Abstract
The interferons (IFNs) are cytokines that play key roles in host defense against viral infections and immune surveillance against cancer. We report that BCR-ABL transformation of hematopoietic cells results in suppression of IFN-dependent responses, including transcription of IFN-inducible genes and generation of IFN-mediated antiviral effects. BCR-ABL transformation suppresses expression of several IFN-regulated genes containing IFN-sensitive response element (ISRE) or GAS elements in their promoters, including Isg15, Irf1, Irf9, and Ifit2 (interferon-induced protein with tetratricopeptide repeats 2). Suppression of transcription of ISRE-containing genes is also seen in cells expressing various BCR-ABL kinase domain mutants, including T315I, H396P, Y253F, and E255K, but not kinase-defective BCR-ABL. Such effects are associated with impaired IFN-dependent phosphorylation of Stat1 on Tyr701 and Stat3 on Tyr705 and defective binding of Stat complexes to ISRE or GAS elements. Beyond suppression of Stat activities, BCR-ABL inhibits IFN-inducible phosphorylation/activation of the p38 MAPK, suggesting a dual mechanism by which this abnormal fusion protein blocks IFN transcriptional responses. The inhibitory activities of BCR-ABL ultimately result in impaired IFNα-mediated protection against encephalomyocarditis virus infection and reversal of IFN-dependent growth suppression. Altogether, our data provide evidence for a novel mechanism by which BCR-ABL impairs host defenses and promotes malignant transformation, involving dual suppression of IFN-activated signaling pathways.
To survive viral infection, cells produce and secrete interferons (IFNs),2 proinflammatory cytokines that inhibit cellular proliferation, block viral infection and replication, and exhibit important immunomodulatory activities (reviewed in Refs. 1–7). One important mechanism by which IFNs mediate their antiviral effects is through the transcriptional regulation of multiple antiviral genes (reviewed in Refs. 8–12). The transcriptional activation of such genes is primarily regulated by the engagement of Jak-Stat signaling cascades, pathways that are rapidly activated after binding of IFNs to their respective cell surface receptor. Type I IFNs (α, β, ω, κ, and ε) utilize the type I IFN receptor, whereas IFNγ binds to a distinct receptor, the type II IFN receptor (reviewed in Refs. 13–15). Following receptor binding of type I IFNs to their receptor, the Jak1 and Tyk2 kinases are activated, leading to induction of a complex network of signaling pathways (reviewed in Refs. 8–12). The classical signaling pathways involve activation of Stat proteins, which form homo- and/or heterodimers, migrate to the nucleus, and bind to the promoters of IFN-regulated genes, leading to their induction or repression (reviewed in Refs. 8–12). Type I IFNs activate the ISGF3 complex, which is composed of IRF9 (interferon regulatory factor 9) and activated Stat1 and Stat2, and other Stat complexes that bind to specific sequences in the promoters of target genes to regulate transcription (8–12, 16–18).
In addition to the classical IFN pathways, various other signaling cascades are engaged by type I IFNs. These include the p38 MAPK signaling cascade (19–25), pathways involving PKC isoforms (26, 27), Crk proteins (reviewed in Refs. 11, 12), and the PI 3′ kinase pathway (9–12, 21), which regulates engagement of mTOR and downstream signals to regulate initiation of mRNA translation (28, 29).
Previous studies have shown that in BCR-ABL (breakpoint cluster region Abelson leukemia fusion protein)-expressing cells, there is deregulation of certain pathways used by IFNs to induce antileukemic effects (20, 30). For instance, in one study, the leukemic cells of a subset of CML patients that did not respond to IFNα were found to lack Stat1 expression (31), whereas approximately half of the IFNα-resistant patients in another study had increased levels of SOCS3 (suppressor of cytokine signaling 3) (32). Thus, although lack of Stat1 expression or overexpression of SOCS3 is associated with IFN resistance in CML, additional mechanisms apparently contribute to IFN resistance in BCR-ABL-expressing cells. Interestingly, other studies have shown that BCR-ABL modestly activates Stat1 and Stat3 through tyrosine phosphorylation of Jak1, Jak2, and Jak3 and strongly phosphorylates and activates Stat5 and the adapter protein CrkL, resulting in the formation of CrkL-Stat5 complexes that bind to GAS elements to regulate transcription of BCR-ABL-regulated genes (33–35). It is intriguing that IFNα also triggers activation of CrkL and formation of IFN-dependent CrkL-Stat5 complexes that regulate transcription of a subset of IFN-sensitive genes (36, 37). In addition, as mentioned above, IFNs activate the phosphatidylinositol 3′-kinase pathway and its effectors (9–12, 21), a pathway well known to promote BCR-ABL-induced leukemogenesis (38). Such findings have raised the possibility that the type I IFN receptor and BCR-ABL compete for the utilization of certain common signaling elements and pathways that are required for the transmission of signals essential for generation of their biological effects.
In the current study, we determined whether the abnormal BCR-ABL protein can activate signals that antagonize the transcriptional activation of IFN-regulated genes. Overexpression of BCR-ABL in IFN-sensitive cells suppressed IFNα-dependent transcriptional activity via ISRE elements in luciferase reporter assays and dramatically decreased the expression of several IFN-inducible genes known to mediate antiviral responses, such as Irf1, Irf9, Isg15 (interferon-stimulated gene 15), and Ifit2. The down-regulation of such genes was also observed in cells expressing various imatinib mesylate resistant BCR-ABL kinase domain mutants, and was associated with decreased levels of IFNα-induced Stat1-Stat1, Stat1-Stat3, and Stat3-Stat3 binding to SIE elements or decreased binding of the ISGF3 complex to ISRE elements in the promoters of IFN-regulated genes. In addition, BCR-ABL suppressed the IFN-dependent phosphorylation/activation of p38 MAPK, whose function is essential for optimal transcription of IFN-regulated genes. Importantly, the generation of IFNα-dependent antiviral and growth-inhibitory responses was suppressed in BCR-ABL-expressing Ba/F3 cells, indicating that the effects of BCR-ABL on IFN transcriptional activity translated to important antagonistic biological responses.
MATERIALS AND METHODS
Antibodies, Cell Lines, Plasmids, and Reagents—Recombinant human IFNβ was obtained from Biogen Inc. (Cambridge, MA). Recombinant mouse interleukin-3 was purchased from Invitrogen. Imatinib mesylate was provided by Novartis (Basel, Switzerland). The antibodies against phosphotyrosine (clone 4G10) and Stat1 were purchased from Upstate Cell Signaling Solutions (Charlottesville, VA). The antibodies against p-Stat1 (Tyr701), p-Stat3 (Tyr705), p38, p-p38 (Thr180/Tyr182), and c-Abl (cellular Abelson murine leukemia virus oncoprotein) were obtained from Cell Signaling Technology (Danvers, MA). Anti-Stat2 was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and anti-p-Stat2 was purchased from Bio-Vision Inc. (Mountain View, CA). U2OS cells were grown in McCoy's medium supplemented with 10% (v/v) fetal bovine serum and gentamycin. K562 cells were grown in RPMI medium supplemented with 10% (v/v) fetal bovine serum and gentamycin. BA/F3 pSRα mock cells, BA/F3 MIGR1, and the BCR-ABL kinase-inactive BA/F3 MIG P210 KI cells were grown in RPMI medium supplemented with 10% (v/v) fetal bovine serum, gentamycin, and recombinant mouse IL3 (10 ng/ml). BA/F3 pSRα BCR-ABL wild-type and kinase domain mutant cells as well as BA/F3 MIG BCR-ABL cells were grown in RPMI medium supplemented with 10% (v/v) fetal bovine serum and antibiotics. BA/F3 cells were starved overnight prior to IFN treatment for Western blot, electrophoretic mobility shift assay, chromatin immunoprecipitation (ChIP), and RNA analysis. The plasmids pcDNA3 and pcDNA3 BCR-ABL, the BA/F3 pSRα cells, and the stably BCR-ABL wild type- and BCR-ABL mutant T315I-, H396P-, E255K-, and Y253F-expressing cells have been previously described (39) (Oregon Health and Sciences University, Portland, OR). The retroviral plasmids MIGR1, MIG P210, and MIG P210 KI (40) were a kind gift from Dr. Rhavi Bhatia (Division of Hematology and Bone Marrow Transplantation, City of Hope National Medical Center, Duarte, CA). The plasmids were used to generate BA/F3 cells stably expressing MIGR1, MIG BCR-ABL wild type, and kinase-inactive MIG BCR-ABL via retroviral infection. Cells expressing the above constructs were GFP+ and were selected by flow cytometry.
Cell Lysis and Immunoblotting—Cells were lysed in phosphorylation lysis buffer as previously described (41–43). Immunoprecipitations and immunoblotting were performed as previously described (41–43).
Cell Proliferation Assays—Cell proliferation was determined using MTT assays, as previously described (25). Equal numbers of cells were plated in 96-well plates and treated with interferon α or STI versus Me2SO as indicated for 5–7 days. BA/F3 pSRα, BA/F3 MIGR1, and BA/F3 MIG BCR-ABL KI cells were grown in the presence of interleukin-3.
Mobility Shift Assays—Actively growing U2OS cells, transfected with pcDNA3 or pcDNA3 BCR-ABL, or BA/F3 pSRα and pSRα BCR-ABL cells were pretreated with Me2SO or STI-571 (1 μm) for 1 h and then treated with IFNβ or IFNα or left untreated, as indicated. 10 μg of nuclear extracts were analyzed using electrophoretic mobility shift assays, as described previously (36). A double-stranded oligonucleotide (ATTTCCCGTAAATCCC), which represents a sis-inducing element (SIE) from the c-fos promoter was synthesized and used to detect Stat1-Stat1, Stat1-Stat3, and Stat3-Stat3 binding in the gel shift assays. ISGF3 complexes were detected using a double-stranded oligonucleotide (CTGTTGGTTTCGTTTCCTCAGA), representing an ISRE element from the Isg15 gene.
Luciferase Reporter Assays—U2OS cells were transfected with an ISRE luciferase construct and a constitutive β-galactosidase expression vector using the superfect transfection reagent according to the protocol of the manufacturer (Qiagen, Hilden, Germany). The same method was used to transfect U2OS cells with pcDNA3 or pcDNA3 BCR-ABL. The ISRE-luciferase construct included the wild-type ISG15 ISRE (TCGGGAAAGGGAAACCGAAACTGAAGCC) cloned via cohesive ends into the BamHI site of the pZtkLuc vector and was provided by Dr. Richard Pine (Public Health Research Institute, New York, NY) (44). Forty-two hours after transfection, triplicate cultures were pretreated with Me2SO or STI-571 (1 or 5 μm) for 1 h and then left untreated or treated with 5 × 103 units/ml of human IFNβ for 6 h, as indicated. In addition, in order to suppress all BCR-ABL effects, Me2SO or STI-571 was added to U2OS cells directly post-transfection, and cells were treated with 5 × 103 units/ml of human IFNβ for 6 h or were left untreated before the luciferase assays were performed.
ChIP Assays—ChIP assays were performed using the ChIP kit from Upstate Cell Signaling Solutions (Charlottesville, VA) according to the manufacturer's instructions. BA/F3 cells were treated with IFNα for 180 min or were left untreated, as indicated. Cells were fixed, and DNA fragments were immunoprecipitated with either an anti-Stat1 antibody or control nonimmune RIgG. Precipitated and input control nonprecipitated DNA fragments were used as templates for real time PCR with the mouse ISRE-ISG15 gene (Mm01705338_s1) probes and primers from Applied Biosystems (Foster City, CA). Values were calculated by subtracting nonspecific binding detected in control RIgG immunoprecipitates from specific ISG15-DNA binding detected in anti-Stat1 immunoprecipitates.
mRNA Isolation and Conversion, Real Time PCR Probes, and Primers—Isolation, purification of mRNA, and conversion into cDNA was performed using the respective kits and oligo(dT)s from Qiagen Inc. (Hilden, Germany) according to the manufacturer's instructions. Probes and primers for real time PCR were purchased from Applied Biosystems (Foster City, CA). The primers and probes used were Isg15 (Mm01705338_s1), Irf9 (Mm0049 2679_m1), Irf1 (Mm00515191_m1), Ifit2 (Mm00492606_m1), and SOCS3 (Mm00545913_s1). Gapdh (Mm99999915_g1) was used as an internal control.
Antiviral Assays—The antiviral effects of mouse IFNα in BA/F3 cells expressing BCR-ABL (wild type) or vector pSRα were determined as previously described (29). Encephalomyocarditis virus (EMCV) was used as the challenge virus.
RESULTS
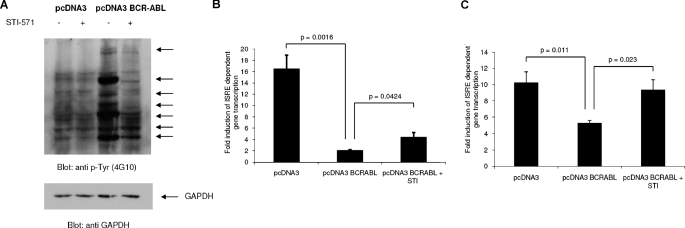
In initial experiments, we examined whether BCR-ABL expression exerts regulatory effects on type I IFN-dependent, ISRE-driven, gene transcription. For this purpose, a system in which BCR-ABL was transiently overexpressed in U2OS cells (Fig. 1A) was initially used. Cells were transfected with either pcDNA3 (empty vector) or a pcDNA3-BCR-ABL construct (Fig. 1A), together with a luciferase reporter plasmid, under the control of an ISRE. As expected, IFNβ treatment led to a robust induction of ISRE-dependent transcription in cells transfected with the empty vector (Fig. 1B). On the other hand, ISRE-driven transcription was significantly suppressed in cells transfected with BCR-ABL (Fig. 1B). Such BCR-ABL-mediated suppression of ISRE-driven transcription was only partially reversed when cells were pretreated with imatinib mesylate (STI-571), added to the cultures 60 min prior to IFN treatment (Fig. 1A). However, if imatinib mesylate was added to the cultures immediately following transfection of cells with the pcDNA3-BCR-ABL construct, there was complete reversal of the suppressive effects of BCR-ABL (Fig. 1C), suggesting that inhibition of the kinase activity of BCR-ABL early in the process is required for reversal of its effects on IFN signals. Thus, BCR-ABL suppresses type I IFN-dependent transcription, and such suppression appears to require the kinase activity of BCR-ABL.
FIGURE 1.
Suppression of ISRE-driven transcription by BCR-ABL. A, U2OS cells were transfected with either pcDNA3 empty vector or a pcDNA3-BCR-ABL construct. 42 h after transfection, the cells were treated for 1 h with imatinib mesylate (STI-571) or left untreated, as indicated. Equal amounts of total cell lysates (100 μg) were resolved by SDS-PAGE and immunoblotted with an antiphosphotyrosine (anti p-Tyr) antibody (4G10) (top). The blots were subsequently stripped and reprobed with an antibody against glyceraldehyde-3-phosphate dehydrogenase to control for protein loading (bottom). The arrows point at proteins that are phosphorylated by BCR-ABL on tyrosine. B, U2OS cells were transfected with either pcDNA3 empty vector or pcDNA3-BCR-ABL. 42 h after transfection, the cells were treated with IFNβ (5000 IU/ml) for 6 h, in the presence or absence of imatinib mesylate (STI-571), and ISRE-driven transcription was determined using a luciferase/β-galactosidase reporter assay. Data are expressed as -fold increases in response to IFN treatment over control untreated samples for each condition. Mean ± S.E. values of six independent experiments are shown. Paired two-tailed t test analysis for ISRE in IFN-treated pcDNA3 versus pcDNA3-BCR-ABL-transfected cells showed a two-tailed p value of 0.016. The paired two-tailed p value for STI-571 treated versus untreated pcDNA3-BCR-ABL cells was 0.0424. C, U2OS cells were transfected with pcDNA3 or pcDNA3-BCR-ABL. Following transfection, cells were incubated in the presence of Me2SO or imatinib mesylate. 42 h after transfection, the cells were treated with IFNβ (5000 IU/ml) for 6 h, and ISRE-driven transcription was determined using a luciferase/β-galactosidase reporter assay. Data are expressed as -fold increases in response to IFN treatment over control untreated samples for each condition. Mean ± S.E. values of five independent experiments are shown. Paired two-tailed t test analysis for pcDNA3- versus pcDNA3-BCR-ABL-transfected cells showed a two-tailed p value of 0.011. When STI-571-treated versus STI-571-untreated pcDNA3-BCR-ABL cells were compared, the two-tailed p value was 0.023.
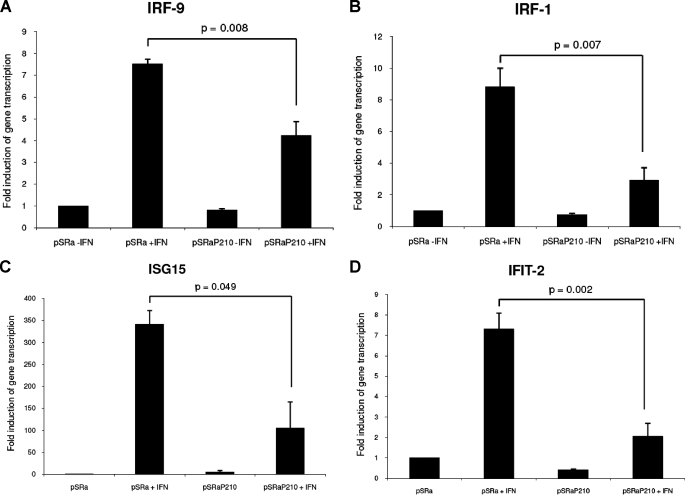
Since ISRE and also SIE sequences are commonly found in the promoters of various type I IFN-inducible genes, including genes that mediate antiviral and/or antiproliferative responses (8–10, 14), we sought to directly examine the effects of BCR-ABL overexpression on the transcription of several genes that are known to play roles in the generation of the biological effects of IFNα and other type I IFNs. For such studies, we used Ba/F3 mouse cells stably expressing BCR-ABL (39). In initial studies, we determined the effects of BCR-ABL on the transcriptional regulation of two gene members of the IRF family, Irf9 (also called ISGF3G or p48) and Irf1 (45, 46). Ba/F3 cells transfected with the empty vector or cells expressing BCR-ABL were treated with mouse IFNα, and expression of Irf1 and Irf9 was analyzed using quantitative real time reverse transcription-PCR. IFNα treatment resulted in a strong induction of Irf9 (Fig. 2A) and Irf1 (Fig. 2B) gene transcription in Ba/F3 cells transfected with pSRα vector alone. On the other hand, the IFN-dependent up-regulation of these genes was severely suppressed in Ba/F3-BCR-ABL-transfected cells (Fig. 2, A and B), and such suppression was statistically significant (paired values were p = 0.008 for Irf9 and p = 0.007 for Irf1) (Fig. 2, A and B). Importantly, expression of BCR-ABL did not alter basal expression of Irf9 and Irf1 (Fig. 2, A and B), and only the IFN-inducible expression of these genes was affected. Two additional genes analyzed were Isg15 (Fig. 2C) and the interferon-induced protein with tetratricopeptide repeats Ifit-2 (Fig. 2D). As in the case of Irf1 and Irf9, Isg15 and Ifit-2 have been previously linked to the antiviral properties of IFNs (47–49). Both genes were fully up-regulated by IFNα treatment of Ba/F3 cells transfected with pSRα vector alone, whereas their expression was suppressed in cells expressing BCR-ABL (Fig. 2, C and D) (paired values p = 0.049 for Isg15 and p = 0.002 for Ifit-2).
FIGURE 2.
IFN-inducible gene transcription in Ba/F3 cells stably transfected with pSRα BCR-ABL or pSRα mock vector. Ba/F3 pSRα BCR-ABL (P210) or Ba/F3 pSRα mock cells were incubated in the presence or absence of mouse IFNα (5 × 103 IU/ml). Cells were lysed, and total mRNA was isolated and used for quantitative reverse transcription-PCR. Gene transcription of Irf9 (A), Irf1 (B), Isg15 (C), and Ifit2 (D) was subsequently assessed by quantitative reverse transcription-PCR. The bars represent the average -fold induction over untreated control BA/F3 pSRα mock cells ± S.E. of three independent experiments. Paired t test analysis comparing interferon-induced transcription in mock vector versus BCR-ABL-expressing cells resulted in p = 0.008 for Irf9 (A), p = 0.007 for Irf1 (B), p = 0.049 for Isg15 (C), and p = 0.002 for Ifit-2 (D).
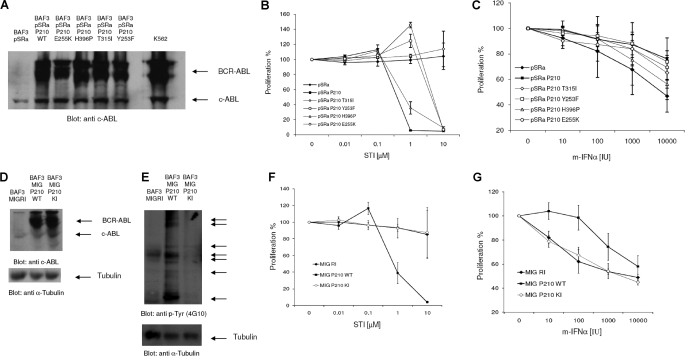
In subsequent studies, we examined the effects of various BCR-ABL kinase point mutations that represent about 60% of kinase domain mutations found in patients with resistance to imatinib mesylate (39) on IFN-dependent gene transcription and responses. In addition to conferring resistance to imatinib mesylate, these mutations also have effects on the intrinsic kinase activity and substrate specificity of BCR-ABL (39). At first we sought to confirm expression of the various mutant proteins in the stably transfected Ba/F3 cells (39). As shown in Fig. 3A, similar levels of expression of wild-type BCR-ABL and the BCR-ABL mutants E255K, H396P, T315I, and Y253F were seen in the various transfectants, and these levels were comparable with the endogenous BCR-ABL levels in the K562 cell line, analyzed in parallel (Fig. 3A). As expected (39), BA/F3 cells expressing the various mutants were insensitive in various degrees to imatinib mesylate, with the T315I mutant being completely refractory, even when a very high concentration (10 μm) of imatinib was used (Fig. 3B). Cells expressing the Y253F and E255K mutants were also resistant, being insensitive to imatinib at a final concentration of 1 μm, whereas the H396P mutant was the least refractory but still clearly less sensitive than wild-type BCR-ABL (Fig. 3B). When the sensitivity of the various transfectants to the growth-inhibitory effects of mouse IFNα was examined, we found that Ba/F3 cells transfected with the empty vector were sensitive to IFNα-mediated growth inhibition, whereas sensitivity to the antiproliferative effects of IFNα was substantially decreased in cells expressing wild-type BCR-ABL (Fig. 3C). Although cells expressing the BCR-ABL T315I and H396P mutants exhibited modestly enhanced sensitivity to IFNα treatment compared with cells expressing wild-type BCR-ABL, such differences were only marginal, suggesting that decreases in the intrinsic kinase activities of these mutants (39) do not alter substantially the ability of BCR-ABL to suppress IFN-dependent growth-inhibitory effects.
FIGURE 3.
Growth-inhibitory effects of IFNα in Ba/F3 cells expressing BCR-ABL and BCR-ABL mutant proteins. A, equal amounts of total cell lysates from the indicated Ba/F3 transfectants were resolved by SDS-PAGE and immunoblotted with anti-ABL antibody. B, Ba/F3-pSRα, Ba/F3-BCR-ABL, and Ba/F3 cells stably expressing the indicated BCR-ABL mutants were incubated with the indicated concentrations (in μmol/liters) of imatinib mesylate for 5 days. Cell proliferation was assessed using MTT assays. Data are expressed as means ± S.E. of four independent experiments. C, Ba/F3-pSRα, Ba/F3-BCR-ABL, and Ba/F3 cells stably expressing the indicated BCR-ABL mutants were incubated with the indicated concentrations (in IU/ml) of mouse IFNα. Cell proliferation was determined with MTT assays after 5 days. Data are expressed as means ± S.E. of three independent experiments. D, equal amounts of total cell lysates from the Ba/F3 MIG R1, MIG BCR-ABL (WT), and kinase-inactive Ba/F3 MIG BCR-ABL (KI) transfectants were resolved by SDS-PAGE and immunoblotted with anti-ABL antibody (top) or anti-tubulin antibody to control for protein loading (bottom). E, Ba/F3 cells stably expressing wild-type and kinase-inactive BCR-ABL were lysed, and equal amounts of total cell lysates (100 μg) were resolved by SDS-PAGE and immunoblotted with an antiphosphotyrosine (anti p-Tyr) antibody (4G10) (top). The blots were subsequently stripped and reprobed with an antibody against α-tubulin to control for protein loading (bottom). The arrows point at proteins that are tyrosine-phosphorylated in BCR-ABL transfected cells. F, Ba/F3-MIG R1, MIG BCR-ABL-transfected, and Ba/F3 MIG BCR-ABL(KI)-transfected cells were incubated with the indicated concentrations (in μmol/liter) of imatinib mesylate for 5 days. Cell proliferation was assessed using MTT assays. Data are expressed as means ± S.E. of four independent experiments. G, Ba/F3-MIG R1, MIG BCR-ABL, and Ba/F3 MIG BCR-ABL(KI) cells were incubated with the indicated concentrations (in IU/ml) of mouse IFNα for 5 days. Cell proliferation was determined by MTT assays. Data are expressed as means ± S.E. of three independent experiments.
To determine whether BCR-ABL tyrosine kinase activity was essential for the suppressive effects of BCR-ABL on the type I IFN-mediated antiproliferative responses, we generated BA/F3 cells stably expressing wild-type BCR-ABL or kinase-inactive BCR-ABL (K1176R) (40), using retroviral transduction. Similar BCR-ABL protein levels were seen in Ba/F3 cells stably transfected with wild-type BCR-ABL or KI-BCR-ABL (Fig. 3D). However, as expected (40), elevated tyrosine kinase activity, as reflected by the phosphorylation of various cellular substrates, was only seen in cells transfected with wild-type BCR-ABL, not in cells expressing the kinase-inactive BCR-ABL mutant (Fig. 3E). Consistent with this, the growth of kinase inactive BA/F3 BCR-ABL cells was not affected by treatment of the cells with imatinib mesylate at concentrations of up to 10 μm (Fig. 3F). Ba/F3-MIG P210 KI cells exhibited enhanced sensitivity to the growth-inhibitory effects of IFNα when compared with Ba/F3 MIG P210 WT cells (Fig. 3G) and followed a similar response pattern to control cells stably transfected with the empty vector (Fig. 3G).
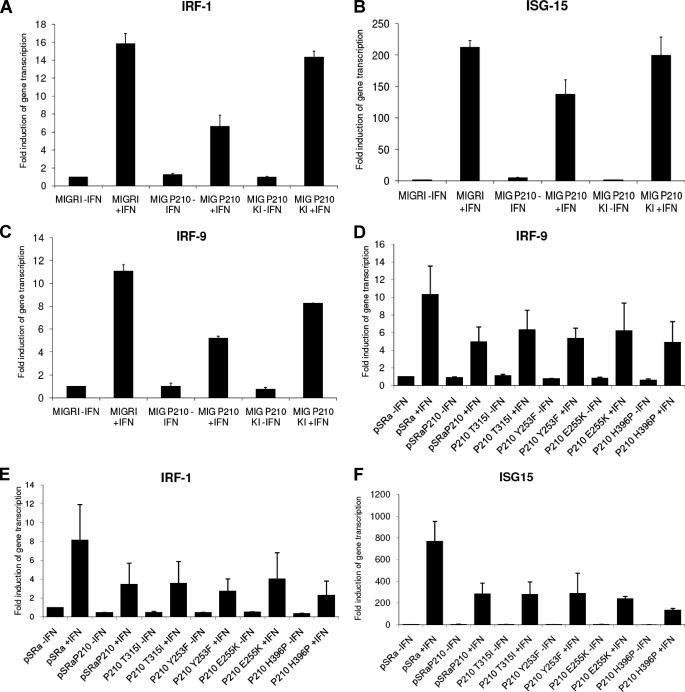
Subsequently, we analyzed the effects of wild-type BCR-ABL expression versus kinase-inactive BCR-ABL expression on the regulation of transcription of ISGs. Although wild-type BCR-ABL significantly suppressed the IFN transcriptional activation of Irf1 (Fig. 4A), Isg15 (Fig. 4B), and Irf9 (Fig. 4C), the kinase-inactive BCR-ABL had minimal or no effects (Fig. 4, A–C). On the other hand, cells expressing the various imatinib mesylate-resistant kinase domain BCR-ABL mutants inhibited IFNα-induced transcription of Irf9 (Fig. 4D), Irf1 (Fig. 4E), Isg15 (Fig. 4F), and Ifit-2 (data not shown) to similar degrees as cells expressing wild-type BCR-ABL. Thus, BCR-ABL tyrosine kinase activity is necessary for suppression of various IFN-sensitive genes known to mediate functional responses, whereas such suppression is unaffected by BCR-ABL mutations known to result to imatinib mesylate resistance.
FIGURE 4.
IFN-inducible gene transcription in cells expressing wild-type BCR-ABL, kinase-inactive BCR-ABL (A–C), or the T315I, Y253F, E255K, and H396P kinase domain BCR-ABL mutants (D–F). The indicated stably transfected cells were incubated in the presence or absence of mouse (IFNα) for 1 h (Irf1 and Irf9) or 3 h (Isg15) and lysed, and total mRNA was isolated. Gene transcription of the indicated genes was assessed by quantitative reverse transcription-PCR. Data are expressed as -fold induction over untreated control Ba/F3 mock cells and represent means ± S.E. of two independent experiments.
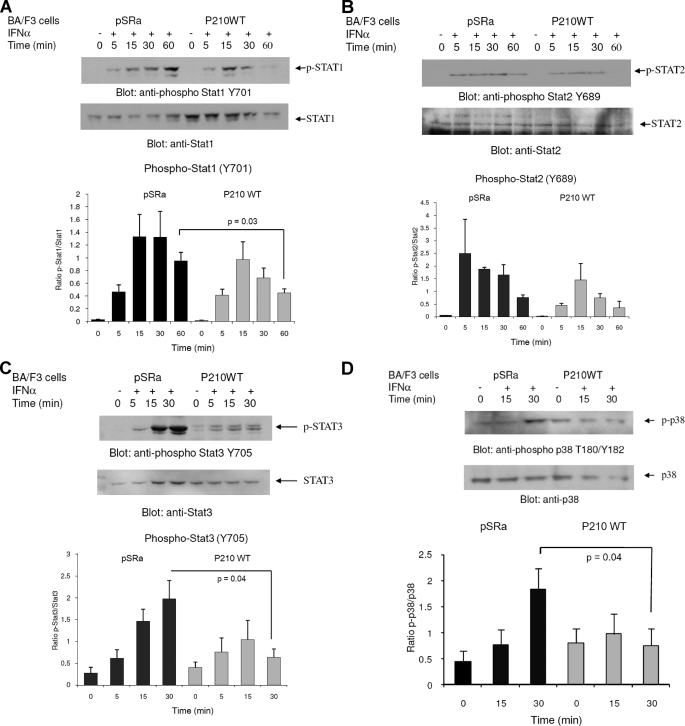
It is well established that transcription of ISGs is controlled by activation of Jak-Stat pathways. Several different Stat proteins are known to be tyrosine-phosphorylated by type I IFN-activated Jak kinases, including Stat1, Stat2, Stat3, and Stat5 (reviewed in Refs. 8–12). It is also well known that phosphorylation of Stat1 and Stat3 proteins on serine 727 is required for their full transcriptional activity. To determine whether the suppressive effects of BCR-ABL on IFN-dependent transcription results from negative regulatory effects on IFN-activated Stats, we examined the effects of BCR-ABL expression on the phosphorylation of Stats. Initially, we analyzed the phosphorylation of Stat1 in Ba/F3-pSRα control cells and Ba/F3-BCR-ABL cells. Although we did not observe any striking differences on the IFN-inducible phosphorylation of Stat1 on Ser727 in empty vector-transfected and BCR-ABL-expressing cells (data not shown), the IFN-inducible phosphorylation of Stat1 on Tyr701 and the duration of the signal were clearly reduced in the BCR-ABL-expressing cells, as compared with the control cells (Fig. 5A). On the other hand, the effects of BCR-ABL expression on Stat2 phosphorylation on Tyr689 were minimal (Fig. 5B), whereas IFN-inducible phosphorylation of Stat3 on Tyr705 (Fig. 5C) was clearly suppressed in BCR-ABL expressing cells. Thus, BCR-ABL expression results in suppression of type I IFN-dependent phosphorylation of Stat1 and Stat3 in Ba/F3 transfectants, suggesting a mechanism that may account for the defective regulation of ISGs and the induction of growth-inhibitory responses by IFNα.
FIGURE 5.
Effects of BCR-ABL on IFNα-dependent tyrosine phosphorylation of Stat proteins and phosphorylation/activation of the p38 MAPK. Ba/F3-pSRα BCR-ABL (P210) or Ba/F3-pSRα mock cells were treated with IFNα (104 IU/ml) for the indicated times or left untreated, as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against the phosphorylated form of Stat1 on Tyr701 (A), the phosphorylated form of Stat2 on Tyr689 (B), the phosphorylated form of Stat3 on Tyr705 (C), and the phosphorylated form of p38 on Thr180/Tyr182 (D). The blots were subsequently stripped and reprobed with antibodies against total Stat1, Stat2, Stat3, or p38, respectively, to control for protein loading. The signals were quantified by densitometry, and the intensities of phosphorylated proteins versus unphosphorylated proteins were calculated. Data are expressed as means ± S.E. of six experiments for A, two experiments for B, four experiments for C, and five experiments for D. Paired t test analysis for Stat1 Tyr701 phosphorylation showed a paired p value of 0.03 comparing IFN-treated (60 min) BA/F3 pSRα versus BA/F3 BCR-ABL cells (A). Paired t test analysis for Stat3 Tyr705 phosphorylation showed a paired p value of 0.04 comparing IFN-treated (30 min) BA/F3 pSRα versus BA/F3 BCR-ABL cells (C). Paired t test for p38 Thr180/Tyr182 phosphorylation comparing IFN-treated (30 min) BA/F3 pSRα versus BA/F3 BCR-ABL cells showed a paired p value of 0.04 (D).
There is accumulating evidence that beyond the function of Stats, engagement of other signaling pathways is required for optimal IFN signaling generation of IFN responses (11, 12, 19). Among them, the p38 MAPK pathway is activated by various type I IFNs, and its function is required for IFN-dependent transcriptional regulation and biological responses (19–25). Interestingly, previous studies have also suggested that the p38 MAPK pathway may be impaired by BCR-ABL transformation (50) and that treatment of cells with imatinib mesylate results in its activation (51). We therefore explored the possibility that activation of p38 MAPK in response to IFNα treatment may be suppressed in BA/F3 BCR-ABL cells compared with empty vector-expressing cells. Although phosphorylation of p38 MAPK on threonine 180/tyrosine 182 was strongly induced in BA/F3 pSRα cells, no significant induction of p38 was seen in BCR-ABL-transfected BA/F3 cells (Fig. 5D). It is therefore possible that inhibition of both Stat1/Stat3 phosphorylation and p38 MAPK activation contributes to the suppressive effects of BCR-ABL on type I IFN-mediated gene transcription.
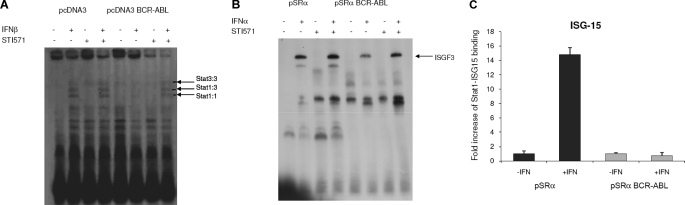
We next examined directly the effects of BCR-ABL expression on the binding of activated Stat complexes to SIE or ISRE sequences. First, we examined the effects of BCR-ABL on the binding of Stat1-Stat1, Stat1-Stat3, and Stat3-Stat3 (SIF) complexes to IFN-specific SIE. Cells transfected with empty vector exhibited normal IFNβ-inducible binding of Stat1 and Stat3 homo- and heterodimers to SIE complexes (Fig. 6A). This binding was strongly suppressed in BCR-ABL-expressing cells, whereas co-treatment of the cells with IFN and imatinib mesylate partially reversed such suppression of Stat protein complex formation (Fig. 6A). Similarly, the IFNα-dependent formation and binding of the ISGF3 complex to ISRE was significantly decreased in BCR-ABL-expressing BA/F3 cells as compared with empty vector-transfected BA/F3 cells (Fig. 6B). Concomitant treatment of BCR-ABL expressing BA/F3 cells with IFN and imatinib mesylate resulted in partial reversal of this suppression (Fig. 6B). In other studies, we determined the binding of ISGF3 complexes to the promoter of the ISG15 gene in Ba/F3-pSRα- and Ba/F3-BCR-ABL-expressing cells by ChIP, using an antibody against Stat1, followed by quantitative PCR analysis. As expected, IFNα treatment of Ba/F3 cells transfected with pSRα vector alone resulted in increased DNA binding of Stat1-containing complexes (ISGF3) to the ISG15 promoter (Fig. 6C). In contrast, we observed no Stat1-ISG15 binding in Ba/F3-BCR-ABL cells whether left untreated or treated with IFNα (Fig. 6C).
FIGURE 6.
Effects of BCR-ABL on IFN-dependent formation and DNA binding of Stat complexes. A, U2OS cells were transfected with pcDNA3 or pcDNA3-BCR-ABL and were incubated in the presence or absence of IFNβ and/or STI-571 or left untreated, as indicated. Nuclear extracts were reacted with 40,000 cpm of a 32P-labeled SIE, and complexes were resolved by native gel electrophoresis and visualized by autoradiography. B, Ba/F3-pSRα mock cells and Ba/F3-pSRα BCR-ABL (P210) were incubated in the presence or absence of mouse IFNα and/or STI-571 or left untreated, as indicated. Nuclear extracts were reacted with 40,000 cpm of a 32P-labeled ISGF3; complexes were resolved by native gel electrophoresis and visualized by autoradiography. C, Ba/F3-pSRα BCR-ABL (P210) or Ba/F3-pSRα mock cells were incubated in the presence or absence of IFNα, as indicated. ChIPs were performed with either anti-Stat1 or control IgG antibodies. Precipitated chromatin DNA fragments were purified and served as a template for quantitative PCR using ISG15-ISRE probes and primers. DNA fragments that were not subject to immunoprecipitation served as input control for each treatment/sample. Bars represent -fold change over input control for each condition and represent means ± S.E. of four independent experiments.
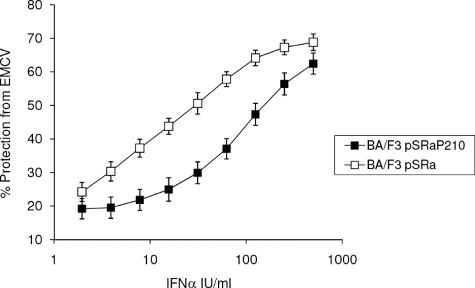
Viewed altogether, our data demonstrate that BCR-ABL regulates the generation of signals that suppress type I IFN-dependent phosphorylation of Stat proteins and formation of DNA binding complexes, resulting in suppressed transcription of several genes known to have antiviral properties. To directly determine whether IFN-induced antiviral effects are diminished in BCR-ABL-expressing cells, we assessed the ability of IFNα to protect from the cytopathic effects of EMCV. Cells were pretreated with IFNα at the indicated concentrations and then challenged with EMCV. Ba/F3 cells expressing BCR-ABL were clearly less sensitive to the antiviral effects of IFNα and less protected from the cytopathic effects of EMCV than BA/F3-pSRα control cells (Fig. 7), strongly suggesting that BCR-ABL decreases the ability of IFNα to generate antiviral responses.
FIGURE 7.
Defective IFN-mediated antiviral responses in cells expressing BCR-ABL. Ba/F3-pSRα BCR-ABL (P210) or Ba/F3-pSRα mock cells were incubated, in triplicate, with the indicated doses of mouse IFNα. The cells were subsequently challenged with EMCV, and CPE was quantified 24 h later. Data are expressed as percentage protection from the CPE of EMCV and are representative of two independent experiments.
DISCUSSION
BCR-ABL, the abnormal product of the fusion of the bcr and c-abl genes (52), transforms hematopoietic cells and generates mitogenic responses via engagement of multiple cellular cascades (30, 53). Several signaling elements and mitogenic signals are activated by BCR-ABL, including Shc (54), Ras-GAP (55), SHP-2 and SHIP (56), c-CBL (57), Vav (58), and the PI 3′ kinase/Akt/mTOR cascade (59–62). In addition, the transcriptional activator Stat5 is engaged in a BCR-ABL-dependent manner and participates in the generation of its effects in target cells (63, 64). The multiplicity of signals and the complexity of pathways activated by BCR-ABL reflect a well coordinated transforming capacity of this oncogene and its ability to overcome the mechanisms of resistance to leukemic transformation that normal cells employ.
Despite the extensive and rapidly accumulating knowledge on the mechanisms by which BCR-ABL transforms cells, very little is known about its effects on IFN-inducible signaling pathways. Understanding the effects of BCR-ABL on IFN signaling is conceivably of importance, since it may uncover unique mechanisms by which the abnormal tyrosine kinase overcomes the normal immune surveillance against cancer and causes malignant transformation. It is of particular interest that IFNα exhibits a selective efficacy in the treatment of chronic myelogenous leukemia, as compared with other tumors and leukemias (30). In fact, prior to the introduction of imatinib mesylate and second generation tyrosine kinase inhibitors in the treatment of CML, IFNα was the treatment of choice for patients not eligible for allogeneic bone marrow transplantation (65). With the emergence of an increasing number of acquired BCR-ABL mutations, rendering BCR-ABL resistant to imatinib mesylate and, in the case of T315I, resistant to second generation BCR-ABL kinase inhibitors, IFNα or combination treatments could re-emerge as important in the treatment of certain cases of CML. Indeed, a recent study found IFN and homoharringtonine to be efficient in the treatment for CML patients with the T315I BCR-ABL mutation that is resistant to all known BCR-ABL kinase inhibitors (66).
In the present work, we examined whether BCR-ABL exerts negative regulatory effects on IFN-inducible transcriptional activation of key target genes. Based on our prior observation that BCR-ABL overexpression impedes PML-gene promoter transcriptional activity (37), we sought to determine its effects on transcriptional activation via ISRE and GAS elements and on specific genes known to play key roles in the induction of IFN-mediated responses. Our data demonstrate that transient or stable BCR-ABL overexpression in target cells blocks IFN-dependent transcriptional activity. Moreover, the expression of several genes known to play key roles in the induction of IFN-induced cellular and antiviral responses is severely suppressed in cells transformed by BCR-ABL. Among them were two well known members of the IRF family of genes, Irf1 and Irf9. Interestingly, the expression of the Irf1 gene has been previously shown to be suppressed in the bone marrows of patients with CML (67, 68), whereas the suppressive effects on Irf9 gene transcription are of particular interest, since its protein-product, IRF9 (also called p48), is a key component of the ISGF3 complex (8–12). Thus, by inhibiting IRF9 expression, BCR-ABL blocks a positive feedback loop for the generation of IFN-inducible responses, involving IRF9-dependent formation of ISGF3 complexes that are in turn required for the transcription of other ISGs. In addition, our data demonstrate that BCR-ABL suppresses the expression of the Isg15 gene, whose protein product plays an important role in IFN signaling by regulating ISGylation (69, 70), and the less characterized Ifit2 gene, which is involved in the generation of antiviral responses (71, 72).
The effects of BCR-ABL on IFN-dependent transcriptional activation of such genes appear to reflect suppressive effects on the phosphorylation/activation of Stat element components of the Jak-Stat pathway, including Stat1, which is required for ISGF3-complex formation and ISRE-driven transcription, and different homo- or heterodimers of Stat1 and Stat3, which are required for SIF complex formation and GAS-driven transcription. Although the precise mechanisms by which such BCR-ABL-mediated suppression occurs remain to be precisely defined, our data establish that the kinase activity of BCR-ABL is essential for such responses. This was shown by studies demonstrating that, in contrast to wild-type BCR-ABL, a kinase-defective mutant did not suppress formation of Stat complexes and IFN-dependent gene transcription. An interesting possibility is that suppression of Isg15 expression by BCR-ABL may lead to a positive feedback loop leading to suppression of Stat phosphorylation, since ISGylation has been previously linked to the regulation of Stat1 phosphorylation (73), whereas cells lacking the ISG15-specific isopeptidase, Ubp43, are more resistant to oncogenic transformation by BCR-ABL (74). However, the validity of such a hypothesis remains to be determined in future studies.
Imatinib mesylate was found to reverse the suppressive effects of BCR-ABL on ISGF3 or SIF complex formation. Interestingly, imatinib mesylate enhances IFN-dependent Stat phosphorylation and formation of Stat-containing DNA-binding complexes in wild-type BCR-ABL-transformed Ba/F3 cells that are resistant to the antiproliferative effects of IFNα, whereas in previous studies, we had found that imatinib mesylate did not exhibit such enhancing effects in the highly IFNα-sensitive KT-1 cell line (51). Thus, it is possible that the enhancing effects of imatinib mesylate correlate with relative levels of expression of BCR-ABL protein and sensitivity of cells to IFNα, being noticeable in the presence of relative IFN resistance. Interestingly, in studies in which we analyzed the antiproliferative effects of IFNα on various Ba/F3 transfectants, we found that Ba/F3 empty vector-transfected cells were relatively sensitive to IFNα, whereas such sensitivity was reversed in Ba/F3-BCR-ABL-transfected cells. Notably, the BCR-ABL kinase domain mutants Y253F and E255K (mutations in the P-loop) exhibited sensitivity to IFNα similar to that of wild-type BCR-ABL-expressing cells, whereas the T315I mutant (mutated hydrogen bond of STI-571 binding site) and, even more so, the BCR-ABL kinase domain mutant H396P (mutation in activation loop) were relatively more sensitive to IFN-inducible growth inhibition.
The fact that IFN-dependent gene transcription and the generation of antiviral effects are antagonized by the abnormal BCR-ABL fusion protein raises questions and issues that may be of clinical and physiological relevance. It is possible that an important mechanism of the transforming capacity of BCR-ABL is suppression of growth-inhibitory pathways. Since IFNs constitute a component of immune surveillance against malignancies, inhibition of IFN signaling by BCR-ABL may contribute to the development of leukemogenesis. Beyond its effects on the activation of classic Stat pathways, our data establish that BCR-ABL suppresses IFN-inducible activation of the p38 MAPK, whose function complements the function of the Stat pathway and is also required for the IFN-dependent transcriptional regulation (22, 23) and generation of growth-inhibitory responses (24, 25). Thus, combined inhibition of more than one growth inhibitory pathway may contribute to the development of the leukemic phenotype by BCR-ABL. The inhibition of expression of IFN-inducible genes with known antiviral properties by BCR-ABL may also contribute in part to the development of an immunity-compromised status and increase the overall sensitivity of leukemic patients to viral infections. Future studies to define the precise mechanisms by which BCR-ABL inhibits activation of Stats and IFN-inducible gene transcription may lead to the identification of novel therapeutic cellular targets and the development of clinical-translational approaches aimed at disrupting the ability of BCR-ABL to suppress signaling pathways engaged by IFNs and, possibly, other cytokines with antitumor properties.
This work was supported by National Institutes of Health Grants CA77816, CA100579, and CA121192 (to L. C. P.), a merit review grant from the Department of Veterans Affairs (to L. C. P.), and Canadian Institutes of Health Research Grant MOP 15094 (to E. N. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IFN, interferon; CML, chronic myeloid leukemia; ISRE, interferon-stimulated response element; GAS, IFNγ-activated sequence; ISG, interferon-stimulated gene; MTT, (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; EMCV, encephalomyocarditis virus; SIE, sis-inducing element; ChIP, chromatin immunoprecipitation; MAPK, mitogen-activated protein kinase; SIF, sis-inducible factor.
References
- 1.Pestka, S., Langer, J. A., Zoon, K. C., and Samuel, C. E. (1987) Annu. Rev. Biochem. 56 727-777 [DOI] [PubMed] [Google Scholar]
- 2.Katze, M. G., He, Y., and Gale, M. (2002) Nat. Rev. Immunol. 2 675-687 [DOI] [PubMed] [Google Scholar]
- 3.Grandvaux, N., tenOever, B. R., Servant, M. J., and Hiscott, J. (2002) Curr. Opin. Infect. Dis. 15 259-267 [DOI] [PubMed] [Google Scholar]
- 4.Galligan, C. L., Murooka, T. T., Rahbar, R., Baig, E., Majchrzak-Kita, B., and Fish, E. N. (2006) Immunol. Res. 35 27-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paun, A., and Pitha, P. M. (2007) Adv. Virus Res. 69 1-66 [DOI] [PubMed] [Google Scholar]
- 6.Sen, G. C. (2001) Annu. Rev. Microbiol. 55 255-281 [DOI] [PubMed] [Google Scholar]
- 7.Theofilopoulos, A. N., Baccala, R., Beutler, B., and Kono, D. H. (2005) Annu. Rev. Immunol. 23 307-336 [DOI] [PubMed] [Google Scholar]
- 8.Darnell, J. E., Jr., Kerr, I. M., and Stark, G. R. (1994) Science 264 1415-1420 [DOI] [PubMed] [Google Scholar]
- 9.Stark, G. R., Kerr, I. M., Williams, B. R., Silverman, R. H., and Schreiber, R. D. (1998) Annu. Rev. Biochem. 67 227-264 [DOI] [PubMed] [Google Scholar]
- 10.van Boxel-Dezaire, A. H., Rani, M. R., and Stark, G. R. (2006) Immunity 25 361-372 [DOI] [PubMed] [Google Scholar]
- 11.Platanias, L. C. (2005) Nat. Rev. Immunol. 5 375-386 [DOI] [PubMed] [Google Scholar]
- 12.Platanias, L. C., and Fish, E. N. (1999) Exp. Hematol. 27 1583-1592 [DOI] [PubMed] [Google Scholar]
- 13.Pestka, S., Krause, C. D., and Walter, M. R. (2004) Immunol. Rev. 202 8-32 [DOI] [PubMed] [Google Scholar]
- 14.Brierley, M. M., and Fish, E. N. (2002) J. Interferon Cytokine Res. 22 835-845 [DOI] [PubMed] [Google Scholar]
- 15.Deonarain, R., Chan, D. C., Platanias, L. C., and Fish, E. N. (2002) Curr. Pharm. Des. 8 2131-2137 [DOI] [PubMed] [Google Scholar]
- 16.Aaronson, D. S., and Horvath, C. M. (2002) Science 296 1653-1655 [DOI] [PubMed] [Google Scholar]
- 17.Ramana, C. V., Gil, M. P., Schreiber, R. D., and Stark, G. R. (2002) Trends Immunol. 23 96-101 [DOI] [PubMed] [Google Scholar]
- 18.Brierley, M. M., and Fish, E. N. (2005) J. Interferon Cytokine Res. 25 733-744 [DOI] [PubMed] [Google Scholar]
- 19.Platanias, L. C. (2003) Pharmacol. Ther. 98 129-142 [DOI] [PubMed] [Google Scholar]
- 20.Katsoulidis, E., Li, Y., Mears, H., and Platanias, L. C. (2005) J. Interferon Cytokine Res. 25 749-756 [DOI] [PubMed] [Google Scholar]
- 21.Kaur, S., Uddin, S., and Platanias, L. C. (2005) J. Interferon Cytokine Res. 25 780-787 [DOI] [PubMed] [Google Scholar]
- 22.Uddin, S., Majchrzak, B., Woodson, J., Arunkumar, P., Alsayed, Y., Pine, R., Young, P. R., Fish, E. N., and Platanias, L. C. (1999) J. Biol. Chem. 274 30127-30131 [DOI] [PubMed] [Google Scholar]
- 23.Uddin, S., Lekmine, F., Sharma, N., Majchrzak, B., Mayer, I., Young, P. R., Bokoch, G. M., Fish, E. N., and Platanias, L. C. (2000) J. Biol. Chem. 275 27634-27640 [DOI] [PubMed] [Google Scholar]
- 24.Verma, A., Deb, D. K., Sassano, A., Uddin, S., Varga, J., Wickrema, A., and Platanias, L. C. (2002) J. Biol. Chem. 277 7726-7735 [DOI] [PubMed] [Google Scholar]
- 25.Mayer, I. A., Verma, A., Grumbach, I. M., Uddin, S., Lekmine, F., Ravandi, F., Majchrzak, B., Fujita, S., Fish, E. N., and Platanias, L. C. (2001) J. Biol. Chem. 276 28570-28577 [DOI] [PubMed] [Google Scholar]
- 26.Uddin, S., Sassano, A., Deb, D. K., Verma, A., Majchrzak, B., Rahman, A., Malik, A. B., Fish, E. N., and Platanias, L. C. (2002) J. Biol. Chem. 277 14408-14416 [DOI] [PubMed] [Google Scholar]
- 27.Srivastava, K. K., Batra, S., Sassano, A., Li, Y., Majchrzak, B., Kiyokawa, H., Altman, A., Fish, E. N., and Platanias, L. C. (2004) J. Biol. Chem. 279 29911-29920 [DOI] [PubMed] [Google Scholar]
- 28.Lekmine, F., Uddin, S., Sassano, A., Parmar, S., Brachmann, S. M., Majchrzak, B., Soneberg, N., Hay, N., Fish, E. N., and Platanias, L. C. (2003) J. Biol. Chem. 278 27772-27780 [DOI] [PubMed] [Google Scholar]
- 29.Kaur, S., Lal, L., Sassano, A., Majchrzak-Kita, B., Srikanth, M., Baker, D. P., Petroulakis, E., Hay, N., Soneberg, N., Fish, E. N., and Platanias, L. C. (2007) J. Biol. Chem. 282 1757-1768 [DOI] [PubMed] [Google Scholar]
- 30.Verma, A., and Platanias, L. C. (2002) Leuk. Lymphoma 43 703-709 [DOI] [PubMed] [Google Scholar]
- 31.Landolfo, S., Guarini, A., Riera, L., Gariglio, M., Gribaudo, G., Cignetti, A., Cordone, I., Montefusco, E., Mandelli, F., and Foa, R. (2000) Hematol. J. 1 7-14 [DOI] [PubMed] [Google Scholar]
- 32.Sakai, I., Takeuchi, K., Yamauchi, H., Narumi, H., and Fujita, S. (2002) Blood 100 2926-2931 [DOI] [PubMed] [Google Scholar]
- 33.Ilaria, R. L., Jr., and Van Etten, R. A. (1996) J. Biol. Chem. 271 31704-31710 [DOI] [PubMed] [Google Scholar]
- 34.Coppo, P., Flamant, S., De Mas, V., Jarrier, P., Guillier, M., Bonnet, M. L., Lacout, C., Guilhot, F., Vainchenker, W., and Turhan, A. G. (2006) Br. J. Haematol. 134 171-179 [DOI] [PubMed] [Google Scholar]
- 35.Rhodes, J., York, R. D., Tara, D., Tajinda, K., and Druker, B. J. (2000) Exp. Hematol. 28 305-310 [DOI] [PubMed] [Google Scholar]
- 36.Fish, E. N., Uddin, S., Korkmaz, M., Majchrzak, B., Druker, B. J., and Platanias, L. C. (1999) J. Biol. Chem. 274 571-573 [DOI] [PubMed] [Google Scholar]
- 37.Grumbach, I. M., Mayer, I. A., Uddin, S., Lekmine, F., Majchrzak, B., Yamuchi, H., Fujita, S., Druker, B., Fish, E. N., and Platanias, L. C. (2001) Br. J. Haematol. 112 327-336 [DOI] [PubMed] [Google Scholar]
- 38.Kharas, M. G., and Fruman, D. A. (2005) Cancer Res. 65 2047-2053 [DOI] [PubMed] [Google Scholar]
- 39.Griswold, I. J., MacPartlin, M., Bumm, T., Goss, V. L., O'Hare, T., Lee, K. A., Corbin, A. S., Stoffregen, E. P., Smith, C., Johnson, K., Moseson, E. M., Wood, L. J., Polakiewicz, R. D., Druker, B. J., and Deininger, M. W. (2006) Mol. Cell. Biol. 26 6082-6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramaraj, P., Singh, H., Niu, N., Chu, S., Holtz, M., Yee, J. K., and Bhatia, R. (2004) Cancer Res. 64 5322-5331 [DOI] [PubMed] [Google Scholar]
- 41.Uddin, S., Yenush, L., Sun, X. J., Sweet, M. E., White, M. F., and Platanias, L. C. (1995) J. Biol. Chem. 270 15938-15941 [DOI] [PubMed] [Google Scholar]
- 42.Platanias, L. C., Uddin, S., Yetter, A., Sun, X. J., and White, M. F. (1996) J. Biol. Chem. 271 278-282 [DOI] [PubMed] [Google Scholar]
- 43.Uddin, S., Fish, E. N., Sher, D. A., Gardziola, C., White, M. F., and Platanias, L. C. (1997) J. Immunol. 158 2390-2397 [PubMed] [Google Scholar]
- 44.Uddin, S., Mazchrzak, B., Woodson, J., Arunkumar, P., Alsayed, Y., Pine, R., Young, P. R., Fish, E. N., and Platanias, L. C. (1999) J. Biol. Chem. 274 30127. [DOI] [PubMed] [Google Scholar]
- 45.Honda, K., and Taniguchi, T. (2006) Nat. Rev. Immunol. 6 644-658 [DOI] [PubMed] [Google Scholar]
- 46.Barnes, B., Lubyova, B., and Pitha, P. M. (2002) J. Interferon Cytokine Res. 22 59-71 [DOI] [PubMed] [Google Scholar]
- 47.Smith, J. B., and Herschman, H. R. (1996) Arch. Biochem. Biophys. 330 290-300 [DOI] [PubMed] [Google Scholar]
- 48.Terenzi, F., Pal, S., and Sen, G. C. (2005) Virology 340 116-124 [DOI] [PubMed] [Google Scholar]
- 49.Martensen, P. M., and Justesen, J. (2004) J. Interferon Cytokine Res. 24 1-19 [DOI] [PubMed] [Google Scholar]
- 50.Wong, S., McLaughlin, J., Cheng, D., and Witte, O. N. (2003) Blood 101 4088-4097 [DOI] [PubMed] [Google Scholar]
- 51.Parmar, S., Katsoulidis, E., Verma, A., Li, Y., Sassano, A., Lal, L., Majchrzak, B., Ravandi, F., Tallman, M. S., Fish, E. N., and Platanias, L. C. (2004) J. Biol. Chem. 279 25345-25352 [DOI] [PubMed] [Google Scholar]
- 52.Ben-Neriah, Y., Daley, G. Q., Mes-Masson, A., Witte, O. N., and Baltimore, D. (1986) Science 233 212-214 [DOI] [PubMed] [Google Scholar]
- 53.Steelman, L. S., Pohnert, S. C., Shelton, J. G., Franklin, R. A., Bertrand, F. E., and McCubrey, J. A. (2004) Leukemia 18 189-218 [DOI] [PubMed] [Google Scholar]
- 54.Matsuguchi, T., Salgia, R., Hallek, M., Eder, M., Druker, B., Ernst, T. D., and Griffin, J. D. (1994) J. Biol. Chem. 269 5016-5021 [PubMed] [Google Scholar]
- 55.Druker, B., Okuda, K., Matulonis, U., Salgia, R., Roberts, T., and Griffin, J. D. (1992) Blood 79 2215-2220 [PubMed] [Google Scholar]
- 56.Sattler, M., Salgia, R., Shrikhande, G., Verma, S., Choi, J. L., Rohrschneider, L. R., and Griffin, J. D. (1997) Oncogene 15 2379-2384 [DOI] [PubMed] [Google Scholar]
- 57.Sattler, M., Salgia, R., Okuda, K., Uemura, N., Durstain, M. A., Pisick, E., Xu, G., Li, J. L., Prasad, K. V., and Griffin, J. D. (1996) Oncogene 12 839-846 [PubMed] [Google Scholar]
- 58.Bassermann, F., Jahn, T., Miething, C., Selpel, P., Bai, R. Y., Coutinho, S., Tybulewicz, V. L., Peschel, C., and Duyster, S. (2002)) J. Biol. Chem. 277 12437-12445 [DOI] [PubMed] [Google Scholar]
- 59.Skorski, T., Bellacosa, A., Nieborowska-Skorska, M., Majewski, M., Martinez, R., Choi, J. K., Trotta, R., Wlodarski, P., Perrotti, D., Chan, T. O., Wasik, M. A., Tsichlis, P. N., and Calabretta, B. (1997) EMBO J. 16 6151-6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ly, C., Arechiga, A. F., Melo, J. V., Walsh, C. M., and Ong, S. T. (2003) Cancer Res. 63 5716-5722 [PubMed] [Google Scholar]
- 61.Mohi, M. G, Boulton, C., Gu, T. L., Sternberg, D. W., Neuberg, D., Griffin, J. D., Gilliland, D. G., and Neel, B. G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 3130-3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parmar, S., Smith, J., Sassano, A., Uddin, S., Katsoulidis, E., Majchrzak, B., Kambhampati, S., Eklund, E. A., Tallman, M. S., Fish, E. N., and Platanias, L. C. (2005) Blood 106 2436-2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gesbert, F., and Griffin, J. D. (2000) Blood 96 2269-2276 [PubMed] [Google Scholar]
- 64.Horita, M., Andreu, E. J., Benito, A., Arbona, C., Sanz, C., Benet, I., Prosper, F., and Fernandez-Luna, J. L. (2000) J. Exp. Med. 191 977-984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sawyers, C. L. (1999) N. Engl. J. Med. 340 1330-1340 [DOI] [PubMed] [Google Scholar]
- 66.de Lavallade, H., Khorashad, J. S., Davis, H. P., Milojkovic, D., Kaeda, J. S., Goldman, J. M., Apperley, J. F., and Marin, D. (2007) Blood 110 2779-2780 [DOI] [PubMed] [Google Scholar]
- 67.Tzoanopoulos, D., Speletas, M., Arvanitidis, K., Veiopoulou, C., Kyriaki, S., Thyphronitis, G., Sideras, P., Kartalis, G., and Ritis, K. (2002) Br. J. Haematol. 119 46-53 [DOI] [PubMed] [Google Scholar]
- 68.Hochhaus, A., Yan, X. H., Willer, A., Hehlmann, R., Gordon, M. Y., Goldman, J. M., and Melo, J. V. (1997) Leukemia 11 933-939 [DOI] [PubMed] [Google Scholar]
- 69.Ritchie, K. J., Hahn, C. S., Kim, K. I., Yan, M., Rosario, D., Li, L., De La Torre, J. C., and Zhang, D. E. (2004) Nat. Med. 10 1374-1378 [DOI] [PubMed] [Google Scholar]
- 70.Ritchie, K. J., and Zhang, D. E. (2004) Semin. Cell Dev. Biol. 15 237-246 [DOI] [PubMed] [Google Scholar]
- 71.Saha, S., Sugumar, P., Bhandari, P., and Rangarajan, P. N. (2006) J. Gen. Virol. 87 3285-3289 [DOI] [PubMed] [Google Scholar]
- 72.Dorn, A., Zhao, H., Granberg, F., HÃsel, M., Webb, D., Svensson, C., Pettersson, U., and Doerfler, W. (2005) J. Virol. 79 2404-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Malakhova, O. A., Yan, M., Malakhov, M. P., Yuan, Y., Ritchie, K. J., Kim, K. I., Peterson, L. F., Shuai, K., and Zhang, D. E. (2003) Genes Dev. 17 455-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan, M., Luo, J. K., Ritchie, K. J., Sakai, I., Takeuchi, K., Ren, R., and Zhang, D. E. (2007) Blood 110 305-312 [DOI] [PMC free article] [PubMed] [Google Scholar]