNA was used as a loading control. (B) Real-time qRT-PCR analysis. RNU43 served as the endogenous control for miRNAs.
The array data obtained with the HPV-18 positive HeLa cell line were also validated by Northern blot and qRT-PCR analyses of three miRNAs, miR-143, miR-145, and miR-218 all of which were found to be downregulated by the various techniques (Supplementary Figure 1).
MicroRNA 218 is also underexpressed in HPV-16 positive cervical lesions and cervical cancer tissues
We also analyzed the expression profile of six representative miRNAs (all of which were also used for the validation of the array data for the cell lines) in three HPV-16 positive cervical intraepithelial neoplasia grade III (CIN III) tissues and five HPV-16 positive cervical cancer tissues (CaCx) by qRT-PCR analysis. Due to the limited amount of RNA available from the tissues, miRNA microarray analysis could not be done. The expression pattern of the above miRNAs in the tissues (Figure 2) was generally consistent with that of the HPV-positive cell lines. Importantly, miR-218 was found to be underexpressed (>2-fold reduction) in all the CIN III and CaCx samples compared to the normal cervix (Figure 2). On average, the CIN III samples showed a more limited underexpression of miR-218 compared to the CaCx tissues. Since miR-218 is encoded by an intron of the SLIT2 tumor suppressor gene (Griffiths-Jones et al., 2006), we tested whether their expression is correlated. Previous analysis of global gene expression (manuscript in preparation) has also shown that SLIT2 was underexpressed in the HPV-positive cell lines. The qRT-PCR results showed that SLIT2 expression paralleled that of miR-218, and both of these were underexpressed in the CIN III and CaCx tissues (Figure 2). MiRNAs 143, 145 and 497 that were underexpressed in the HPV-16 positive cell lines were also underexpressed in the HPV-positive tissues compared to the normal cervical tissue (Figure 2), although the relative levels of various miRNAs varied between the individual samples. In the case of miR-368, 5 out of 8 cervical cancer and CINIII lesions showed downregulation as compared to the normal cervix (Figure 2). Overall, the results obtained with the tissues provide further validation of the data obtained with the cervical cell lines.
Figure 2.
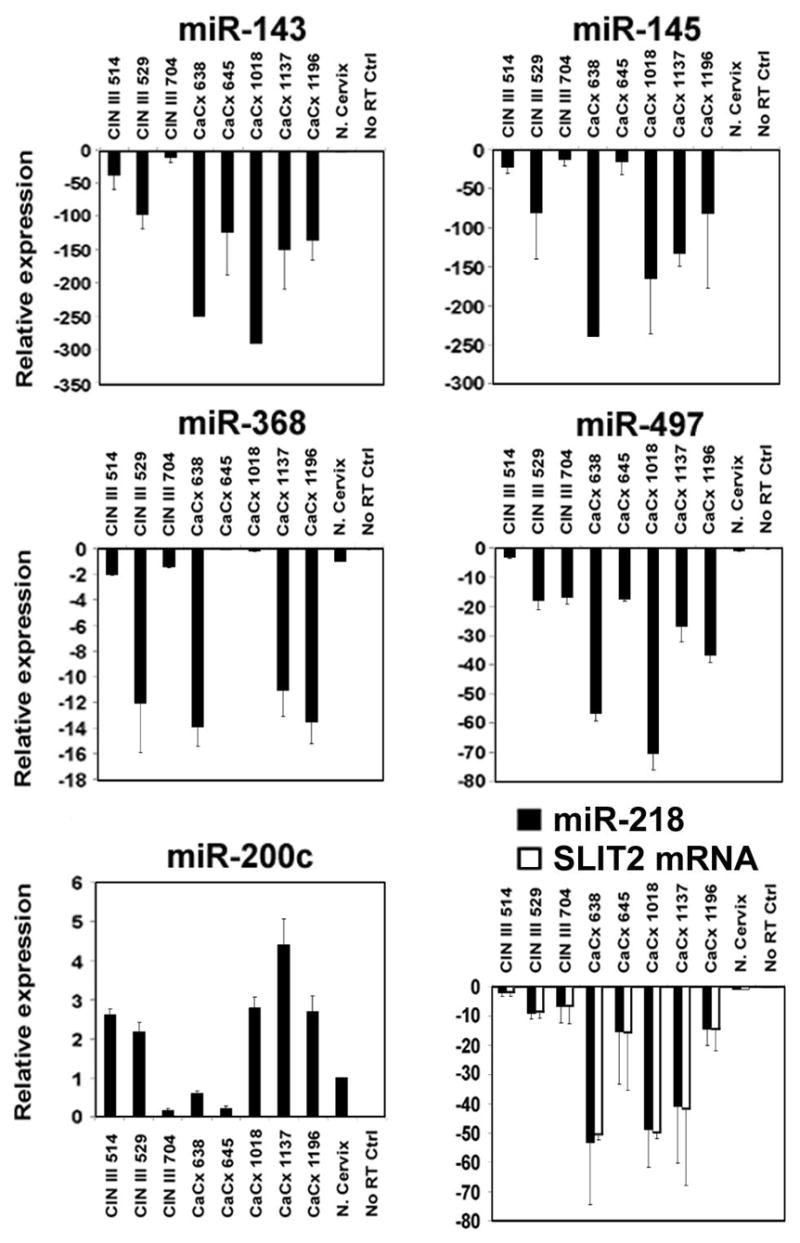
Expression of miRNAs and the SLIT2 gene in cervical tissues. qRT-PCR analysis of three cervical intraepithelial neoplasias type III (CIN III) and five cervical carcinomas (CaCx). The normal cervix sample was obtained from Stratagene. G3PDH served as the endogenous control for SLIT2.
HPV-16 E6 oncogene downregulates miR-218
To test whether E6 and/or E7 expression is directly correlated with reduced expression of miR-218, we utilized the osteosarcoma cell line U2OS either expressing the HPV-16 E6 or E7 gene, or the control neomycin resistance gene. The qRT-PCR results showed that both miR-218 and SLIT2 were underexpressed in the U2OS-E6 cell line compared to U2OS-E7 and the control U2OS-Neo cell line (Figure 3A). In another approach, the 20861 cell line containing integrated HPV-16 was transfected with HPV-16 E6/E7 siRNAs. Since E6 and E7 are derived from alternative splicing of the same RNA, a specific siRNA for E6 alone could not be used. The E6/E7 siRNAs reduced expression of these genes while increasing the expression of both miR-218 and the SLIT2 gene in 20861 cells (Figure 3B). These results indicate that the HPV-16 E6 gene is involved in the downregulation of miR-218 and the SLIT2 gene in HPV-16 positive cell lines. Since a U2OS derivative expressing the E6 gene of a low-risk HPV is not available, we utilized normal oral keratinocytes (NOK) expressing the HPV-6 E6 gene to study whether the E6 gene of a low-risk HPV also affects miR-218 expression. The qRT-PCR analysis showed that NOK-16E6 cells had reduced expression of miR-218 compared to both NOK-NEO and NOK-6E6 (Figure 3C). These results suggest that the E6 gene of the high-risk HPV-16, but not the low-risk HPV-6, reduces miR-218 expression.
Figure 3.
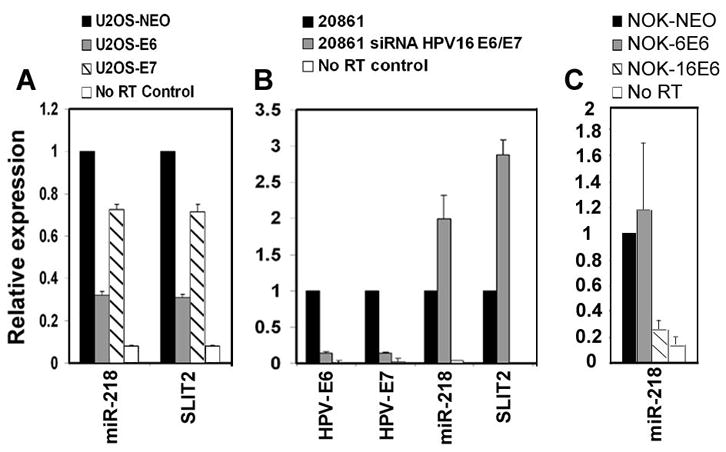
HPV-16 E6 oncogene reduces the expression of miR-218. (A) qRT-PCR analysis of miR-218 and SLIT2 in U2OS-NEO, U2OS-16E6, and U2OS-16E7. (B) Expression of HPV-16 E6 and E7, miR-218 and SLIT2 in the 20861 cell line with or without RNAi against HPV-16 E6/E7. (C) qRT-PCR analysis of miR-218 in NOK-NEO, NOK cell line expressing the E6 gene of either the low-risk HPV-6 (NOK-6E6) or high-risk HPV-16 (NOK-16E6). RNU43 served as the endogenous control for miRNAs, while G3PDH served as the endogenous control for E6, E7 and SLIT2.
Laminin 5 β3 is a transcriptional target of miR-218
To identify possible miR-218 targets, we compared computationally predicted targets in the miRBase Registry (Griffith-Jones et al., 2006) with our gene expression data (manuscript in preparation), and then used the program rna22 (http://cbcsrv.watson.ibm.com/rna22.html). This analysis revealed six possible targets of miR-218: emopamil binding protein (EBP), mitochondrial ribosomal protein S27 (MRPS27), nucleoporin 93kDa (NUP93), ephrin-A1 (EFNA1), laminin 5 β3 (LAMB3) and muscleblind-like 2 (MBNL2). The expression of these genes was analyzed by qRT-PCR in SiHa and 20861 cell lines transfected with an artificial miR-218 precursor molecule. After confirming the expression of mature miR-218 (Figure 4A), we found that only the LAMB3 transcript was significantly underexpressed in miR-218 expressing cells (Figure 4B and data not shown). Furthermore, Western blot analysis showed that miR-218 expression also greatly reduced the levels of the LAMB3 protein in SiHa cells (Figure 4C). We also found that LAMB3 was underexpressed in the 20861 cell line in the presence of the E6/E7 siRNAs compared to a control oligo (Figure 4D). Furthermore, U2OS-16E6 cells showed an increase in the levels of LAMB3 mRNA as compared to the U2OS-NEO cells (Figure 4D). Taken together, these results demonstrate that miR-218 reduces LAMB3 expression at the transcriptional level.
Figure 4.
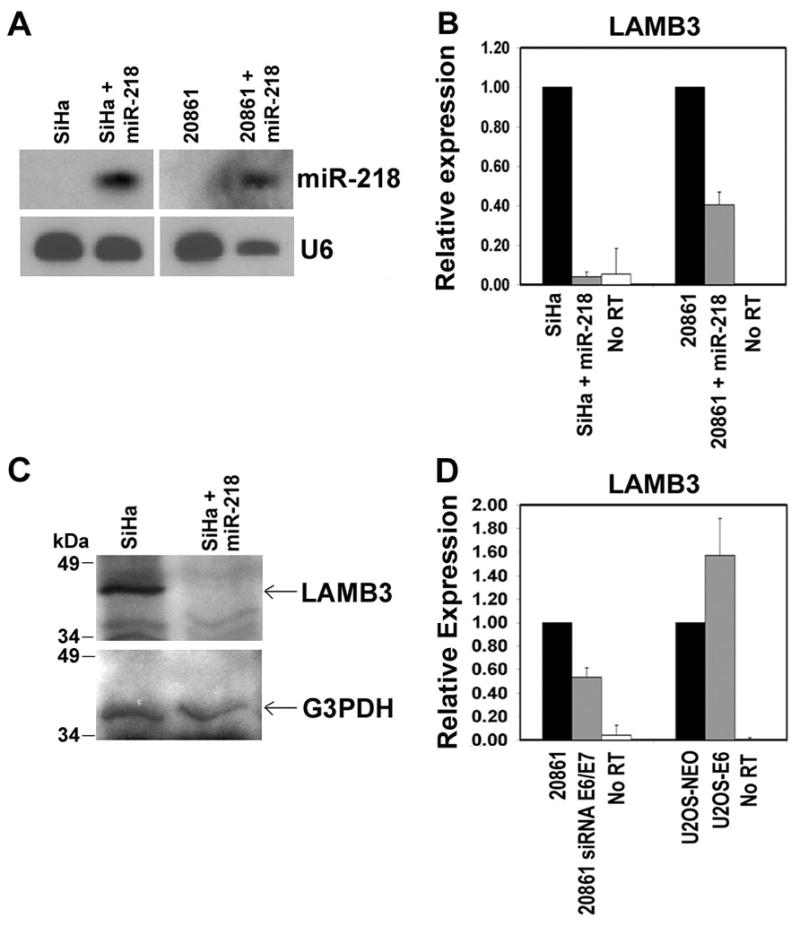
Expression of LAMB3 is reduced in the presence of miR-218. (A) Northern blot analysis of miR-218 after transfection of a precursor of miR-218 in SiHa and 20861 cell lines. U6 RNA was used as a loading control. (B) qRT-PCR analysis of LAMB3 in SiHa and 20861 cell lines transfected with a pre-miR-218. (C) Western blot analysis of LAMB3 protein in SiHa cells transfected with pre-miR-218. Location of the 40-kDa LAMB3 protein is indicated. G3PDH was used as a control. (D) Expression of LAMB3 mRNA in the 20861 cell line with or without RNAi against HPV-16 E6/E7, and in the U2OS-NEO and U2OS-16E6 cell lines. G3PDH served as the endogenous control for LAMB3.
Discussion
The microarray data showed that 24 miRNAs were underexpressed and 3 overexpressed in integrated HPV-16 cell lines compared to the normal cervix (Table 1). We also validated these data by Northern blot and qRT-PCR analyses of five miRNAs that were underexpressed (miR-143, miR-145, miR-218, miR-368 and miR-497) and three that were overexpressed (miR-193b, miR-200c and miR-205) in the HPV-16 positive cell lines compared to the normal cervix or the HPV-negative cervical carcinoma cell line C-33A (Figure 1). The probable targets of the above eight miRNAs are shown in Supplementary Table 7. The observed differences in the levels of differentially expressed miRNAs were generally much greater in qRT-PCR and Northern blot studies as compared to the array data. Thus, the array data likely underestimate the fold-changes in the levels of differentially expressed miRNAs. Our studies identified 10 underexpressed miRNAs in integrated HPV-16 cell lines that are not known to be altered in any cancers. These included 7 known human miRNAs, miR-1, miR-126-AS, miR-133a, miR-376a, miR-451, miR-487b, miR-495, and 3 predicted miRNAs, ambi-miR-5021, ambi-miR-7029, and ambi-miR-7070. MiR-143 and miR-145, which are underexpressed in colon and breast cancers (Calin and Croce, 2006), were also underexpressed in all cervical cell lines including C-33A (Tables 1 and 3), suggesting that they may be important in cervical carcinogenesis independent of HPV infection. MiR-368, miR-497 and miR-193b which are underexpressed in integrated HPV-16 cell lines are known to be underexpressed in colon cancer cell lines (Bandres et al., 2006), papillary thyroid carcinomas (Calin and Croce, 2006), and colon cancer (Cummins et al., 2006), respectively. Interestingly, miR-218 was the only miRNA found to be underexpressed in integrated HPV-16 cell lines compared to both the normal cervix and C-33A cells (Tables 1 and 2, and Figure 1). Northern blot and qRT-PCR analysis showed that miR-218 is also underexpressed in HeLa cells containing integrated HPV-18 DNA and the 20863 cell line containing episomal HPV-16 DNA (Figure 1, and Supplementary Figure 1). These results suggest that miR-218 may be a specific cellular target of high-risk HPVs. MiR-218 is known to be underexpressed in several cancers and the DNA encoding miR-218 is also deleted in ovarian, breast, and melanoma cancers (Calin and Croce, 2006; Zhang et al., 2006). Of the miRNAs overexpressed in integrated HPV-16 cell lines, miR-210 is overexpressed in many epithelial cancers (Calin and Croce, 2006), while miR-182 and miR-183 are overexpressed in colon cancer cell lines (Bandres et al., 2006). We also found that 6 miRNAs have reduced expression in the episomal HPV-16 cell line 20863 compared to the normal cervix (Supplementary Table 2). These miRNAs may represent early targets of HPV-16 infection. A comparison of the miRNA expression profile of HPV-16 and HPV-18 positive cell lines with that of the normal cervix showed that while some miRNAs are similarly affected in the HPV-16 and HPV-18 positive cells, others are not. This may indicate the presence of both common as well as unique pathways that are altered in cells containing HPV-16 and HPV-18 DNA.
Analysis of 6 representative miRNAs, miR-143, miR-145, miR-200c, miR-218, miR-368 and miR-497 (whose expression levels in various cell lines were also analyzed by a variety of methods) in CIN III and CaCx tissues by qRT-PCR showed expression patterns that were generally similar to those of the HPV-positive cell lines (Figures 1 and 2). Importantly, miR-218 was also underexpressed (>2-fold difference) in all the CIN III and CaCx tissues (Figure 2). These results suggest that miR-218 underexpression is likely linked to the process of HPV-associated carcinogenesis in vivo. The parallel expression of miR-218 and the SLIT2 gene (which encodes miR-218) in all the tumor tissues (Figure 2) suggests that they may be coordinately regulated. Previous studies have shown that the SLIT2 tumor suppressor gene is frequently inactivated in lung and breast cancers (which have reduced miR-218 levels) (Dallol et al., 2002). Whether regulation of SLIT2 plays a role in the pathogenesis of HPV-associated malignancies is currently unknown.
Our results showed that both miR-218 and SLIT2 were underexpressed in the U2OS-16E6 cell line (Figure 3A). Similarly, miR-218 was also underexpressed in NOK-16E6 but not in the NOK-6E6 cell line (Figure 3C). These results suggest that E6 of the high-risk HPV-16, but not the low-risk HPV-6, may contribute to the downregulation of miR-218. This possibility is also supported by our observations that expression of E6/E7 siRNAs in 20861 cells increases miR-218 and SLIT2 mRNA levels (Figure 3B). Interestingly, SLIT2 is a possible target of miR-200c (overexpressed in the HPV-positive cell lines and tissues) (Supplementary Table 7). One study has shown that the absence of the p53 gene increases miR-200c expression (Xi et al., 2006). Thus, it is possible that E6-dependent degradation of the p53 protein results in miR-200c overexpression which in turn reduces the levels of miR-218 and SLIT2 mRNA.
Our studies showed that introduction of miR-218 into SiHa and 20861 cell lines reduced the levels of Laminin 5 β3 (LAMB3) mRNA as well as the protein (Figure 4A, B, C). This strongly supports implies that miR-218 can downregulate LAMB3 expression independent of SLIT2. Furthermore, introduction of E6/E7 siRNA into the 20861 cell line also reduced LAMB3 expression (Figure 4D). Also, E6 expression in U2OS cells increased LAMB3 expression (Figure 4D) presumably by reducing miR-218 expression. Taken together, the above studies suggest that miR-218 may regulate LAMB3 expression at the transcriptional level. LAMB3 protein is part of the polymeric cell surface receptor laminin 5 that is expressed in the basal lamina of the epithelium and is overexpressed in cervical cancers (Skyldberg et al., 1999; Kohlberger et al., 2003). LAMB3 increases cell migration and tumorigenicity in SCID mice, and in collaboration with its ligand α6β4-integrin promotes tumorigenesis in human keratinocytes (Dajee et al., 2003; Calaluce et al., 2004). A recent study suggests that secreted laminin 5 can be used by HPV as a transient receptor to aid the virus in the infection of basal cells that express α6β4-integrin (Culp et al., 2006). Thus, downregulation of miR-218 by E6 and the consequent overexpression of LAMB3 may promote viral infection of the surrounding tissue and contribute to eventual tumorigenesis.
Materials and methods
Cell lines
Cervical cancer cell lines CaSki, SiHa (HPV-16 positive), HeLa (HPV-18 positive), and C-33A (HPV-negative) and their growth conditions have been described (Meissner, 1999). Three clonal populations of the HPV-16 cervical cell line W12, 20863 (episomal HPV-16), 20861 and 201402 (integrated HPV-16), were obtained from the laboratories of Drs. Margaret Stanley and Paul Lambert (Medical Research Council, UK and University of Wisconsin, USA, respectively). The cell lines U2OS-Neo, U2OS-E6, U2OS-E7 (Duensing et al., 2000) were obtained from Dr. Stefan Duensing (University of Pittsburgh, USA). Normal oral keratinocytes (NOK) cell lines immortalized by hTERT expression and expressing the E6 gene of HPV-6 or HPV-16 (Piboonniyom et al., 2003) were obtained from Drs. Stefan Duensing and Karl Munger (Harvard University).
Cervical tissues characteristics
Human cervical tissue samples were collected under an IRB approved protocol from patients with cervical preinvasive neoplasia undergoing leep excision or radical hysterectomy for invasive cervical cancer. Informed consent was obtained from all subjects. Parallel specimens of the same site were stained (Hemotoxylin and Eosin) to confirm the diagnosis. HPV-16 positivity was confirmed by RT-PCR. Total RNA from normal human cervix was obtained from a pool of 2 different donors (Stratagene). Prior to further RNA purification, these preparations contained limited levels of cellular DNA and were used to confirm the absence of HPV DNA. PCR amplification showed positive results for G3PDH and negative results for several HPV types using the degenerate MY09/MY11 primer pair (Manos et al., 1994). Furthermore, the preparations also did not give any PCR signals for the HPV-16 E7 and E2 genes. CaSki cells were used as a positive control and gave positive signals for the G3PDH gene as well as for the HPV L1 gene and HPV-16 E7 and E2 genes (data not shown).
MicroRNA microarray analysis
Total RNA was extracted using the Ultraspec™ RNA isolation system (BIOTECX, USA) according to the manufacturer’s instructions. Total RNA from cell lines and normal cervix was used to isolate small RNAs (<200 nt) which includes miRNAs using the RNeasy Mini Kit and the MinElute Cleanup Kit (Qiagen, USA). MirVana miRNA Bioarray (Ambion, USA) was used to analyze miRNA expression in cervical cell lines. The Bioarray consist of 662 probes (~22 nt long antisense oligonucleotides) that include known human miRNAs (328), theoretical human microarrays known as ambi-miRs (152) as well as unique miRNAs from mouse (114) and rat (46). Each array contains probes for all miRNAs in quadruplicate, and the signals obtained for each miRNA is represented as an average of these four values. The enriched small RNA fractions obtained from 25 μg of total RNA were fluorescently labeled and hybridized to the Bioarrays according to the manufacturer’s instructions (Ambion, USA). Each experiment was done twice. The Bioarrays were scanned using GenePix 4000B Scanner and the median fluorescent intensity was obtained after subtracting the background using the GenePix Pro 6.0 software. To identify differential miRNA expression between samples, the median fluorescent intensities were log2 transformed and normalized using the median within the array and the global mean adjustment among arrays using the GEDA program (http://bioinformatics.upmc.edu/Help/GEDADescription.html). After transformation and normalization, we used the Significance Analysis of Microarray (SAM) program version 1.21 (www-stat.stanford.edu/~tibs/SAM/) to obtain the differential expression profiles.
MicroRNA Northern blot analysis
Enriched miRNA fractions obtained from 25 μg of total RNA were separated on 15% urea-containing polyacrylamide gels, transferred onto GeneScreen Plus membranes and hybridized to 32P-labeled oligonucleotide probes complementary to various cellular miRNAs. We hybridized a probe complementary to the housekeeping splicing-related small U6 RNA for loading control. Hybridization was carried out overnight at 50°C and the membranes were subjected to autoradiography at −80°C.
MicroRNA and mRNA real-time quantitative RT-PCR analysis
DNase I-treated total RNA (10 ng) was subjected to qRT-PCR analysis using the TaqMan® miRNA Reverse Transcription Kit and miRNA Assays (Applied Biosystems, USA), and the Real-Time thermocycler iQ5 (BioRad, USA). The small nucleolar RNU43 was used as the housekeeping small RNA reference gene. For qRT-PCR analysis of SLIT2 and LAMB3 mRNAs, the following primers were used: (SLIT2, forward 5′-CTGTGAATGCAGCAGTGGAT-3′ and reverse 5′-TTGTTTGGCAAGCAGCATAG-3′ (116-bp product); LAMB3, forward 5′-GGGAGACCATGGAGATGATG-3′and reverse 5′-ACACGCTTCTCCAGTCCTGT-3′ (112-bp product). Also, 500 ng of total RNA from the cervical samples was amplified using the one step QuantiTect SYBR Green RT-PCR Master Mix (Qiagen, USA). The primer sequences for the control housekeeping glyceraldehyde-3-phosphate dehydrogenase gene (G3PDH) have been described previously (Martinez et al., 2007). All reactions were done in triplicate and relative expression of RNAs was calculated using the 2 delta CT method (Livak et al., 2001).
HPV-16 E6 and E7 siRNA and transfection assays
Double-stranded small interfering RNAs (siRNAs) against HPV-16 E6 (siRNA 209, 5′-UCCAUAUGCUGUAUGUGAUTT-3′; complementary to HPV-16 positions 277 to 298) and E7 (E7 siRNA, 5′-CCAUCUAUUUCAUCCUCCUTT-3′, complementary to HPV-16 positions 662 to 682) (Jiang and Milner, 2002; Tang et al., 2006) were obtained from Dharmacon, USA. BLOCK-iT™ Fluorescent double-stranded oligo (with no human homologous sequence) was used as a negative control as well as to measure the transfection efficiency (Invitrogen, USA). MiR-218 was expressed in cell lines by transfecting with a Pre-miR™ miR-218 precursor molecule (Ambion, USA). Cell lines were seeded (1.5 × 105) into 6-well plates, and after 24 hr, transfected (125 nM per well of HPV siRNAs or 100 nM of Pre-miR™ miR-218 precursor Molecule) using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions. Cells were harvested after 72 hr, and RNA and protein extractions were performed.
Western blot analysis
Twenty micrograms of total cellular proteins were separated on 7.5% SDS-polyacrylamide gels, transferred to Immobilon-P PVDF membranes (Millipore), and incubated with primary murine monoclonal antibody (Calaluce et al., 2004) against the •3 chain of laminin-5 (Kalinin B1; BD Biosciences, USA). The membranes were subsequently incubated with a 1:5,000 dilution of the secondary anti-mouse horseradish peroxidase antibody (Amersham Biosciences, USA). Specific proteins were detected using chemiluminescence with ECL Plus Western Blotting Detection Reagents (Amersham Biosciences, USA). Murine monoclonal antibody against G3PDH (Chemicon) was used to demonstrate equal loading.
Supplementary Material
Supplementary information is available at Oncogene’s web site.
Acknowledgments
We thank Stefan Duensing and Karl Munger for U2OS and NOK cell lines, Margaret Stanley and Paul Lambert for W12 derived cell lines, Cathy Ma for help with standardization of miRNA microarrays, Naftali Kaminski for the use of the GenePix Scanner and Yugandhar Reddy for helpful discussions. This work was supported in part by National Institutes of Health Grant DC016406 to S.A.K. ASG was supported by NIH training grant 5T32GM065100 (Biotechnology Training Grant) and by NIH Ruth Kirschstein predoctoral fellowship DE019028.
Abbreviations
- miRNA
microRNA
- HPVs
human papillomaviruses
- CaCx
cancer of the cervix
References
- Alazawi W, Pett M, Arch B, Scott L, Freeman T, Stanley MA, et al. Changes in cervical keratinocyte gene expression associated with integration of human papillomavirus 16. Cancer Res. 2002;62:6959–6965. [PubMed] [Google Scholar]
- Band V, Dalal S, Delmolino L, Androphy EJ. Enhanced degradation of p53 protein in HPV-6 and BPV-1 E6-immortalized human mammary epithelial cells. EMBO J. 1993;12:1847–1852. doi: 10.1002/j.1460-2075.1993.tb05833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29–38. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calaluce R, Bearss DJ, Barrera J, Zhao Y, Han H, Beck SK, et al. Laminin-5 beta3A expression in LNCaP human prostate carcinoma cells increases cell migration and tumorigenicity. Neoplasia. 2004;6:468–479. doi: 10.1593/neo.03499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- Culp TD, Budgeon LR, Marinkovich MP, Meneguzzi G, Christensen ND. Keratinocyte-secreted laminin 5 can function as a transient receptor for human papillomaviruses by binding virions and transferring them to adjacent cells. J Virol. 2006;80:8940–8950. doi: 10.1128/JVI.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajee M, Lazarov M, Zhang JY, Cai T, Green CL, Russell AJ, et al. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- Dallol A, Da Silva NF, Viacava P, Minna JD, Bieche I, Maher ER, et al. SLIT2, a human homologue of the Drosophila Slit2 gene, has tumor suppressor activity and is frequently inactivated in lung and breast cancers. Cancer Res. 2002;62:5874–5880. [PubMed] [Google Scholar]
- Duensing S, Lee LY, Duensing A, Basile J, Piboonniyom S, Gonzalez S, et al. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc Natl Acad Sci U S A. 2000;97:10002–10007. doi: 10.1073/pnas.170093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiol D, Kuhne C, Glaunsinger B, Lee SS, Javier R, Banks L. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene. 1999;18:5487–5496. doi: 10.1038/sj.onc.1202920. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebner CM, Laimins LA. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev Med Virol. 2006;16:83–97. doi: 10.1002/rmv.488. [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Milner J. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene. 2002;21:6041–6048. doi: 10.1038/sj.onc.1205878. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kohlberger P, Beneder C, Horvat R, Leodolter S, Breitenecker G. Immunohistochemical expression of laminin-5 in cervical intraepithelial neoplasia. Gynecol Oncol. 2003;89:391–394. doi: 10.1016/s0090-8258(03)00086-6. [DOI] [PubMed] [Google Scholar]
- Lechner MS, Mack DH, Finicle AB, Crook T, Vousden KH, Laimins LA. Human papillomavirus E6 proteins bind p53 in vivo and abrogate p53-mediated repression of transcription. EMBO J. 1992;11:3045–3052. doi: 10.1002/j.1460-2075.1992.tb05375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Manos MM, Waldman J, Zhang TY, Greer CE, Eichinger G, Schiffman MH, et al. Epidemiology and partial nucleotide sequence of four novel genital human papillomaviruses. J Infect Dis. 1994;170:1096–1099. doi: 10.1093/infdis/170.5.1096. [DOI] [PubMed] [Google Scholar]
- Martinez I, Wang J, Hobson KF, Ferris RL, Khan SA. Identification of differentially expressed genes in HPV-positive and HPV-negative oropharyngeal squamous cell carcinomas. Eur J Cancer. 2007;43:415–432. doi: 10.1016/j.ejca.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massimi P, Pim D, Storey A, Banks L. HPV-16 E7 and adenovirus E1a complex formation with TATA box binding protein is enhanced by casein kinase II phosphorylation. Oncogene. 1996;12:2325–2330. [PubMed] [Google Scholar]
- Meissner JD. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J Gen Virol. 1999;80:1725–1733. doi: 10.1099/0022-1317-80-7-1725. [DOI] [PubMed] [Google Scholar]
- Munger K, Howley PM. Human papillomavirus immortalization and transformation functions. Virus Res. 2002;89:213–228. doi: 10.1016/s0168-1702(02)00190-9. [DOI] [PubMed] [Google Scholar]
- Piboonniyom SO, Duensing S, Swilling NW, Hasskarl J, Hinds PW, Munger K. Abrogation of the retinoblastoma tumor suppressor checkpoint during keratinocyte immortalization is not sufficient for induction of centrosome-mediated genomic instability. Cancer Res. 2003;63:476–483. [PubMed] [Google Scholar]
- Skyldberg B, Salo S, Eriksson E, Aspenblad U, Moberger B, Tryggvason K, et al. Laminin-5 as a marker of invasiveness in cervical lesions. J Natl Cancer Inst. 1999;91:1882–1887. doi: 10.1093/jnci/91.21.1882. [DOI] [PubMed] [Google Scholar]
- Stanley MA, Browne HM, Appleby M, Minson AC. Properties of a non-tumorigenic human cervical keratinocyte cell line. Int J Cancer. 1989;43:672–676. doi: 10.1002/ijc.2910430422. [DOI] [PubMed] [Google Scholar]
- Tang S, Tao M, McCoy JP, Jr, Zheng ZM. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J Virol. 2006;80:4249–4263. doi: 10.1128/JVI.80.9.4249-4263.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, Pim D, Banks L. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene. 1999;18:7690–7700. doi: 10.1038/sj.onc.1202953. [DOI] [PubMed] [Google Scholar]
- Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res. 2006;12:2014–2024. doi: 10.1158/1078-0432.CCR-05-1853. [DOI] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information is available at Oncogene’s web site.