Abstract
Eisosomes are recently described fungal structures that play roles in the organization of the plasma membrane and endocytosis. Their major protein components are Pil1 and Lsp1, and previous studies showed that these proteins are phosphorylated by the sphingolipid long-chain base-activated Pkh1 and Pkh2 protein kinases in vitro. We show that Pkh1 and Pkh2 phosphorylate Pil1 and Lsp1 in vivo to produce species B, and that heat stress, which activates Pkh1 and Pkh2, generates a more highly phosphorylated species, C. Cells with low Pkh activity lack species B and C and contain abnormally organized eisosomes. To verify that Pil1 phosphorylation is essential for correct eisosome organization, phosphorylated serine and threonine residues were identified and changed to alanines. A variant Pil1 protein lacking five phosphorylation sites did not form eisosomes during log phase growth, indicating that phosphorylation is critical for eisosome organization. We also found that eisosomes are dynamic structures and disassemble when the Ypk protein kinases, which are activated by the sphingolipid-Pkh signaling pathway, are inactivated or when the sphingolipid signal is pharmacologically blocked with myriocin. We conclude that eisosome formation and turnover are regulated by the sphingolipid-Pkh1/2-Ypk1/2 signaling pathway. These data and previous data showing that endocytosis is regulated by the sphingolipid-Pkh1/2-Ypk1/2 signaling pathway suggest that Pkh1 and -2 respond to changes in membrane sphingolipids and transmit this information to eisosomes via Pil1 phosphorylation. Eisosomes then control endocytosis to align the composition and function of the plasma membrane to match demand.
The plasma membrane of eukaryotic cells is a dynamic assembly of lipids and proteins that modulates and integrates a complex spectrum of cellular processes in ways that remain unclear. Recently, two related Saccharomyces cerevisiae proteins, Pil1 and Lsp1, were shown to comprise novel structures on the cytoplasmic face of the plasma membrane termed eisosomes (1). Eisosomes mark sites for endocytosis of proteins and lipids, and they are sequestered on patches in the plasma membrane rich in sterols and specific protein transporters (2). We showed previously that Pil1 and Lsp1 are phosphorylated in vitro by the Pkh1 and Pkh2 protein kinases (3), which are structural and functional homologs of mammalian phosphoinositide-dependent protein kinase 1 (4, 5). Phosphorylation by Pkh1/2 suggests that some function of Pil1, Lsp1, and eisosomes is regulated by phosphorylation. Here we show that both proteins are highly phosphorylated in vivo by Pkh1/2 and that phosphorylation of Pil1 regulates eisosome organization.
Pil1 and Lsp1 were found in a complex with Pkh1/2 (6, 7). Because Pkh1/2 activate protein kinase C, Pkc1, in yeast (8, 5) and Pkc1 controls a mitogen-activated protein kinase cascade culminating with the Slt2 kinase, a controller of cell wall integrity (reviewed in Ref. 9), we determined if Pil1 and Lsp1 play a role in activating Slt2 during heat stress. Heat stress was examined, because heat induces a transient increase in sphingolipid long-chain bases (LCBs),6 the only known activators of Pkh1/2 (3, 8, 10). Deletion of either PIL1 or LSP1 led to an increase in the level of activated Slt2 during a heat stress, suggesting that Pil1 and Lsp1 play a role in regulating the Pkc1-mitogen-activated protein kinase cascade (3). In addition, both deletion strains were more resistant to heat stress. Several other results implied that Pil1 and Lsp1 regulate the activity of Ypk1. Together these data suggested that Pil1 and Lsp1 modulate Pkh1/2 and downstream kinases to control cell wall integrity during non-stressful and stressful circumstances.
Pkh1 and Pkh2 have partially overlapping functions that play incompletely characterized roles in endocytosis (8, 11, 12), cell wall integrity (5, 13, 14), growth (4, 5), actin dynamics (15, 5, 16), and exocytosis (17). Recently, Pkh1, but not Pkh2, has been implicated in the regulation of yeast life-span (18).
To further understand how Pil1 and Lsp1 regulate cell wall integrity, we examined their phosphorylation in vivo and now show that there are two species, A and B, of each protein in unstressed, log phase cells and that these species differ in their level of phosphorylation. Heat stress generates a third, more highly phosphorylated species, C. Phosphorylation of species B and C is mediated by Pkh1/2, and these kinases may also be responsible for phosphorylation of species A. In cells with reduced Pkh activity, phosphorylation of Pil1 and Lsp1 is reduced, there are fewer eisosomes, the eisosomes are not spaced as evenly on the plasma membrane as in wild-type cells, and eisosomes are less homogeneous in size, suggesting that phosphorylation plays roles in eisosome organization. In addition, we find that the Ypk1/2 kinases, which are regulated by the LCB-Pkh1/2 pathway, are necessary for maintaining eisosomes. Together our data suggest that phosphorylation of Pil1 is important both for assembly and for disassembly of eisosomes, which have not previously been recognized as dynamic structures whose number, size, and spatial organization changes. Given that eisosomes mark sites of endocytosis, we speculate that Pil1 phosphorylation plays roles in regulating endocytosis thereby modulating the composition and functions of the plasma membrane.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Media—Yeast strains used in these studies are described in Table 1. For heat shock experiments, cells were grown in YPD (1% yeast extract, 2% peptone, 2% glucose), and for overproducing Pil1 they were grown in a defined medium (19). Synthetic medium lacking uracil (0.34% yeast nitrogen base (Difco), 1% ammonium sulfate, 2% glucose, 30 mg/liter each of adenine, isoleucine, and tyrosine, and 20 mg/liter each of histidine, leucine, lysine, methionine, and tryptophan) was used to grow cells for fluorescence microscopy and for selecting cells transformed with a plasmid carrying URA3. Solid media contained 2% agar. For microscopy cells were grown in YPD supplemented with 150 mg/liter of adenine sulfate (YPAD).
TABLE 1.
Strains used in this work
| Strain | Genotype | Reference |
|---|---|---|
| 15 Dau | MATa ura3Δ0 leu2Δ0 trp1Δ his2Δ ade1Δ | (5) |
| INA106-3B | MATa ura3Δ0 leu2Δ0 trp1Δ his2Δ ade1Δ pkh1D398G pkh2::LEU2 | (5) |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | (41) |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | (41) |
| RCD364 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pil1::KAN | (3) |
| RCD368 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 lsp1::KAN | (3) |
| RCD775 | MATa ura3Δ0 leu2Δ0 trp1Δ his2Δ ade1Δ LSP1-YFP::KAN (made in the 15 Dau strain background) | This study |
| RCD778 | MATa ura3Δ0 leu2Δ0 trp1Δ his2Δ ade1Δ pkh1D398G pkh2::LEU2 PIL1-YFP::KAN (made in the INA106-3B strain background) | This study |
| RCD783 | MATa ura3Δ0 leu2Δ0 trp1Δ his2Δ ade1Δ LSP1-YFP::KAN (made in the 15 Dau strain background) | This study |
| RCD786 | MATa ura3Δ0 leu2Δ0 trp1Δ his2Δ ade1Δ pkh1D398G pkh2::LEU2 LSP1-YFP::KAN (made in the INA106-3B strain background) | This study |
| W303-1B | MATa ade2-1 can1-100 ura3-1 his3-11,15 trp1-1 leu2-3,112 LSP1-GFP::HIS3 | (1) |
| W303-1B | MATa ade2-1 can1-100 ura3-1 his3-11,15 trp1-1 leu2-3,112 pil1::KAN PIL1-GFP::HIS3 | (1) |
| DBY746 | MATα leu2-3,112 his3Δ1 trp1-289 ura3-52 GAL+ | (42) |
| YPH499 | MATa ura3-52 lys2-801amber ade2-101ochre leu2Δ1 his3Δ200 trp1Δ63 | (20) |
| YPT40 | MATa ura3-52 lys2-801amber ade2-101ochre leu2Δ1 his3Δ200 trp1Δ63 ypk1-1ts::HIS3, ypk2-Δ1::TRP1 | (4) |
| RCD520 | MATa ura3 leu2 trp1 his2 ade1 pkh1D398G pkh2::LEU2, lsp1::KAN | This study |
| RCD521 | MATa ura3 leu2 trp1 his2 ade1 pkh1D398G pkh2::LEU2 pil1::KAN | This study |
Six histidine residues (His6) were added to the N terminus of Pil1 by cloning the PIL1 coding region into pYES2/NTA (Invitrogen) so that the galactose-inducible GAL1 promoter drove gene expression. PKH1, PKH2, and wild-type or mutant forms of PIL1 were transformed into yeast cells by using pRS316 (20). A wild-type allele of PKH1 containing 197 bp upstream of start codon and an added BamH1 site, and 298 bp downstream of stop codon plus an added SalI site, was synthesized by PCR and cloned into the corresponding sites of pRS316 to give pRS316-PKH1. The same strategy was used to clone a wild-type allele of PKH2 containing 698 bp upstream of start codon and 380 bp downstream of stop codon into pRS316 to give pRS316-PKH2. A wild-type allele of PIL1 containing 520 bp upstream of start codon and 458 bp downstream of stop codon was cloned into pRS316 in an identical manner to give pRS316-PIL1, which served as a template for site-directed mutagenesis (GeneEditor, Promega, Madison, WI).
Protein Extraction and Analysis—Cell-free yeast extracts were prepared by using glass beads and vortexing as described previously (5). The published Hot-SDS protein extraction protocol was modified slightly (21). Ten A600 units of yeast cells were collected by centrifugation, washed once with water, and then suspended in 400 μl of cold distilled water, followed by the addition of 400 μl of 0.2 m NaOH. Samples were mixed and incubated for 5 min at room temperature. Cells were concentrated by centrifugation in a microcentrifuge, suspended in 200 μl of SDS sample buffer (0.06 m Tris-HCl, pH 8.6, 5% glycerol, 1% SDS, 10 mm β-mercaptoethanol), and boiled for 3–5 min. Samples were centrifuged, and the protein concentration of the supernatant fluid was determined by using the DC protein assay kit (Bio-Rad, Richmond, CA).
Species A, B, and C of Pil1 were prepared for mass spectrometry (MS) by growing DBY746 cells transformed with pYES2/NTA-PIL1 in 1 liter of defined medium lacking uracil and containing 2% sucrose and 0.1% glucose to an A600 of 0.3 at 30 °C. Powdered galactose (2%, w/v), peptone (1%), and yeast extract (1%) were added to induce production of His6-Pil1, and the culture was incubated for 10 h (A and B species) or for 48 h (C species). The A and B species were isolated by nickel affinity chromatography as described previous (10) except that Triton X-100 and sodium deoxycholate were omitted from the lysis buffer, the concentration of EDTA was reduced to 0.02 mm, and 0.5% glycerol and 0.033 mm dithiothreitol were included. After lysis with glass beads, n-octyl-β-glucoside (0.5% w/v) and NaCl (final concentration 300 mm) were added to the lysate, which was vortexed for 10 s and incubated for 30 min at 4 °C before centrifugation at 100,000 × g for 15 min at 4 °C. The supernatant was loaded onto a 0.5-ml nickel affinity column and washed with 30 ml of buffer I (50 mm Tris-HCl, ph7.5, 300 mm NaCl, 5% glycerol, 10 mm imidazole, and 0.05% n-octyl-β-glucoside) followed by washing with 50 ml of buffer II (same as buffer I except 50 mm NaCl and 20 mm imidazole). Following another wash with 10 ml of buffer III (50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 20 mm imidazole), His6-Pil1 was eluted with 15 ml of elution buffer (50 mm Tris-HCl, pH 7.5, 200 mm imidazole). Fractions were analyzed on a 10% SDS-PAGE, and the Coomassie Blue-stained A and B bands were excised from the gel and stored at –80 °C.
The C species of Pil1 was prepared from 30–40 A600 units of cells by scaling up the Hot-SDS procedure. The extract was diluted 10-fold with immunoprecipitation buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm sodium fluoride, 1 mm sodium pyrophosphate) and rotated for 2 h at 4 °C with 0.1 ml of a 50% solution of nickel-nitrilotriacetic acid-agarose (Sigma-Aldrich), which had been washed with and suspended in immunoprecipitation buffer. Agarose-bound His6-Pil1 was washed three times with immunoprecipitation buffer, suspended in SDS sample buffer, heated 5 min at 95 °C, and run on a 9% SDS-PAGE (20 cm long gel). The C band was excised from a Coomassie Blue-stained gel and stored at –80 °C.
Samples were treated with lambda protein phosphatase as described by the supplier (New England BioLabs) with small modifications. For analysis of Pil1 (100 μg of cell-free extract) the total reaction volume was 0.6 ml and for analysis of Lsp1 (50 μg cell-free extract) the reaction volume was 0.3 ml. EDTA, final concentration of 50 mm, and sodium orthovanadate, final concentration of 5 mm, were used to inhibit phosphatase activity. Proteins were concentrated by acetone precipitation,7 and the pellet was dissolved in 20 μl of 1× SDS sample loading buffer and immunoblotted.
Immunoblotting—Protein extracts were electrophoresed on a 9% SDS-PAGE. Proteins were transferred (transfer buffer: 48 mm Tris, 39 mm glycine, 20% methanol) for 1 h at 12 V to a polyvinylidene fluoride membrane (Millipore, Billerica, MA). Membranes were blocked in 2% nonfat milk for 1 h, washed with TBS (50 mm Tris-HCl, 150 mm NaCl, pH 7.5) for 10 min, and incubated 1 h with anti-Vma2 mouse antibody (Molecular Probes, Eugene, OR, diluted 1:1,000) and polyclonal rabbit antibody against Lsp1 or Pil1 (diluted 1:2,500) diluted in TBS buffer. The membrane was washed twice with TBS containing 0.1% Tween 20 (TBS-T), twice with TBS and then incubated 1 h with alkaline phosphatase-linked anti-rabbit antibody (Chemicon, Temecula, CA, diluted 1:3,000 in TBS) and anti-mouse antibody (diluted 1:5,000, Sigma). The membrane was washed with TBS-T three times and exposed to an ECF substrate (Amersham Biosciences). Fluorescent signals were analyzed by using a PhosphorImager (Storm 860, Amersham Biosciences) and ImageQuaNT software. Rabbit polyclonal antibodies were raised against the C terminus of Pil1 (CVGHQQSESLPQQTTA) and Lsp1 (CHHVSQNGHTSGSENI, Genemed Synthesis Inc., San Francisco, CA).
Microscopy—A Nikon Eclipse E600 fluorescence microscope equipped with a Plan Apo 100×, 1.40 oil immersion objective was used for fluorescence microscopy. Room temperature samples were photographed with a SPOT RT 9.0 Monochrome-6 camera using the MetaMorph (Version 6.3.0) acquisition software. A Leica TSC SP5 broadband microscope with an HCX PL Apo, 100×, 1.40 oil immersion objective was used for confocal microscopy. Photographs were taken at room temperature using LAS AF 1.5.1 acquisition software. Images were processed by using Adobe Photoshop (version 7.0).
Mass Spectrometry—Coomassie-stained bands containing Pil1 were excised from SDS-PAGE gels and cut in small cubes. Gel pieces were destained by alternating washes with H2O and 50% acetonitrile. Reduction and alkylation was not performed, because Pil1 does not contain any cysteines. Gel pieces were incubated with acetonitrile for 10 min, which was then removed. Proteases were dissolved in 50 mm NH4HCO3 at a concentration of 12.5 ng/μl. Gel pieces were covered with protease solution and incubated for 30 min on ice. Remaining protease solution was removed, 30 μl of 50 mm NH4HCO3 was added, and samples were incubated overnight at 37 °C. Proteolysis was stopped by making the samples 0.2% (v/v) trifluoroacetic acid.
For MALDI-MS analysis, samples were prepared by the dried droplet method (22), and 0.5–1 μl of sample was spotted on the target together with matrix, either α-cyano-hydroxy cinnemaic acid (10 mg/ml in 70% acetonitrile, 1% trifluoroacetic acid) or 2,5-dihydroxy benzoic acid (in 50% acetonitrile, 1% phosphoric acid). Samples were measured either on Ultraflex (Bruker) or MALDI-Q-Tof (Micromass, Manchester, UK) mass spectrometers.
LC-MS analysis was performed on a nano-LC system (Proxeon Biosystems) coupled to a LTQ-FT mass spectrometer (ThermoFinnigan). Digested protein samples were either loaded directly onto the high-performance liquid chromatograph or after concentration on C18 STAGE tips (23). Peptides were loaded onto a 1.5-cm-long trapping column (75-μm inner diameter) and separated on a 9-cm-long analytical column (50-μm inner diameter). Both columns were prepared by pressure-packing ReproSil Pur C18-AQ 3-μm reversed-phase beads (Dr. Maisch, HPLC GmbH, Tuebingen, Germany) into fused silica capillaries containing Kasil frits (PQ Corp.). Peptides were eluted with a 30-min linear gradient of 4–32% acetonitrile in 0.5% acetic acid at a flow of 250 nl/min. Analysis of peptides on the LTQ-FT was performed as described previously (24).
For nano-ESI-MS, peptides were bound to a C18 STAGE tip and eluted with 2 μl of 50% MeOH and 0.5% acetic acid into a nano-electrospray emitter (Proxeon Biosystems). Peptides were measured on a Q-Tof Ultima (Micromass) without back pressure at ∼900 V in positive ion mode and fragmented with low energy collision-induced decomposition. Data analysis and MaxEnt3 processing was performed with MassLynx 4 (Micromass).
All spectra were searched with MASCOT using the SGD data base of translated yeast open-reading frames, which also contained mammalian keratins and trypsin. The following search parameters were used. (a) For MALDI: mass accuracy, 50 ppm; modifications, carbamidomethyl (fixed), oxidation (M), and deamidation (NQ); two missed cleavages; cleavage specificity, either trypsin AspN or LysC. (b) For LC-MS: mass accuracy, 20 ppm (parent ion) and 0.8 Da (fragment ions); modifications, carbamidomethyl (fixed), oxidation (M), and deamidation (NQ); two missed cleavages; cleavage specificity, either trypsin AspN or LysC.
Analysis of Species A Made in pkh1/2 Mutant Cells—Strains RCD520 (transformed with pYES2-NTA-LSP1), RCD521 (transformed with pYES2-NTA-LSP1), and 15 Dau (transformed with pYE2S-NTA-LSP1 or with pYES2-NTA-PIL1) were grown in defined medium lacking uracil and containing 2% galactose and 1% sucrose for 12 h at 25 °C to an A600 of 0.4. 50 A600 units of cells were lysed by using the Hot-SDS protein extraction protocol described above. His6-tagged proteins were purified by combining 60 μl of nickel-nitrilotriacetic acid-agarose beads (Qiagen), 200 μl of protein extract, and 4.5 ml of lysis buffer (Qiagen: 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm NaF, 1 mm sodium pyrophosphate, 5 mm imidazole) and incubating 3 h at 4 °C followed by centrifugation. The beads were washed twice with 0.5 ml of buffer I (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5% glycerol, 10 mm imidazole) and once with wash buffer II (50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 5% glycerol, 20 mm imidazole, 0.05% Nonidet P-40). Beads were treated with λ-phosphatase (New England Biolabs) according to the manufacturer's instructions. Proteins stained with Pro-Q Diamond (Molecular Probes) were analyzed on a Typhoon 9400 scanner with the following settings: Emission filter = 560 LP/Green, PMT = 400, Laser = Green 532 nm, Sensitivity = normal.
RESULTS
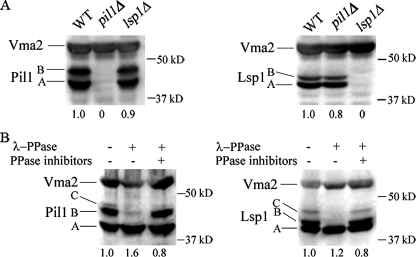
Pil1 and Lsp1 Are Phosphoproteins in Vivo—Because Pil1 and Lsp1 each complex with (6, 7) and are phosphorylated by (3) Pkh1 and Pkh2 in vitro, we determined if Pil1 and Lsp1 are phosphorylated in vivo and if these protein kinases are responsible for phosphorylation. Polyclonal rabbit antibodies generated against the C terminus of Pil1 and Lsp1 each detect two electrophoretic species (A and B) on denaturing gels of about equal concentration in cell-free yeast extracts (Fig. 1A), and the proteins are of the expected size (Pil1 = 38,349 Da; Lsp1 = 38,071 Da). The antibodies are specific, because they do not detect these or any other proteins in extracts made from pil1-or lsp1-deleted cells.
FIGURE 1.
Phosphorylated forms of Lsp1 and Pil1 detected by immunoblotting. A, cell-free extracts (50 μg of protein) from log phase wild-type (BY4742), pil1Δ (RCD364), or lsp1Δ (RCD368) cells were immunoblotted with anti-Pil1 (left panel) or anti-Lsp1 (right panel) rabbit antibodies. The antibodies are specific, because they do not react with proteins in extracts of lsp1Δ or pil1Δ cells. The vacuolar protein Vma2 served as an internal loading control. B, cell-free extracts made from log phase 15 Dau cells were treated or not treated with lambda protein phosphatase, and equal amounts of total protein (100 μg for Pil1 and 50 μg for Lsp1) were immunoblotted to determine if the slower migrating species (B and C) of Lsp1 are phosphorylated.
The concentration of the two species was examined during heat shock, because LCBs, Pkh1/2, Pil1, and Lsp1 all play roles in heat stress tolerance (reviewed in Ref. 25). Initially, log phase cells were broken by vortexing with glass beads, and the cell-free protein extracts were used for immunoblotting. After 40 min of heat shock at 39 °C a third, slower migrating species of Lsp1, termed C, appeared while a similar species with a reduced concentration appeared in a Pil1 immunoblot (Fig. 1B, and see below). We speculated that the C species was a hyperphosphorylated form of Lsp1 and Pil1 that might be susceptible to dephosphorylation by phosphatases. To reduce the risk of dephosphorylation, proteins were extracted by using high temperature and SDS (21). This alternative procedure enhanced the level of species A, B, and C of both Lsp1 and Pil1 and enabled the C species to be detected at earlier times after heat stress (see below).
To establish that the B and C species are phosphorylated forms of species A, we treated cell-free protein extracts with λ-protein phosphatase. Both B and C disappear and the intensity of species A increases following phosphatase treatment (Fig. 1B), demonstrating that B and C are phosphorylated proteins. Because these multiple species have not been seen before, we examined several common strain backgrounds and found that all strains (BY4742, DBY746, YPH499, 15 Dau, and W303, Table 1) contain species A and B and produce species C upon heat shock (data not shown).
Additional evidence for phosphorylation was obtained by overproducing each protein with an N-terminal tag of six histidines, His6-Lsp1 and His6-Pil1, in yeast and purifying them. Proteins were subjected to SDS-PAGE and stained with Pro-Q Diamond (Molecular Probes) to detect phosphoproteins. Both species A and B of Lsp1 and Pil1 stained before but not after treatment with lambda phosphatase (data not shown). We conclude that all forms of Lsp1 and Pil1 are phosphorylated and suggest that A, B, and C are a spectrum that goes from the least to the most phosphorylated species.
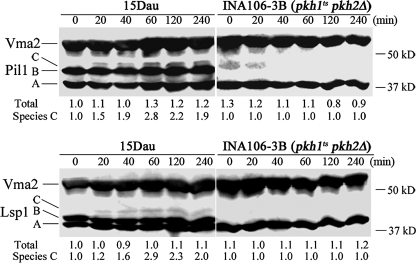
Pkh1/2 Phosphorylate Pil1 and Lsp1 in Vivo—To determine if Pkh1 and Pkh2 phosphorylate Pil1 and Lsp1 in vivo, we compared a strain (INA106-3B) (5) having pkh2 deleted and PKH1 replaced with a temperature-sensitive allele (pkh1D398G) to its parental strain, 15 Dau. At the permissive temperature of 25 °C (Fig. 2, time 0) parental cells contain an equal amount of the A and B species of Pil1 (Fig. 2, top panel) and Lsp1 (Fig. 2, bottom panel) and no species C. The concentration of the A and B species do not change during the heat shock, but the concentration of species C increases 2.8- to 2.9-fold after 60 min of heat stress at 39 °C and then starts to decrease. The mutant cells behave very differently. They contain mostly the A species. There is no detectable Lsp1 B species and a very small amount of Pil1 B species, which disappears. Heat stress does not induce species C of either protein. These data demonstrate that Pkh1 and Pkh2 are responsible for generating the phosphorylated B and C species of Pil1 and Lsp1 in vivo.
FIGURE 2.
Phosphorylation of Lsp1 and Pil1 in log phase cells requires Pkh1 and Pkh2. Log phase wild-type 15 Dau and INA106-3B (pkh1ts pkh2Δ) mutant cells were grown in YPD medium at 25 °C and then switched to 39 °C at time 0. Proteins were extracted at the indicated times, and 50 μg of total protein was immunoblotted. The numbers below each lane represent the total of species A, B, and C or just the concentration the C species of Lsp1 and Pil1, normalized first to the Vma2 signal and then divided by the value of the 0 time wild-type sample.
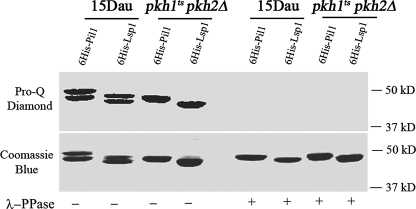
To determine if species A of Pil1 and Lsp1 present in pkh1ts pkh2Δ cells contains phosphates, His6-tagged Pil1 and Lsp1 were isolated from RCD521 (pil1Δ pkh1ts pkh2Δ) and RCD520 (lsp1Δ pkh1ts pkh2Δ) cells, respectively, and from wild-type 15 Dau cells as a positive control. Isolated proteins were treated or not treated with λ-protein phosphatase followed by staining with Pro-Q Diamond. His6-Pil1 and His6-Lsp1 species A isolated from pkh1ts pkh2Δ cells stained before but not after phosphatase treatment, indicating that species A is phosphorylated in pkh1ts pkh2Δ cells grown at the permissive temperature (Fig. 3).
FIGURE 3.
The A species of Pil1 and Lsp1 made in pkh1t pkh2Δ mutant cells is phosphorylated. His6-Pil1 and His6-Lsp1 were produced in and purified from RCD521 (pYES2-NTA-PIL1) and RCD520 (pYES2-NTA-LSP1) cells, respectively, and from wild-type 15 Dau cells. Purified proteins were treated or not treated with lambda phosphatase as indicated and displayed on SDS-PAGE followed by staining with Pro-Q-Diamond to detect phosphorylation followed by staining with Coomassie Blue to measure total protein.
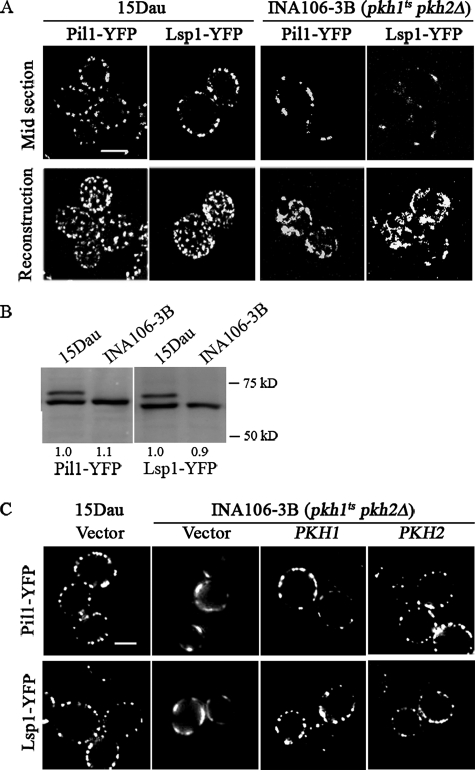
Cells with Reduced Pkh Activity Make Abnormal Eisosomes—To begin to understand why Pil1 and Lsp1 are phosphorylated, eisosomes were examined in parental 15 Dau (PKH1 PKH2) and mutant INA106-3B (pkh1ts pkh2Δ) cells having a sequence encoding the yellow fluorescent protein (YFP) added to the chromosomal PIL1 or LSP1 gene so that eisosome formation could be examined by confocal or regular light fluorescence microscopy. Eisosomes in mutant cells were strikingly different than those in parental cells. Both mid-sections and whole cells image reconstructions from Z-sections show that mutant cells contain fewer eisosomes, the eisosomes are not as evenly distributed throughout the cell, and they are less uniform in size with some seeming to fuse to form fluorescent masses that are larger than eisosomes in wild-type 15 Dau cells (Fig. 4A, supplemental Fig. S1). These phenotypes were present in 96% of the 209 mutant cells expressing Pil1-YFP and 98% of 234 mutant cells expressing Lsp1-YFP compared with wild-type cells in which 99% of the >240 cells had normal looking eisosomes. The observed changes in eisosomes are not due to a reduction in the concentration of Pil1-YFP or Lsp1-YFP (Fig. 4B).
FIGURE 4.
Eisosomes are disrupted in cells with low Pkh protein kinase activity. A, parental 15 Dau (PKH1 PKH2) and mutant INA106-3B (pkh1ts pkh2Δ) cells carrying chromosomal PIL1 or LSP1 tagged at their C terminus with YFP were grown in YPAD at a permissive temperature (25 °C) and examined by confocal microscopy. Z-sections of 250 nm thickness were used to make the whole cell reconstructions. Bar is 5 μm. B, immunoblotting of Pil1-YFP or Lsp1-YFP in 15 Dau or INA106-3B cells with anti-GFP antibody. Numbers below each lane indicate the concentration of YFP-tagged protein relative to the concentration in 15 Dau cells. C, INA106-3B cells expressing Pil1-YFP or Lsp1-YFP were transformed with a vector (pRS316) or the vector carrying PKH1 or PKH2 and examined by regular fluorescence microscopy. 15 Dau cells expressing Pil1-YFP or Lsp1-YFP and transformed with the vector are a positive control for eisosome formation. Bar, 2.5 μm.
A normal eisosomes pattern reappeared when either PKH1 or PKH2 were expressed in INA106-3B mutant cells (Fig. 4C), showing that the eisosome defects are due to a low level of Pkh activity and consequent loss of phosphorylation of Pil1 and Lsp1 (Fig. 2). These data, when combined with in vitro (3) and in vivo (Fig. 2) phosphorylation studies, support the hypothesis that phosphorylation of Pil1 and Lsp1 by Pkh1/2 plays a role in the assembly and distribution of eisosomes.
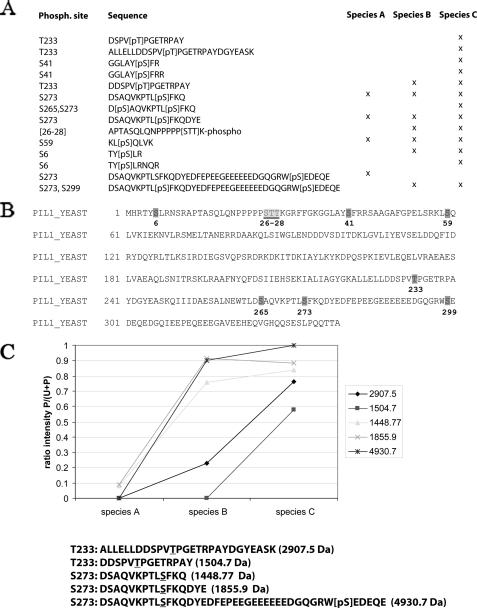
Phosphorylation Sites in the A, B, and C Species of Pil1 Identified by MS—Although the results with INA106-3B (pkh1ts pkh2Δ) cells support the idea that phosphorylation of Pil1 and Lsp1 is vital for assembly of normal eisosomes, they do not rule out the possibility that the eisosome defects are due to Pkh1/2 phosphorylating another protein that is essential for eisosome assembly. To exclude this possibility and to show that phosphorylation of Pil1 is necessary for eisosome assembly, we first identified phosphorylation sites in the A, B, and C species of Pil1 by MS and then mutated phosphorylation sites to determine if eisosome formation was disrupted. Pil1 was chosen for detailed analyses, because it is essential for eisosome formation whereas Lsp1 is not.
Mapping of phosphorylation sites in proteins requires high amino acid sequence coverage to detect all potential phosphorylation sites. High sequence coverage is achieved by digesting the protein with several different proteases, in combination with MS analysis and identification of the produced peptide and phosphopeptide species (26). The N terminus of Pil1 is largely basic, and the C terminus contains a long stretch of acidic residues lacking lysines and arginines (supplemental Fig. S2). Therefore, to maximize the amount of sequence analyzed by MS, three proteases, trypsin, which cleaves C-terminal to arginine and lysine residues, LysC, which cleaves after lysine, and AspN, which cleaves N-terminal to aspartate, were used to digest the A, B, and C species of Pil1 produced with an N-terminal His6 tag and isolated as described under “Experimental Procedures.” Peptides identified by LC-MS typically have masses between 700 and 4000 Da, and a theoretical digest of Pil1 with each of the three proteases produced a combined amino acid sequence coverage of 93%, considering only fully cleaved peptides in this mass range.
Peptides generated from species A of Pil1 by each of the three proteases were analyzed by MALDI mass spectrometry (supplemental Fig. S2A). Calibrated mass spectra were used for sequence data base searching, using MASCOT and a data base of translated yeast open-reading frames. Sequence coverage varied from 44% to 60% for the individual enzymes, and the total combined sequence coverage was 91% (supplemental Fig. S2B), demonstrating that our approach covers most of the potential phosphorylation sites in Pil1.
Detection and analysis of phosphopeptides by MS is often challenging due to the low abundance of many phosphorylation events in biological samples, ionization bias of phosphopeptides during MS analysis, and the lability of phosphoserine and phosphothreonine residues that leads to loss of phosphoric acid and difficulties in assigning the precise phosphorylation site. However, sequencing of phosphopeptides can by improved in ion trap mass spectrometers by neutral loss-directed MS/MS/MS (MS3) analysis. The often prominent product ion formed by the loss of phosphoric acid from the precursor phosphopeptide ion is fragmented by another round of MS/MS. This technique has been reported to increase the number of identified phosphopeptides by 40% in a complex samples from yeast cells (24). Because Pil1 was affinity-purified and the corresponding in-gel digests did not contain a large number of peptides, aliquots of species A, B, and C were directly analyzed by LC-MS/MS on a LTQ-FT-ICR-MS instrument with neutral loss-directed MS3. A number of phosphopeptides were identified in the trypsin and AspN digests (supplemental Fig. S2A).
Affinity purification of phosphopeptides is a commonly used technique to enhance their detection by MS (27). We investigated, therefore, whether the enrichment of phosphopeptides with either immobilized metal affinity chromatography (Fe(III)-IMAC) or TiO2 (28–31) would allow the identification of additional phosphorylation sites. One additional peptide, D[pS]AQVKPTL[pS]FKQ, with two phosphate groups was identified in this manner.
Initially, phosphopeptides were identified by sequence data base searching using MASCOT. To localize the exact phosphorylation sites, the MS spectra were manually validated and annotated (supplemental Fig. S3). All phosphorylation events occurred on serine or threonine residues and most of the MSMS spectra contained b- or y-ion series of both the phosphorylated peptide ion fragments and the corresponding ions without phosphoric acid, thereby allowing the exact and unambiguous determination of the phosphorylated residue. MALDI TOF MS is a highly sensitive technique for peptide mass mapping and it allows analysis of large peptides and phosphopeptides (>4 kDa). MALDI MS analysis of Pil1 species A revealed a peptide at 4930.7 Da, which was shifted by +80 Da in samples from species B and C (supplemental Fig. S4). These peptides were not detected by LC-MS analyses and, therefore, sequenced by static nanoelectrospray ionization QTOF MS/MS. Peptides were observed at m/z 1253.27 for species A and at m/z 1253.21 for species B, corresponding to the quadruply charged (quadruply protonated) precursor ions. Both of these peptides were subjected to MS/MS analysis for sequence determination. Complex MS/MS fragmentation spectra with good mass resolution and high signal to background ratios were recorded (supplemental Figs. S5 and S6). The tandem mass spectra were processed with the MaxEnt3 algorithm, which detects the charge of peptide peaks and generates a spectrum where all fragments are converted to charge state +1 (supplemental Figs. S5 and S6). The peak lists generated from these MS/MS spectra were searched with MASCOT against a yeast data base and the phosphopeptide 264DSAQVKPTLSFKQDYEDFEPEEGEEEEEEDGQGRWSEDEQE304, containing either one or two phosphorylation sites, was identified. Manual validation assigned the phosphorylation to Ser-299 for species A and to Ser-273 and Ser-299 for species B (and also species C; data not shown). These two sites are also predicted with high probability by NetPhosYeast (32).
Only one phosphopeptide, APTASQLQNPPPPP[STT]K (Fig. 5A), failed to give sufficient sequence information to determine the exact site of phosphorylation in either the MS/MS or the MS3 spectra. This ambiguity is due to the five prolines preceding the phosphorylation site, which are preferred MS/MS-induced cleavage sites and prevent the generation of a comprehensive series of y-ions (33). Therefore, we conclude that phosphorylation occurs on Ser-26, Thr-27, or Thr-28. Prediction of potential phosphorylation sites in Pil1 by NetPhosYeast suggests that Ser-26 is the most likely site for phosphorylation.
FIGURE 5.
Summary of phosphorylated peptides identified by MS in Pil1 species A, B, and C. A, peptides in the A, B, and C species of Pil1 that were identified by MS. B, sequence of Pil1 showing the eight identified sites of phosphorylation. C, plot showing the relative intensity of phosphorylated Thr-233 and Ser-273 in specific phosphopeptides found in the A, B, and C species of Pil1. Peak intensities were determined for the unphosphorylated and phosphorylated (mono- and diphosphorylated in the case of the peptide of 4930.7 Da) forms. The ratio of the intensities of the phosphorylated form divided by the sum of unphosphorylated plus phosphorylated forms is blotted for each species. The graphs show a clear increase in phosphorylation from species A to C. The underlined amino acids in the peptides at the bottom of the figure are ones whose phosphorylation increases in the B and C species, and [pS] indicates a phosphorylated residue whose concentration remains relatively constant.
A total of 14 distinct phosphopeptides were identified in the three species of Pil1 by MS (Fig. 5A), and these establish that species A is phosphorylated on Ser-59, Ser-273, and Ser-299. Species B contains these three sites plus phosphates on Ser-6, [26STT28], and Thr-233. Species C contains the six phosphates present in species B plus phosphorylation on Ser-41 and Ser-265 (Fig. 5B). These data corroborate the hypothesis that phosphorylation increases from species A to B to C, which contain 3, 6, and 8 phosphorylation sites, respectively.
Quantitative analysis by MS generally requires stable isotope labeling or external standards. As a consequence, the specific level of phosphorylation in species A, B, and C could not be determined. However, the shift of the mono-phosphorylated Pil1 peptide at 4930.7 Da in species A to the double phosphorylated form in species B and C was observed in a MALDI analysis (supplemental Fig. S4). The signal intensities of both peptides change drastically form species A to B, indicating that almost no monophosphorylated form is present in species B. This is only an estimate, because the response factors of the two different peptides are not known, and, consequently, precise quantitative measurements are not possible from these spectra. Nevertheless, these results prompted us to look for other phosphorylated peptides and quantitative changes between physiological species in MALDI spectra. To have the highest sensitivity for phosphopeptides, the MALDI analysis was preformed with a 2,5-dihydrobenzoic acid matrix in the presence of phosphoric acid, which has been reported to increase the intensity of phosphopeptides (34). Several peptides with a mass shift of 80 Da between the different physiological species were identified. Comparison of their masses with the identified phosphopeptides revealed five phosphopeptides and two phosphorylation sites. The intensity ratios of the phosphopeptides divided by the sum of intensities of both forms were determined for each physiological species (Fig. 5C). Three different Pil1 peptides contained phosphorylation on Ser-273, and their curves correlate well showing a strong increase in phosphorylation between species A and B. In contrast, the correlation for phosphorylation on Thr-233 was not as good. This is mainly due to the fact that the phosphopeptide DSPV[pT]-PGETRPAY, generated in an AspN digest, is not or only very weakly detected in samples from species B, whereas the tryptic peptides show high intensities. However, in species C all peptides reveal a high degree of phosphorylation. These data show the general trends, but more precise measurements will require stable isotope labeling techniques such as SILAC or chemical modification with e.g. iTRAQ.
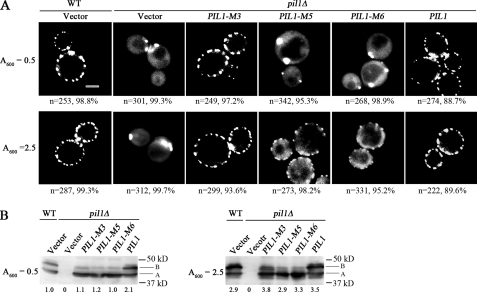
Eisosome Organization Is Disrupted by Pil1 Variant Proteins Lacking Phosphorylation Sites—To directly examine the role of Pil1 phosphorylation in eisosome formation, phosphorylated serine and threonine residues were changed to alanines by site-directed mutagenesis, and cells producing a variant Pil1 protein in place of the wild type were examined by fluorescence microscopy. One Pil1 variant, M3, lacks the phosphorylation sites present in species A (S59A, S273A, and S299A), another variant, M5, lacks five of the six phosphorylation sites present in the B species (S6A, S59A, T233A, S273A, and S299A), and a third variant, M6, that is the same as M5 but has Thr-27 is mutated to alanine. Wild-type and variant Pil1 proteins were produced by using the PIL1 promoter and a single-copy vector (pRS316) in pil1Δ cells that express Lsp1-GFP. Cells were grown to mid-log phase (A600 = 0.5) in defined medium. Eisosomes in cells expressing the M3 variant of Pil1 look like eisosomes in wild-type cells and pil1Δ cells carrying wild-type PIL1 on a plasmid (Fig. 6A). In contrast, eisosomes are not observed in cells making the M5 or the M6 variant proteins, and instead these cells make one or two large fluorescent bodies that look like the eisosome remnants of Lsp1-GFP seen in pil1Δ cells transformed with just the vector (Fig. 6A). Mutant cells expressing the vector show diffuse cytoplasmic fluorescence, which was suggested previously (1) to be Lsp1-GFP that had not assembled into eisosomes. This same cytoplasmic fluorescence is seen in mutant cells producing the M5 or M6 variant of Pil1 that fail to assemble into eisosomes, a further indication that these variant Pil1 proteins do not assemble into eisosomes in log phase cells.
FIGURE 6.
Eisosome formation requires phosphorylation of Pil1. A, wild-type W303 (WT) or a pil1Δ mutant transformed with the vector (pRS316) or the vector carrying PIL1-M3, PIL1-M5, PIL1-M 6, or wild-type PIL1 were grown to early log phase (A600 = 0.5) or almost to stationary phase (A600 = 2.5) in SD medium containing 40 mg/liter adenine sulfate and lacking uracil and examined by fluorescence microscopy. All strains carry chromosomal LSP1-GFP. Bar, 2.5 μm. B, immunoblots showing that the concentration of the M3, M5, and M6 variants of Pil1 produced in pil1Δ cells is the same as the concentration of Pil1 in wild-type cells grown to an A600 of 0.5 (left panel) or 2.5 (right panel).
The lack of eisosomes in cells making the M5 and M6 variant proteins is not due to a reduced level of Pil1, because these cells make as much total Pil1 as do wild-type cells (Fig. 6B, A600 = 0.5). We expected there would be no protein that migrated like species B in cells making any of the variant proteins. However, there is a small amount of a band that migrates like species B. For the M3 variant this band could be due to phosphorylation at Ser-6, Ser/Thr[26–28], Thr-233, and Ser-265 that are phosphorylated in species B and C. For the M5 and M6 variants this B-like band could be due to phosphorylation of sites that define species C (Ser-41 and Ser-265). Another explanation is that when the normal phosphorylation sites are mutated, phosphorylation occurs at alternative sites. In any case, it appears that the number of phosphates is too low or they are in the wrong position for the M5 and M6 variants to form stable eisosomes in log phase cells. We conclude from the data shown in Fig. 6 (A and B) that phosphorylation of Pil1 plays an essential role in eisosome assembly and organization. Furthermore, the data eliminate the possibility that eisosome assembly in INA106-3B (pkh1ts pkh2Δ) cells (Fig. 4) is abnormal, because Pkh1/2 phosphorylate some unknown protein, in addition to Pil1 and Lsp1, that is essential for eisosome organization.
Occasionally cells making the M5 variant of Pil1 contained eisosomes, and further examination revealed that these cells and cells making the M6 variant were able to make some eisosomes but not a normal complement of eisosomes when grown to a higher density (Fig. 6A, A600 = 2.5), which was slightly short of their maximal density (A600 ∼ 3). The fluorescence images shown in Fig. 6 represent the eisosome pattern observed in the majority of cells, and the numbers below each panel indicate the number of cells examined and the percentage with the eisosome pattern shown in the panel. To understand why eisosomes appeared at the higher cell density, the concentration and species of Pil1 were quantified by immunoblotting. Total Pil1 increased ∼3-fold in all strains grown to a higher density (Fig. 6B, A600 = 2.5). In addition, in all strains there was a small increase in bands migrating faster than species A, which could be intermediates with fewer phosphates than species A. There were also slower migrating species that could be more phosphorylated than species A, but not as phosphorylated as species B. Eisosomes may be present in cells containing the M5 and M6 variants of Pil1 when grown to a high density, because the concentration of Pil1 increases and possibly because the variant proteins become slightly more phosphorylated.
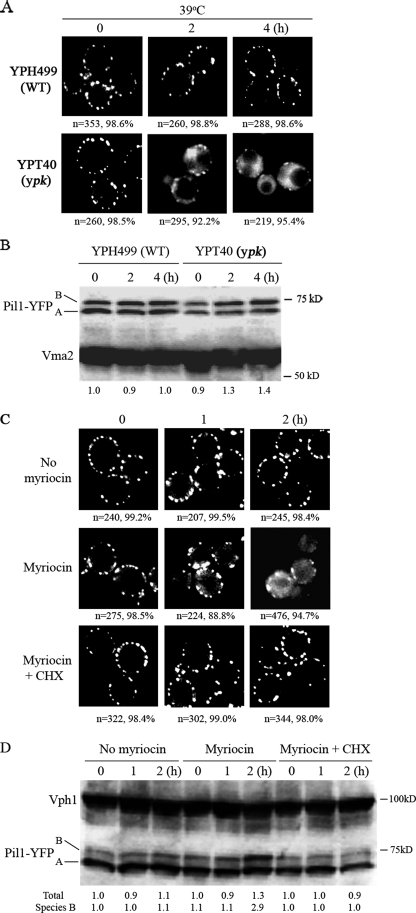
The Ypk Kinases Prevent Disassembly of Eisosomes—Because the LCB-Pkh1/2 signaling pathway regulates the activity of the Ypk1/2, Pkc1, and Sch9 protein kinases, we determined if any of these kinases play a role in eisosome organization. Ypk1 and -2 do play a role in eisosome organization: Cells (YPT40) with a ypk1 temperature-sensitive allele and YPK2 deleted make normal looking eisosomes when grown at a permissive temperature (25 °C), but, upon shifting to a restrictive temperature (39 °C), the number of eisosomes per cell decreases over a 4-h time frame, whereas eisosomes do not change in wild-type cells (YPH499, Fig. 7A). YPT40 cells lyse when grown for >1 h at 39 °C, because Ypk1 and -2 play roles in cell wall integrity (13, 14). To prevent lysis, 1 m sorbitol was included in the culture medium. The numbers below each panel indicate the numbers of cells examined, and the percentage with the eisosome pattern is shown in the panel. About 2 h after the temperature shift, fluorescence appears in the cytoplasm of YPT40 mutant cells and becomes more pronounced with time. Such cytoplasmic fluorescence is not seen in YPH499 wild-type cells. Cytoplasmic fluorescence probably represents Pil1-YFP that has not assembled into eisosomes. Sch9 and Pkc1 do not seem to play a role in eisosome organization, because eisosomes remain stable in sch9Δ cells or in cells carrying a temperature-sensitive pkc1 allele following a shift to 39 °C (data not shown).
FIGURE 7.
Eisosome stability is controlled by the Ypk1/2 kinases and LCBs. A, wild-type YPH499 or Ypk mutant cells (YPT40, ypk1ts ypk2Δ) with chromosomal PIL1 carrying a C-terminal YFP tag, were grown in YPAD containing 1 m sorbitol to an A600 of 0.5 and shifted to 39 °C. Eisosomes were examined by fluorescence microscopy at the indicated times. B, cells used in panel A were immunoblotted for Pil1-YFP. The relative intensity of the combined Pil1-YFP bands (species A and B) divided by the intensity of the Vma2 loading control and normalized to the 0 time value gives the relative amounts of Pil1-YFP shown below each lane. C, YPH499 cells (PIL1-YFP) were grown at 30 °C in YPAD to an A600 of 0.5 and either treated with the ethanol vehicle (1% final concentration, No myriocin), or with 5 μm myriocin or myriocin plus 100 μg/ml cycloheximide (CHX) for the indicated times and analyzed by fluorescence microscopy for eisosomes. D, cells used in panel C immunoblotted for Pil1-YFP with the normalized signal intensity calculated as for panel B and shown below each lane.
The decrease in eisosomes following a shift of YPT40 cells to a restrictive temperature is not due to dephosphorylation of Pil1 or Lsp1 or a decrease in their abundance, because the concentration of the A and B species remains the same in mutant and wild-type cells (Fig. 7B). However, our data do not rule out dephosphorylation of one or more residues that have no affect upon the electrophoretic mobility of species A or B. These results demonstrate that eisosomes are dynamic structures whose stability and disassembly are controlled by the Ypk kinases.
Because Ypk1/2 are activated by the LCB-Pkh1/2 pathway, we reduced the concentration of LCBs by treating cells with myriocin to determine if eisosomes disassembled. Myriocin inhibits serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis, and has been widely used to reduce LCBs (reviewed in Ref. 25). Myriocin treatment did trigger a loss of eisosomes as early as 1 h after treatment, and the effect is even more evident by 2 h when cytoplasmic fluorescence appears in most cells (Fig. 7C). The concentration of the A species of Pil1 did not differ significantly between myriocin-treated and untreated cells, but the concentration of the B species increased 2.9-fold after 2 h in two separate experiments (Fig. 7D). These results suggest that increased phosphorylation of the B species of Pil1 may play a role in eisosome disassembly. To ensure that the myriocin affect on eisosomes was due to reduced LCBs, phytosphingosine was added to the culture medium during myriocin treatment. Phytosphingosine prevented eisosome disassembly (supplemental Fig. S7). To determine if eisosome disassembly following myriocin treatment required de novo protein synthesis, cells were treated with cycloheximide along with myriocin. Cycloheximide treatment blocked eisosome disassembly, suggesting that disassembly requires new protein synthesis (Fig. 7C).
DISCUSSION
The yeast phosphoinositide-dependent protein kinase 1 homologs, Pkh1 and Pkh2, control cellular processes primarily by phosphorylating and activating protein kinases, including Pkc1, Ypk1/2, and Sch9. The in vivo phosphorylation results presented here along with our previous in vitro phosphorylation data establish Pil1 and Lsp1 as the first physiological substrates of Pkh1/2 that are not protein kinases (Figs. 1 and 2). Furthermore, our data show that phosphorylation controls eisosome organization (Figs. 4 and 6) and that eisosomes are dynamic structures whose disassembly is regulated by the Ypk1/2 kinases and by sphingolipid LCBs (Fig. 7). These data when combined with previous data (reviewed in Ref. 25) establish the LCB-Pkh1/2-Ypk1/2 signaling pathway as a regulator of eisosome assembly and disassembly. The location of eisosomes on the inner surface of the plasma membrane places them in a strategic position to control endocytosis and align the functions of the plasma membrane with the changing needs of cells as they encounter environmental stresses. Because LCBs are found in membranes, they and the Pkh1/2-Ypk1/2 kinase serve as links between cellular stresses and eisosomes.
We found two species, A and B, of Pil1 and Lsp1 in unstressed, log phase cells and a third more highly phosphorylated species C, generated during heat shock (Fig. 2). MS analyses show that these species are due to differences in phosphorylation with species A, B, and C of Pil1 having 3, 6, and 8 phosphorylation sites, respectively (Fig. 7). Species A is phosphorylated on Ser-59, Ser-273, and Ser-299, and species B has three additional phosphorylation sites, Ser-6, [26STT28], and Thr-233. Species B is converted to C by additional phosphorylation at Ser-41 and Ser-265. A recently described neural network-based method for prediction of phosphorylation sites in yeast proteins (32) correctly predicted 7 of the 8 phosphorylation sites in Pil1 that we identified (Fig. 5B), the only site that was not strongly predicted was Ser-41, and this site fell slightly below the cut-off score. Further refinement of the method should lead to enhanced predictability of phosphorylation sites. Our data show that Pkh1/2 are responsible for generating the B and C species of Pil1 by phosphorylating it on Ser-6, [26STT28], Thr-233, Ser-41, and Ser-265. Because the A species of Pil1 is phosphorylated in pkh mutant cells (Fig. 3), we cannot be sure if Pkh1D398G or some other protein kinase is responsible. There may be additional phosphorylation sites that were not detected or that are generated by different growth or stress conditions. Investigations are under way to determine the function of species C, which is generated by other stresses besides heat shock, and may play a role in stress resistance.
Our observation, that cells with reduced Pkh activity (INA106-3B, grown at 25 °C) lack the more phosphorylated B species of Pil1 and contain fewer eisosomes that are not as evenly distributed as in wild-type cells (Fig. 4A), provides one avenue of support for the importance of Pil1 phosphorylation in assembly of eisosomes. The need for Pil1 phosphorylation in eisosome assembly is also supported by the M5 and M6 phosphorylation site mutants that do not assembly into eisosomes in log phase cells. Failure to assemble eisosomes is due to reduced phosphorylation of Pil1 as evidenced by the low concentration of species B (Fig. 6B, A600 = 0.5). By the time the cell density increases 5-fold and cells enter the diauxic shift, eisosomes begin to appear, demonstrating that the M5 and M6 variant proteins can form eisosomes (Fig. 6A, A600 = 2.5) and eliminating the possibility that replacement of serine and threonine residues with alanines causes the proteins to misfold into one or more conformations that cannot assemble or that block assembly. Even at the higher cell density there are fewer eisosomes and a large fraction of Lsp1-YFP remains unassembled as indicated by cytoplasmic fluorescence. At the higher cell density the concentration of the variant Pil1 proteins increases 3-fold, and they become slightly more phosphorylated but still not as phosphorylated as wild-type Pil1 (Fig. 6B). One or both of these factors probably work to overcome a rate-limiting step in eisosome formation that is normally promoted in wild-type cells by the highly phosphorylated B species of Pil1.
It is not clear if all five of the phosphorylation sites mutated in the M5 variant must be present to block assembly. Further work will be required to identify the specific phosphorylation sites necessary for eisosome assembly and correct distribution on the plasma membrane. The distribution of eisosomes is probably due to their localization to patches on the plasma membrane that are enriched for sterols, proton symporters and the Sur7 protein, which is thought to anchor eisosomes (2). Our data suggest that Pil1 phosphorylation plays a role in localizing eisosomes to these patches.
Because Pkh1 and -2 activate the Ypk1/2, Pkc1, and Sch9 protein kinases, we determined if any of these kinases play roles in eisosome formation or organization. We found that Ypk1/2 kinase activity is required to maintain eisosomes and prevent their disassembly (Fig. 7A). Ypk1/2 may regulate a protein that inhibits disassembly or that promotes disassembly when Ypk1/2 activity is reduced. Disassembly is not due to a reduction in phosphorylation of Pil1 that changes protein mobility on denaturing gels or to a decrease in protein concentration, because the concentration of the A and B species is the same in mutant Ypk1/2 and parental cells (Fig. 7B). However, disassembly could involve dephosphorylation of one or more specific residues in Pil1 that do not change the mobility of the A and B species on a denaturing gel. Because Ypk1/2 are activated by the LCB-Pkh1/2 pathway, we determined if lowering the concentration of LCBs by treating cells with myriocin also triggered eisosome disassembly. This hypothesis proved to be correct (Fig. 7C). Unexpectedly, cycloheximide treatment prevented myriocin from promoting eisosome disassembly (Fig. 7C), indicating that the disassembly process requires synthesis of one or more proteins. Because cycloheximide also prevented the nearly 3-fold increase in the concentration of Pil1 species B induced by myriocin treatment (Fig. 7D), the newly made protein could be required to promote phosphorylation or inhibit dephosphorylation of Pil1. Our results with Ypk mutant cells and myriocin treatment indicate for the first time that eisosomes are dynamic structures whose assembly and maintenance requires an active LCB-Pkh1/2-Ypk1/2 pathway.
How might the LCB-Pkh1/2-Ypk1/2 signaling pathway control eisosome assembly and disassembly? We suggest that the basal level of LCBs activates some fraction of Pkh1/2, which phosphorylate Pil1 to generate electrophoretic species B that is necessary for assemble into eisosomes. Ypk1/2 activity, on the other hand, is required to maintain eisosomes and prevent them from dissembling. It is not clear how Ypk1/2 maintain eisosomes, but the most direct way would be for them to phosphorylate the Pil1 present in eisosomes. A less direct way would be through modulation of the activity of a protein phosphatase that dephosphorylates Pil1. One potential phosphatase that lies downstream of the LCB-Pkh1/2 pathway is calcineurin, a calcium/calmodulin-regulated protein phosphatase (11, 17, 16). Alternatively, Ypk1/2 could maintain eisosomes in more indirect ways.
Mammalian phosphoinositide-dependent protein kinase 1 phosphorylates the first threonine in the consensus sequence TFCGTXGY (where X is any amino acid) that is found in the activation loop in all of its known protein kinase substrates (4, 35). The four known yeast protein kinases that are substrates of Pkh1/2 contain part of this consensus sequence, TFCGTXX(Y/F) (4). Surprisingly, none of the seven phosphorylation sites we identified in Pil1 have a sequence similar to the mammalian phosphoinositide-dependent protein kinase 1 or yeast Pkh1/2 consensus sequence, and, in fact, the sequences around the seven phosphorylation sites in Pil1 show no similarity (Fig. 7A). Thus, our data suggest that Yeast Pkh1/2 probably rely more on protein conformation than on amino acid sequence to identify cognate phosphorylation sites, at least when Pil1 is the substrate. Conservation of the sequence in the activation loop of protein kinases phosphorylated by Pkh1/2 may promote a conformation that is required for enzyme activity rather than being a strict sequence required for substrate recognition.
The increase in Pil1 concentration that occurred as cells entered the diauxic shift on their way to stationary phase (Fig. 6B) may be due to increased transcription mediated by the stress response elements located in the PIL1 promoter (36–39). The functional role of this increase remains to be uncovered, but it suggests that Pil1 and perhaps eisosomes play roles in stress protection. In addition to an increased concentration, there was also an increase in the B species relative to the A species (Fig. 6B, WT lane), which could play unique roles in stationary phase cells.
The sequence of Pil1 is highly conserved up to about residue 275 in six other fungal species (Aspergillus fumigatu, Candida glabrata, Kluyveromyces lactis, Neurospora crassa, Schizosaccharomyces pombe, and Yersinia lipolyti, from the Saccharomyces Genome Data base). Seven of eight phosphorylation sites we identified lie within this region, the only exception being Ser-299, found in the unconserved C-terminal tail region of Pil1. The Ser-6, [26STT28] and Thr-233 phosphorylation sites are conserved in all seven fungal Pil1 sequences, and Ser-273 is found in all but the S. pombe sequence, which lacks sequence similarity around Ser-273. We predict that these conserved residues are likely to be phosphorylated in other fungi and to play a role in eisosome organization.
A recent publication comes to many of the same conclusions about the role of the LCB-Pkh1/2 pathway in phosphorylating Pil1 and controlling eisosome organization (40) as we do. However, there are differences, and the most significant is the function of Pil1 phosphorylation. Our data support a model in which phosphorylation of Pil1 promotes eisosome assembly, whereas Walther et al. propose that phosphorylation promotes disassembly. This difference seems to be due to three experimental results of Walther et al. First, overproduction of Pkh1 or Pkh2, which increased the amount of Pil1 species B, caused eisosomes to disassemble. Second, production of a quadruple phosphorylation site mutant of Pil1 (S45A, S59A, S230A, and T233A) as a GFP fusion protein in pil1Δ cell, resulted in eisosomes with increased fluorescence (larger eisosomes). Third, when the same phosphorylation sites were changed to aspartic acids to try and mimic phosphorylation, very few eisosomes were observed. It is difficult to know if overproduction of protein kinases enhances a normal physiological process or whether it produces a non-physiological response. The phosphorylation site mutants of Walther et al. are different from ours and only share the S59A and T233A changes, so it is hard to directly compare them. In addition, there are differences in the phosphorylation sites identified by us and by Walther et al. Some of these could be due to the way cells were grown while others could be due to a variation in the level of phosphorylation at particular sites, reflecting differences in cell growth and purification procedures. Resolving the differences between our interpretation of the function of Pil1 phosphorylation and that of Walther et al. will probably require knowledge of the structure of Pil1 and a more detailed analysis of phosphorylation sites and their affect upon eisosome assembly and disassembly.
Supplementary Material
Acknowledgments
We thank Drs. Hainning Zhu and Carol Beach for initial analyses of phosphorylation sites in Pil1 and Jens Andersen for providing access to and Lene Jakobsen for measuring samples on the LTQ-FT instrument.
This work was supported in part by Grant GM41302 from the National Institutes of Health (NIH) and by core facilities supported by a Grant P20-RR020171 from the National Center for Research Resources, a component of NIH. The LTQ-FT ICR-MS instrument was financed by a generous grant from the Danish Basic Research Foundation to the Center for Experimental Bioinformatics. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
Footnotes
The abbreviations used are: LCB, long-chain base; ESI, electrospray ionization; GFP, green fluorescent protein; LC, liquid chromatography; LTQ-FT, ion trap-Fourier transform; MALDI, matrix-assisted laser desorption/ionization; MS, mass spectrometry; QTOF, quadrupole time of flight; YFP, yellow fluorescent protein; YPD, yeast extract, peptone, dextrose; TBS, Tris-buffered saline; MS/MS, tandem MS; MS3, MS/MS/MS.
Pierce Chemicals Technical Resource brochure Acetone Precipitation of Proteins.
References
- 1.Walther, T. C., Brickner, J. H., Aguilar, P. S., Bernales, S., Pantoja, C., and Walter, P. (2006) Nature 439 998–1003 [DOI] [PubMed] [Google Scholar]
- 2.Grossmann, G., Opekarova, M., Malinsky, J., Weig-Meckl, I., and Tanner, W. (2007) EMBO J. 26 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang, X., Lester, R. L., and Dickson, R. C. (2004) J. Biol. Chem. 279 22030–22038 [DOI] [PubMed] [Google Scholar]
- 4.Casamayor, A., Torrance, P. D., Kobayashi, T., Thorner, J., and Alessi, D. R. (1999) Curr. Biol. 9 18697. [DOI] [PubMed] [Google Scholar]
- 5.Inagaki, M., Schmelzle, T., Yamaguchi, K., Irie, K., Hall, M. N., and Matsumoto, K. (1999) Mol. Cell. Biol. 19 8344–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., Yang, L., Wolting, C., Donaldson, I., Schandorff, S., Shewnarane, J., Vo, M., Taggart, J., Goudreault, M., Muskat, B., Alfarano, C., Dewar, D., Lin, Z., Michalickova, K., Willems, A. R., Sassi, H., Nielsen, P. A., Rasmussen, K. J., Andersen, J. R., Johansen, L. E., Hansen, L. H., Jespersen, H., Podtelejnikov, A., Nielsen, E., Crawford, J., Poulsen, V., Sorensen, B. D., Matthiesen, J., Hendrickson, R. C., Gleeson, F., Pawson, T., Moran, M. F., Durocher, D., Mann, M., Hogue, C. W., Figeys, D., and Tyers, M. (2002) Nature 415 180–183 [DOI] [PubMed] [Google Scholar]
- 7.Krogan, N. J., Cagney, G., Yu, H., Zhong, G., Guo, X., Ignatchenko, A., Li, J., Pu, S., Datta, N., Tikuisis, A. P., Punna, T., Peregrin-Alvarez, J. M., Shales, M., Zhang, X., Davey, M., Robinson, M. D., Paccanaro, A., Bray, J. E., Sheung, A., Beattie, B., Richards, D. P., Canadien, V., Lalev, A., Mena, F., Wong, P., Starostine, A., Canete, M. M., Vlasblom, J., Wu, S., Orsi, C., Collins, S. R., Chandran, S., Haw, R., Rilstone, J. J., Gandi, K., Thompson, N. J., Musso, G., St Onge, P., Ghanny, S., Lam, M. H., Butland, G., Altaf-Ul, A. M., Kanaya, S., Shilatifard, A., O'Shea, E., Weissman, J. S., Ingles, C. J., Hughes, T. R., Parkinson, J., Gerstein, M., Wodak, S. J., Emili, A., and Greenblatt, J. F. (2006) Nature 440 637–643 [DOI] [PubMed] [Google Scholar]
- 8.Friant, S., Lombardi, R., Schmelzle, T., Hall, M. N., and Riezman, H. (2001) EMBO J. 20 6783–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levin, D. E. (2005) Microbiol. Mol. Biol. Rev. 69 262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu, K., Zhang, X., Lester, R. L., and Dickson, R. C. (2005) J. Biol. Chem. 280 22679–22987 [DOI] [PubMed] [Google Scholar]
- 11.Bultynck, G., Heath, V. L., Majeed, A. P., Galan, J. M., Haguenauer-Tsapis, R., and Cyert, M. S. (2006) Mol. Cell. Biol. 26 4729–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.deHart, A. K., Schnell, J. D., Allen, D. A., and Hicke, L. (2002) J. Cell Biol. 156 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roelants, F. M., Torrance, P. D., Bezman, N., and Thorner, J. (2002) Mol. Biol. Cell 13 3005–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmelzle, T., Helliwell, S. B., and Hall, M. N. (2002) Mol. Cell. Biol. 22 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fadri, M., Daquinag, A., Wang, S., Xue, T., and Kunz, J. (2005) Mol. Biol. Cell 16 1883–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabuchi, M., Audhya, A., Parsons, A. B., Boone, C., and Emr, S. D. (2006) Mol. Cell. Biol. 26 5861–5875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daquinag, A., Fadri, M., Jung, S. Y., Qin, J., and Kunz, J. (2007) Mol. Cell. Biol. 27 633–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powers, R. W., 3rd, Kaeberlein, M., Caldwell, S. D., Kennedy, B. K., and Fields, S. (2006) Genes Dev. 20 174–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabrizio, P., Liou, L. L., Moy, V. N., Diaspro, A., SelverstoneValentine, J., Gralla, E. B., and Longo, V. D. (2003) Genetics 163 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sikorski, R. S., and Hieter, P. (1989) Genetics 122 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kushnirov, V. V. (2000) Yeast 16 857–860 [DOI] [PubMed] [Google Scholar]
- 22.Karas, M., and Hillenkamp, F. (1988) Anal. Chem. 60 2299–2301 [DOI] [PubMed] [Google Scholar]
- 23.Rappsilber, J., Ishihama, Y., and Mann, M. (2003) Anal. Chem. 75 663–670 [DOI] [PubMed] [Google Scholar]
- 24.Gruhler, A., Olsen, J. V., Mohammed, S., Mortensen, P., Faergeman, N. J., Mann, M., and Jensen, O. N. (2005) Mol. Cell Proteomics 4 310–327 [DOI] [PubMed] [Google Scholar]
- 25.Dickson, R. C., Sumanasekera, C., and Lester, R. L. (2006) Prog. Lipid Res. 45 447–465 [DOI] [PubMed] [Google Scholar]
- 26.Mann, M., and Jensen, O. N. (2003) Nat. Biotechnol. 21 25561. [DOI] [PubMed] [Google Scholar]
- 27.Morandell, S., Stasyk, T., Grosstessner-Hain, K., Roitinger, E., Mechtler, K., Bonn, G. K., and Huber, L. A. (2006) Proteomics 6 4047–4056 [DOI] [PubMed] [Google Scholar]
- 28.Larsen, M. R., Thingholm, T. E., Jensen, O. N., Roepstorff, P., and Jorgensen, T. J. (2005) Mol. Cell Proteomics 4 873–886 [DOI] [PubMed] [Google Scholar]
- 29.Neville, D. C., Rozanas, C. R., Price, E. M., Gruis, D. B., Verkman, A. S., and Townsend, R. R. (1997) Protein Sci 6 2436–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinkse, M. W., Uitto, P. M., Hilhorst, M. J., Ooms, B., and Heck, A. J. (2004) Anal. Chem. 76 3935–3943 [DOI] [PubMed] [Google Scholar]
- 31.Stensballe, A., Andersen, S., and Jensen, O. N. (2001) Proteomics 1 207–222 [DOI] [PubMed] [Google Scholar]
- 32.Ingrell, C. R., Miller, M. L., Jensen, O. N., and Blom, N. (2007) Bioinformatics 23 895–897 [DOI] [PubMed] [Google Scholar]
- 33.Yu, W., Vath, J. E., Huberty, M. C., and Martin, S. A. (1993) Anal. Chem. 65 3015–3023 [DOI] [PubMed] [Google Scholar]
- 34.Kjellstrom, S., and Jensen, O. N. (2004) Anal. Chem. 76 5109–5117 [DOI] [PubMed] [Google Scholar]
- 35.Hanks, S. K., and Hunter, T. (1995) FASEB J. 9 576–596 [PubMed] [Google Scholar]
- 36.Causton, H. C., Ren, B., Koh, S. S., Harbison, C. T., Kanin, E., Jennings, E. G., Lee, T. I., True, H. L., Lander, E. S., and Young, R. A. (2001) Mol. Biol. Cell 12 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jelinsky, S. A., Estep, P., Church, G. M., and Samson, L. D. (2000) Mol. Cell. Biol. 20 8157–8167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rep, M., Krantz, M., Thevelein, J. M., and Hohmann, S. (2000) J. Biol. Chem. 275 8290–8300 [DOI] [PubMed] [Google Scholar]
- 39.Yale, J., and Bohnert, H. J. (2001) J. Biol. Chem. 276 15996–16007 [DOI] [PubMed] [Google Scholar]
- 40.Walther, T. C., Aguilar, P. S., Frohlich, F., Chu, F., Moreira, K., Burlingame, A. L., and Walter, P. (2007) EMBO J. 26 4946–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., and Boeke, J. D. (1998) Yeast 14 115–132 [DOI] [PubMed] [Google Scholar]
- 42.Fabrizio, P., Pozza, F., Pletcher, S. D., Gendron, C. M., and Longo, V. D. (2001) Science 292 288–290 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.