Abstract
The lumen of endosomal organelles becomes increasingly acidic when going from the cell surface to lysosomes. Luminal pH thereby regulates important processes such as the release of internalized ligands from their receptor or the activation of lysosomal enzymes. The main player in endosomal acidification is the vacuolar ATPase (V-ATPase), a multi-subunit transmembrane complex that pumps protons from the cytoplasm to the lumen of organelles, or to the outside of the cell. The active V-ATPase is composed of two multi-subunit domains, the transmembrane V0 and the cytoplasmic V1. Here we found that the ratio of membrane associated V1/Vo varies along the endocytic pathway, the relative abundance of V1 being higher on late endosomes than on early endosomes, providing an explanation for the higher acidity of late endosomes. We also found that all membrane-bound V-ATPase subunits were associated with detergent resistant membranes (DRM) isolated from late endosomes, raising the possibility that association with lipid-raft like domains also plays a role in regulating the activity of the proton pump. In support of this, we found that treatment of cells with U18666A, a drug that leads to the accumulation of cholesterol in late endosomes, affected acidification of late endosome. Altogether our findings indicate that the activity of the vATPase in the endocytic pathway is regulated both by reversible association/dissociation and the interaction with specific lipid environments.
Introduction
During evolution, compartmentalization of the intracellular space has allowed to spatially restrict and optimize certain biochemical reactions and pathways. Therefore the lumens of different organelles have different properties in terms of ion concentrations, redox states and also pH. Organellar pH is tightly regulated ranging from neutral in the endoplasmic reticulum, to mildly acidic in early endosomes and highly acidic in late endosomes/lysosomes [1]. Acidic pH affects a number of biological events such as membrane trafficking, dissociation of ligand-receptor complexes after internalization and activation of lysosomal enzymes [1]. Although pH regulation can be modulated by a variety of factors such as proton leak, ClC chloride channels or Na,K-ATPases [2], [3], [4], [5], it is primarily determined by the activity of the vacuolar ATPase (V-ATPase), which is expressed in all eukaryotes from yeast to mammals [6], [7].
The V-ATPase is a multi-subunit complex composed of two domains, a peripheral V1 domain containing the ATPase activity and a membrane bound V0 domain responsible for translocation of protons across the membrane [6], [8]. The central function of the V-ATPase is to pump protons from the cytoplasm to the lumen of organelles. In some specialized cells, the V-ATPase can also be found at the plasma membrane. Its role is then either to acidify the extracellular medium such as around osteoclasts and renal cells [9], [10], [11], or to control the cytoplasmic pH as in neutrophiles and macrophages [12], [13]. The proton pump also appears to be involved in cancer through the promotion of metastasis and tumor progression and it is therefore considered as a potential drug target [6], [13].
Despite the importance of the V-ATPase in physiological and pathological processes, the exact mechanisms that control the activity of the V-ATPase remain to be fully elucidated. Four regulatory mechanisms have been described to date. The first is the reversible dissociation of the catalytic V1 domain from the membrane-associated V0, which was observed in S. cerevisiae and Manduca sexta upon glucose deprivation and starvation respectively [14], [15]. Change in V1/V0 association was also reported during maturation of murine dendritic cells [16]. The second mechanism involves the abundance of the proton pump at a given site. In cells such as renal cells and osteoclasts, where the proton pump is at the cell surface, acid secretion was indeed found to be modulated by a differential surface expression of the V-ATPase through reversible exocytosis and endocytosis of the pump [17]. The third mechanism by which the activity of the V-ATPase could be modulated is by changing of the coupling efficiency between ATP hydrolysis and proton translocation, attributable to different isoforms of subunit V0a. This differential coupling efficiency was proposed to explain that lysosomes are more acidic than the Golgi [1], [6], [18] [19]. Finally, it has been proposed that specific lipids can affect the activity of the V-ATPase. More specifically it was shown in yeast that sphingolipids with a C26 acyl group are required for generating V1 domains with ATPase activity [20]. Interestingly, several V-ATPase inhibitors where shown to incorporate into the lipid bilayer and affect the V-ATPase structural flexibility [21], [22].
In this study, our interest was to get a better understanding of how pH is controlled along the endocytic pathway in mammalian cells, since most studies on V-ATPase regulation where performed in yeast. We found that the increased acidity of late endosomes is not due to a higher density of proton pumps but rather to an increase in the V1/V0 ratio when compared to early endosomes. Thus regulation would occur via reversible association/dissociation, reminiscent of what occurs found in yeast. Additionally, we found that the lipid environment of the V-ATPase is of essential importance for its activity, suggesting a second mode of regulation and highlighting that the V-ATPase activity is modulated by multiple, simultaneously operating, mechanisms.
Results
DRM association of all the subunits of the V-ATPase
Endosomes, and in particular late endosomes, are well known to be composed of mosaics of domains [23], [24]. We have shown that late endosomes, in particular, contain raft-like domains that are rich in cholesterol and sphingomyelin and resistant to extraction in non-ionic detergents at 4°C [25], [26]. To further characterize late endosomal DRMs, we have here performed a proteomic analysis. Proteomics analysis of DRM fractions have been reported in numerous studies [27], [28], [29], [30], [31], but this is the first that proteomics was performed on DRMs of a purified endocytic organelle. As previously, late endosomes were obtained from Baby Hamster Kidney cells, since a well-established subcellular fractionation protocol to purify late endosomes is available for this cell line [25], [32]. We have previously shown using surface biotinylation of proteins that this fraction does not contain detectable amounts of plasma membrane [33]. Moreover this late endosomal fraction is not contaminated by Golgi, endoplasmic reticulum, early endosomal or caveolar membranes, as shown by western blotting using marker proteins [34]. It is likely that the high purity of this fraction is due to the fact that the late endosomal fraction has a very low buoyancy and thus migrates through the entire gradient to the top of the tube, far away from all other cellular compartments. The llate endosomal fraction was subsequently submitted to solubilization in triton X-100 at 4°C and the Detergent-Resistant-Membranes (DRMs, fractions 1 and 2 from the top of the gradient) were separated from the Detergent-Soluble-Membranes (DSMs, fractions 5 and 6) using floatation gradients. The proteomes of both the DRMs and DSMs were determined by mass spectrometry after appropriate trypsinization and sample preparation.
Some 126 and 161 proteins were identified in the DRM and DSM fractions respectively, 44 of which were found in both fractions (Table 1 and 2), which jointly provide a global proteome of the organelle. Reassuringly, known late endosome specific proteins such as the small GTPase Rab7, the lysosomal glycoprotein LAMP2, or the NPC1 protein involved in Niemann Pick type C disease [35] were found. Also the DRM fraction contained most of the well-documented rafts markers such as caveolin-1 and 2 –involved in the formation of caveolae, in signaling and in lipid regulation [36]–, erlin 2, –a DRM associated ER protein [37], flotillins 1 and 2 [38], which were also found in the DSM in agreement with detectable amounts by western blotting [25], stomatin and the hyaluronic acid receptor CD44 [39].
Table 1. Proteins identified in the late endosomal DRM fraction.
| Numb | Uniprot Numb | Protein name | Protein family | Mascot score | Mw | Cov (%) | Nb pep |
| 1 | P56564 | Excitatory amino acid transporter 1 | AA transport | 150.46 | 59584 | 7.21 | 2 |
| 2 | Q3UFR4 | Neutral amino acid transporter ASCT2 | AA transport | 186.95 | 58327 | 6.42 | 3 |
| 3 | Q9JKY1 | Peroxiredoxin-1 | Cell redox homeostasis | 45.56 | 22248 | 5.45 | 1 |
| 4 | Q71FK5 | Actin, cytoplasmic 1 (Beta-actin) | Cytoskeleton | 1034.51 | 41710 | 54.35 | 15 |
| 5 | P63260 | Actin, cytoplasmic 2 (Gamma-actin) | Cytoskeleton | 1071.66 | 41766 | 54.35 | 15 |
| 6 | Q7TSH1 | Actr3b protein | Cytoskeleton | 55.1 | 48539 | 2.49 | 1 |
| 7 | Q8BMK4 | Cytoskeleton-associated protein 4 | Cytoskeleton | 74.92 | 60655 | 2.9 | 1 |
| 8 | P47757 | F-actin-capping protein subunit beta | Cytoskeleton | 44.23 | 31195 | 3.53 | 1 |
| 9 | Q5M810 | Myo1g protein | Cytoskeleton | 114.37 | 58945 | 7.1 | 2 |
| 10 | Q3UFT0 | myosin heavy chain IX | Cytoskeleton | 108.31 | 110908 | 4.27 | 2 |
| 11 | Q63355 | Myosin I heavy chain | Cytoskeleton | 1530.65 | 118017 | 27.33 | 23 |
| 12 | P14869 | Myosin regulatory light chain 2-A | Cytoskeleton | 80.24 | 34195 | 3.87 | 1 |
| 13 | P46735 | Myosin-Ib | Cytoskeleton | 45.16 | 128447 | 0.77 | 1 |
| 14 | Q9ERB6 | Nuclear myosin I beta | Cytoskeleton | 1508.15 | 119802 | 27.09 | 23 |
| 15 | P30427 | Plectin-1 | Cytoskeleton | 42.29 | 533214 | 0.23 | 1 |
| 16 | Q9EPK2 | Protein XRP2 | Cytoskeleton | 39.92 | 39220 | 3.93 | 1 |
| 17 | P54116 | Stomatin | Cytoskeleton | 192.32 | 31384 | 15.79 | 3 |
| 18 | P68372 | Tubulin beta-2C chain | Cytoskeleton | 144.93 | 49799 | 6.42 | 3 |
| 19 | Q3TWV0 | vimentin | Cytoskeleton | 97.46 | 53633 | 4.72 | 2 |
| 20 | Q8K3H8 | Calnexin | ER chaperone | 48.48 | 67422 | 2.61 | 1 |
| 21 | Q8BFZ9 | Erlin-2 precursor | ERAD pathway | 129.07 | 37849 | 10.76 | 3 |
| 22 | O08917 | Flotillin-1 | Flotillin complex | 900.61 | 47484 | 42.92 | 14 |
| 23 | Q9Z2S9 | Flotillin-2 | Flotillin complex | 782.94 | 47009 | 34.43 | 12 |
| 24 | P17809 | Glucose transporter type 1 (GLUT-1) | Glucose transporter | 105.89 | 53899 | 3.68 | 2 |
| 25 | Q3UAZ6 | ATP-binding cassette, sub-family B (MDR/TAP), member 6 | Heme homeostasis | 41.06 | 84765 | 1.95 | 1 |
| 26 | O88783 | Coagulation factor V | Ion binding | 43.16 | 247076 | 0.45 | 1 |
| 27 | Q6PFA8 | Moxd1 protein | Ion binding | 89.33 | 69734 | 3.63 | 2 |
| 28 | Q3TVE3 | similar to S100 calcium-binding protein A16 | Ion binding | 49.01 | 14287 | 8.53 | 1 |
| 29 | Q7TMC7 | Ab2-417 (Cc1-8) | Ion transport | 97.43 | 107343 | 3.08 | 2 |
| 30 | P51881 | ADP/ATP translocase 2 | Ion transport | 92.37 | 32779 | 7.74 | 2 |
| 31 | Q8VDN2 | Na(+)/K(+) ATPase alpha-1 subunit | Ion transport | 634.88 | 112910 | 14.91 | 10 |
| 32 | Q3TX38 | voltage-dependent anion channel 3 | Ion transport | 143.97 | 30733 | 12.19 | 3 |
| 33 | Q9Z2L0 | Voltage-dependent anion-selective channel protein 1 (VDAC-1) | Ion transport | 107.19 | 30606 | 7.55 | 2 |
| 34 | Q60930 | Voltage-dependent anion-selective channel protein 2 (VDAC-2) | Ion transport | 49.07 | 31713 | 4.07 | 1 |
| 35 | O08532 | Voltage-dependent calcium channel subunit alpha-2/delta-1 | Ion transport | 508.19 | 124551 | 12.37 | 9 |
| 36 | P51909 | Apolipoprotein D | Lipid biogenesis | 58.56 | 21596 | 6.12 | 1 |
| 37 | Q9JKU9 | Sigma 1 receptor beta variant | Lipid biogenesis | 42.74 | 21589 | 6.63 | 1 |
| 38 | P00762 | Anionic trypsin-1 precursor | Peptidase | 81.74 | 25943 | 8.51 | 1 |
| 39 | P07356 | Annexin A2 | Phospholipid binding/modifying | 339.59 | 38521 | 24 | 6 |
| 40 | Q8BG11 | cadherin 13 | Protein binding | 43.41 | 78137 | 2.39 | 1 |
| 41 | Q8BFU4 | GAIP/RGS19 short isoform | Protein binding | 48.57 | 22240 | 6.44 | 1 |
| 42 | Q4FCR4 | Intercellular adhesion molecule 1 | Protein binding | 72.74 | 53809 | 2.86 | 1 |
| 43 | P70490 | Lactadherin precursor | Protein binding | 43.23 | 47382 | 2.56 | 1 |
| 44 | Q6DI58 | Rpl12 protein | Protein binding | 53.03 | 22973 | 7.21 | 1 |
| 45 | Q6PDW1 | 40S ribosomal protein S12 | Protein biosynthesis | 108.17 | 14505 | 16.79 | 1 |
| 46 | P62629 | Elongation factor 1-alpha 1 | Protein biosynthesis | 50.37 | 50082 | 5.27 | 1 |
| 47 | Q3U561 | ribosomal protein L10A | Protein biosynthesis | 70.68 | 24800 | 5.78 | 1 |
| 48 | Q3UBI6 | ribosomal protein L7 | Protein biosynthesis | 53.16 | 31331 | 3.87 | 1 |
| 49 | Q9QWC2 | Ribosomal protein P2 | Protein biosynthesis | 40.29 | 5063 | 23.08 | 1 |
| 50 | Q3UCL7 | ribosomal protein S3 | Protein biosynthesis | 136.27 | 26658 | 16.12 | 3 |
| 51 | Q62186 | Translocon-associated protein subunit delta precursor | Protein biosynthesis | 61.03 | 18924 | 6.4 | 1 |
| 52 | Q3S4T7 | 78 kDa glucose-regulated protein precursor | Protein folding | 268.69 | 72284 | 8.68 | 5 |
| 53 | P24369 | Cyclophilin B | Protein folding | 38.8 | 22699 | 6.31 | 1 |
| 54 | P30412 | Cyclophilin C | Protein folding | 253.35 | 22780 | 22.71 | 4 |
| 55 | Q3KQJ4 | Hspa8 protein (Fragment) | Protein folding | 252.63 | 62117 | 8.87 | 5 |
| 56 | Q6AYQ9 | Peptidyl-prolyl cis-trans isomerase | Protein folding | 211.16 | 22995 | 20.1 | 4 |
| 57 | P62858 | 40S ribosomal protein S28 | Protein processing | 60.75 | 7836 | 16.9 | 1 |
| 58 | Q9R0Y5 | Adenylate kinase isoenzyme 1 | Protein processing | 51.75 | 21526 | 6.67 | 1 |
| 59 | Q9WU83 | Dolichol-phosphate mannosyltransferase | Protein processing | 42.88 | 29636 | 4.09 | 1 |
| 60 | P57716 | Nicastrin precursor | Protein processing | 43.83 | 78441 | 2.24 | 1 |
| 61 | Q6AXQ3 | Viral oncogene yes-1 homolog 1 | Protein processing | 249.57 | 54236 | 14 | 5 |
| 62 | P25286 | V0 subunit a1 | Proton transport | 419.99 | 96265 | 9.03 | 7 |
| 63 | Q5XK06 | V0 subunit C | Proton transport | 45.38 | 20434 | 9.73 | 1 |
| 64 | P51863 | V0 subunit d | Proton transport | 210.04 | 40275 | 7.92 | 3 |
| 65 | Q3U5W3 | V1 subunit A, isoform 1 | Proton transport | 790.08 | 68211 | 30.97 | 13 |
| 66 | P62814 | V1 subunit B2 (brain isoform) | Proton transport | 639.65 | 56515 | 29.82 | 12 |
| 67 | Q9Z1G3 | V1 subunit C | Proton transport | 176.41 | 43702 | 10.08 | 3 |
| 68 | P57746 | V1 subunit D | Proton transport | 47.54 | 28351 | 6.23 | 1 |
| 69 | Q3UK59 | V1 subunit E isoform 1 | Proton transport | 357.23 | 26145 | 27 | 5 |
| 70 | Q9D1K2 | V1 subunit F | Proton transport | 91.08 | 13362 | 17.36 | 2 |
| 71 | Q8R2H0 | V1 subunit G2 | Proton transport | 165.4 | 13659 | 12.9 | 2 |
| 72 | Q8BVE3 | V1 subunit H | Proton transport | 38.7 | 55819 | 3.16 | 1 |
| 73 | P63001 | Ras-related C3 botulinum toxin substrate 1 | Ras-related protein | 345.83 | 21436 | 32.47 | 6 |
| 74 | Q99JI6 | Ras-related protein Rap-1b precursor | Ras-related protein | 291.41 | 20812 | 32.28 | 5 |
| 75 | P61226 | Ras-related protein Rap-2b precursor | Ras-related protein | 142.64 | 20491 | 20.43 | 2 |
| 76 | P62071 | Ras-related protein R-Ras2 precursor | Ras-related protein | 125.1 | 23385 | 13.21 | 2 |
| 77 | Q60522 | CD44 antigen precursor | Receptor | 127.99 | 46778 | 5.41 | 2 |
| 78 | P17852 | Integrin alpha-3 precursor | Receptor | 150.39 | 118475 | 2.79 | 2 |
| 79 | Q9ERD4 | Ankyrin repeat-rich membrane-spanning protein | Signalling | 45.27 | 190414 | 1.21 | 1 |
| 80 | P32261 | Antithrombin-III precursor (ATIII) | Signalling | 68.51 | 51971 | 2.54 | 1 |
| 81 | Q7TMA5 | Apolipoprotein B-100 | Signalling | 276.81 | 535688 | 1.36 | 5 |
| 82 | P61022 | Calcium-binding protein p22 | Signalling | 93.05 | 22287 | 13.86 | 2 |
| 83 | P62204 | Calmodulin | Signalling | 331.96 | 16696 | 54.97 | 4 |
| 84 | Q9DAS9 | G protein G(I)/G(S)/G(O) subunit gamma-12 precursor | Signalling | 40.48 | 7861 | 22.54 | 1 |
| 85 | P62874 | G protein G(I)/G(S)/G(T) subunit beta-1 | Signalling | 195.17 | 37222 | 10.06 | 2 |
| 86 | P62880 | G protein G(I)/G(S)/G(T) subunit beta-2 | Signalling | 171.96 | 37176 | 10.09 | 2 |
| 87 | P38403 | G protein G(k) subunit alpha (G(i) alpha-3) | Signalling | 177.74 | 40447 | 11.44 | 3 |
| 88 | P08752 | Guanine nucleotide-binding protein G(i) | Signalling | 264.43 | 40314 | 14.21 | 4 |
| 89 | Q6R0H7 | Guanine nucleotide-binding protein G(s) subunit alpha isoforms | Signalling | 206.68 | 121429 | 3.26 | 4 |
| 90 | Q3U2W7 | Kirsten rat sarcoma oncogene 2 | Signalling | 67.25 | 21400 | 6.19 | 1 |
| 91 | P62977 | Ubiquitin | Signalling | 112.42 | 8560 | 32.47 | 2 |
| 92 | Q6IN24 | Galectin 8 | sugar binding | 96.51 | 36050 | 5.81 | 2 |
| 93 | Q9CQW2 | ADP-ribosylation factor-like protein 8B | Trafficking | 77.66 | 21525 | 19.49 | 2 |
| 94 | P17426 | AP-2 complex subunit alpha-1 | Trafficking | 110.59 | 107596 | 2.45 | 2 |
| 95 | Q9DBG3 | AP-2 complex subunit beta-1 | Trafficking | 92.4 | 104516 | 4.95 | 2 |
| 96 | P62743 | AP-2 complex subunit sigma-1 | Trafficking | 50.54 | 17007 | 9.74 | 1 |
| 97 | P49817 | Caveolin-1 | Trafficking | 289.28 | 20525 | 40.32 | 6 |
| 98 | Q8VIK7 | Caveolin-2 | Trafficking | 46.57 | 18163 | 9.7 | 1 |
| 99 | P08712 | Endoplasmin | Trafficking | 41.48 | 46765 | 3.29 | 1 |
| 100 | Q6WRU0 | GPI-anchored protein GREG | Trafficking | 49.7 | 22812 | 5.31 | 1 |
| 101 | Q5SW87 | RAB1 | Trafficking | 144.33 | 15016 | 27.94 | 3 |
| 102 | Q4FJL0 | RAB10 | Trafficking | 129.62 | 22527 | 16.67 | 3 |
| 103 | Q91V41 | RAB14 | Trafficking | 51.95 | 23751 | 6.05 | 1 |
| 104 | P35293 | RAB18 | Trafficking | 48.64 | 23021 | 5.26 | 1 |
| 105 | P53994 | RAB2A | Trafficking | 177.51 | 23533 | 20.19 | 3 |
| 106 | P61021 | RAB5B | Trafficking | 36.86 | 23692 | 6.51 | 1 |
| 107 | Q3TCT9 | RAB5C | Trafficking | 132.96 | 23397 | 21.23 | 3 |
| 108 | Q3U4W5 | RAB6 | Trafficking | 41.42 | 23531 | 5.63 | 1 |
| 109 | Q4AEF6 | RAB7 | Trafficking | 163.59 | 23489 | 17.37 | 3 |
| 110 | P13596 | RAB7a | Trafficking | 140.11 | 94599 | 3.49 | 2 |
| 111 | P63321 | Ral-A precursor | Trafficking | 121.59 | 23538 | 12.68 | 3 |
| 112 | Q3UZ06 | SEC22 vesicle trafficking protein-like 1 | Trafficking | 67.84 | 18935 | 6.98 | 1 |
| 113 | O70377 | SNAP-23 | Trafficking | 84.91 | 23220 | 18.48 | 2 |
| 114 | Q5F234 | Syntaxin 8 | Trafficking | 66.07 | 26908 | 7.38 | 1 |
| 115 | O35587 | Transmembrane protein Tmp21 precursor | Trafficking | 90.42 | 24805 | 8.89 | 2 |
| 116 | Q9WV55 | VAMP-associated protein A (VAMP-A) | Trafficking | 93.96 | 27262 | 5.67 | 2 |
| 117 | O88384 | Vesicle transport v-SNARE protein Vti1-like 1 (Vti1-rp1) | Trafficking | 53.28 | 26697 | 4.96 | 1 |
| 118 | Q791V5 | Mitochondrial carrier homolog 2 | Tranporter | 52.55 | 33477 | 7.89 | 1 |
| 119 | O54724 | Polymerase I and transcript release factor | Transcription | 138.19 | 43927 | 7.27 | 2 |
| 120 | Q9Z239 | Phospholemman precursor | Unknown cellular process | 52.05 | 10316 | 12.9 | 1 |
| 121 | Q3TH64 | Q3TH64 | Unknown cellular process | 40.29 | 16236 | 23.13 | 1 |
| 122 | Q6UL10 | Q6UL10 | Unknown cellular process | 45.96 | 223986 | 0.59 | 1 |
| 123 | Q9QUR8 | Semaphorin-7A precursor | Unknown cellular process | 60.59 | 74946 | 1.91 | 1 |
| 124 | Q3TJK3 | serine (or cysteine) proteinase inhibitor | Unknown cellular process | 119.34 | 46505 | 9 | 2 |
| 125 | Q3TIP1 | solute carrier family 3 | Unknown cellular process | 40.75 | 10997 | 17.17 | 1 |
| 126 | Q6P791 | UPF0404 protein C11orf59 homolog | Unknown cellular process | 137.22 | 17710 | 16.25 | 2 |
Mw: theoretical molecular weight (kDa). Cov (%), protein sequence coverage; Nb pep, number of peptides assigned. In bold are the proteins found in both DRM and DSM fractions.
Table 2. Proteins identified in the late endosomal DSM fraction.
| Numb | Uniprot Numb | Protein name | Protein family | Mascot score | Mw | Cov (%) | Nb pep |
| 1 | Q60458 | Cationic amino acid transporter-1 | AA transport | 97.13 | 35185 | 11.6 | 2 |
| 2 | Q8VII6 | Choline transporter-like protein 1 | AA transport | 47.02 | 73043 | 1.81 | 1 |
| 3 | Q3UFR4 | Neutral amino acid transporter ASCT2 | AA transport | 174.62 | 58327 | 6.42 | 3 |
| 4 | P17897 | Lysozyme C type P precursor | Bacteriolytic | 49.06 | 16783 | 7.89 | 1 |
| 5 | Q920L2 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit | Carbohydrate metabolism | 55.31 | 71570 | 2.15 | 1 |
| 6 | Q9Z0G9 | Claudin-3 | Cell adhesion | 85.27 | 23269 | 13.27 | 2 |
| 7 | Q3T1H3 | Ncam1 protein | Cell adhesion | 400.3 | 93515 | 11.65 | 6 |
| 8 | Q91Z81 | ERP57 protein | Cell redox homeostasis | 493.44 | 56761 | 20.93 | 9 |
| 9 | P20070 | NADH-cytochrome b5 reductase 3 | Cell redox homeostasis | 243.98 | 34022 | 12.94 | 4 |
| 10 | Q60451 | NADPH-cytochrome P450 oxidoreductase | Cell redox homeostasis | 46.89 | 75803 | 1.6 | 1 |
| 11 | Q52KJ9 | Thioredoxin domain containing 1 | Cell redox homeostasis | 120.38 | 31415 | 10.53 | 2 |
| 12 | Q3THH1 | thioredoxin domain containing 5 | Cell redox homeostasis | 326.36 | 48627 | 11.54 | 4 |
| 13 | Q71FK5 | Actin, cytoplasmic 1 (Beta-actin) | Cytoskeleton | 583.76 | 41710 | 37.2 | 10 |
| 14 | P18760 | Cofilin-1 | Cytoskeleton | 111.27 | 18417 | 13.77 | 2 |
| 15 | P47753 | F-actin-capping protein subunit alpha-1 | Cytoskeleton | 54.64 | 32788 | 3.36 | 1 |
| 16 | P62962 | Profilin-1 | Cytoskeleton | 50.02 | 14816 | 10.45 | 1 |
| 17 | P54116 | Stomatin | Cytoskeleton | 48.34 | 31384 | 4.21 | 1 |
| 18 | P68361 | Tubulin alpha-1B chain | Cytoskeleton | 55.57 | 50120 | 1.98 | 1 |
| 19 | P69893 | Tubulin beta-5 chain | Cytoskeleton | 115.73 | 49639 | 4.43 | 2 |
| 20 | P02544 | Vimentin | Cytoskeleton | 305.99 | 53566 | 11.93 | 5 |
| 21 | Q8K3H8 | Calnexin | ER chaperone | 119.25 | 67224 | 5.07 | 2 |
| 22 | O08917 | Flotillin-1 | Flotillin complex | 383.61 | 47484 | 20.19 | 6 |
| 23 | Q9Z2S9 | Flotillin-2 | Flotillin complex | 445.33 | 47009 | 20.84 | 7 |
| 24 | Q9Z1W7 | GP50 | Folding and Transport regulator | 264.19 | 46569 | 14.89 | 4 |
| 25 | Q3TWF2 | heat shock 70kD protein 5 | Folding and Transport regulator | 909.11 | 72419 | 27.66 | 15 |
| 26 | P19378 | Heat shock cognate 71 kDa protein | Folding and Transport regulator | 500.69 | 70761 | 16.49 | 9 |
| 27 | P35293 | Heat shock protein HSP 90-beta | Folding and Transport regulator | 44.7 | 23021 | 5.26 | 1 |
| 28 | P17809 | Glucose transporter type 1 (GLUT-1) | Glucose transporter | 51.34 | 53899 | 1.64 | 1 |
| 29 | P05064 | Fructose-bisphosphate aldolase A | Glycolysis | 64.07 | 39200 | 5.62 | 1 |
| 30 | Q3THM2 | glyceraldehyde-3-phosphate dehydrogenase | Glycolysis | 91.15 | 35789 | 8.11 | 2 |
| 31 | O70252 | Heme oxygenase 2 | Heme homeostasis | 85.89 | 35716 | 12.35 | 2 |
| 32 | Q1MWP8 | EH-domain containing 4-KJR | Ion binding | 38.62 | 61613 | 1.96 | 1 |
| 33 | Q3U7R1 | Extended-synaptotagmin-1 (E-Syt1) | Ion binding | 114.58 | 121478 | 2.63 | 2 |
| 34 | Q6PFA8 | N-acylsphingosine amidohydrolase 1 | Ion binding | 45.67 | 69734 | 1.58 | 1 |
| 35 | Q05186 | Reticulocalbin-1 precursor | Ion binding | 42.58 | 38090 | 4.34 | 1 |
| 36 | Q7TMC7 | Ab2-417 (Cc1-8) | Ion transport | 110.61 | 107343 | 2.97 | 2 |
| 37 | Q3V132 | ADP/ATP translocase 4 | Ion transport | 47.67 | 35235 | 3.75 | 1 |
| 38 | P15999 | ATP synthase subunit alpha | Ion transport | 58.99 | 59717 | 2.21 | 1 |
| 39 | P56480 | ATP synthase subunit beta | Ion transport | 140.84 | 56265 | 5.09 | 2 |
| 40 | O55143 | endoplasmic reticulum calcium ATPase 2 | Ion transport | 107.98 | 114784 | 3.45 | 2 |
| 41 | Q09143 | High affinity cationic amino acid transporter 1 | Ion transport | 98.32 | 67048 | 3.94 | 2 |
| 42 | Q91VS7 | Microsomal glutathione S-transferase 1 | Ion transport | 83.61 | 17409 | 7.59 | 1 |
| 43 | Q8VDN2 | Na(+)/K(+) ATPase alpha-1 subunit | Ion transport | 621.11 | 112910 | 16.37 | 10 |
| 44 | P11505 | Plasma membrane calcium-transporting ATPase 1 | Ion transport | 522.49 | 138632 | 10.32 | 11 |
| 45 | Q3TX38 | voltage-dependent anion channel 3 | Ion transport | 232.58 | 30733 | 19.35 | 4 |
| 46 | Q9Z2L0 | Voltage-dependent anion-selective channel protein 1 (VDAC-1) | Ion transport | 245.29 | 30606 | 23.38 | 5 |
| 47 | Q60930 | Voltage-dependent anion-selective channel protein 2 (VDAC-2) | Ion transport | 177.68 | 31713 | 14.93 | 3 |
| 48 | P51909 | Apolipoprotein D | Lipid biogenesis | 62.59 | 21596 | 6.12 | 1 |
| 49 | Q8BLF1 | Arylacetamide deacetylase-like 1 | Lipid biogenesis | 122.47 | 45711 | 5.06 | 2 |
| 50 | O88531 | Palmitoyl-protein thioesterase 1 precursor | Lipid biogenesis | 80.12 | 34467 | 4.79 | 1 |
| 51 | Q9JKU9 | Sigma 1 receptor beta variant | Lipid biogenesis | 52.54 | 21589 | 6.63 | 1 |
| 52 | Q9R1J0 | Sterol-4-alpha-carboxylate 3-dehydrogenase | Lipid biogenesis | 42.14 | 40660 | 3.52 | 1 |
| 53 | Q69ZN7 | Myoferlin (Fer-1-like protein 3) | Membrane repair | 152.49 | 236053 | 2.38 | 3 |
| 54 | Q99KV1 | DnaJ homolog subfamily B member 11 precursor | mRNA modification | 41.04 | 40530 | 2.99 | 1 |
| 55 | P00762 | Anionic trypsin-1 precursor | Peptidase | 81.51 | 25943 | 8.51 | 1 |
| 56 | P07150 | Annexin A1 | Phospholipid binding/modifying | 223.58 | 38674 | 7.12 | 3 |
| 57 | P07356 | Annexin A2 | Phospholipid binding/modifying | 934.99 | 38521 | 40.57 | 14 |
| 58 | Q3UCL0 | annexin A4 | Phospholipid binding/modifying | 177.72 | 35893 | 9.51 | 3 |
| 59 | P48037 | Annexin A6 | Phospholipid binding/modifying | 367.61 | 75575 | 14.26 | 8 |
| 60 | P14824 | Annexin A6 | Phospholipid binding/modifying | 359.86 | 75707 | 13.66 | 8 |
| 61 | Q8VDP6 | Phosphatidylinositol synthase | Phospholipid binding/modifying | 37.88 | 23583 | 5.14 | 1 |
| 62 | Q8R366 | Immunoglobulin superfamily member 8 precursor | Protein binding | 115.51 | 64970 | 3.73 | 2 |
| 63 | Q4FCR4 | Intercellular adhesion molecule 1 | Protein binding | 186.97 | 53809 | 8.38 | 3 |
| 64 | P21956 | Lactadherin precursor | Protein binding | 47.39 | 51236 | 2.37 | 1 |
| 65 | P70117 | Pancreas cancer-associated protein 4 | Protein binding | 166.84 | 64358 | 6.67 | 3 |
| 66 | P70206 | Plexin-A1 precursor | Protein binding | 75.36 | 210965 | 1.3 | 2 |
| 67 | Q9WV91 | Prostaglandin F2 receptor negative regulator precursor | Protein binding | 91.15 | 98646 | 3.46 | 2 |
| 68 | Q5SS40 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein | Protein binding | 50.26 | 29155 | 4.91 | 1 |
| 69 | P62629 | Elongation factor 1-alpha 1 | Protein biosynthesis | 208.12 | 50082 | 12.97 | 4 |
| 70 | P10630 | Eukaryotic initiation factor 4A-II | Protein biosynthesis | 40.71 | 46373 | 2.38 | 1 |
| 71 | Q66H94 | Peptidyl-prolyl cis-trans isomerase (PPIase) | Protein biosynthesis | 44.5 | 63086 | 2.09 | 1 |
| 72 | Q8R2Y8 | Peptidyl-tRNA hydrolase 2 (PTH 2) | Protein biosynthesis | 50.9 | 19514 | 7.82 | 1 |
| 73 | Q9CY50 | Translocon-associated protein subunit alpha precursor | Protein biosynthesis | 44.77 | 32045 | 5.15 | 1 |
| 74 | Q62186 | Translocon-associated protein subunit delta precursor | Protein biosynthesis | 58.73 | 18924 | 6.4 | 1 |
| 75 | Q60432 | 150 kDa oxygen-regulated protein | Protein folding | 111.56 | 111202 | 3.56 | 2 |
| 76 | Q1PSW2 | 84 kDa heat shock protein | Protein folding | 268.13 | 83230 | 5.29 | 4 |
| 77 | P24369 | Cyclophilin B | Protein folding | 159.73 | 22699 | 17.96 | 3 |
| 78 | P30412 | Cyclophilin C | Protein folding | 111.37 | 22780 | 12.08 | 2 |
| 79 | Q8R180 | ERO1-like protein alpha precursor (ERO1-Lalpha) | Protein folding | 40.76 | 54004 | 2.45 | 1 |
| 80 | Q3U5T8 | t-complex protein 1 | Protein folding | 97.92 | 55438 | 4.17 | 2 |
| 81 | P80318 | T-complex protein 1 subunit gamma | Protein folding | 82.19 | 60591 | 2 | 1 |
| 82 | O70152 | Dolichol-phosphate mannosyltransferase | Protein processing | 41.77 | 29156 | 8.3 | 1 |
| 83 | Q3UC51 | dolichyl-di-phosphooligosaccharide-protein glycotransferase | Protein processing | 197.03 | 49011 | 9.44 | 4 |
| 84 | Q03145 | Ephrin type-A receptor 2 precursor | Protein processing | 77.94 | 108753 | 2.73 | 2 |
| 85 | O08795 | Glucosidase 2 subunit beta precursor | Protein processing | 42.18 | 58756 | 1.87 | 1 |
| 86 | P17439 | Glucosylceramidase precursor | Protein processing | 81.57 | 57585 | 4.59 | 2 |
| 87 | P57716 | Nicastrin precursor | Protein processing | 48.9 | 78441 | 1.54 | 1 |
| 88 | P27773 | Protein disulfide-isomerase A3 precursor | Protein processing | 421.29 | 56586 | 18.29 | 8 |
| 89 | P38660 | Protein disulfide-isomerase A6 precursor | Protein processing | 264.65 | 48131 | 11.67 | 3 |
| 90 | Q8R4U2 | Protein disulfide-isomerase precursor (PDI) | Protein processing | 516.21 | 56975 | 27.85 | 9 |
| 91 | P52480 | Pyruvate kinase isozymes M1/M2 | Protein processing | 257.65 | 57719 | 14.89 | 5 |
| 92 | Q91YQ5 | Ribophorin I | Protein processing | 47.47 | 68486 | 1.93 | 1 |
| 93 | Q9DBG6 | Ribophorin II | Protein processing | 97.57 | 69020 | 3.67 | 2 |
| 94 | Q00993 | Tyrosine-protein kinase receptor UFO precursor | Protein processing | 70.28 | 98188 | 1.23 | 1 |
| 95 | Q6AXQ3 | Viral oncogene yes-1 homolog 1 | Protein processing | 36.55 | 54236 | 4.46 | 1 |
| 96 | Q3TFU8 | Glycoprotein 25L2 homolog | Protein transport | 89.06 | 31239 | 7.42 | 2 |
| 97 | P62835 | Rap-1A precursor | Ras-related protein | 240.69 | 20974 | 18.42 | 4 |
| 98 | P80236 | Ras-related C3 botulinum toxin substrate 1 (p21-Rac1) | Ras-related protein | 47.41 | 8805 | 17.5 | 1 |
| 99 | P61226 | Ras-related protein Rap-2b precursor | Ras-related protein | 69.2 | 20491 | 6.45 | 1 |
| 100 | P62071 | Ras-related protein R-Ras2 precursor | Ras-related protein | 79.46 | 23385 | 7.55 | 1 |
| 101 | Q61411 | Transforming protein p21 (p21ras) | Ras-related protein | 49.31 | 21335 | 6.22 | 1 |
| 102 | P17852 | Integrin alpha-3 precursor | Receptor | 329.02 | 118475 | 6.59 | 5 |
| 103 | P43406 | Integrin alpha-V precursor | Receptor | 120.21 | 115205 | 2.87 | 3 |
| 104 | P09055 | Integrin beta-1 precursor | Receptor | 90.71 | 88173 | 2.5 | 2 |
| 105 | P61022 | Calcium-binding protein p22 | Signalling | 58.15 | 22287 | 5.94 | 1 |
| 106 | P62204 | Calmodulin | Signalling | 187.09 | 16696 | 30.46 | 3 |
| 107 | P62874 | G protein G(I)/G(S)/G(T) subunit beta-1 | Signalling | 186.51 | 37222 | 13.02 | 3 |
| 108 | P62880 | G protein G(I)/G(S)/G(T) subunit beta-2 | Signalling | 151 | 37176 | 13.06 | 3 |
| 109 | Q3HR13 | Guanine nucleotide binding protein alpha inhibiting 2 | Signalling | 235.3 | 40419 | 14.17 | 4 |
| 110 | Q5EAP4 | Guanine nucleotide binding protein, alpha 14 | Signalling | 227.12 | 41415 | 12.5 | 4 |
| 111 | P21279 | Guanine nucleotide-binding protein G(q) subunit alpha | Signalling | 273.99 | 41457 | 20.21 | 5 |
| 112 | Q8R4A8 | Guanine nucleotide-binding protein G(s) subunit alpha | Signalling | 232.48 | 45621 | 13.04 | 5 |
| 113 | P62977 | Ubiquitin | Signalling | 114.62 | 8560 | 32.47 | 2 |
| 114 | O88736 | 3-keto-steroid reductase | Steroid metabolism | 58.55 | 37293 | 3.54 | 1 |
| 115 | P47953 | Galectin-3 | Sugar binding | 50.42 | 25592 | 4.31 | 1 |
| 116 | Q3U0D7 | ADP-ribosylation factor 6 | Trafficking | 37.23 | 20069 | 6.04 | 1 |
| 117 | Q9CQW2 | ADP-ribosylation factor-like protein 8B | Trafficking | 205.81 | 21525 | 17.44 | 4 |
| 118 | Q9DB05 | Alpha-soluble NSF attachment protein (SNAP-alpha) | Trafficking | 173.24 | 33168 | 12.62 | 3 |
| 119 | P49020 | COPI-coated vesicle membrane protein p24 | Trafficking | 66.59 | 22175 | 6.47 | 1 |
| 120 | P08113 | Endoplasmin precursor | Trafficking | 334.23 | 92418 | 5.6 | 5 |
| 121 | P49130 | LAMP-2 | Trafficking | 41.5 | 44999 | 2.44 | 1 |
| 122 | O35604 | Niemann-Pick C1 protein precursor | Trafficking | 36.07 | 142795 | 1 | 1 |
| 123 | Q91ZX7 | Prolow-density lipoprotein receptor-related protein 1 precursor (LRP) | Trafficking | 107.05 | 504411 | 0.68 | 2 |
| 124 | O35074 | Prostacyclin synthase | Trafficking | 170.13 | 57011 | 7.34 | 3 |
| 125 | Q9D0F3 | Protein ERGIC-53 precursor | Trafficking | 52.6 | 57753 | 2.1 | 1 |
| 126 | Q4FJL0 | RAB10 | Trafficking | 174.32 | 22559 | 16.1 | 3 |
| 127 | P62492 | RAB11A | Trafficking | 51.03 | 24247 | 5.91 | 1 |
| 128 | Q91V41 | RAB14 | Trafficking | 99.99 | 23751 | 11.16 | 2 |
| 129 | P62821 | RAB1A | Trafficking | 187.04 | 22532 | 19.12 | 3 |
| 130 | P53994 | RAB2A | Trafficking | 263.69 | 23533 | 30.52 | 5 |
| 131 | Q3UCX7 | RAB5A | Trafficking | 80.78 | 23585 | 11.68 | 2 |
| 132 | P61021 | RAB5B | Trafficking | 141.52 | 23692 | 20.47 | 3 |
| 133 | Q3TCT9 | RAB5C | Trafficking | 200.16 | 23398 | 23.11 | 4 |
| 134 | Q3U4W5 | RAB6 | Trafficking | 269.71 | 23531 | 28.64 | 5 |
| 135 | Q4AEF6 | RAB7 | Trafficking | 40.92 | 28557 | 3.47 | 1 |
| 136 | Q3UZ06 | SEC22 vesicle trafficking protein-like 1 | Trafficking | 97.82 | 18935 | 6.98 | 1 |
| 137 | Q8K021 | Secretory carrier membrane protein 1 | Trafficking | 40.66 | 38004 | 6.67 | 1 |
| 138 | Q62465 | Synaptic vesicle membrane protein VAT-1 homolog | Trafficking | 77.87 | 43069 | 3.58 | 1 |
| 139 | O88385 | Syntaxin 12 | Trafficking | 55.46 | 31090 | 5.32 | 1 |
| 140 | Q3TSL5 | syntaxin 4A | Trafficking | 76.27 | 34204 | 7.1 | 2 |
| 141 | Q9JI92 | Syntenin-1 | Trafficking | 109.68 | 32403 | 11.33 | 2 |
| 142 | Q61235 | Syntrophin-3 (SNT3) | Trafficking | 162.55 | 56346 | 7.42 | 3 |
| 143 | Q07891 | Transferrin receptor protein 1 (TfR) | Trafficking | 51.95 | 85027 | 1.42 | 1 |
| 144 | O35587 | Transmembrane protein Tmp21 precursor | Trafficking | 152.47 | 24805 | 16.44 | 3 |
| 145 | P63024 | Vesicle-associated membrane protein 3 (VAMP-3) | Trafficking | 55.54 | 11473 | 15.38 | 1 |
| 146 | Q791V5 | Mitochondrial carrier homolog 2 | Transporter | 71.58 | 33477 | 7.89 | 1 |
| 147 | Q7TP91 | Ab1-205 | Unknown cellular process | 50.91 | 83209 | 1.72 | 1 |
| 148 | Q9D7N9 | Adipocyte plasma membrane-associated protein | Unknown cellular process | 94.98 | 46405 | 5.46 | 2 |
| 149 | P20944 | CD44 antigen precursor | Unknown cellular process | 142.8 | 39750 | 4.99 | 2 |
| 150 | Q9QV38 | ERP6 | Unknown cellular process | 54.19 | 2112 | 73.68 | 1 |
| 151 | Q8BLN5 | Lanosterol synthase (EC 5.4.99.7) | Unknown cellular process | 68.6 | 83088 | 1.85 | 1 |
| 152 | Q6P7S1 | N-acylsphingosine amidohydrolase 1 | Unknown cellular process | 150.07 | 44415 | 10.42 | 3 |
| 153 | P97300 | Neuroplastin precursor | Unknown cellular process | 60.1 | 31258 | 3.52 | 1 |
| 154 | Q9Z239 | Phospholemman precursor | Unknown cellular process | 38.49 | 10316 | 12.9 | 1 |
| 155 | O55221 | Putative CD98 protein | Unknown cellular process | 63.88 | 58889 | 3.74 | 1 |
| 156 | Q9JK11 | Reticulon-4 | Unknown cellular process | 59.01 | 126310 | 1.13 | 1 |
| 157 | Q9CWD1 | similar to DB83 PROTEIN | Unknown cellular process | 66.14 | 19857 | 6.67 | 1 |
| 158 | Q3TIP1 | solute carrier family 3 | Unknown cellular process | 55.26 | 10997 | 17.17 | 1 |
| 159 | Q5XIK2 | Thioredoxin domain-containing protein 14 precursor | Unknown cellular process | 39.08 | 33844 | 3.91 | 1 |
| 160 | Q2TBF8 | Transmembrane 9 superfamily protein member 4 | Unknown cellular process | 41.68 | 74584 | 1.33 | 1 |
| 161 | Q6P791 | UPF0404 protein C11orf59 homolog | Unknown cellular process | 116.65 | 17710 | 16.25 | 2 |
Mw: theoretical molecular weight (kDa). Cov (%), protein sequence coverage; Nb pep, number of peptides assigned. In bold are the proteins found in both DRM and DSM fractions.
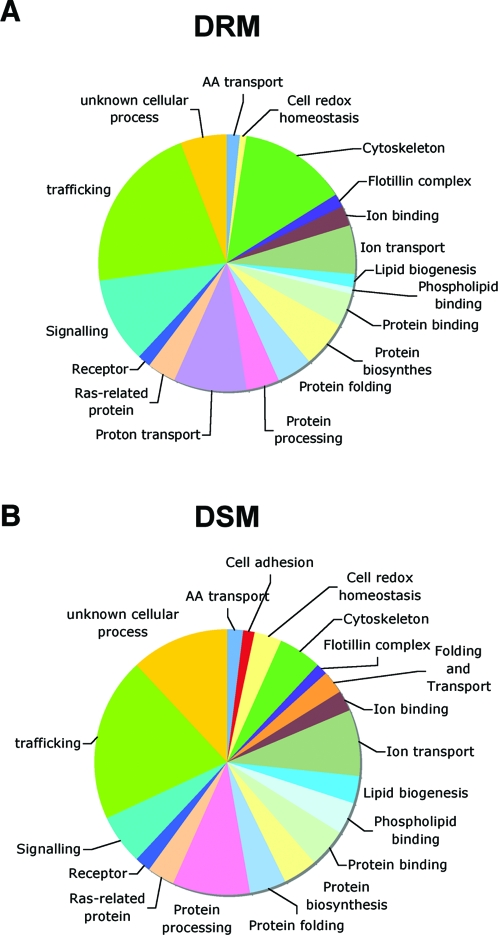
As illustrated in Fig. 1, proteins forming the largest group, in both fractions, were involved in trafficking, confirming that late endosomes are implicated in intensive communication with other compartments. In particular, several small GTPases of the Rab family, in addition to Rab7 well known to localize to late endosomes, were found, in agreement with other proteomic studies [40] and suggesting a more complex role/localization of Rabs than expected.
Figure 1. Graphic representation of the main functional categories found in the proteomic studies.
A: DRM fraction, B: DSM fraction.
Interestingly many of the identified proteins were involved in signaling, in particular in the DRMs, such as the α and ß subunits of several G proteins, as also reported by others reported [40], [41]. The presence of these proteins in late endosomes supports the notion that endosomes are active signaling sites [42] and that cholesterol rich domains are important in organizing signaling platforms [43].
However, the most striking finding to us was the identification of all the subunits of the V-ATPase (a1, c and d from the V0 subunit and A, B2, C, D, E, F, G2 and H from the V1 subunit) exclusively in the DRM fraction. This observation suggested that the V-ATPase may localize to specific membrane domains within late endosomes and that this could be important for its function or the regulation of its activity.
There is increasing evidence that the V-ATPase can interact with numerous regulatory proteins, and thus we investigated whether some of these proteins came up in our proteomics analysis. The H-subunit, which shows homology to ß-adaptins [44], was found to interact with the m2-chain of AP2 adaptor and promotes clathrin-coated vesicle (CCV) formation [44]. Interestingly we found three of the subunits of the adaptor complex AP-2, in the late endosomal DRM fraction. Functional AP-2 complex was previously observed in lysosomes and found to support clathrin assembly and bud formation [45], [46]. We also found the small GTPase Arf6, albeit in DSM fraction. Arf6 was found to interact with the Vo c subunit and it was proposed that through this interaction, the vATPase could modulate membrane trafficking in the endosomal system [47]. In addition to mediating proton transport, it has been proposed that the Vo subunit is involved in vesicles fusion both in yeast [48] and in higher eukaryotes [49], [50]. Numerous proteins involved in membrane fusion were found in the DRM fraction including SNAP-23, syntaxin 8, VAMP-A and the v-SNARE like protein Vtil-rp1.
Finally the dissociation of the V1/vo complex, in yeast, has been shown to require an intact microtubule network [51] and well aldolase, which would act as a glucose sensor and signal for the dissociation of the V-ATPase [52]. Both tubulin and aldolase were detected in our proteomic analysis. Interestingly aldolase was only found in the DSM fraction and it is tempting to speculate that spatial segregation between the V-ATPase and aldolase is necessary to prevent uncontrolled disassembly of the complex. Clearly, non transmembrane proteins could also be removed from the detergent resistant domains during the solubilization.
DRM association of the V-ATPase
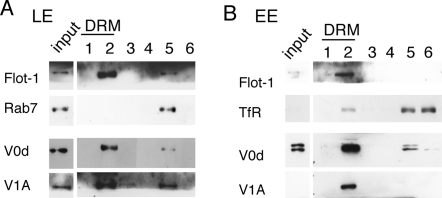
To validate the DRM association of the V-ATPase revealed by the proteomic analysis, we performed a Western blot analysis probing for V-ATPase subunits of the V1 and V0 domains. Distribution of the V0 domain was monitored by following the d subunit and distribution of the V1 domain by following the A subunit. Both V0d and V1A subunits were found in the DRM fractions of both late endosomes and early endosomes (Fig. 2). Note that the total protein content of each fraction was analyzed in Fig. 2, as opposed to the same amount of protein. Fig. 2 thus shows that the majority of the V-ATPase localized to DRMs, showing a distribution similar to that of Flotillin-1, a well-established marker of DRM fractions [38].
Figure 2. Association of V1 and V0 domains of the V-ATPase with the DRMs of early and late endosomes.
Late (A) and early (B) endosomes were purified from BHK cells and the fractions were submitted to solubilization in 1% triton X-100 at 4°C. The Triton X-100 treated fractions were subsequently loaded at the bottom of an OptiPrep gradient. After centrifugation, 6 fractions were collected from the top and analyzed by SDS-PAGE followed by Western blotting to detect flotilin-1, Rab7, the vATPas subunits V0d and V1A.
Effects of U18666A treatment on the acidification of late endosomes
The above experiments show that all along the endocytic pathway, from early to late endosomes, the V-ATPase exhibits a strong affinity for DRMs suggesting its preference for cholesterol rich, ordered membrane domains. To evaluate the physiological relevance of these findings, we investigated whether perturbing cholesterol rich domains in late endosomes would affect the function of the V-ATPase, by monitoring the acidity of late endosomes.
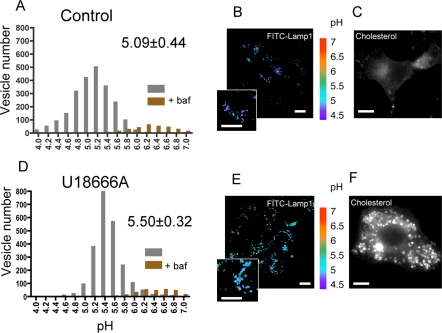
We first investigated whether extraction of cholesterol from cells using ß-methylcyclodextrin (ß-MCD) would lead to a change in late endosome acidification. Cells were treated with 10 mM ß-MCD for 55 min leading to an ≈50% decrease in total cholesterol content [53]. As illustrated in Fig. S1, ß-MCD treatment led to a significant increase in late endosomal pH, supporting the notion that alteration of cholesterol rich domains affects the function of the V-ATPase. These findings however need to be taken with a word of caution, since we have no evidence that extraction of cholesterol from the plasma membrane actually leads to a significant reduction of cholesterol in late endosomes. Indeed when purifying late endosomes from ß-MCD treated cells, we could not detect a significant change in cholesterol, as analyzed by thin layer chromatography, a technique that might not be sensitive enough to detect small changes (10–20%). We therefore decided to induce perturbation of late endosomal membrane domains by treating cells with the negatively charged amine 3beta-(2-diethylaminoethoxy)-androstenone HCl (U18666A) [54], which leads to the accumulation of cholesterol in late endosomes, through unknown mechanisms, and to alterations in the membrane dynamics of this compartment [34]. Treatment for 18 hrs with U18666A mimics the Niemann Pick type C phenotype [34], [54], [55] showing a drastic cholesterol accumulation in late endosomes (Fig. 3CF and 4CF).
Figure 3. Effects of U18666A treatment on the pH at the limiting membrane (FITC-Lamp1) of late endosomes.
Lamp1-FITC was internalized overnight at 37°C by control cells (A, B, C) and cells treated by U18666A (D, E, F). Typical examples of the respective labels are shown (B and E). The pH of individual organelles was measured by fluorescence ratio imaging of internalized Lamp1-FITC. The histograms show the pH distribution of 2304 and 2325 endosomes for the upper and the lower panels, respectively, with the mean±SD given in the left panels (A and D). Pseudocolor pH scale is on the side. Histograms of vesicles of cells treated with bafilomycin are shown as a control. The observed difference is significant according to a paired t-test with p<0.001. Cells were checked for their phenotype of cholesterol accumulation with filipin staining (C and F). Bars, 10 µm.
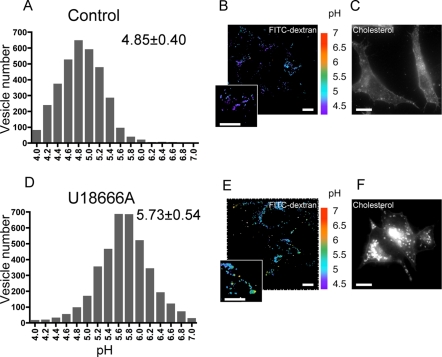
Figure 4. Effects of U18666A treatment on the pH of late endosomal lumen (FITC-dextran).
FITC-dextran was internalized for 15 minutes and chased for 40 minutes at 37°C in control cells (A, B, C) and cells treated by U18666A (D, E, F) to allow it to reach late endosomes. Typical examples of the respective labels are shown (B and E). The pH of individual organelles was measured by fluorescence ratio imaging of internalized FITC-Dextran. Pseudocolor pH scale is on the side. The histograms show the pH distribution of 3346 and 3880 endosomes for the upper and the lower panels, respectively, with the mean±SD given in the left panels (A and D). The observed difference is significant according to a paired t-test with p<0.001. Cells were checked for their phenotype of cholesterol accumulation with filipin staining (C and F). Bars, 10 µm.
The pH measurements were performed by fluorescence ratio imaging, using the pH sensitive florescent probe FITC (fluorescein isothiocyanate) [56]. We used two different probes: FITC coupled to dextran, which was endocytosed by fluid phase and thus provides information on the bulk of the endosomal lumen, and FITC coupled to a monoclonal antibody, 4A1, against the luminal domain of the hamster late endosomal protein Lamp1. Lamp1 localizes specifically to the limiting membrane of late endosomes [57] –this organelle being multivesicular [23]–, and thus FITC anti-lamp1 allows the measurement of the pH in the vicinity of the limiting membrane. We find that the 4A1 anti-Lamp1 antibody internalized overnight (Fig. 3B, 3E), reaches late endosomes where it binds to its antigen (co-localizing with the late endosomal lipid LBPA, not shown) and is not degraded, in agreement with observations by others [58], [59]. In contrast, a none specific antibody endocytosed at the same concentration was degraded in late endosomes/lysosomes, as documented by others [59]. The fluid phase fluorescent probe, FITC-Dextran was internalized for 15′ and chased for 40′ in order to reach late endosomes (Fig. 4B, 4E). Importantly we have previously shown that U18666A treatment does not prevent transport to late endosomes [34]. Series of images were taken at both 440 nm and 490 nm, prior to calibration with different pH solutions, which allows the translation of ratio values into pH values. As illustrated by the pseudocolor images (Fig. 3B, 4B) and quantified in the distribution histograms (Fig. 3A, 4A), the pH of late endosomes in control cells varied between 4.85±0.40 and 5.09±0.44 depending on the probe used, a pH that was neutralized upon treatment with the V-ATPase specific inhibitor bafilomycin (Fig. 3A). Late endosomes of U18666A cells were however 0.41 (using anti-lamp1 FITC) to 0.88 (using FITC dextran) pH units less acidic than late endosomes from control cells, yet pH was still sensitive to bafilomycin, suggesting that acidity was still mainly due to the V-ATPase.
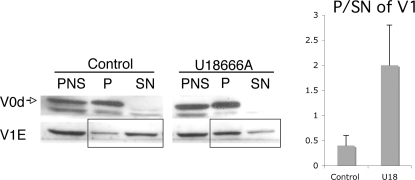
We next analyzed the effect of U18666A biochemically, monitoring the association of the V-ATPase subunits with membranes. Post nuclear supernatant (PNS) from control and U18666A treated BHK cells were submitted to high-speed centrifugation to separate the membranes (P: pellet) from the cytosol (SN: supernatant). As expected for a V0 domain subunit, V0d was entirely membranes associated, irrespective of U18666A treatment. In contrast, the majority of V1E was in the cytosolic fraction of control cells with only a minor fraction associated with membranes. Interestingly, this distribution was reversed upon U18666A treatment, leading a two-fold increase in the membrane to cytosol ratio of the V1E subunit (Fig. 5). This result indicates that the lipid composition of the membrane, and possibly the degree of order of the membrane, affects the V1-V0 association/dissociation of the V-ATPase. The observations are compatible with a higher V1 off rate in a more fluid membrane. The transport of protons by the V-ATPase requires the association of the V1 sector with the V0. Therefore an increased association of V1 with membranes as observed upon U18666A would a priori be expected to lead to an increased activity and thus more acidic endosomes. Yet U18666A led to a decrease in acidity. Together these observations suggest that U18666A altered the dynamics of association/dissociation, which appear to be essential for proper V-ATPase function, leading to a locked configuration of the V-ATPase.
Figure 5. Increase of V1 association to membrane upon U18666A treatment.
PNS was submitted to high speed spinning and pellet (P) and supernatant (SN) were loaded on a gel. Western blotting was revealed with V0d and V1E subunit antibody. Quantification reveals that upon U18666A treatment, V1E is 4-fold more associated with the membrane (pellet) than in control cells.
Increase of the V1/V0 ratio along the endocytic pathway
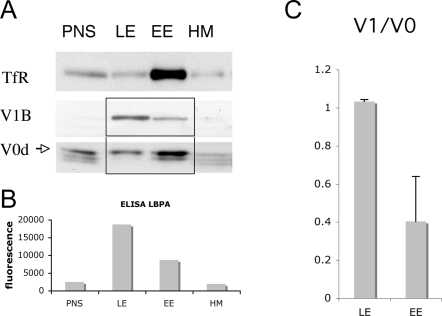
The above observation suggests that the membrane environment influences the activity of the V-ATPase. This does not exclude other mechanisms of regulation and in particular we found that the V-ATPase was associated with DRMs in both early and late endosomes, rendering it unlikely that membrane localization is the sole responsible for the different pH in early and late endosomes. We therefore investigate whether association/dissociation of the V1 domain form the V0 domain, the regulatory mechanism found in yeast, could contribute to the increase in acidity along the endocytic pathway. Early and late endosomes fractions, enriched in the transferrin receptor and the late endosomal lipid lysobisphosphatidic acid (LBPA, used a marker of late endosomes) respectively (Fig. 6A and 6B), were obtained by differential centrifugation from BHK cells and separated from heavy membranes containing the endoplasmic reticulum and the Golgi (Fig. 6). As expected, the V0 and V1 subunits, revealed with anti- V0d and anti-V1B respectively, were detected in both early and late endosomes (Fig. 6A). As observed through the analysis of multiple experiments, the enrichment of the V0 subunit d was quite similar in early and late endosomes indicating that the density of the proton translocator subunit was the same in both compartments (not shown). What was strikingly different between early and late endosomes was however the relative abundance of the V1 domain, as witness by following the B (Fig. 6A), the A or E subunits (data not shown): the V1/V0 ratio was always higher in late than in early endosomes, as quantified from several experiments in Fig. 6B (n = 3, note that the ratios only have a relative meaning and do not provide stochiometric information because the antibodies against the two subunits are by definition different). The higher V1/V0 ratio in late vs. early endosomes correlates with the higher acidity of this compartment, suggesting that an increase in V-ATPase activity through an increase in V1-V0 association is the underlying mechanism and that this mechanism of regulation is not restricted to yeast [14], [60] and maturing dendritic cells [16].
Figure 6. Increased assembly of the two sectors of the V-ATPase in late endosome.
Subcellular fractionation of postnuclear supernatants (PNS) was performed to separate early (EE) from late endosomes (LE) and heavy membranes (HM) A: 20 µg of protein from each fraction were separated in 12.5% SDS-PAGE and blotted for the presence of TfR (early endosomal marker), V0d and V1B. B: The late endosomal lipid LBPA was used to follow the distribution of late endosomes on the gradient. An equal amount of protein was used for LBPA detection by ELISA with the LBPA antibody (6C4). C: Western blot was quantified using a Phosphoimager (Bio-Rad Laboratories) and the ratio V1/V0 was plotted. Note that the ratio LE/EE is higher for the V1B subunit than for the V0d subunit suggesting a higher assembly of the V-ATPase in the late endosomes compared to early endosomes.
Discussion
Endosomes are the most acidic organelles in mammalian cells, yet little is known about the exact mechanisms that regulate their pH and how acidity increases along the endocytic pathway. Although the V-ATPase is the main contributor to this acidity, how its activity is regulated is to a large extent unknown. The main mechanism described to date is the reversible dissociation of the V0 and V1 domains of the V-ATPase. It was first discovered in yeasts and insects. In these organisms however, dissociation appears as a survival mechanism to conserve the ATP stock upon conditions of energy limitation by glucose deprivation or starvation [7], [14], [15]. The known glucose-induced signaling pathways do not seem to be involved since mutants in the ras-cyclic AMP pathway or in the protein kinase C pathway, continue to assemble in the presence of glucose [7], [61]. Regulation by reversible dissociation has also been described in specialized mammalian cells, such as renal epithelial cells, where reversible dissociation is, as in yeast, coupled to glucose levels [62], [63]. The only situation in which changes in V1-V0 association were aimed at controlling endosomal pH was during maturation of dendritic cells, where increased acidification was shown to increase the activity of hydrolytic enzymes and thus the efficiency of antigen processing [16]. Our work suggests that, as in maturing dendritic cells, increased assembly of the V-ATPase sectors along the endocytic pathway of fibroblast-like cells contributes to increasing the acidity of the organelles.
Changes in V1-V0 association raise the questions of the mechanisms that regulate this process. Our striking finding that all the subunits of the V-ATPase localized to the DRM fraction and that treatment of cells with U18666A, which affects late endosomal cholesterol levels, lead to an increase in V1-V0 association, together point to a role of the membrane composition in controlling the rate of V1-V0 association-dissociation. This could be either through the direct effect of specific lipids on the V-ATPase mediated by a lipid-protein interaction, or/and through an effect of the membrane fluidity. High fluidity, as possibly found at the plasma membrane, would lead to V1 dissociation and a drop in V-ATPase mediated acidification. Similarly low membrane fluidity, as promoted by U18666A dependent cholesterol accumulation, would lead to a too strong V1-V0 interaction preventing proper function of the proton pump, and thus again reduction in activity. The effect of U18666A is reminiscent of the marine-derived V-ATPase inhibitor salicylihalamide A. This metabolite binds to the Vo complex, as do Bafilomycin A or concanamycin, but to a different site. More strikingly, salicylihalamide A triggers a dramatic redistribution of the V1 complex from the cytoplasm to the endosomes, a redistribution not observed with the other inhibitors [64]. The paradoxical effects of U18666A and salicylihalamide A suggest that excessive stabilization of the V1-V0 complexes is detrimental to the pumps activity.
The hypothesis that raft-like domains are involved in regulating the activity of the mammalian V-ATPase is further supported by findings by others, in yeast, that sphingomyelin [20], and possibly ceramide [65] modulate the function of the V-ATPase. Interestingly, ceramide was also found to be required for oligomerization of the plasma membrane proton ATPase in yeast [66].
We wish to mention that the V-ATPase has previously been found in a raft fraction of the plasma membrane of neurons [67]. However only components of the V0 domain were identified. In contrast, we here found all eight V1 subunits (table 1). Yoshinaka et al. [43] hypothesize that the V-ATPase may disassemble during sample preparation or that the V0 sector selectively associates with rafts. Our results show that both V1 and V0 sectors can be recovered in the DRM fraction. The apparent discrepancy between the two studies could be due to the fact that plasma membrane fractions were analyzed in the first study and that the V-ATPase is mostly disassembled at that cellular site as opposed to endosomes studied here. This interpretation is supported by our finding that assembly of V1 and V0 increases along the endocytic pathway and would thus be lowest at the cell surface.
The here identified effect of membrane composition on the activity of the V-ATPase suggests that diseases that affect lipid distribution or composition, in particular in the endocytic pathway, may lead to alterations in endosomal pH. It will therefore be of interest to determine if endosomal pH is affected in cells from patients suffering from lipid storage diseases such as Niemann Pick type C and how this affects endosomal function.
Materials and Methods
Cell, reagents and drug treatment
Monolayers of Baby hamster kidney (BHK) cells were grown and maintained as described previously [68], [69]. The monoclonal antibodies against LBPA (6C4) and Lamp 1 (4A1) have been described [58], [70]. Monoclonal anti-transferrin receptor antibodies were from Zymed Laboratories Inc. Rabbit polyclonal anti-flotillin antibodies were produced by our laboratory [25]. Polyclonal antibody against the 39-kDa subunit of the V-ATPase (V0d) was previously described [71]. Rabbit polyclonal antibodies against V1 subunit A and V1 subunit E were kind gifts from I. Schultz and antibodies against V1 subunit B were produced by Eurogentech. Rhodamine-dextran (10,000 Da) was from Molecular Probes. For pH measurement, Lamp1 antibody was labeled with FITC (Fluo reporter labeling kit) from Molecular probes. Cells were treated with 3beta-(2-diethylaminoethoxy)-androsterone HCl (U18666A) as described [34]. Briefly, the cell culture medium was removed 4 hours after plating and fresh medium containing U18666A at 3 µg/ml was added for 18 hours.
Subcellular fractionation and Immunoblotting
Early and late endosomal fractions were prepared as described [32], [34]. Briefly, BHK cells were homogenized, a post nuclear supernatant (PNS) was prepared, adjusted to 40.6% sucrose, loaded on the bottom of a SW41 tube and overlaid sequentially with 35%, 25% and 8.5% sucrose solution in 3mM imidazole, pH 7.4. The gradient was centrifuged for 90 minutes at 35000 rpm. Heavy membranes, early and late endosomal fractions were collected at the 40.6/35%, 35/25% and 25/8.5% interfaces respectively. Proteins were separated by SDS-PAGE using 12.5% acrylamide gels and transferred onto a nitrocellulose membrane. Western blots were revealed with SuperSignal Chemiluminescence (Pierce) and quantified by densitometry.
Isolation of DRMs from early and late endosomal fraction
DRMs were prepared from late endosomes as described [25]. Early or late endosomes were diluted four times, sedimented by centrifugation (TLS55 Beckman Rotor, 30 min, 55000 rpm) and resuspended in 200 µl of lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA) in the presence of Complete, a cocktail of protease inhibitors (Roche) and 1% Triton X-100. After 20 min of incubation at 4°C, the lysat was adjusted to 40% OptiPrep (Nycodenz), overlaid with 30% and 0% OptiPrep cushions and centrifuged for two hours at 55000 rpm (4°C) using a TSL55 rotor. Six fractions were collected and precipitated with 6% trichloroacetic acid in the presence of sodium deoxycholate as a carrier.
Sample preparation for mass spectrometry analysis
DRMs and DSMs fractions were prepared from late endosomes (200 µg) as explained in precedent paragraph. 30 µg proteins from recovered DRMs and DSMs fractions were precipitated using methanol/chloroform. Proteins were then run on 2 cm in one-dimensional SDS-PAGE (15% acrylamide gel, 0.75 mm thick). Protein bands (12, and 14 for DRMs and soluble fraction, respectively) were excised and destained by repeated cycles of incubation in 25mM NH4HCO3 for 15 min and then with 50% (v/v) ACN in the same buffer (25mM NH4HCO3) for 15 min. After drying by vacuum centrifugation, the gel pieces were incubated with an oxidizing solution (7% H2O2) for 15 min [72]. Gel pieces were then washed in HPLC grade water (Sigma-Aldrich) for 15 min before being dehydrated with 100% ACN. In-gel digestion was performed (0.5 µg trypsin/band; sequencing grade modified trypsin, Promega) in 25mM NH4HCO3 overnight at 37°C. Peptides were extracted from the gel using passive diffusion in the following solutions: 50% ACN, then 5% formic acid, and finally 100% ACN. The extracts were dried by vacuum centrifugation and peptides were resolubilized in 5% ACN, 0.2% formic acid.
MS/MS analysis
The different sample fractions were injected into a CapLC nanoLC system (Waters) and first preconcentrated on a 300 µm×5 mm precolumn (PepMap C18; Dionex). The peptides were then eluted onto a C18 column (75 µm×150 mm; Dionex). Chromatographic separation used a gradient transition from solution A (2% acetonitrile, 98% water and 0.1% formic acid) to solution B (80% acetonitrile, 20% water and 0.08% formic acid) over 60 min at a flow rate of 200 nl/min. The LC system was directly coupled to a mass spectrometer (QTOF; Waters). MS and MS/MS data were acquired and processed automatically using MassLynx software (Waters). Database searching was performed using MASCOT 2.2 software (Matrix Science) using SwissProt_Trembl as the database and Rodent as the taxonomy. Variable modifications accorded were: Acetyl N-terminal of protein, simple and dioxidation of Methionine and cysteic acid on cysteine. Precisions on both MS and MS/MS data were set to 0.3 Da. The .dat files obtained through Mascot were further filtered using an “in-house” parsing solution (Irma, to be published: Bioinformatics Application Notes) build from Mascot Parser. The two sets of data were checked for false positive but none were found. Peptides whose score were > = to query identity threshold (p<0.05) and rank < = 1 were marked as significant through a first filtering and at final, a manual validation was done on proteins containing only one or two peptides. The peptide sequences were considered as validated if they contained at least 3 consecutives Y or B ions with a S/N threshold >3. Classical MS contaminants such as trypsin and keratin proteins were removed manually.
Fluorescence microscopy
Cells grown on cover slips were fixed with 3% paraformaldehyde for 20 min at room temperature and saturated with 10% phosphate-buffered saline-fetal calf serum (PBS-FCS) for 20 min. Cholesterol was labeled with 50 µg/ml Filipin in 10% PBS-FCS.
pH measurements
BHK cells grown on glass cover slips, were incubated with a FITC conjugated Lamp1 antibody overnight at 37°C or a FITC dextran for 15 minutes at 37°C and washed for 40 minutes. Late endosomal pH was measured by ratio fluorescence imaging as described [73], [74] with the use of a Nipkow dual spinning disk confocal laser imaging system with a QLC module (Visitron systems GmbH, Switzerland). Cover slips were inserted into a perfusion chamber (Medical Systems, Green- vale, NY) at 37°C in 1 ml of IM medium and imaged with a video/CCD camera controlled by MetaMorph/Metafluor imaging software. Images were acquired for 500 ms at two different wavelengths, using the two lasers 490 and 440 nm. Calibration and image processing were performed as described previously [73]. Data were graphed using the Prism software.
Supporting Information
Effects of β−MCD treatment on the pH of late endosomal lumen (FITC-dextran). FITC-dextran was internalized for 15 minutes and chased for 40 minutes at 37°C in control cells (A) and cells treated by β−MCD (B) to allow it to reach late endosomes. The histograms show the pH distribution of 3700 and 2194 endosomes for the upper and the lower panels. Values represent the mean±SD. The observed difference is significant according to a paired t-test with p<0.001.
(1.24 MB TIF)
Acknowledgments
We warmly thank M. Moniatte and J. Rougemont for their help in analyzing the proteomics data, J. Gruenberg for providing antibodies against LBPA and hamster lamp1, S. Grinstein for antibodies against V0d, I. Schultz for antibodies against V1A and V1E.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the Swiss National Science foundation and from a grant from Novartis Consumer Health Foundation to G.v.d.G. C.L is a recipient of a Roche Research Foundation post-doctoral fellowship. G.v.d.G is an international Fellow of the Howard Hughes Medical Institute.
References
- 1.Nishi T, Forgac M. The vacuolar (H+)-ATPases–nature's most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- 2.Faundez V, Hartzell HC. Intracellular chloride channels: determinants of function in the endosomal pathway. Sci STKE. 2004;2004:re8. doi: 10.1126/stke.2332004re8. [DOI] [PubMed] [Google Scholar]
- 3.Grabe M, Oster G. Regulation of organelle acidity. J Gen Physiol. 2001;117:329–344. doi: 10.1085/jgp.117.4.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jentsch TJ. Chloride and the endosomal-lysosomal pathway: emerging roles of CLC chloride transporters. J Physiol. 2007;578:633–640. doi: 10.1113/jphysiol.2006.124719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheel O, Zdebik AA, Lourdel S, Jentsch TJ. Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature. 2005;436:424–427. doi: 10.1038/nature03860. [DOI] [PubMed] [Google Scholar]
- 6.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 7.Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev. 2006;70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshansky V, Futai M. The V-type H(+)-ATPase in vesicular trafficking: targeting, regulation and function. Curr Opin Cell Biol. 2008 doi: 10.1016/j.ceb.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown D, Breton S. H(+)V-ATPase-dependent luminal acidification in the kidney collecting duct and the epididymis/vas deferens: vesicle recycling and transcytotic pathways. J Exp Biol. 2000;203:137–145. doi: 10.1242/jeb.203.1.137. [DOI] [PubMed] [Google Scholar]
- 10.Frattini A, Orchard PJ, Sobacchi C, Giliani S, Abinun M, et al. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat Genet. 2000;25:343–346. doi: 10.1038/77131. [DOI] [PubMed] [Google Scholar]
- 11.Li YP, Chen W, Liang Y, Li E, Stashenko P. Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat Genet. 1999;23:447–451. doi: 10.1038/70563. [DOI] [PubMed] [Google Scholar]
- 12.Nanda A, Brumell JH, Nordstrom T, Kjeldsen L, Sengelov H, et al. Activation of proton pumping in human neutrophils occurs by exocytosis of vesicles bearing vacuolar-type H+-ATPases. J Biol Chem. 1996;271:15963–15970. doi: 10.1074/jbc.271.27.15963. [DOI] [PubMed] [Google Scholar]
- 13.Hinton A, Bond S, Forgac M. V-ATPase functions in normal and disease processes. Pflugers Arch. 2007 doi: 10.1007/s00424-007-0382-4. [DOI] [PubMed] [Google Scholar]
- 14.Kane PM. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J Biol Chem. 1995;270:17025–17032. [PubMed] [Google Scholar]
- 15.Sumner JP, Dow JA, Earley FG, Klein U, Jager D, et al. Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits. J Biol Chem. 1995;270:5649–5653. doi: 10.1074/jbc.270.10.5649. [DOI] [PubMed] [Google Scholar]
- 16.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299:1400–1403. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- 17.Wagner CA, Finberg KE, Breton S, Marshansky V, Brown D, et al. Renal vacuolar H+-ATPase. Physiol Rev. 2004;84:1263–1314. doi: 10.1152/physrev.00045.2003. [DOI] [PubMed] [Google Scholar]
- 18.Nelson N. A journey from mammals to yeast with vacuolar H+-ATPase (V-ATPase). J Bioenerg Biomembr. 2003;35:281–289. doi: 10.1023/a:1025768529677. [DOI] [PubMed] [Google Scholar]
- 19.Kawasaki-Nishi S, Bowers K, Nishi T, Forgac M, Stevens TH. The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. J Biol Chem. 2001;276:47411–47420. doi: 10.1074/jbc.M108310200. [DOI] [PubMed] [Google Scholar]
- 20.Chen SH, Bubb MR, Yarmola EG, Zuo J, Jiang J, et al. Vacuolar H+-ATPase binding to microfilaments: regulation in response to phosphatidylinositol 3-kinase activity and detailed characterization of the actin-binding site in subunit B. J Biol Chem. 2004;279:7988–7998. doi: 10.1074/jbc.M305351200. [DOI] [PubMed] [Google Scholar]
- 21.Dixon N, Pali T, Kee TP, Ball S, Harrison MA, et al. Interaction of spin-labeled inhibitors of the vacuolar H+-ATPase with the transmembrane Vo-sector. Biophys J. 2008;94:506–514. doi: 10.1529/biophysj.107.111781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon N, Pali T, Kee TP, Marsh D. Spin-labelled vacuolar-ATPase inhibitors in lipid membranes. Biochim Biophys Acta. 2004;1665:177–183. doi: 10.1016/j.bbamem.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Gruenberg J. The endocytic pathway: a mosaic of domains. Nat Rev Mol Cell Biol. 2001;2:721–730. doi: 10.1038/35096054. [DOI] [PubMed] [Google Scholar]
- 24.Miaczynska M, Zerial M. Mosaic organization of the endocytic pathway. Exp Cell Res. 2002;272:8–14. doi: 10.1006/excr.2001.5401. [DOI] [PubMed] [Google Scholar]
- 25.Fivaz M, Vilbois F, Thurnheer S, Pasquali C, Abrami L, et al. Differential sorting and fate of endocytosed GPI-anchored proteins. Embo J. 2002;21:3989–4000. doi: 10.1093/emboj/cdf398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobo K, Chevallier J, Parton RG, Gruenberg J, van der Goot FG. Diversity of raft-like domains in late endosomes. PLoS ONE. 2007;2:e391. doi: 10.1371/journal.pone.0000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blonder J, Hale ML, Lucas DA, Schaefer CF, Yu LR, et al. Proteomic analysis of detergent-resistant membrane rafts. Electrophoresis. 2004;25:1307–1318. doi: 10.1002/elps.200405891. [DOI] [PubMed] [Google Scholar]
- 28.Foster LJ, De Hoog CL, Mann M. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc Natl Acad Sci U S A. 2003;100:5813–5818. doi: 10.1073/pnas.0631608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLellan DL, Steen H, Adam RM, Garlick M, Zurakowski D, et al. A quantitative proteomic analysis of growth factor-induced compositional changes in lipid rafts of human smooth muscle cells. Proteomics. 2005;5:4733–4742. doi: 10.1002/pmic.200500044. [DOI] [PubMed] [Google Scholar]
- 30.Karsan A, Blonder J, Law J, Yaquian E, Lucas DA, et al. Proteomic analysis of lipid microdomains from lipopolysaccharide-activated human endothelial cells. J Proteome Res. 2005;4:349–357. doi: 10.1021/pr049824w. [DOI] [PubMed] [Google Scholar]
- 31.Adam RM, Yang W, Di Vizio D, Mukhopadhyay NK, Steen H. Rapid preparation of nuclei-depleted detergent-resistant membrane fractions suitable for proteomics analysis. BMC Cell Biol. 2008;9:30. doi: 10.1186/1471-2121-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aniento F, Emans N, Griffiths G, Gruenberg J. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J Cell Biol. 1993;123:1373–1387. doi: 10.1083/jcb.123.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Goot FG. Separation of early steps in endocytic membrane transport. Electrophoresis. 1997;18:2689–2693. doi: 10.1002/elps.1150181426. [DOI] [PubMed] [Google Scholar]
- 34.Sobo K, Le Blanc I, Luyet PP, Fivaz M, Ferguson C, et al. Late endosomal cholesterol accumulation leads to impaired intra-endosomal trafficking. PLoS ONE. 2007;2:e851. doi: 10.1371/journal.pone.0000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- 36.Parton RG, Simons K. The multiple faces of caveolae. Nat Rev Mol Cell Biol. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- 37.Browman DT, Resek ME, Zajchowski LD, Robbins SM. Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J Cell Sci. 2006;119:3149–3160. doi: 10.1242/jcs.03060. [DOI] [PubMed] [Google Scholar]
- 38.Morrow IC, Parton RG. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic. 2005;6:725–740. doi: 10.1111/j.1600-0854.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 39.Oliferenko S, Paiha K, Harder T, Gerke V, Schwarzler C, et al. Analysis of CD44-containing lipid rafts: Recruitment of annexin II and stabilization by the actin cytoskeleton. J Cell Biol. 1999;146:843–854. doi: 10.1083/jcb.146.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schroder B, Wrocklage C, Pan C, Jager R, Kosters B, et al. Integral and associated lysosomal membrane proteins. Traffic. 2007;8:1676–1686. doi: 10.1111/j.1600-0854.2007.00643.x. [DOI] [PubMed] [Google Scholar]
- 41.Van Dyke RW. Heterotrimeric G protein subunits are located on rat liver endosomes. BMC Physiol. 2004;4:1. doi: 10.1186/1472-6793-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez-Gaitan M. Signal dispersal and transduction through the endocytic pathway. Nat Rev Mol Cell Biol. 2003;4:213–224. doi: 10.1038/nrm1053. [DOI] [PubMed] [Google Scholar]
- 43.Roy CR. Trimming the fat: a Brucella abortus survival strategy. Nat Immunol. 2005;6:546–548. doi: 10.1038/ni0605-546. [DOI] [PubMed] [Google Scholar]
- 44.Geyer M, Yu H, Mandic R, Linnemann T, Zheng YH, et al. Subunit H of the V-ATPase binds to the medium chain of adaptor protein complex 2 and connects Nef to the endocytic machinery. J Biol Chem. 2002;277:28521–28529. doi: 10.1074/jbc.M200522200. [DOI] [PubMed] [Google Scholar]
- 45.Keyel PA, Watkins SC, Traub LM. Endocytic adaptor molecules reveal an endosomal population of clathrin by total internal reflection fluorescence microscopy. J Biol Chem. 2004;279:13190–13204. doi: 10.1074/jbc.M312717200. [DOI] [PubMed] [Google Scholar]
- 46.Traub LM, Bannykh SI, Rodel JE, Aridor M, Balch WE, et al. AP-2-containing clathrin coats assemble on mature lysosomes. J Cell Biol. 1996;135:1801–1814. doi: 10.1083/jcb.135.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurtado-Lorenzo A, Skinner M, El Annan J, Futai M, Sun-Wada GH, et al. V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat Cell Biol. 2006;8:124–136. doi: 10.1038/ncb1348. [DOI] [PubMed] [Google Scholar]
- 48.Peters C, Bayer MJ, Buhler S, Andersen JS, Mann M, et al. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature. 2001;409:581–588. doi: 10.1038/35054500. [DOI] [PubMed] [Google Scholar]
- 49.Hiesinger PR, Fayyazuddin A, Mehta SQ, Rosenmund T, Schulze KL, et al. The v-ATPase V0 subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila. Cell. 2005;121:607–620. doi: 10.1016/j.cell.2005.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liegeois S, Benedetto A, Garnier JM, Schwab Y, Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J Cell Biol. 2006;173:949–961. doi: 10.1083/jcb.200511072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu T, Forgac M. Microtubules are involved in glucose-dependent dissociation of the yeast vacuolar [H+]-ATPase in vivo. J Biol Chem. 2001;276:24855–24861. doi: 10.1074/jbc.M100637200. [DOI] [PubMed] [Google Scholar]
- 52.Lu M, Sautin YY, Holliday LS, Gluck SL. The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J Biol Chem. 2004;279:8732–8739. doi: 10.1074/jbc.M303871200. [DOI] [PubMed] [Google Scholar]
- 53.Abrami L, van der Goot FG. Plasma membrane microdomains act as concentration platforms to facilitate intoxication by aerolysin. J Cell Biol. 1999;147:175–184. doi: 10.1083/jcb.147.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koh CH, Cheung NS. Cellular mechanism of U18666A-mediated apoptosis in cultured murine cortical neurons: bridging Niemann-Pick disease type C and Alzheimer's disease. Cell Signal. 2006;18:1844–1853. doi: 10.1016/j.cellsig.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 55.Liscum L, Faust JR. The intracellular transport of low density lipoprotein-derived cholesterol is inhibited in Chinese hamster ovary cells cultured with 3-beta-[2-(diethylamino)ethoxy]androst-5-en-17-one. J Biol Chem. 1989;264:11796–11806. [PubMed] [Google Scholar]
- 56.Demaurex N. pH Homeostasis of cellular organelles. News Physiol Sci. 2002;17:1–5. doi: 10.1152/physiologyonline.2002.17.1.1. [DOI] [PubMed] [Google Scholar]
- 57.Kobayashi T, Beuchat MH, Chevallier J, Makino A, Mayran N, et al. Separation and characterization of late endosomal membrane domains. J Biol Chem. 2002;277:32157–32164. doi: 10.1074/jbc.M202838200. [DOI] [PubMed] [Google Scholar]
- 58.Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG, et al. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- 59.Petiot A, Faure J, Stenmark H, Gruenberg J. PI3P signaling regulates receptor sorting but not transport in the endosomal pathway. J Cell Biol. 2003;162:971–979. doi: 10.1083/jcb.200303018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kane PM. The long physiological reach of the yeast vacuolar H(+)-ATPase. J Bioenerg Biomembr. 2007 doi: 10.1007/s10863-007-9112-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parra KJ, Kane PM. Reversible association between the V1 and V0 domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol Cell Biol. 1998;18:7064–7074. doi: 10.1128/mcb.18.12.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sautin YY, Lu M, Gaugler A, Zhang L, Gluck SL. Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+-ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol Cell Biol. 2005;25:575–589. doi: 10.1128/MCB.25.2.575-589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Breton S, Brown D. New insights into the regulation of V-ATPase-dependent proton secretion. Am J Physiol Renal Physiol. 2007;292:F1–10. doi: 10.1152/ajprenal.00340.2006. [DOI] [PubMed] [Google Scholar]
- 64.Xie XS, Padron D, Liao X, Wang J, Roth MG, et al. Salicylihalamide A inhibits the V0 sector of the V-ATPase through a mechanism distinct from bafilomycin A1. J Biol Chem. 2004;279:19755–19763. doi: 10.1074/jbc.M313796200. [DOI] [PubMed] [Google Scholar]
- 65.Dawson K, Toone WM, Jones N, Wilkinson CR. Loss of regulators of vacuolar ATPase function and ceramide synthesis results in multidrug sensitivity in Schizosaccharomyces pombe. Eukaryot Cell. 2008;7:926–937. doi: 10.1128/EC.00037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee MC, Hamamoto S, Schekman R. Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. J Biol Chem. 2002;277:22395–22401. doi: 10.1074/jbc.M200450200. [DOI] [PubMed] [Google Scholar]
- 67.Yoshinaka K, Kumanogoh H, Nakamura S, Maekawa S. Identification of V-ATPase as a major component in the raft fraction prepared from the synaptic plasma membrane and the synaptic vesicle of rat brain. Neurosci Lett. 2004;363:168–172. doi: 10.1016/j.neulet.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 68.Abrami L, Fivaz M, Glauser PE, Parton RG, van der Goot FG. A pore-forming toxin interacts with a GPI-anchored protein and causes vacuolation of the endoplasmic reticulum. J Cell Biol. 1998;140:525–540. doi: 10.1083/jcb.140.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gruenberg J, Howell KE. Membrane traffic in endocytosis: insights from cell-free assays. Annu Rev Cell Biol. 1989;5:453–481. doi: 10.1146/annurev.cb.05.110189.002321. [DOI] [PubMed] [Google Scholar]
- 70.Kobayashi T, Beuchat MH, Lindsay M, Frias S, Palmiter RD, et al. Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat Cell Biol. 1999;1:113–118. doi: 10.1038/10084. [DOI] [PubMed] [Google Scholar]
- 71.Schapiro F, Sparkowski J, Adduci A, Suprynowicz F, Schlegel R, et al. Golgi alkalinization by the papillomavirus E5 oncoprotein. J Cell Biol. 2000;148:305–315. doi: 10.1083/jcb.148.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jaquinod M, Villiers F, Kieffer-Jaquinod S, Hugouvieux V, Bruley C, et al. A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol Cell Proteomics. 2007;6:394–412. doi: 10.1074/mcp.M600250-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Demaurex N, Furuya W, D'Souza S, Bonifacino JS, Grinstein S. Mechanism of acidification of the trans-Golgi network (TGN). In situ measurements of pH using retrieval of TGN38 and furin from the cell surface. J Biol Chem. 1998;273:2044–2051. doi: 10.1074/jbc.273.4.2044. [DOI] [PubMed] [Google Scholar]
- 74.Piguet V, Gu F, Foti M, Demaurex N, Gruenberg J, et al. Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of beta-COP in endosomes. Cell. 1999;97:63–73. doi: 10.1016/s0092-8674(00)80715-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects of β−MCD treatment on the pH of late endosomal lumen (FITC-dextran). FITC-dextran was internalized for 15 minutes and chased for 40 minutes at 37°C in control cells (A) and cells treated by β−MCD (B) to allow it to reach late endosomes. The histograms show the pH distribution of 3700 and 2194 endosomes for the upper and the lower panels. Values represent the mean±SD. The observed difference is significant according to a paired t-test with p<0.001.
(1.24 MB TIF)