Abstract
Mitochondrial NAD+-specific isocitrate dehydrogenases (IDHs) are key regulators of flux through biosynthetic and oxidative pathways in response to cellular energy levels. Here we present the first structures of a eukaryotic member of this enzyme family, the allosteric, hetero-octameric, NAD+-specific IDH from yeast in three forms: 1) without ligands, 2) with bound analog citrate, and 3) with bound citrate + AMP. The structures reveal the molecular basis for ligand binding to homologous but distinct regulatory and catalytic sites positioned at the interfaces between IDH1 and IDH2 subunits and define pathways of communication between heterodimers and heterotetramers in the hetero-octamer. Disulfide bonds observed at the heterotetrameric interfaces in the unliganded IDH hetero-octamer are reduced in the ligand-bound forms, suggesting a redox regulatory mechanism that may be analogous to the “on-off” regulation of non-allosteric bacterial IDHs via phosphorylation. The results strongly suggest that eukaryotic IDH enzymes are exquisitely tuned to ensure that allosteric activation occurs only when concentrations of isocitrate are elevated.
The oxidative decarboxylation of isocitrate to α-ketoglutarate catalyzed by mitochondrial NAD+-specific isocitrate dehydrogenases is a rate-limiting step in the tricarboxylic acid cycle. The affinities of the yeast and human enzymes (IDH,5 EC 1.1.1.41) for isocitrate are allosterically regulated positively by AMP and ADP, respectively, and the human enzyme is also regulated negatively by ATP (1, 2). Both enzymes are negatively regulated by NADH (3, 4). This control has been proposed to contribute to inverse regulation of rates of energy production by oxidative pathways and by glycolysis (5). Under conditions of energy sufficiency, i.e. when relative cellular ratios of [ATP]/[AMP] and [NADH]/[NAD+] are high, flux through the tricarboxylic acid cycle would be attenuated at the level of IDH, and tricarboxylic acids, citrate and isocitrate, would be diverted into biosynthetic pathways.
Yeast IDH is a hetero-octamer composed of four IDH1 (Mr = 38,001) and four IDH2 (Mr = 37,755) subunits with 42% sequence identity (6-8) (supplemental Fig. 1). Results of targeted mutagenesis studies (9-13) suggest that IDH2 contains a catalytic isocitrate/Mg2+- and NAD+-binding site, whereas the homologous site in IDH1 functions in cooperative binding of isocitrate and in binding of the allosteric activator AMP. Yeast two-hybrid assays and other mutagenesis studies (13, 14) strongly suggest that a heterodimer of IDH1 and IDH2 is the fundamental structural building block of the enzyme and that IDH1 contributes a few residues to catalytic sites in IDH2, while the analogous residues of IDH2 function in the regulatory ligand-binding sites in IDH1. Visualization of subunit interactions within and between heterodimers in the holoenzyme is essential to define the molecular basis for allosteric communication between such sites and to provide information prerequisite for an understanding of the evolution of the highly regulated allosteric IDH hetero-octamer from the more ancient nonallosteric, homodimeric bacterial IDH enzymes. Mammalian NAD+-specific isocitrate dehydrogenases are structurally more complex octamers than yeast IDH, containing three subunits in a ratio of α2:β:γ, with the α-subunit being more similar in sequence to the catalytic yeast IDH2, and the β- and γ-subunits apparently contributing regulatory functions similar to those provided by IDH1 (15-19).
Because structural information on mitochondrial eukaryotic NAD+-specific isocitrate dehydrogenases has remained elusive, analyses of these multimeric, allosteric enzymes have thus far relied upon modeling based on the known structures of homodimeric homologues of bacterial origin (20-22) and of non-allosteric homodimeric mammalian NADP+-specific enzymes (23, 24). Here, we present the first structures of hetero-octameric yeast IDH determined using the well established tools of single crystal x-ray diffraction. Together, the unliganded, citrate-, and citrate + AMP-bound structures reveal: 1) the molecular details of interactions between IDH1 and IDH2 in the heterodimer; 2) the overall spatial arrangement and subunit orientation of the IDH1/IDH2 components in the hetero-octameric holoenzyme; 3) structural transitions at the regulatory isocitrate/AMP-binding site of IDH1 upon ligand binding; 4) evidence for communication of these structural transitions to another IDH heterodimer through the oxidation and reduction of a disulfide bond at the “clasp” β-barrel motif found at the 2-fold symmetric heterotetrameric interface; and 5) a pathway of communication between the two heterotetramers through the N-terminal residues of IDH1 subunits. These structures provide a platform for understanding the evolution of IDH from a nonallosteric, homodimeric enzyme to a highly regulated, allosteric, hetero-octameric enzyme and highlight the increasingly fine levels of metabolic regulation in moving from prokaryotic to eukaryotic organisms.
EXPERIMENTAL PROCEDURES
Protein Expression and Crystallization—Selenomethionine-substituted IDH (SeMet-IDH) was generated by the method of Hendrickson et al. (25) using the methionine auxotrophic Escherichia coli strain B834(DE3) (Novagen). The strain was transformed with pET-15bIDH1His/IDH2. Cells were grown to an A600 of 0.5-0.8, and expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were harvested, resuspended, and sonicated on ice in 40 mm Tris-HCl (pH 7.4) containing 300 mm NaCl, 10 mm imidazole, 10 mm sodium citrate, and 4 mm MgCl2. This buffer and all solutions used in purification were supplemented with 2 mm Tris(2-carboxyethyl)phosphine to prevent selenomethionine oxidation. The histidine-tagged yeast IDH protein was purified using Ni2+-nitrilotriacetic acid chromatography as described previously (13). IDH eluted from this column was dialyzed overnight in buffer A (10 mm Tris-HCl (pH 7.4), 40 mm NaCl, 10 mm sodium citrate, and 4 mm MgCl2) and further purified using Affi-Gel blue chromatography (Bio-Rad). After extensive washing, IDH was eluted off the column with buffer A containing 1.0 m NaCl, dialyzed overnight at 4 °C in buffer A, and concentrated to ∼80 mg/ml (ε = 168,820 m-1cm-1) for use in crystallization trials. Purified SeMet-IDH subjected to matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry revealed increases in mass of ∼330 and ∼144 Da for IDH1 and IDH2, respectively, indicating that on average 7 of 8 methionine residues in IDH1 and 3 of 4 methionine residues in IDH2 were substituted by SeMet (∼83% total for the IDH1/IDH2 heterodimer).
Native IDH lacking a histidine tag was expressed in E. coli BL21(DE3) cells and purified using conventional column chromatography essentially as described previously (26). Protein concentrations of ∼15 mg/ml were used for crystallization screens.
Initial crystals of both forms of yeast IDH were obtained using the sitting-drop vapor diffusion method with commercially available crystal screens and optimized with the hanging-drop vapor diffusion method. Crystals of SeMet-IDH with bound citrate were grown from drops containing 3 μl of protein solution and 3 μl of 0.1 m CHES (pH 9.5), 1.1 m sodium citrate, and 15% ethylene glycol (reservoir solution pH 9.7). Ligand-free native IDH crystals were grown from drops containing 3 μl of protein solution and 3 μl of 16% polyethylene glycol 3350, 0.13 m K2SO4, 0.2 m trimethylamineoxide, and 15% propanediol (reservoir solution, pH 8.1). Crystals of IDH with citrate and AMP were grown from drops containing 3 μl of protein solution (with additions of 10 mm sodium citrate and 4 mm AMP) and 3 μl of 18% polyethylene glycol 3350, 0.2 m K2SO4, and 10% ethylene glycol (reservoir solution, pH 6.6). SeMet-IDH crystals suitable for x-ray diffraction work were obtained in space group C2 with a = 256.9 Å, b = 112.9 Å, c = 125.6 Å, β = 106.7° with one hetero-octamer per asymmetric unit. Ligand-free native IDH crystals were obtained in space group P1 with a = 112.4 Å, b = 115.2 Å, c = 159.2 Å, α = 111.0°, β = 96.1°, γ = 107.1° with two hetero-octamers per asymmetric unit. The crystal form with added citrate and AMP crystallized in space group P1 with a = 113.2 Å, b = 116.3 Å, c = 163.6 Å, α = 99.0°, β = 110.2°, γ = 106.6° with two hetero-octamers per asymmetric unit. The packing of IDH in the two crystal forms is similar in that robust protein-protein interactions result in the formation of “columns” of hetero-octamers. However, the way these columns pack with respect to each other in the two crystal forms is different.
Data Collection, Structure Determination, and Refinement—IDH crystals were flash-cooled in liquid nitrogen prior to data collection at the Berkeley Advanced Light Source beamline 5.0.2 (citrate-bound SeMet-IDH and citrate/AMP-bound native IDH) and the Brookhaven National Synchrotron Light Source beamline X29 (unliganded native IDH). Both crystal forms demonstrated sensitivity to synchrotron radiation, limiting the multiwavelength anomalous diffraction (MAD (27)) experiment to energies corresponding only to the selenium absorption peak and inflection point. All diffraction data were processed with HKL-2000 (28) and are summarized in Table 1.
TABLE 1.
Data collection, phasing, and refinement statistics
| Native IDH (unliganded) | Native IDH (citrate + AMP) | SeMet IDH (citrate) | |||
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P1 | P1 | C2 | ||
| Cell dimensions | |||||
| a, b, c (Å) | 112.4, 115.2, 159.2 | 113.2, 116.3, 163.6 | 256.9, 112.9, 125.6 | ||
| α, β, γ (°) | 111.0, 96.1, 107.1 | 99.0, 110.2, 106.6 |
90, 106.7, 90
|
||
|
Peak
|
Inflection
|
||||
| Wavelength | 1.1000 | 1.0000 | 0.9791 | 0.9796 | |
| Resolution (Å) | 50-2.7 | 50-4.3 | 50-3.2 | 50-3.3 | |
| Rsyma | 0.078 (0.554) | 0.130 (0.334) | 0.101 (0.556) | 0.128 (0.511) | |
| I/σI | 13.7 (2.0) | 7.9 (2.4) | 11.0 (2.1) | 10.2 (2.3) | |
| Completeness (%) | 96.6 (94.1) | 90.1 (66.7) | 96.8 (97.9) | 98.8 (99.3) | |
| Redundancy | 3.6 (2.7) | 3.2 (2.3) | 3.1 (3.0) | 3.0 (2.8) | |
| Refinement | |||||
| Resolution (Å) | 50-2.7 | 50-4.3 | 50-3.2 | ||
| No. reflections | 183,432 | 44,278 | 107,063 | ||
| Rwork/Rfree | 0.239/0.264 | 0.274/0.311 | 0.221/0.265 | ||
| No. atoms | |||||
| Protein | 41,336 | 41,406 | 20,828 | ||
| Ligand/ion | 288 | 52 | |||
| B-factors | |||||
| Protein | 58.2 | 191.4 | 74.0 | ||
| Ligand/ion | 175.2 | 82.4 | |||
| Root-mean-square deviations | |||||
| Bond lengths (Å) | 0.011 | 0.012 | 0.008 | ||
| Bond angles (°) | 1.275 | 1.145 | 1.149 | ||
Values in parentheses are for highest resolution shell.
The citrate-bound SeMet IDH structure in space group C2 was determined first using a combination of phases derived from molecular replacement (MR) (29) and MAD. Initial MR phases were calculated from the structure of homodimeric IMDH from Sulfolobus tokodaii (Protein Data Bank entry 1WPW)6 as the search model using the program PHASER (30). These phases were used to locate 40 selenium positions in an anomalous difference Fourier map calculated from the selenium absorption peak data set. MR and MAD phases were combined in the program SHARP (31) to yield a mean figure of merit to 0.53 calculated at 3.2 Å resolution. Maximum likelihood density modification including 4-fold averaging in the program RESOLVE (32) resulted in an interpretable electron density map. The selenium atom positions distinguished IDH1 (8 SeMet) from IDH2 (4 SeMet). The molecular model for the IDH heterodimer was built using the program COOT (33). The coordinates of a single IDH1/IDH2 citrate-bound heterodimer were subsequently used as the search model to determine the native unliganded yeast IDH structure in space group P1 to 2.7 Å and the citrate + AMP-bound structure to 4.3 Å resolution. The low resolution of the latter structure permitted only rigid body refinement, but because AMP was not in the search model, Fo - Fc electron density map calculations clearly illuminated the positions of the AMP-binding sites. For the native and citrate-bound structures, non-crystallographic symmetry restraints were invoked in PHENIX (34) during refinement. Crystallographic refinement statistics for both crystal systems are shown in Table 1.
Structure Analysis and Figure Preparation—All figures were prepared using the program PyMol (Delano Scientific). Coordinates have been deposited in the Protein Data Bank with accession codes 3BLX, 3BLV, and 3BLW for the unliganded, citrate-bound, and citrate + AMP bound structures, respectively.
RESULTS
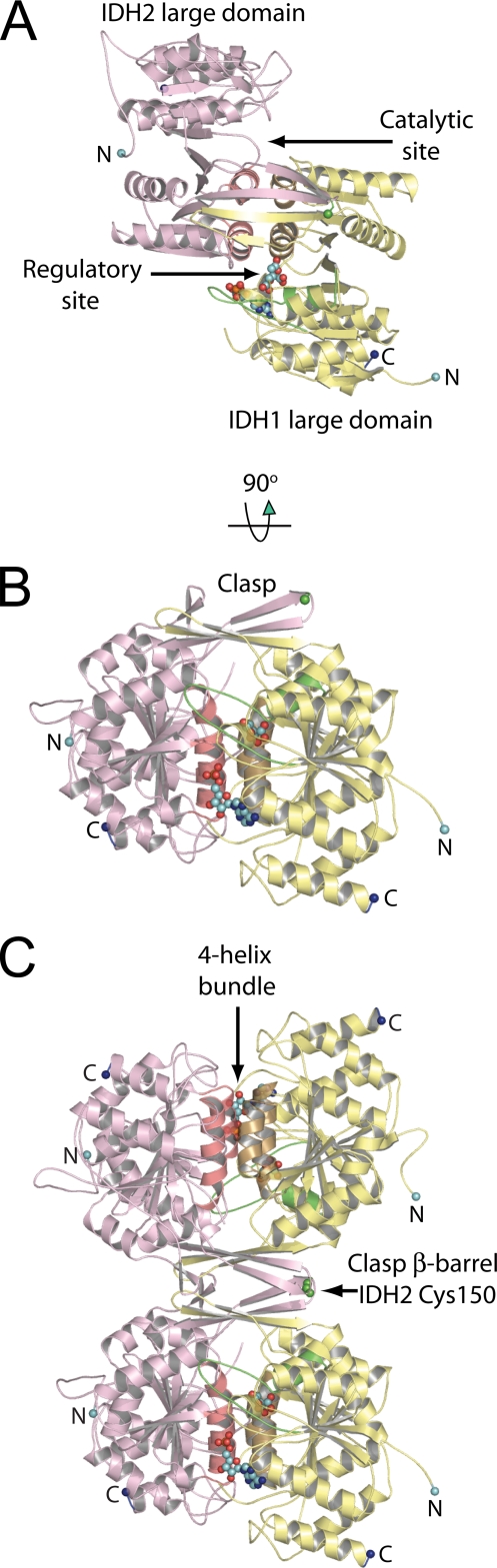
The IDH1/IDH2 Heterodimer—A structure-based multiple sequence alignment of yeast IDH1 and IDH2 subunits with selected bacterial dehydrogenases is shown in supplemental Fig. 1. Supplemental Fig. 2 shows the similarities and differences in the packing of yeast IDH hetero-octamers in the C2 and P1 crystals forms. Table 1 shows the x-ray diffraction data, MAD phasing, and protein structure refinement statistics. The overall architecture of the IDH heterodimer is shown in Fig. 1A. IDH1 and IDH2 both contain 12 β-strands, designated A-L, and 11 α-helices, designated a-k (supplemental Fig. 3). Ten of the β-strands (A-J) form a continuous, twisted, mixed β-sheet that serves as the core of both the “large” and “small” domains in each subunit. A pseudo 2-fold axis brings the g and h helices from the small domains of IDH1 and IDH2 together to form a four-helix bundle at the heterodimer interface. The ligand-binding sites are recessed into prominent clefts positioned on opposite sides of the four-helix bundle. The binding site formed in part from the large domain of IDH1 is designated as the “regulatory” site, and the site formed in part by the large domain of IDH2 is designated as the “catalytic” site. Fig. 1B shows that strands K and L protrude from the small domains of each subunit at the heterodimer interface to form a concave clasp-like structure (see below). Overall, the citrate-bound IDH structure reveals many differences relative to the unliganded IDH structure, and these differences define the molecular basis for allosteric regulation in the hetero-octamer as described below.
FIGURE 1.
The yeast IDH heterodimer and heterotetramer. The regulatory IDH1 subunits are shown in yellow and the catalytic IDH2 subunits in pink. The N and C termini are shown as cyan and dark blue spheres, respectively. Helices g and h, which form the four-helix bundle at the heterodimer interface, are shown in gold and salmon coming from IDH1 and IDH2, respectively. Citrate and AMP are shown as spheres bound to IDH1. Residues 78-92 of IDH1, which undergo the helix-loop structural transition upon ligand binding, are shown in green. A, the IDH heterodimer shown looking down the pseudo-2-fold axis of rotation. The regulatory and catalytic sites and the large domains in each subunit are labeled. B, the IDH heterodimer shown looking perpendicular to the pseudo-2-fold axis of rotation. The β-hairpins formed by β-strands K and L and comprising the clasp region are indicated. The sulfur atom of Cys-150 coming from IDH2 in the turn between β-strands K and L is shown as a green sphere. C, the yeast IDH heterotetramer. The lower heterodimer is shown in the same orientation as in B. The 2-fold axis of rotation formed at the clasp β-barrel heterotetrameric interface is horizontal in the plane of the page. The disulfide bond formed in the unliganded IDH structure by IDH2 Cys-150 residues is indicated.
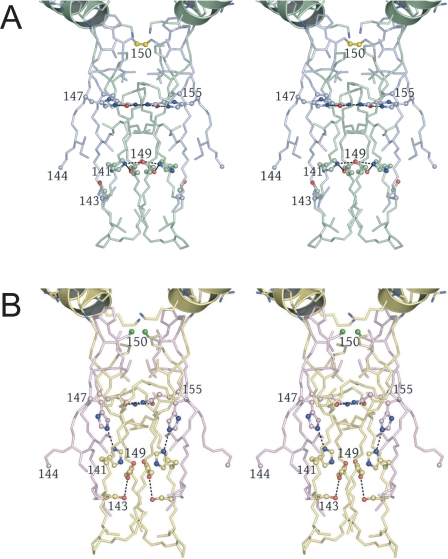
The Heterotetrameric Interface—Fig. 1C shows that the clasp regions of two IDH heterodimers associate around a molecular 2-fold axis to form a heterotetrameric interface consisting of an unusual open-ended eight-stranded β-barrel. Fig. 2A shows that the interior of the clasp β-barrel in the unliganded IDH structure contains alternating layers of polar and apolar side chains oriented approximately normal to the long axis of the barrel. The polar IDH2 His-147 and Gln-155 side chains form hydrogen bonds with their counterparts across the 2-fold of the barrel. A layer of Ile and Leu residues separate these polar side chains from a second hydrogen-bonded polar layer consisting of symmetry-related IDH1 His-141, Ser-143, and Glu-149 side chains. A disulfide bond, observed in the unliganded structure near the entrance to the clasp β-barrel, is formed by Cys-150 residues coming from the β-hairpins located between strands K and L in the IDH2 subunits. The disulfide covalently links the two IDH2 components of the heterodimers across the heterotetrameric interface in the absence of ligand binding to the regulatory site. In both the unliganded and citrate-bound IDH structures, the Sγ atom of each IDH2 Cys-150 residue is centered on the long axis of an uncapped five-turn α-helix consisting of IDH1 residues 156-174. Fig. 2B shows that in the citrate-bound structure, the disulfide bond between Cys-150 residues is reduced, hydrogen bonds between polar side chains in the clasp β-barrel are broken, and the 2-fold-related IDH2 His-147 side chains adopt different rotameric conformations that disrupt the Leu- and Ile-containing apolar layer intervening between the two polar layers in the unliganded structure.
FIGURE 2.
Alterations at the clasp β-barrel formed at the heterotetrameric interface in the unliganded and citrate-bound yeast IDH structures. The interior of the barrel is formed by alternating layers of apolar and polar residues (see “Results”). Hydrogen bonds formed by polar side chains are shown as dotted lines. A, the clasp β-barrel in the unliganded yeast IDH structure. IDH1 and IDH2 subunits are shown in green and slate blue, respectively. IDH2 Cys-150 residues form a disulfide bond (bright yellow). The five-turn helices formed by IDH1 residues 157-174 are positioned such that the uncapped N termini are adjacent to the Cys-150 Sγ atoms, which are centered on the long helix axes (see “Results” and supplemental Figs. 6 and 7). B, the clasp β-barrel in the citrate-bound yeast IDH structure shown in the same orientation as in A. IDH1 and IDH2 subunits are shown in yellow and pink, respectively. The disulfide bond between IDH2 Cys-150 residues (bright green) is reduced, the side chain of IDH2 His-147 disrupts the lower adjacent apolar layer of side chains, and the remaining polar side chains form fewer hydrogen bonds than in the unliganded IDH structure.
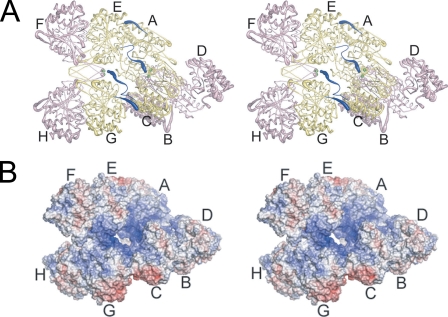
The IDH Hetero-octamer—Fig. 3A shows that two IDH heterotetramers associate via contacts in their IDH1 subunits to form the hetero-octamer. However, the hetero-octamer does not demonstrate pseudo-222 symmetry as might be expected. The relationship between heterotetramers and the deviation from pseudo-222 symmetry involve an ∼24° offset of the heterotetrameric molecular 2-fold axes from a line connecting the center of masses of the heterotetramers plus a rotation of ∼296° around this line. These deviations permit the N termini from IDH1 subunits C and E to be positioned on the inside of the solvent-filled “hole” in the hetero-octamer, whereas the N termini from IDH1 subunits A and G are positioned on the exterior of the hetero-octamer exposed to solvent. Thus, the “twist” of one heterotetramer relative to the other in the hetero-octamer permits two types of interactions between IDH1 subunits, and these interactions underlie the allosteric communication between heterotetramers (see below).
FIGURE 3.
The yeast IDH hetero-octamer does not obey pseudo-222 symmetry. Regulatory IDH1 subunits are shown in yellow and catalytic IDH2 subunits in pink. The N termini of IDH1 subunits are shown in dark blue, and the Sγ atoms of IDH2 Cys-150 residues are shown as bright green spheres. The orientation of the leftmost heterotetramer in each panel is in the same orientation as in Fig. 1C. The thickness of the tubes is proportional to the thermal parameters for the backbone atoms, with thicker tubes representing higher atomic displacement parameters. A, the yeast IDH hetero-octamer with citrate bound. Note that the N termini of two IDH1 subunits run along the interior of the central solvent-filled cavity of the hetero-octamer and the remaining two IDH1 N termini are positioned on the exterior of the hetero-octamer. The interior IDH1 N termini from each heterotetramer interact reciprocally with the Cys-150 residues of theβ-barrel clasp of the other heterotetramer in the ligand-bound but not in the unliganded IDH structures (see “Results”). The 2-fold axis of rotation relating heterotetramers runs normal to a line running from ∼1:00 to 7:00 o'clock in the plane of the page. B, electrostatic surface potential calculated at ±10 kT for the citrate-bound yeast IDH hetero-octamer. The orientation is the same as shown in panel A and in Fig. 1C. Basic amino acid side chains coming from the uncapped IDH1 helices consisting of residues 156-173 and the proximity of the N-terminal amino group of IDH1 chain C and E contribute to a substantial positive electrostatic potential (blue shading) at IDH2 residues Cys-150 (see “Results”).
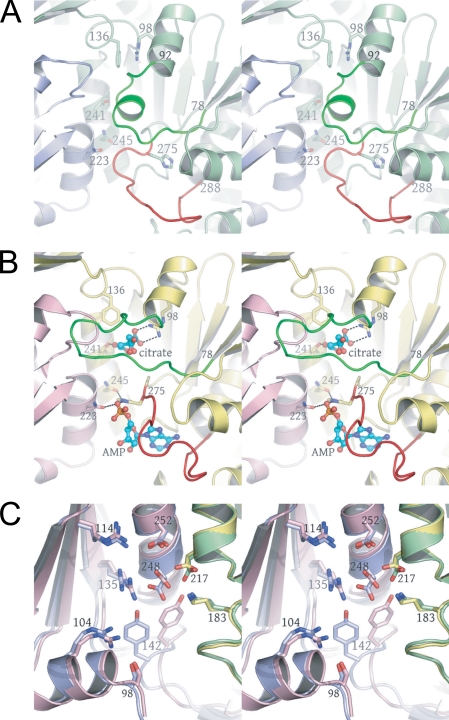
Structural Transitions upon Ligand Binding in the Heterodimer—Fig. 4, A and B, shows the unliganded and citrate-bound structures in the IDH1 regulatory site, revealing that residues 78-92 and 275-283 in IDH1 change conformation upon ligand binding. Fig. 5A shows a difference map contoured at 3σ after rigid body refinement of the citrate-bound structure against the 4.3-Å citrate + AMP diffraction data, revealing the AMP-binding site. In the absence of citrate, residues 78-92 adopt a helical conformation such that these residues physically block the citrate-binding site. IDH1 residues 275-283 pack against this helix, blocking the AMP-binding site. Upon citrate binding, residues 78-92 “unwind” to form an extended loop positioned out of the citrate-binding site in a conformation similar to that observed for the structurally equivalent residues in the active site of the homologous homodimeric E. coli enzyme (20, 22) (see supplemental Fig. 4). This movement permits residues 275-283 to move partially into the space formerly occupied by the 78-92 helix, which concomitantly unblocks the AMP-binding site. Thus, it appears that the binding of citrate induces the largest conformational changes, and the subsequent binding of AMP acts to “prop” these sites open, preventing the reverse loop-helix transition that would again obstruct the ligand-binding sites. As shown in Fig. 4B and in supplemental Fig. 5, the binding of ligand affects several residues coming from IDH1 helix h and IDH2 helix g of the four-helix bundle, which form part of the regulatory binding site. IDH1 Thr-241 is directly involved in citrate binding, and IDH1 Asn-245 forms a hydrogen bond to IDH1 His-275 in the liganded structure, but not in the unliganded structures. Both Thr-241 and Asn-245 are positioned on IDH1 helix h, which is in direct contact with the three other helices of the four-helix bundle. Also in the regulatory site, IDH2 residue Asp-222 forms a hydrogen bond to IDH1 Thr-88, a residue in the 78-92 helix, and IDH2 residue Asn-223 forms a hydrogen bond to the phosphate group of the AMP. These latter two residues are contributed by IDH2 helix g. The binding of citrate and AMP in the regulatory site therefore is likely communicated through the four-helix bundle at the heterodimer interface to the active site. Indeed, previous mutagenesis and kinetic studies (35) have demonstrated that the four-helix bundle plays a role in allostery in yeast IDH.
FIGURE 4.
The regulatory and catalytic sites in the unliganded (green and slate blue) and citrate-bound (yellow and pink) yeast IDH heterodimers. Amino acid residues undergoing a conformational change upon citrate binding are shown in bright green and red. Citrate and AMP are shown as ball-and-stick, and the side chains of residues involved in ligand binding are shown as sticks alone. The AMP-binding site was identified in difference electron density maps after rigid body refinement of the citrate-bound IDH structure against the 4.3 Å citrate + AMP diffraction data (see “Experimental Procedures” and Fig. 5A). A, regulatory IDH1-binding site in the unliganded IDH structure. Note the position of the green helix corresponding to residues 78-92 and the large red loop corresponding to residues 275-285. These residues block both the citrate- and AMP-binding sites, respectively. B, regulatory IDH1-binding site in the citrate-bound IDH structure. Upon citrate binding, the green residues undergo a helix-loop transition that causes a shift in the position of the red loop that in turn opens the AMP-binding site (see “Results”). C, catalytic IDH2-binding sites with the same color coding as shown in A. The side chains of catalytically invariant Arg-104, Arg-114, Arg-135, Tyr-142, Lys-183, Asp-217, Asp-248, and Asp-252 are shown as ball-and stick.
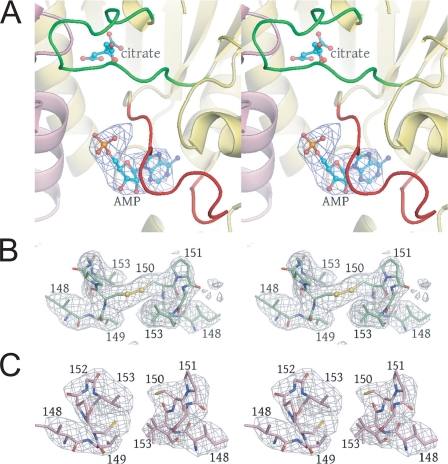
FIGURE 5.
Difference and annealed omit electron density superimposed on key regions of IDH. A, the AMP-binding site is revealed by Fo - Fc electron density contoured at 3σ after rigid body refinement of the citrate-bound IDH structure against the 4.3 Å citrate + AMP diffraction data. The orientation is approximately the same as that shown in Fig. 4, A and B. B, annealed omit map contoured at 3σ calculated for the unliganded IDH structure in which IDH2 residues 148-153 were removed from the phase calculations. The electron density reveals an oxidized disulfide bond between symmetry-related IDH2 Cys-150 residues. C, annealed omit map contoured at 4σ calculated for the citrate-bound IDH structure in which IDH2 residues 148-153 were removed from the phase calculations. The electron density reveals a reduced disulfide bond between symmetry-related IDH2 Cys-150 residues (see “Results”).
Fig. 4C shows the superposition of catalytic sites from the unliganded and citrate-bound yeast IDH2 structures. The most significant difference in the unliganded and citrate-bound yeast IDH catalytic sites is found in Tyr-142, where the α-carbon atom positions differ by 3.1 Å and the side chain hydroxyl oxygen atom positions differ by 5.6 Å. In the E. coli IDH structure, the analogous residue is Tyr-160, the side chain hydroxyl group of which makes a hydrogen bond to one of the substrate carboxylate groups and to the guanidinium nitrogen of Arg-153, the arginine residue equivalent to Arg-135 in IDH2. The ability to form these hydrogen bonds has been shown to be critical for proper substrate binding, as a Y160F mutant is inactive (36). It should be noted that citrate is not bound in the catalytic site in these structures. The absence of citrate in the catalytic site is presumably due to an alternate hydrogen bonding pattern with active site amino acid side chains relative to isocitrate, the low concentrations of Mg2+ (which is required for (iso)citrate binding at the catalytic site) (11), the absence of NAD+ during crystallization, or a combination of the above factors. The presence of isocitrate, Mg2+, and NAD+ in the catalytic site would likely have an influence on the position of Tyr-142.
Communication of Ligand Binding to the Clasp β-Barrel—As shown in Fig. 4, A and B, the loop-helix transition of IDH1 residues 78-92 causes a shift in the connected helix comprising residues 92-102. The Cα of Arg-98, a key residue in the binding of citrate (and isocitrate), shifts 3 Å, such that in the unliganded structure, the guanidinium moiety of Arg-98 makes a stacking interaction with the aromatic ring of Phe-136. These shifts are propagated from Phe-136 through Thr-155, which form strands K and L of the IDH1 contribution to the clasp β-barrel. Because these residues are intimately associated with the other β-hairpins in the clasp β-barrel, they result in the alterations in the heterotetrameric interface shown in Fig. 2.
Communication between IDH Heterotetramers—Fig. 3A shows that in the citrate-bound form, residues 12-18 of IDH1 chains C and E interact extensively with IDH1 Chains G and A, respectively. These N-terminal residues in chains C and E form a tight turn that inserts into a pocket in chains G and A formed by residues 74-75, 165-172, 262-266, and 301. The N-terminal amino groups of IDH1 subunits C and E bridge the distance between the two heterotetramers of the hetero-octamer, positioning the N-terminal amino group of these chains ∼4 Å from the Sγ atoms of IDH2 Cys-150 residues. In contrast, the first seven N-terminal residues of IDH1 chains C and E in the unliganded IDH structure are disordered, leaving the Cys-150 residues exposed to solvent. Fig. 3B shows that there exists a significant positive electrostatic potential near Cys-150 residues of the clasp β-barrel. This positive electrostatic potential arises in part from the uncapped helix dipoles coming from the helices shown in Fig. 2, the presence of several nearby Arg and Lys residues, and the presence of the N-terminal amino group of IDH1 chains C and E positioned ∼4 Å away. The possible significance of these differences in the local environment near Cys-150 in these structures is discussed below.
DISCUSSION
As predicted from genetic and site-directed mutagenesis studies (13, 14), IDH1 and IDH2 associate to form a heterodimer that acts as the basic building block of the heterooctameric enzyme. Because of its heteromeric composition, the yeast IDH structure is distinct at every level from those described for homodimeric or homotetrameric microbial enzymes (20, 22, 37).
The crystallographic observation that four IDH1/IDH2 heterodimers assemble to form a hetero-octamer is in agreement with the oligomeric state predicted by analytical ultracentrifugation studies (26, 38). The regulatory IDH1 subunits form the core of the hetero-octamer, whereas the IDH2 subunits are positioned on the periphery of the enzyme. As shown in Fig. 3A, the thermal parameters for the IDH2 subunits are considerably higher than the IDH1 subunits. The unexpected deviation from pseudo-222 symmetry upon assembly of heterotetramers to form the hetero-octamer results in IDH1 subunits A and G being in different local environments than IDH1 subunits C and E. The N-terminal 20 residues of IDH1 subunits C and E mediate the assembly of two heterotetramers, forming one surface of the central “hole” in the enzyme where they make extensive protein-protein interactions with IDH1 subunits A and G. In contrast, the N termini of IDH1 subunits A and G are positioned on the exterior of the enzyme, forming more tenuous protein-protein interactions with IDH1 subunits C and E at the “hinge” region between the two types of IDH1 subunits. This deviation from pseudo-222 symmetry may underlie the molecular basis for previous perplexing binding data indicating that, although there are theoretically eight isocitrate, four AMP, and four NAD+-binding sites in the hetero-octamer, the enzyme exhibits binding to only half of the sites for each ligand when saturated (10, 11, 39). In this context, the observed binding of AMP to all four regulatory sites in the citrate + AMP structure could be an artifact of crystallization, as only two AMP-binding sites in the IDH holoenzyme are predicted by ligand-binding analyses of the soluble enzyme.
We note that the mammalian enzyme contains one type of catalytic subunit and two types of regulatory subunits (18). We speculate that the two types of regulatory subunits found in the mammalian enzyme reflect the two distinct local environments of the IDH1 subunits in yeast IDH. However, it is unclear whether the mammalian enzyme also exhibits the property of half-site ligand binding, because original studies were performed under the assumption of a tetrameric structure for the enzyme (40). More recent studies have confirmed that the human enzyme is also a hetero-octamer (15, 19).
The structural transitions revealed in Fig. 4, A and B, are consistent with the previous observation that the binding of isocitrate must precede the binding of AMP in the regulatory IDH1 subunit (10, 11). The binding of isocitrate results in a helix-loop structural transition that opens the AMP-binding site. The binding of AMP stabilizes this “open” conformation of the enzyme, preventing the reverse loop-helix structural transition. The structural changes in the IDH1 regulatory binding site are transmitted to the clasp β-barrel at the heterotetrameric interface, where the positions of IDH2 Cys-150 and His-147 and IDH1 His-141 and Ser-143 side chains are altered substantially from their positions in the unliganded IDH structure. Our observation of an oxidized disulfide bond between IDH2 Cys-150 residues in the unliganded IDH structure and a reduced disulfide bond in the citrate-bound structure suggests that oxidation and reduction of this disulfide bond may be associated with ligand binding by the enzyme. Alternatively, the presence of the disulfide bond may stabilize a form of IDH with very low affinity for isocitrate, as well as for other ligands, because isocitrate is a prerequisite for binding of both AMP and NAD+ (10, 11). Such a scenario may represent an “on-off” switch for yeast IDH enzymatic activity analogous to the on-off behavior of prokaryotic IDH enzymes in which a critical active site serine residue is reversibly phosphorylated (21).
The Sγ atom of each IDH2 Cys-150 residue sits on the long axis of the five-turn α-helix e coming from its cognate IDH1 subunit in the heterodimer. α-Helices are typically “capped” in protein structures by the side chains of acidic residues at helix N termini and by the side chains of basic residues at helix C termini (42). However, as shown in Fig. 2 and in supplemental Figs. 6 and 7, the IDH1 e-helices are uncapped and begin with an unusual trio of residues: Arg-156, Pro-157, and Lys-158. The positive electrostatic potential coming from these basic amino acid residues is predicted to substantially lower the pKa of the Cys-150 side chain and to stabilize the thiolate.
Supplemental Figs. 6 and 7 also show that, in the unliganded structure, the Cys-150 residues are “sunken” into one end of the symmetrical clasp barrel and are surrounded on three sides by apolar side chains, including those from IDH2 Val-149, Val-153, and Pro-151 and IDH1 Val-153. The N-terminal positions of IDH2 Pro-157 in the e-helices also partially shield the Cys-150 residues from solvent. The partially apolar environment may help to stabilize the disulfide bond. This spatial arrangement of structural elements leaves a single unidirectional avenue of access to these Cys-150 residues, which is along the heterotetramer 2-fold facing the other heterotetramer of the hetero-octamer. As shown in supplemental Fig. 7, in the citrate-bound structure, these single avenues of access to Cys-150 residues in each heterotetramer are occupied by the N termini of IDH1 chains C and E with the nitrogen of the NH+3 group residing only ∼4 Å from one of the Cys-150 Sγ atoms, placing yet an additional positive charge adjacent to this atom. This arrangement likely stabilizes the thiolate and disfavors the disulfide bond. In test helical peptides, cysteine thiolates proximal to the N terminus have been reported to be stabilized through both charge and hydrogen bond interactions (43). Taking all of the above into consideration, it appears that the local environment of these Cys-150 residues is such that the redox potential of the disulfide bond may be tuned to go either to the oxidized or the reduced form. The exact mechanism of how Cys-150 is oxidized and reduced in this context is unclear, as the source of electrons for the reduction of the disulfide bond is unknown.
Support for the notion that this disulfide bond is part of the allosteric regulation of yeast IDH enzymatic activity comes from kinetic analyses of C150S and C150A mutants of IDH and of the wild type enzyme in the presence and absence of dithiothreitol. The reduced wild type and the Cys-150 mutant enzymes exhibit a 30-40% increase in activity and an approximate 50% decrease in cooperativity with respect to isocitrate (i.e. a reduction in Hill coefficients from ∼4 to 2). Dithiothreitol and the residue substitutions have no substantial effect on the overall affinity of the enzyme for isocitrate, nor do they affect allosteric properties with respect to AMP activation.7 Thus, it appears that the disulfide bond is important for limiting maximal catalytic activity and for cooperative interactions between half of the isocitrate-binding sites. This scenario seems realistic, because both kinetic and structural data suggest that loss or reduction of the disulfide bond would result in a more active state of the enzyme and would likely reduce or eliminate cooperative communication between regulatory isocitrate-binding sites. The existence of allosteric disulfide bonds has been predicted (44), but there are currently few examples. Thus, yeast IDH may be a model for examining mechanisms for oxidation and reduction of such bonds and the subsequent effects on metabolism and allostery.
The Cys-150 residue of IDH2 appears to be unique among most eukaryotic homologues. In fact, even among other yeast species for which genome sequence information is available, only those closely related to Saccharomyces cerevisiae (in a group denoted Saccharomyces sensu stricto) (45, 46) contain a cysteine at the residue position equivalent to Cys-150 in IDH2. Interestingly, this group is metabolically unique in that these yeasts exhibit a strong repression of respiratory metabolism in the presence of hexoses even if oxygen is available (47). Thus, it is tempting to speculate that formation of a disulfide bond in S. cerevisiae IDH, which would substantially reduce ligand binding, might be a cellular mechanism for rapidly reducing flux through the tricarboxylic acid cycle when a hexose is made available for degradation. Such regulation would be analogous to phosphorylation and inactivation of E. coli IDH, which occurs upon introduction of acetate to the medium and results in a redirection of net carbon flux from the tricarboxylic acid cycle into the biosynthetic glyoxylate cycle (41).
Several other aspects of the regulation of yeast IDH remain to be elucidated. For example, binding of isocitrate to catalytic sites requires Mg2+, and binding of NAD+ requires isocitrate plus Mg2+ (10, 11). Also unclear is the structural basis for AMP activation by increasing apparent affinity for isocitrate. These elements of ordered ligand binding and allostery are currently being studied in our laboratories via structural analyses of the enzyme in complexes with various ligands.
Supplementary Material
Acknowledgments
We thank Dr. Stephen Holloway for help with the figures and the staff at the Advanced Light Source beamline 5.0.2, the staff at the National Synchrotron Light Source beamline X29, and Dr. Duilio Cascio for valuable discussions. Support for the X-ray Crystallography Core Laboratory by the Executive Research Council at the University of Texas Health Science Center at San Antonio is also gratefully acknowledged.
The atomic coordinates and structure factors (codes 3BLX, 3BLV, and 3BLW) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by National Institutes of Health Grant 5R01GM051265 (to L. M.-H.) and Robert A. Welch Foundation Grant AQ-1399 (to P. J. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1-7.
Footnotes
The abbreviations used are: IDH, isocitrate dehydrogenase; MAD, multiwavelength anomalous diffraction; MR, molecular replacement; CHES, 2-(cyclohexylamino)ethanesulfonic acid.
R. Hirose, M. Sakurai, T. Suzuki, H. Moriyama, T. Sato, A. Yamagishi, T. Oshima, and N. Tanaka, unpublished observations.
A.-P. Lin, J. Garcia, A. B. Taylor, P. J. Hart, and L. McAlister-Henn, unpublished observations.
References
- 1.Chen, R. F., and Plaut, G. W. (1963) Biochemistry 2 1023-1032 [DOI] [PubMed] [Google Scholar]
- 2.Plaut, G. W. (1970) Curr. Top. Cell. Regul. 2 1-27 [Google Scholar]
- 3.Barnes, L. D., McGuire, J. J., and Atkinson, D. E. (1972) Biochemistry 11 4322-4329 [DOI] [PubMed] [Google Scholar]
- 4.Gabriel, J. L., Zervos, P. R., and Plaut, G. W. (1986) Metabolism 35 661-667 [DOI] [PubMed] [Google Scholar]
- 5.Sols, A., Gancedo, C., and DelaFuente, G. (1971) in The Yeasts, (Rose, A. H., and Harrison, J. S., eds) 1st Ed., Vol. 2, pp. 271-307, Academic Press, London [Google Scholar]
- 6.Cupp, J. R., and McAlister-Henn, L. (1991) J. Biol. Chem. 266 22199-22205 [PubMed] [Google Scholar]
- 7.Cupp, J. R., and McAlister-Henn, L. (1992) J. Biol. Chem. 267 16417-16423 [PubMed] [Google Scholar]
- 8.Keys, D. A., and McAlister-Henn, L. (1990) J. Bacteriol. 172 4280-4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cupp, J. R., and McAlister-Henn, L. (1993) Biochemistry 32 9323-9328 [DOI] [PubMed] [Google Scholar]
- 10.Lin, A. P., and McAlister-Henn, L. (2002) J. Biol. Chem. 277 22475-22483 [DOI] [PubMed] [Google Scholar]
- 11.Lin, A. P., and McAlister-Henn, L. (2003) J. Biol. Chem. 278 12864-12872 [DOI] [PubMed] [Google Scholar]
- 12.Lin, A. P., McCammon, M. T., and McAlister-Henn, L. (2001) Biochemistry 40 14291-14301 [DOI] [PubMed] [Google Scholar]
- 13.Zhao, W. N., and McAlister-Henn, L. (1997) J. Biol. Chem. 272 21811-21817 [DOI] [PubMed] [Google Scholar]
- 14.Panisko, E. A., and McAlister-Henn, L. (2001) J. Biol. Chem. 276 1204-1210 [DOI] [PubMed] [Google Scholar]
- 15.Kim, Y. O., Oh, I. U., Park, H. S., Jeng, J., Song, B. J., and Huh, T. L. (1995) Biochem. J. 308 63-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nichols, B. J., Hall, L., Perry, A. C., and Denton, R. M. (1993) Biochem. J. 295 347-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols, B. J., Perry, A. C., Hall, L., and Denton, R. M. (1995) Biochem. J. 310 917-922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramachandran, N., and Colman, R. F. (1980) J. Biol. Chem. 255 8859-8864 [PubMed] [Google Scholar]
- 19.Soundar, S., Park, J. H., Huh, T. L., and Colman, R. F. (2003) J. Biol. Chem. 278 52146-52153 [DOI] [PubMed] [Google Scholar]
- 20.Hurley, J. H., Dean, A. M., Koshland, D. E., Jr., and Stroud, R. M. (1991) Biochemistry 30 8671-8678 [DOI] [PubMed] [Google Scholar]
- 21.Hurley, J. H., Dean, A. M., Sohl, J. L., Koshland, D. E., Jr., and Stroud, R. M. (1990) Science 249 1012-1016 [DOI] [PubMed] [Google Scholar]
- 22.Imada, K., Sato, M., Tanaka, N., Katsube, Y., Matsuura, Y., and Oshima, T. (1991) J. Mol. Biol. 222 725-738 [DOI] [PubMed] [Google Scholar]
- 23.Ceccarelli, C., Grodsky, N. B., Ariyaratne, N., Colman, R. F., and Bahnson, B. J. (2002) J. Biol. Chem. 277 43454-43462 [DOI] [PubMed] [Google Scholar]
- 24.Xu, X., Zhao, J., Xu, Z., Peng, B., Huang, Q., Arnold, E., and Ding, J. (2004) J. Biol. Chem. 279 33946-33957 [DOI] [PubMed] [Google Scholar]
- 25.Hendrickson, W. A., Horton, J. R., and LeMaster, D. M. (1990) EMBO J. 9 1665-1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnes, L. D., Kuehn, G. D., and Atkinson, D. E. (1971) Biochemistry 10 3939-3944 [DOI] [PubMed] [Google Scholar]
- 27.Hendrickson, W. A. (1991) Science 254 51-58 [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski, Z., and Minor, W. (1997) in Methods in Enzymology (Carter, C. W., Jr., and Sweet, R. M., eds) Vol. 276, pp. 306-326, Academic Press, New York [Google Scholar]
- 29.Rossmann, M. G., and Blow, D. M. (1962) Acta Crystallogr. 15 24-31 [Google Scholar]
- 30.McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C., and Read, R. J. (2005) Acta Crystallogr. Sect. D Biol. Crystallogr. 61 458-464 [DOI] [PubMed] [Google Scholar]
- 31.de La Fortelle, E., and Bricogne, G. (1997) in Methods in Enzymology (Carter, C. W., Jr., and Sweet, R. M., eds) Vol. 276, pp. 472-494, Academic Press, New York [DOI] [PubMed] [Google Scholar]
- 32.Terwilliger, T. C. (2000) Acta Crystallogr. Sect. D Biol. Crystallogr. 56 965-972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 34.Adams, P. D., Grosse-Kunstleve, R. W., Hung, L. W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K., and Terwilliger, T. C. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1948-1954 [DOI] [PubMed] [Google Scholar]
- 35.Hu, G., and McAlister-Henn, L. (2006) Arch. Biochem. Biophys. 453 207-216 [DOI] [PubMed] [Google Scholar]
- 36.Lee, M. E., Dyer, D. H., Klein, O. D., Bolduc, J. M., Stoddard, B. L., and Koshland, D. E., Jr. (1995) Biochemistry 34 378-384 [DOI] [PubMed] [Google Scholar]
- 37.Miyazaki, J., Asada, K., Fushinobu, S., Kuzuyama, T., and Nishiyama, M. (2005) J. Bacteriol. 187 6779-6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson, S. L., Schirf, V., and McAlister-Henn, L. (2002) Biochemistry 41 7065-7073 [DOI] [PubMed] [Google Scholar]
- 39.Kuehn, G. D., Barnes, L. D., and Atkinson, D. E. (1971) Biochemistry 10 3945-3951 [DOI] [PubMed] [Google Scholar]
- 40.Ehrlich, R. S., and Colman, R. F. (1981) J. Biol. Chem. 256 1276-1282 [PubMed] [Google Scholar]
- 41.Walsh, K., and Koshland, D. E., Jr. (1985) J. Biol. Chem. 260 8430-8843 [PubMed] [Google Scholar]
- 42.Richardson, J. S., and Richardson, D. C. (1988) Science 240 1648-1652 [DOI] [PubMed] [Google Scholar]
- 43.Kortemme, T., and Creighton, T. E. (1995) J. Mol. Biol. 253 799-812 [DOI] [PubMed] [Google Scholar]
- 44.Schmidt, B., Ho, L., and Hogg, P. J. (2006) Biochemistry 45 7429-7433 [DOI] [PubMed] [Google Scholar]
- 45.Piskur, J., and Langkjaer, R. B. (2004) Mol. Microbiol. 53 381-389 [DOI] [PubMed] [Google Scholar]
- 46.Dujon, B. (2005) Curr. Opin. Genet. Dev. 15 614-620 [DOI] [PubMed] [Google Scholar]
- 47.Pronk, J. T., Steensma, H. Y., and Van Dijken, J. P. (1996) Yeast 12 1607-1633 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.