Abstract
Molecular, sequence-based environmental surveys of microorganisms have revealed a large degree of previously uncharacterized diversity. However, nearly all studies of the human endogenous bacterial flora have relied on cultivation and biochemical characterization of the resident organisms. We used molecular methods to characterize the breadth of bacterial diversity within the human subgingival crevice by comparing 264 small subunit rDNA sequences from 21 clone libraries created with products amplified directly from subgingival plaque, with sequences obtained from bacteria that were cultivated from the same specimen, as well as with sequences available in public databases. The majority (52.5%) of the directly amplified 16S rRNA sequences were <99% identical to sequences within public databases. In contrast, only 21.4% of the sequences recovered from cultivated bacteria showed this degree of variability. The 16S rDNA sequences recovered by direct amplification were also more deeply divergent; 13.5% of the amplified sequences were more than 5% nonidentical to any known sequence, a level of dissimilarity that is often found between members of different genera. None of the cultivated sequences exhibited this degree of sequence dissimilarity. Finally, direct amplification of 16S rDNA yielded a more diverse view of the subgingival bacterial flora than did cultivation. Our data suggest that a significant proportion of the resident human bacterial flora remain poorly characterized, even within this well studied and familiar microbial environment.
The endogenous bacterial flora in man is thought to play a role in nutrition, carcinogenesis, and resistance to colonization by pathogens, and to harbor potential opportunistic pathogens (1). An accurate understanding of these roles, and the nature of the interactions among and between individual members and the host, requires as a first step knowledge of the composition of the microbial community. Many studies of environmental microbial communities have demonstrated the limitations of cultivation-dependent methods in determining community composition. Environmental surveys based on acquisition of phylogenetically useful microbial sequences (2–4) such as that of the 16S rRNA gene (16S rDNA) have revealed a great deal of previously unsuspected bacterial and archaeal diversity. In most instances, the cultivated members represent <1% of the total extant population. Broad range small subunit rDNA PCR methods have also revealed cultivation-resistant pathogens in disease settings (5).
Despite the limitations of this approach, most surveys of the human endogenous bacterial flora have relied on cultivation. Nonetheless, nearly 500 bacterial strains have been recovered from the subgingival crevice, a particularly well studied microbial niche. Many of these strains are thought to be commensals, and a smaller number, opportunistic pathogens (6, 7). Local disease, including dental caries, gingivitis, and periodontitis, has prompted most examinations of the oral bacterial flora. These diseases are associated with changes in both local bacterial density and species composition (8).
Few efforts have been undertaken to characterize human endogenous microbial communities using broad-based molecular methods. A small number of 16S rDNA sequences have been characterized after direct amplification from oral specimens associated with disease (9–11). However, a direct comparison between cultivation-dependent and -independent methods has not been described. In this study, we characterized bacterial diversity within a specimen from the human subgingival crevice by using both methods. Our results reveal a significantly broader diversity of bacterial 16S rDNA sequence types (phylotypes) by using the cultivation-independent approach, although each method identified previously uncharacterized phylotypes and should be viewed as complementary.
Materials and Methods
Specimen Collection.
Subgingival plaque was collected from the mesial surface of tooth 3 and the mesial and distal surfaces of tooth 30 of a 39-year-old Caucasian male, in accordance with a protocol approved by the Stanford Administrative Panel on Human Subjects in Medical Research. The subject was a nonsmoker with mild gingivitis and no record of antibiotic use during the previous 3 months. The plaque material was dispersed in 1.3 ml of reduced Treponeme broth (Anaerobe Systems, San Jose, CA). The vial was flushed with an anaerobic gas mixture (80% N/10% H2/10% CO2) and was maintained on ice for 1 hr before processing.
Cultivation and Phenotypic Analysis of Bacteria from Subgingival Specimen.
The plaque suspension was diluted 1:2 and 1:20 in reduced Treponeme broth. A 100-μl aliquot of the 1:2 dilution was examined by dark field microscopy immediately and after 24-hr incubation for the presence of spirochetes. The diluted samples were used to inoculate a wide range of routine clinical microbiological media (see supplemental material on the PNAS web site, www.pnas.org). Isolates were chosen for further characterization based on differences in colony morphology and were identified by using routine clinical microbiological assays (see supplemental material).
Specimen Digestion and DNA Extraction.
An undiluted 100-μl aliquot of the suspended plaque material was added to an equal volume of lysis buffer [400 μg/ml proteinase K (Boehringer Mannheim)/2% Laureth-12 (Mazer Chemicals, Gurnee, IL)/100 mM Tris⋅HCl, pH 8.5/2 mM EDTA] and was incubated overnight at 55°C. The digest was then sonicated for 2 min at 120 W in a bath sonicator (cell disrupter W185; Branson). One half of the digested sample was purified and precipitated with phenol, chloroform, and ethanol by using standard protocols. The remainder of the digest was centrifuged at 1,200 × g for 2 min, and the supernatant was used directly in PCR.
PCR Amplification.
PCR was performed with broadly conserved bacterial 16S rDNA primers or spirochete-specific 16S rDNA primers (Table 1). Broad range primer pairs were (“A”) S-D-Bact-0338-a-A-18 + S-D-Bact-1525-a-S-17; (“B”) S-D-Bact-0515-a-A-19 + S-D-Bact-1371-a-S-20; (“C”) S-D-Bact-0515-a-A-19 + S-D-Bact-1525-a-S-17; and (“D”) S-D-Bact-0515-a-A-19 + S-D-Bact-1529-a-S-17. A 1-μl aliquot of extracted DNA or digest supernatant was added to 99 μl of PCR reaction mix (10 mM Tris⋅HCl, pH 8.5/2 mM MgCl2/50 mM KCl/200 μM of each dNTP/20 pmol of each primer). The reaction was heated at 94°C for 3 min before the addition of 2.5 units of AmpliTaq DNA polymerase (Perkin–Elmer). 16S rDNA genes were amplified for 30 or 40 cycles; each cycle consisted of 94°C (30 s), 55°C (30 s), and 72°C (30 s), with the exception of PCRs using S-D-Bact-1529-a-S-17, for which an annealing temperature of 50°C was used. The sensitivity of PCRs with primer pair “B” was determined by amplifying serial dilutions of cloned phylotype A-38 16S rDNA in water.
Table 1.
Oligonucleotide primers and probe
| Primers/probe* | Sequence (5′ → 3′) | Ref. |
|---|---|---|
| SDBact0338aA18† | ACTCCTACGGGAGGCAGC | 12 |
| SDBact0515aA19 | GTGCCSGCMGCCGCGGTAA | 13‡ |
| SDBact0515aS19 | TTACCGCGGCMGCSGGCAC | 13‡ |
| SDBact1371aS20 | AGGCCCGGGAACGTATTCAC | 14 |
| SDBact1525aS17 | AAGGAGGTGATCCAGCC | 15‡ |
| SDBact1529aS17 | YAKAAAGGAGGTGWTCC | This study |
| SGTrep0008aA20 | AGAGTTTGATCMTGGCTCAG | 9 |
| SGStrp1264aS22§ | AGAGATTAGCTTGCCGTCACCG | This study |
Primers and probes are named according to Oligonucleotide Probe Database convention (16). The names incorporate the 5′ terminal position (E. coli 16S rRNA numbering convention) and primer length in nucleotides.
† Orientation indicates PCR primer sequence. Reverse complement sequence used for in situ probing.
‡ Modified from indicated reference.
§ Streptococcus-specific probe.
Cloning of Amplified 16S rRNA Genes and Sequencing.
Amplicons were ligated into pCR 2.1 (Invitrogen). One half of the product from each ligation reaction was restricted with Bsu36I (New England BioLabs) (see Results). Escherichia coli Top10F′ (Invitrogen) was transformed with either restricted or unrestricted plasmid DNA. 16S rDNA was amplified and purified as previously described (17) from randomly chosen E. coli Top10F′ clones and all cultivated isolates with unique phenotypes. 16S rDNA was sequenced by using the Prism AmpliTaq FS DyeDeoxy terminator cycle sequencing kit and a 373A sequencer (Applied Biosystems). GenBank database accession numbers are AF201965–AF202029.
Phylogenetic Analysis.
After determining preliminary phylogenetic associations, the integrity of amplified sequences displaying <99% identity to database sequences was confirmed by using the check_chimera program of the Ribosomal Database Project. Chimeras were defined as sequences in which subfragments possessed a higher similarity to database sequences other than the database sequence most similar to the entire query sequence.
Initial alignment of amplified sequences was performed by using the automated 16S rRNA sequence aligner of the arb software package (Technical Univ. of Munich, Munich, Germany) against a database of 7,916 complete and partial rRNA sequences (March 1997 arb small subunit rRNA database release). Ambiguously and incorrectly aligned positions were aligned manually on the basis of conserved primary sequence and secondary structure. Similarity matrices were generated from 516 to 961 masked (unambiguously aligned) positions. Sequences with ≥99% identity were considered as a single phylotype. A single representative clone from each phylotype was chosen for further phylogenetic analysis.
The phylogenetic associations of all representative sequences were determined from 100 bootstrapped replicates (18) by using a maximum-likelihood algorithm (19). These associations were confirmed by using a least-squares fit (20) of evolutionary distances (with Jukes-Cantor correction) and by using parsimony algorithms. More detailed phylogenetic analyses based on 864 and 894 masked positions of Low GC Gram Positive and Actinobacteria sequences, respectively, were performed by using the same methods. Coverage of clone libraries was calculated according to the method of Good (21), and an estimate of the number of unseen species was obtained by using a parametric model (22).
In Situ Hybridization and Enumeration.
A 50-μl aliquot of subgingival plaque suspension was fixed and simultaneously hybridized with 33 ng/μl fluorescently labeled S-G-Strp-1264-a-S-22 and 1.6 ng/μl fluorescently labeled S-D-Bact-0338-a-S-18 as described (23), but omitting sodium borohydride. Probe S-G-Strp-1264-a-S-22 was designed from conserved regions of Streptococcus spp. 16S rDNA sequences in the RDP small subunit aligned database v3.0 (24) and GenEMBL database, as well as from Streptococcus spp. 16S rDNA sequences found from this study. Probe S-G-Strp-1264-a-S-22 was synthesized with 5′-Aminolink 2 (Applied Biosystems) and was labeled by using the FluoReporter Texas Red-X Oligonucleotide Amine Labeling Kit (Molecular Probes). The conserved bacterial probe S-D-Bact-0338-a-S-18 was synthesized directly with the FITC-labeled phosphoramidite 6-FAM (401527, Applied Biosystems). Samples were examined on an Olympus (New Hyde Park, NY) IX-70 microscope, and images were collected by using the DeltaVision deconvolution system (Applied Precision, Seattle). A series of 32 slices were collected every 0.2 μm along the z axis and was projected by using the maximum intensity algorithm to form a single image. Streptococcus spp. were enumerated in 12 representative fields at ×1,000 magnification.
Results
Direct Amplification of 16S rDNA from Subgingival Plaque.
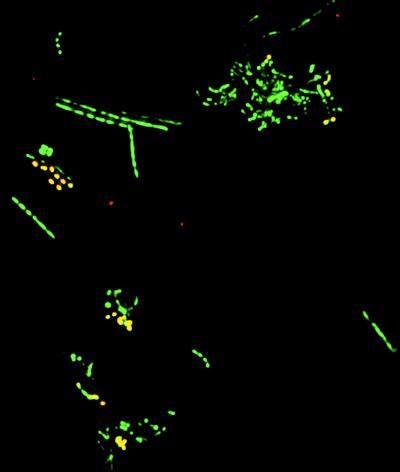
Initial amplification and cloning with four broad range bacterial primer pairs (see supplemental material) revealed that 46 of 50 amplified 16S rRNA genes were of streptococcal origin. This high percentage of streptococcal 16S rDNA appeared to conflict with reported microscopic counts in which streptococci comprised ≈30% of plaque-associated bacterial communities (25). The relative abundance of Streptococcus species to total bacteria in the original sample was evaluated by using a fluorescently labeled Streptococcus-specific probe and a nonspecific bacterial probe (S-D-Bact-0338-a-S-18). Streptococcus spp. comprised 79 of 268 (29.5%) bacteria in this specimen as determined by in situ hybridization (Fig. 1).
Figure 1.
Streptococcus spp. (yellow) in a thin smear of the gingival specimen revealed by in situ hybridization with a Streptococcus-specific Texas Red-labeled oligonucleotide probe (red) and a broad range bacterial FITC-labeled oligonucleotide probe (green). (×500.)
The over-representation of Streptococcus spp. was reduced in subsequent clone libraries by restriction of plasmids with Bsu36I before transformation. This restriction endonuclease recognizes a site present in the 16S rDNA of nearly all Streptococcus spp. Transformant colony counts were reduced by >7-fold after Bsu36I restriction of equivalent amounts of DNA. Having solved the problem of over-representation of the Streptococcus spp., we created 21 clone libraries from 15 separate PCR amplifications of rDNA from digested subgingival plaque (see supplemental material); 245 randomly chosen clones from 14 Bsu36I-restricted libraries and 25 clones from 7 unrestricted libraries were sequenced. Six of the 270 amplified sequences were chimeras and were removed from subsequent analyses.
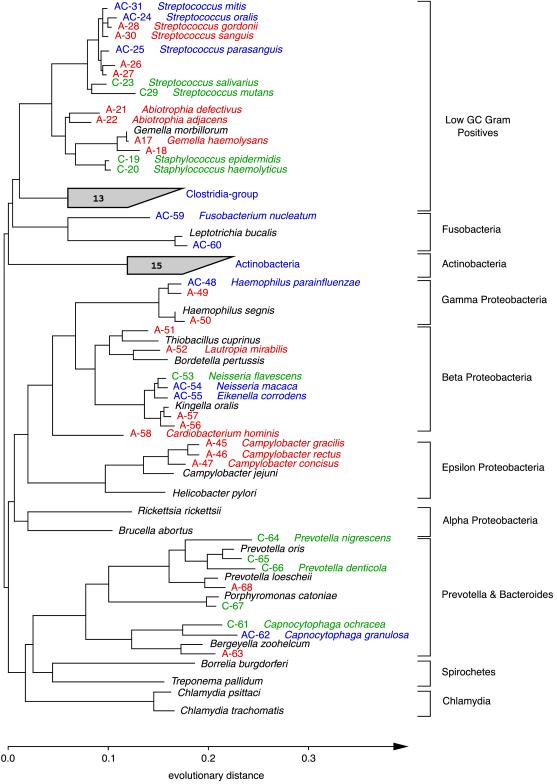
The percent sequence identity among the 264 remaining clones was determined by comparison of 489–690 homologous 16S rDNA positions between members of monophyletic groups. Sequences differing by ≤1% were considered to be representative of a single phylotype. This phylotype definition was chosen after evaluating the heterogeneity among multiple copies of 16S rRNA genes in 6 fully sequenced bacterial genomes containing multiple rrn operon copies: Synechocystis PCC 6803, 0% among 2 operons; Haemophilus influenzae, 0% among 6 operons; Helicobacter pylori, ≤0.1% sequence dissimilarity among 3 operons; Treponema pallidum, 0.06% among 2 operons; Bacillus subtilis, ≤0.6% among 10 operons; and E. coli, ≤1.1% among 7 operons. This definition may in fact under-represent the diversity found among closely related taxa. Based on our definition, the 264 clones represented 59 unique phylotypes (Figs. 2–4). These 59 phylotypes constitute ≈88% of all sequences within the cloned population [(number of phylotypes observed once/total number of observations) ×100] (21). The remaining 12% of the cloned population could potentially be composed of hundreds of additional unique phylotypes. The number of unique unseen species may be estimated by using a parametric model that examines the number of phylotypes observed only once versus those observed multiple times (Δt = n1t − n2t2 + n3t3… ; where Δt is the expected number of new phylotypes, t is the number of additional sample sets and nx is the number of phylotypes appearing x times in the original sample set). Based on methods for estimating the number of unseen species (22), these 59 phylotypes constitute <70% of the total predicted number of unique phylotypes within the clone libraries.
Figure 2.
Phylogenetic relationships of bacteria detected in a subgingival specimen, inferred from 16S rDNA sequence analysis. This unrooted tree was constructed by using 489 homologous sequence positions (534–1,050; E. coli numbering) and a maximum-likelihood algorithm. Sequences were given a colored alphanumeric designation according to whether they were amplified directly from the specimen (A; red), obtained from organisms cultivated from the specimen (C; green), or found by both methods (AC; blue). An organism name is provided adjacent to an alphanumeric designation when a sequence generated from this study was ≥99.0% identical to a sequence in a public database. Sequences selected from a public database for purposes of reference (not detected in the specimen) are indicated with the organism name in black. The number within each shaded fan indicates the total number of unique (amplified and cultivated) phylotypes within the group.
Figure 4.
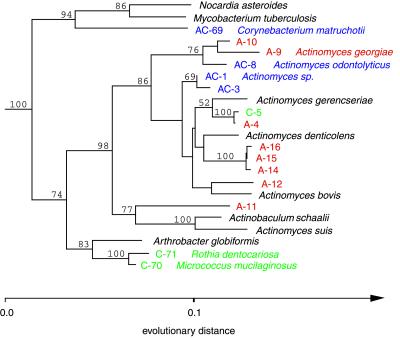
Phylogenetic relationships of bacteria that belong within the Actinobacteria division, inferred from 16S rDNA sequence analysis. This tree was constructed by using 894 homologous positions (534–1,438; E. coli numbering) and a maximum-likelihood algorithm; it was rooted by using the sequence of Streptomyces coelicolor. Confidence values >50% from bootstrapped data are displayed. For sequence designation scheme, see Fig. 2 legend.
The creation of 15 individual libraries by using different methods allowed us to examine the effect of DNA purification and primer pairs on library composition. Phenolchloroform extraction of DNA had no significant effect. Primer selection was associated with several notable differences: Capnocytophaga were represented by only 1 of 86 clones (1.2%) in libraries created by using primer pair “C” whereas they were represented by 14 of 124 clones (11.3%) in libraries created with primer pair “A” and 5 of 52 clones (9.6%) in libraries created by using primer pair “D.” None of the Capnocytophaga sequences within the arb 16S rDNA database contained mismatches with the S-D-Bact-0515-a-A-19 primer. Similarly, Actinomyces spp. were observed frequently by using primer pair “C” (20 of 86 clones) but not with primer pair “D” (1 of 52 clones). There were no mismatches between the Actinomyces sequences and the S-D-Bact-1529-a-S-17 primer.
Comparison of Cultivated and Amplified Phylotypes.
Cultivation methods yielded 56 isolates from subgingival plaque that were distinct based on antigenic or biochemical traits; 37.5% of these isolates could not be definitively identified at the genus level, and 55% could not be identified at the species level, using conventional phenotypic methods. Sequence analysis of 489–690 homologous positions from the 16S rDNA of these 56 isolates indicated 28 unique phylotypes based on our 1% identity definition (Figs. 2–4).
The phylotypes recovered by amplification and cloning exhibited greater divergence and diversity than phylotypes recovered by cultivation. The use of a strict definition of species boundaries (≤97% sequence identity) (36) did not alter this finding. The 16S rDNA from 22 of the 28 cultivated isolates showed ≥99% identity to sequences within public databases. Conversely, 52.5% of the 59 amplified phylotypes had not been previously characterized (<99% identity) by 16S rDNA analysis (Table 2). Fifty percent of the cultivated phylotypes were also present within the 59 phylotypes recovered by direct, broad-range amplification whereas only 24% of the directly amplified phylotypes were recovered by cultivation. Although cultivation did not reveal the same degree of divergence as amplification, selected groups of bacteria were more readily recovered by cultivation. Cultivation recovered four Prevotella phylotypes and two phylotypes of Staphylococcus, Rothia dentocariosa and Micrococcus luteus, whereas only a single Prevotella phylotype and none of the other organisms were detected by direct amplification.
Table 2.
Distribution of similarity scores of sequences to the most closely related public database entry
| Percent identity* | Isolates
|
Amplicons
|
||
|---|---|---|---|---|
| No. | Percent of total | No. | Percent of total | |
| ≥99.0% | 22 | 78.6 | 28 | 47.5 |
| 97.0–98.9% | 3 | 10.7 | 13 | 22.0 |
| 95.0–96.9% | 3 | 10.7 | 10 | 17.0 |
| 90.0–94.9% | 0 | 0.0 | 6 | 10.1 |
| ≤89.9% | 0 | 0.0 | 2 | 3.4 |
Values are based on an analysis of 489 or more homologous positions (see text).
Degree of identity with most closely related database sequence.
Approximately 48% of the 77 phylotypes (59 amplified and 28 cultivated) were previously uncharacterized sequences (<99% identical to any of the nearly 5,000 bacterial 16S rRNA sequences >1,000 nucleotides in length within the arb database). Sequences obtained by direct amplification and cloning represented ≈83% of these novel phylotypes whereas ≈10.5% were discovered only by cultivation; the remaining 6.5% were revealed by both approaches (Figs. 2–4). Many of the novel phylotypes with the lowest similarity to public database sequences fell within the Low GC Gram Positive Clostridia and Actinobacteria divisions (Figs. 3 and 4).
Figure 3.
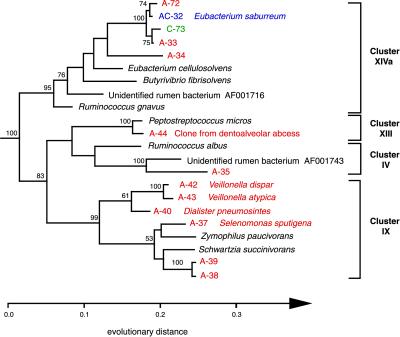
Phylogenetic relationships of bacteria that belong within the Low GC Gram Positive division, inferred from 16S rDNA sequence analysis. This tree was constructed by using 860 homologous sequence positions (534–1,400; E. coli numbering) and a maximum-likelihood algorithm; it was rooted by using the sequence of Peptococcus niger. Confidence values >50% from bootstrapped data are displayed. For sequence designation scheme, see Fig. 2 legend. Cluster designations are based on the proposed phylogenetic scheme of Collins et al. (26).
The Clostridia group of Low GC Gram Positives displays extensive phenotypic and phylogenetic diversity (26). In our study, the majority of phylotypes within this group were recovered only by amplification (Fig. 3). Among the 15 phylotypes identified, 9 were previously uncharacterized by 16S rDNA analysis, and 5 of these displayed <93% sequence identity with organisms within the arb database and to each other. Phylotype A-35 exhibited the greatest divergence among the clone libraries: 89.0% sequence identity with its closest relative, a ruminal bacterium sequence (GenBank accession no. AF001743). Similarly, A-36 (not shown) displayed 89.4% sequence identity with a different ruminal bacterium sequence (GenBank accession no. AF001716). A-38 and A-39 were 98% identical to each other and 93% identical to their closest relative, Schwartzia succinivorans. S. succinivorans is an obligate succinate fermenter that has been isolated from the rumen of a cow. Veillonella spp. are early plaque colonizers (27) and are instrumental in the formation of the matrix to which late colonizers such as Selenomonas and Eubacterium adhere. The A-44 sequence is identical to one recently cloned from a dentoalveolar abcess (11). Finally, A-33, A-34, A-72, and C-73 are related to Eubacterium saburreum, a common oral isolate. Many of the Eubacterium species have been isolated from the rumen and/or are capable of degrading various plant polymers (saccharolytic, xylanotic, or cellulolytic).
Phylogenetic analysis revealed 18 phylotypes within the Actinobacteria group, 12 of which were previously unidentified. Nine of these twelve novel phylotypes displayed <93% sequence identity with organisms within the arb database (Fig. 4). Of these, A-11 had the lowest similarity value to its closest relative (≈90%). This phylotype clustered consistently with Actinobaculum schaalii and Actinomyces suis. A. schaalii has been isolated from human blood and urine (28) whereas A. suis has been associated with porcine cystitis and pyelonephritis.
Several interesting phylotypes were not assigned to the Clostridia or Actinobacteria groups and are listed in Table 3. For example, the deeply divergent phylotype A-63 was most similar to Weeksella zoohelcum, a normal inhabitant of the canine upper respiratory tract that has been cultured from human dog bite wounds (29). However, drawing conclusions about an organism whose existence is inferred from only sequence is problematic when the nearest-known relative is so distant. Amplification reactions using Treponema-specific 16S rDNA primers (9) failed to reveal treponemal sequences. The sensitivity of detection using conserved bacterial primers and cloned bacterial (phylotype A-38) 16S rDNA was 4–40 gene copies.
Table 3.
Phylotypes with <99.0% identity to a public database sequence, excluding the Actinobacteria and the Low GC Gram Positive Clostridia groups
| Phylotypes* | Related organism(s) | Percent Identity | Nucleotides |
|---|---|---|---|
| A50 | Haemophilus segnis | 98.9 | 541 |
| A57 | Kingella oralis | 98.5 | 545 |
| A27 | Streptococcus parasanguis | 98.3 | 529 |
| A49 | Haemophilus parainfluenzae | 97.6 | 541 |
| A26 | Streptococcus parasanguis | 97.5 | 529 |
| A68 | Prevotella loescheii | 97.2 | 702 |
| A56 | K. oralis | 97.1 | 545 |
| AC60 | Leptotrichia bucallis | 97.1 | 1,370 |
| C65 | Prevotella oris | 96.3 | 814 |
| C67 | Porphyromonas catoniae | 96.1 | 818 |
| A63 | W. zoohelcum | 95.5 | 663 |
| A18 | Gemella morbillorum | 95.3 | 953 |
| A51 | T. cuprinus | 93.9 | 933 |
Phylotypes were obtained by either direct amplification (A), cultivation (C), or both (AC).
Discussion
Sequence-based environmental microbial surveys have taught us that cultivation methods woefully under-represent the true extent of bacterial diversity. The gross outlines of this diversity have begun to emerge in recent years from comparative analyses of 16S rRNAs. From these surveys, 36–40 higher order bacterial divisions can be discerned (30). Our study sought to examine the extent of bacterial diversity within a well studied human endogenous niche, the subgingival crevice, and to compare the representations of diversity provided by two approaches, that based on cultivation and that based on amplification of rDNA directly from the same specimen. A priori, one might postulate that, within a relatively familiar ecological niche that has been subjected to numerous cultivation procedures—prompted in part by the search for causative agents of local and systemic disease—relatively few previously unrecognized bacteria would be identified. Some of the phylotypes recovered by rDNA amplification directly from the plaque specimen conformed well with previous descriptions of the bacterial flora of the subgingival crevice, including members associated with local and systemic disease such as bacteremia, and endocarditis. However, we found that the diversity revealed by direct amplification of rDNA directly from a subgingival plaque specimen substantively exceeded that revealed by cultivation from the same specimen. Furthermore, many of the more divergent phylotypes have not been detected previously within humans.
Overall, we detected members of five of the bacterial divisions proposed by Hugenholtz et al. (30) within subgingival plaque. These five divisions (Low GC Gram Positives, Fusobacteria, Actinobacteria, Proteobacteria, and Cytophagales) all appear to be cosmopolitan in nature and are well represented among cultivated microorganisms. Initially, most of the amplified 16S rDNA was of streptococcal origin, a finding that was contradicted by previously published data (25) and by our in situ hybridization data. This discrepancy pointed to a bias in PCR amplification. Previously reported PCR biases have been attributed to variation in bacterial genome size, 16S rDNA allele copy number, G+C content, rDNA secondary structure, and template concentration (31–33). Our subsequent strategy to reduce the number of streptococcal 16S rDNA clones by using Bsu36I was successful; however, it probably reduced the representation of other bacterial sequences within the libraries. In fact, there are Bsu36I endonuclease restriction sites within 16S rDNAs from cyanobacteria and spirochetes. Another obvious limitation in our study was the use of just one specimen from one individual. High-throughput sequencing methods and the possibility of high density microarrays for microbial surveying should make analysis of multiple specimens from numerous individuals increasingly practicable.
Typically, large numbers of Treponema have been associated with periodontitis (9, 34, 35). Choi et al. found that only 7% of 16S rDNA clones obtained by amplification using broad-range bacterial primers were of treponemal origin whereas phase-contrast microscopy indicated that spirochetes comprised ≈20% of the bacterial population (9). Attempts to amplify Treponema 16S rDNA from our specimen were unsuccessful. This failure does not appear to be explained by the choice of primers because the predicted annealing sites are well conserved among the Treponema sequences within the arb 16S rRNA database. It is probable that treponemes, if present in our specimen, were present in numbers below the detection limits of our PCR. The absence of spirochetes detected by darkfield microscopy supports this presumption.
A crude estimate of the population coverage by the 59 unique phylotypes we identified by direct amplification may be obtained by using the estimator proposed by Good (21). By this estimate, the 59 phylotypes accounted for 88% of all cloned sequences. Using a parametric model for estimating the number of “unseen” species, the 59 unique phylotypes represented <70% of the total number of unique phylotypes within the clone population (22). By inference, an additional 25 [(59/0.7) − 59] unique but undetected phylotypes were present within the clone libraries and comprised ≈12% of the clones. In ecological terms, the clone libraries, which are assumed to represent the subgingival microbial flora from this specimen, are uneven (a small number of species dominate the population) but contain a large degree of richness (a large number of unique phylotypes). This conclusion is consistent with the cultivation-based findings of Moore (8) in which 10–16 species accounted for 60–71% of the cultivatable oral bacteria in adult gingivitis. The transfer of mathematical constructs, such as the Good coverage estimator or the Shannon-Weaver index, from macroecology to human microbial ecology should facilitate a more meaningful and quantifiable comparison of the richness and evenness between different hosts and sites over time.
Overall, direct amplification of 16S rDNA yielded a more diverse view of the bacterial flora associated with subgingival plaque than that found by cultivation. Our findings raise the possibility that previously unidentified higher order taxonomic divisions still exist within the human ecosystem. Direct amplification and cultivation may be viewed as complementary techniques for the study of microbial diversity and identification of potential opportunistic pathogens. Obviously, the physiological and clinical relevance of the organisms detected in this study remain unclear. At one extreme, some of the phylotypes may be transient residents acquired from the environment. For example, A-51 is related to metal-oxidizing γ-proteobacteria, such as the sulfur-oxidizer Thiobacillus cuprinus, a bacterium typically isolated from lake sediments. A-51 may be a transient oral resident obtained from drinking water. Other organisms may be critical for maintenance of ecosystem stability and oral health or may be occasional pathogens. Large-scale ecosystem analyses at multiple intraoral sites in multiple hosts and at multiple times should help to clarify some of these issues.
Supplementary Material
Acknowledgments
We thank Pat Mickelsen and Arlene Morton from the Stanford University Hospital Clinical Microbiology Laboratory for bacterial cultivation; Victor T. Lee and Penny Asami Wong for specimen collection; the Stanford University Digestive Diseases Center; and Doug Smith and Deborah Dodge from Perkin–Elmer for DNA sequencing technology support. This work was supported by the Donald E. and Delia B. Baxter Foundation and the Lucille P. Markey Charitable Trust.
Footnotes
References
- 1.Mackowiak P A. N Engl J Med. 1982;307:83–93. doi: 10.1056/NEJM198207083070203. [DOI] [PubMed] [Google Scholar]
- 2.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Nature (London) 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 4.Stackebrandt E, Liesack W, Goebel B M. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 5.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 6.Thoden van Velzen S K, Abraham-Inpijn L, Moorer W R. J Clin Periodontol. 1984;11:209–220. doi: 10.1111/j.1600-051x.1984.tb02211.x. [DOI] [PubMed] [Google Scholar]
- 7.Meyer D H, Fives-Taylor P M. Curr Opin Microbiol. 1998;1:88–95. doi: 10.1016/s1369-5274(98)80147-1. [DOI] [PubMed] [Google Scholar]
- 8.Moore W E, Burmeister J A, Brooks C N, Ranney R R, Hinkelmann K H, Schieken R M, Moore L V. Infect Immun. 1993;61:2891–2898. doi: 10.1128/iai.61.7.2891-2898.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi B K, Paster B J, Dewhirst F E, Gobel U B. Infect Immun. 1994;62:1889–1895. doi: 10.1128/iai.62.5.1889-1895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashimoto A, Chen C, Bakker I, Slots J. Oral Microbiol Immunol. 1996;11:266–273. doi: 10.1111/j.1399-302x.1996.tb00180.x. [DOI] [PubMed] [Google Scholar]
- 11.Dymock D, Weightman A J, Scully C, Wade W G. J Clin Microbiol. 1996;34:537–542. doi: 10.1128/jcm.34.3.537-542.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amann R I, Krumholz L, Stahl D A. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fredricks D N, Relman D A. J Clin Microbiol. 1998;36:2810–2816. doi: 10.1128/jcm.36.10.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards U, Rogall T, Blocker H, Emde M, Bottger E C. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Relman D A. In: Diagnostic Molecular Microbiology: Principles and Applications. Persing D H, Smith T F, Tenover F C, White T J, editors. Washington, DC: Am. Soc. Microbiol.; 1993. pp. 489–495. [Google Scholar]
- 18.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 19.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 20.DeSoete G. Psychometrika. 1983;48:621–626. [Google Scholar]
- 21.Good I J. Biometrika. 1953;40:237–264. [Google Scholar]
- 22.Efron B, Thisted R. Biometrika. 1976;63:435–447. [Google Scholar]
- 23.DeLong E F, Wickham G S, Pace N R. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 24.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Socransky S S, Gibbons R J, Dale A C, Bortnick L, Rosenthal E, Macdonald J B. Arch Oral Biol. 1963;8:275–280. doi: 10.1016/0003-9969(63)90019-0. [DOI] [PubMed] [Google Scholar]
- 26.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez G J, Garcia P, Cai J, Hippe H, Farrow J A. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 27.Kolenbrander P E, London J. J Bacteriol. 1993;175:3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawson P A, Falsen E, Akervall E, Vandamme P, Collins M D. Int J Syst Bacteriol. 1997;47:899–903. doi: 10.1099/00207713-47-3-899. [DOI] [PubMed] [Google Scholar]
- 29.Talan D A, Citron D M, Abrahamian F M, Moran G J, Goldstein E J C. N Engl J Med. 1999;340:85–92. doi: 10.1056/NEJM199901143400202. [DOI] [PubMed] [Google Scholar]
- 30.Hugenholtz P, Goebel B M, Pace N R. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farrelly V, Rainey F A, Stackebrandt E. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki M T, Giovannoni S J. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandler D P, Fredrickson J K, Brockman F J. Mol Ecol. 1997;6:475–482. doi: 10.1046/j.1365-294x.1997.00205.x. [DOI] [PubMed] [Google Scholar]
- 34.Riviere G R, Smith K S, Carranza N J, Tzagaroulaki E, Kay S L, Dock M. J Periodontol. 1995;66:829–837. doi: 10.1902/jop.1995.66.10.829. [DOI] [PubMed] [Google Scholar]
- 35.Kigure T, Saito A, Seida K, Yamada S, Ishihara K, Okuda K. J Periodontal Res. 1995;30:332–341. doi: 10.1111/j.1600-0765.1995.tb01284.x. [DOI] [PubMed] [Google Scholar]
- 36.Stackenbrandt E, Groebel B M. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.