Abstract
Oxidation of low density lipoprotein (LDL) occurs in vivo and significantly contributes to the development of atherosclerosis. An important mechanism of LDL oxidation in vivo is its modification with 12/15-lipoxygenase (LO). We have developed a model of minimally oxidized LDL (mmLDL) in which native LDL is modified by cells expressing 12/15LO. This mmLDL activates macrophages inducing membrane ruffling and cell spreading, activation of ERK1/2 and Akt signaling, and secretion of proinflammatory cytokines. In this study, we found that many of the biological activities of mmLDL were associated with cholesteryl ester (CE) hydroperoxides and were diminished by ebselen, a reducing agent. Liquid chromatography coupled with mass spectroscopy demonstrated the presence of many mono- and polyoxygenated CE species in mmLDL but not in native LDL. Nonpolar lipid extracts of mmLDL activated macrophages, although to a lesser degree than intact mmLDL. The macrophage responses were also induced by LDL directly modified with immobilized 12/15LO, and the nonpolar lipids extracted from 12/15LO-modified LDL contained a similar set of oxidized CE. Cholesteryl arachidonate modified with 12/15LO also activated macrophages and contained a similar collection of oxidized CE molecules. Remarkably, many of these oxidized CE were found in the extracts of atherosclerotic lesions isolated from hyperlipidemic apoE–/– mice. These results suggest that CE hydroperoxides constitute a class of biologically active components of mmLDL that may be relevant to proinflammatory activation of macrophages in atherosclerotic lesions.
There is now considerable evidence that oxidized low density lipoprotein (LDL)2 exists in vivo and that LDL oxidation enhances the atherogenicity of LDL (1). Among several mechanisms suggested to explain how LDL is oxidized in vivo, 12/15-lipoxygenase (LO) has been proposed to play a major role (2–7). The family of 12/15LO enzymes is conserved between various animal and plant species and includes human 15LO, mouse 12/15LO (both expressed in macrophages), soybean lipoxygenase, and others (8, 9). The classic in vitro reaction of 12/15LO is the oxygenation of arachidonic acid at carbons 12 and/or 15 (hence the name of the LO enzyme). 12/15LO is capable of oxygenating not only free fatty acids (FFA) but also polyunsaturated acyl chains in phospholipids (PL) and cholesteryl esters (CE) (8). Thus, an incubation of LDL with isolated 12/15LO or with the cells expressing 12/15LO produces hydroperoxides of three classes, FFA-OOH, PL-OOH, and CE-OOH (8, 10, 11).
The importance of 12/15LO in the development of diet-induced atherosclerosis has been established in several murine models, including 12/15LO knock-out and transgenic mice (3–7). The 12/15LO, apoE double knock-out mice on a high fat diet have less atherosclerosis, significantly lower titers of autoantibodies against oxidized LDL in plasma and lower isoprostane levels in urine as compared with apoE–/– mice, indicating that 12/15LO is important in LDL oxidation in vivo (5). To model 12/15LO-induced LDL oxidation in vivo, we developed a method to modify LDL by incubating it with murine fibroblasts stably overexpressing human 15LO (15LO cells) (11–13). In this model, LDL is incubated with 15LO cells in DMEM, which minimizes any extracellular oxidation of the LDL, resulting in the generation of a minimally oxidized LDL, termed minimally modified LDL (mmLDL). mmLDL is enriched mainly with early lipid peroxidation products, such as hydroperoxides and hydroxides, as opposed to more complex and advanced products typically found in more extensively oxidized LDL. This mmLDL is biologically active; for example, it induces endothelial cells to have enhanced binding of monocytes (14, 15). It induces membrane ruffling, actin polymerization and cell spreading in macrophages (13, 16, 17). In addition, mmLDL activates phosphoinositide 3-kinase/Akt and extracellular signal regulated kinase (ERK) signaling pathways in macrophages, further resulting in secretion of proinflammatory cytokines and chemokines. Remarkably, some of the biological effects of mmLDL are mediated by toll-like receptor-4 signaling (17, 18), implying cross-talk between the innate immune response to bacterial pathogens and to modified LDL (19). We have also demonstrated that mmLDL, via activation of the Akt signaling pathway, offsets the proapoptotic effects of oxidized LDL and of free cholesterol accumulation in macrophages (20).
It has been reported that isolated 12/15LO preferentially oxygenates CE in LDL (10, 21). Accumulation of CE hydroperoxides has been documented in human atherosclerotic lesions and in the lesions of apoE–/– mice fed a high fat diet (22–24). We now demonstrate that complex CE hydroperoxides are a biologically active component of mmLDL and that similar polyoxygenated CE, as found in mmLDL, are also present in murine atherosclerotic lesions.
EXPERIMENTAL PROCEDURES
Reagents—All free fatty acid, eicosanoid, and cholesteryl ester standards for mass spectrometry were purchased from Cayman Chemical (Ann Arbor, MI). Hepoxilin B3 was purchased from Biomol (Plymouth Meeting, PA). Solvents used for liquid chromatography were of chromatography grade and purchased from OmniSolv (Gibbstown, NJ). Ammonium acetate and triethylamine used as liquid chromatography additives as well as cholesteryl arachidonate were purchased from Sigma-Aldrich. Ebselen and a borohydride resin were from Calbiochem.
LDL Isolation and Modification—LDL (density = 1.019–1.063 g/ml) was isolated from plasma of normolipidemic donors by sequential ultracentrifugation (25). Contamination of native and modified LDL preparations by endotoxin was assessed with a Limulus Amoebocyte Assay (Cambrex, Walk-ersville, MD). LDL preparations with lipopolysaccharide levels higher than 50 pg/mg protein were discarded.
To produce mmLDL, we incubated 50 μg/ml LDL in serum-free DMEM for 18 h with a murine fibroblast cell line overexpressing 15LO (12). The conditioned medium containing mmLDL was centrifuged, filtered through a 0.22-μm filter, concentrated to 1 mg/ml using a 100-kDa cut-off centrifugal concentrator (Millipore, Billerica, MA), and sterile filtered (0.22 μm). We previously documented that this procedure generates mmLDL, i.e. it binds to native LDL receptors but not to scavenger receptors (11–14, 17). mmLDL contains early lipid peroxidation products (11, 17), but it does not contain any measurable thiobarbituric acid reactive substances or EO6-reactive phospholipid oxidation products above that of “native LDL” (13). The mmLDL modification appeared to be very reproducible, and a successful modification was documented in a biological assay in which mmLDL induced membrane ruffling of J774 macrophages in cell culture (13, 17).
LDL modification with isolated 12/15LO enzymes (not cellular) has been reported previously (26). However, to avoid direct effects of the enzyme on the plasma membrane of macrophages, we immobilized commercial (Cayman Chemical, Ann Arbor, MI) 15LO (soybean) and 5LO (potato) on carboxylated magnetic beads (MagnaBind Beads from Pierce) using the manufacturer's protocol. The 15LO and 5LO beads were thoroughly washed to remove unbound enzymes and used to modify native LDL (200 μg/ml). The 15LO reaction was conducted in 2 mm borate buffer (pH 9.0) for 2 h at room temperature. The 5LO reaction was conducted in phosphate-buffered saline (pH 7.4) for 2 h at room temperature. The magnetic beads were removed, and the resulting modified LDL was diluted with DMEM to 50 μg/ml, sterile filtered (0.22 μm), tested for endotoxin, and kept at 4 °C for up to 5 days.
Oxidation of Cholesteryl Arachidonate—Arachidonic acid cholesteryl ester (AA-CE; purchased from Sigma) was reconstituted in hexane at 2.5 mg/ml and kept at –80 °C. Fifty μgof AA-CE was incubated with or without 24,000 units of 15LO (soybean) in 1 ml of buffer (20 mm Tris-HCl, 0.2 m NaCl, 20 mm deoxycholate, pH 8.5) for 24 h at room temperature. The reaction mixture was extracted with one volume of methanol and 2 volumes of chloroform supplemented with 0.01% butylated hydroxytoluene. The chloroform layer was collected and dried under argon, and the AA-CE was reconstituted in n-hexane.
LDL and Tissue Lipid Extraction—Nonpolar lipids were isolated from LDL preparations or mouse aorta homogenates (see below) using a procedure described earlier (27). In brief, 0.3 ml of mmLDL (0.05–0.5 mg/ml) was vigorously vortexed with 7.5 ml of ice-cold methanol for 15 s in a 50 ml glass tube, and then 30 ml of ice-cold n-hexane was added, and the mixture was vigorously vortexed for 1 min. Organic phases were separated by centrifugation for 5 min at 1000 rpm, and the hexane phase was collected and dried under argon. Dry lipid was immediately resuspended in absolute ethanol and stored at –80 °C no longer than 48 h. We confirmed that this storage did not affect the stability nor resulted in de novo formation of oxidized CE moieties. In some experiments, after collecting the hexane layer, we also collected the aqueous/methanol layer and extracted it with 2 volumes of chloroform. This fraction of LDL contained polar lipids. Independently, FFA were analyzed in LDL total lipid extracts, obtained with acidified chloroform/methanol extraction as described (28).
Reverse Phase Liquid Chromatography of CE—High performance liquid chromatography was carried out using two Shimadzu (Columbia, MD) LC-10AD high performance pumps interfaced with a Shimadzu SCL-10A controller. For cholesteryl ester analyses, separation was performed using a 2.1 mm × 250 mm Vydac (Hysperia, CA) reverse phase C18 column (catalog number 201TP52) equipped with a guard column (Vydac; catalog number 201GD52T) held at 35 °C. LC buffer A was water/tetrahydrofuran (50/50, v/v) containing 5 mm ammonium acetate; buffer B was tetrahydrofuran. Gradient elution was achieved using 100/0 A/B at 0 min and linearly ramped to 55/45 A/B by 15 min. A/B was linearly ramped back to 100/0 by 17 min and held there until 25 min to achieve column re-equilibration. The buffer flow rate was 0.3 ml/min. Fifteen μl of sample was injected onto the column using a Leap Technologies (Carrboro, NC) PAL autosampler. The liquid chromatography effluent was coupled to a mass spectrometer (see below) for further analysis.
Reverse Phase Liquid Chromatography of FFA—The same pump/controller described above was used. Separation was performed using a 2.1 mm × 100 mm Waters (Taunton, MA) XTerra MS reverse phase C18 column (catalog number 186000404) equipped with a guard column (Waters; catalog number 186000632) held at 35 °C. LC buffer A was water/acetonitrile (90/10, v/v) containing 10 mm triethylamine; buffer B was acetonitrile/isopropyl alcohol (50/50, v/v) containing 10 mm triethylamine. Gradient elution was achieved using 100/0 A/B at 0 min and linearly ramped to 40/60 A/B by 16 min. A/B was linearly ramped back to 100/0 by 18 min and held there until 28 min to achieve column re-equilibration. The buffer flow rate was 0.2 ml/min. Five μl of sample was injected onto the column using the same autosampler described above. The liquid chromatography effluent was coupled to a mass spectrometer (see below) for further analysis.
Mass Spectrometry—All of the mass spectral analyses were performed using an Applied Bioscience (Foster City, CA) 4000 QTRAP hybrid triple quadrupole linear ion trap mass spectrometer operated in multiple reaction monitoring mode (see below for more detail) and equipped with a Turbo V ion source. For cholesteryl ester analyses, the Turbo V ion source was operated in positive electrospray mode using the following mass spectrometer settings: CUR, 10 p.s.i.; GS1, 50 p.s.i.; GS2, 20 p.s.i.; IS, 5500 V; CAD, high; temperature, 525 °C; ihe, ON; DP, 60 V; EP, 15 V; and CXP, 10 V. The voltage used for collisional activated dissociation (collisional energy) for all cholesteryl ester molecular species was 25 V. For FFA analyses, the Turbo V ion source was operated in negative electrospray mode using the following mass spectrometer settings: CUR, 10 p.s.i.; GS1, 40 p.s.i.; GS2, 0 p.s.i.; IS, –4200 V; CAD, high; temperature, 425 °C; ihe, ON; DP, –30 V; EP, –15 V; and CXP, –15 V. The voltage used for collisional activated dissociation (collisional energy) for all FFA molecular species was –30 V.
CE cations were formed through molecular ammonium adduction (CE+NH4)+. Fragmentation of the CE cations yielded the same fragment ion regardless of the CE moiety, the cholesterol portion of the molecule minus its OH group located on carbon number 3 (C3-OH) having a mass-to-charge ratio (m/z) of 369 (C27 H45)+; no other fragment ions were observed. A specialized mode of tandem mass spectrometry, which was used in this work, is multiple reaction monitoring (MRM). In MRM mode the first filtering section of the mass spectrometer (Q1) is set to pass ions of a specific m/z; these ions are fragmented in a collision cell (q2), and the second filtering section of the mass spectrometer (Q3) is set to pass fragment ions having a specific m/z. An instrument method is created to detect specified MRM pairs (e.g. 691/369 for cholesteryl arachidonate, 667/369 for cholesteryl linoleate, 723/369 for HpETE-CEs, etc.). The first member of the MRM pair is the m/z of the CE cation, and the second member is the m/z of their corresponding fragment ion, in these examples always the cholesterol cation. An MRM method can monitor very many species, according to their specific MRM pair, in a single relatively short analysis. The mass spectrometer cycles through all the MRM pairs repeatedly from the beginning of the analysis to the end (one complete cycle for as many as 100 MRM pairs can be accomplished in as little as 3 s, so a 16-min analysis would repeat through more than 300 cycles). By coupling a liquid chromatographic separation to the mass spectrometer and employing the MRM mode, the mass spectrometer is utilized as a highly selective and highly sensitive HPLC detector. A similar MRM method was created for FFA analysis, as we previously reported (29).
Testing Biological Activity of mmLDL, Lipid Extracts, and 15LO-modified LDL and AA-CE—A mouse macrophage cell line J774A.1 was used to test biological effects of mmLDL and other compounds. The J774 cells were maintained in 10% heat-inactivated fetal bovine serum/DMEM supplemented with 50 μg/ml gentamicin. Native LDL or mmLDL were added to J774 macrophages at a concentration of 50 μg/ml in serum-free DMEM. When extracted LDL lipid components were tested, native LDL was used as a carrier, and the results were compared with the samples containing the native LDL alone. In the case of 15LO-modified AA-CE, the lipid in hexane (no more than 0.5% hexane in aqueous solution) was added directly to the cell growth media. Cytoskeletal changes were observed 15 min after the addition of mmLDL or other compounds. The cells were fixed with 3.7% formaldehyde and stained for F-actin with 1 μm TRITC-phalloidin (Sigma) and for nucleus with 2 μg/ml Hoechst 33342 (Sigma). The images were captured using a DeltaVision deconvolution microscopic system operated by SoftWorx software (Applied Precision, Issaquah, WA) as described (16). Phosphorylation of signaling proteins was tested 15 min following the stimulation of J774 cells. The cell lysates were subjected to SDS-PAGE and blotted to a polyvinylidene difluoride membrane and probed with the antibodies against phospho-Akt (Ser473) and phospho-ERK1/2 (Thr202/Tyr204) (Cell Signaling Technology, Danvers, MA). The concentration of MIP-2 in cell culture media was assayed 5 h after the stimulation using a DuoSet enzyme-linked immunosorbent assay kit from R & D Systems (Minneapolis, MN), as we reported previously (18). The data shown are means ± standard deviation of quadruplicates (technical duplicates of two biological replicates). The experiments were repeated two to five times, and representative images and graphs are shown.
Isolation of Mouse Aorta—All of the animal experiments were performed according to National Institutes of Health guidelines and were approved by the University of California, San Diego Animal Subjects Committee. The mice were euthanized with CO2, chilled on ice, opened from abdomen to thorax, and bled via vena cava. Immediately, aortas were perfused via a canula inserted into the left ventricle, with ice-cold phosphate-buffered saline containing 2 μm EDTA at pH 7.4 until the eluate became clear (30). The entire aorta was dissected from the proximal ascending aorta, including the three carotids, to the bifurcation of the iliac artery using a dissecting microscope. Minor branching arteries (e.g. intercostal arteries) were cut off, and the adventitia was thoroughly and carefully removed in situ. The aorta was divided into three subsections: the arch section, defined as the ascending aorta to the third rib including visual parts of the three carotids in the chest cavity; the abdominal section, defined as from the diaphragm to 3-mm distal to the iliac bifurcation; and the thoracic section, defined as between the arch and abdominal sections without including the renal arteries. The average wet weight of the arch, thoracic, and abdominal sections for the apoE–/– mice (n = 2; 50-week old males fed a 1.25% cholesterol and 21% milk fat atherogenic diet for the last 9 weeks) were 7.5, 3.8, and 4.6 mg, respectively. The arch and thoracic segments from the age-matched C57BL/6 control mice (n = 3; 51 weeks old) weighed on average 3.1 mg. The C57BL/6 aorta segments were combined prior to lipid extraction to make one arch (9.4 mg) and one thoracic (9.4 mg) pool. Aorta segments were homogenized on ice in 100 μlof ice-cold distilled H2O(3 × 5 s; PowerGen 125 equipped with a Generator 7 × 95 mm probe; Fisher), and lipids were extracted with methanol and n-hexane as described above. The harvest and extraction of aortic material was carried out quickly in the cold, and the resulting lipid extracts were stored at –80 °C and analyzed within 24 h after extraction.
RESULTS
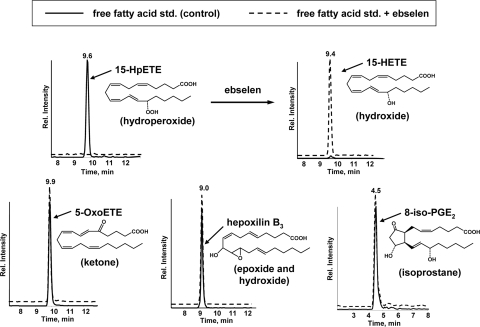
Ebselen Specifically Reduces Lipid Hydroperoxides—Because the mmLDL was modified by exposure to cells overexpressing 15LO, we initially examined the hypothesis that biologically active components of mmLDL were nonreduced lipid hydroperoxides, originating from the lipoxygenase activity. To provide evidence for this, we examined whether the biological effects of mmLDL could be diminished by reducing the lipid hydroperoxides present in mmLDL. Borohydride is a commonly used reducing agent but is known to reduce a wide variety of oxidized groups. Ebselen is a seleno-organic mimetic of glutathione peroxidase (31) that has been widely used as a hydroperoxide reducing agent. However, we were unable to find literature exploring whether ebselen is capable of also reducing other oxidation products that might be formed on unsaturated fatty acids, such as epoxides or isoprostanes. Thus, we tested whether ebselen can reduce 15-HpETE, 5-OxoETE, hepoxilin B3, and 8-iso-PGE2 (Fig. 1). Only the hydroperoxide, 15-HpETE, was reduced by ebselen, and the reaction produced the corresponding hydroxide, 15-HETE. The ketone, epoxide, hydroxide, and isoprostane were resistant to ebselen. Thus, ebselen appears to specifically reduce hydroperoxides under the conditions used.
FIGURE 1.
Ebselen sensitivity of oxygenated fatty acids. Chromatographs of FFAs were monitored using the mass spectrometer in MRM mode (the x axis shows chromatographic retention time). For each FFA analysis, ∼15 pmol of standard (5 μl of a 3 μm solution) was loaded onto the column. To test for ebselen sensitivity, ebselen (10 μm) was added to the FFA solutions for 1 h at 22 °C. Note that for the two chromatographs in the top of the figure, MRM pairs allowed both 15-HpETE and 15-HETE to be observed, should either compound be present. 15-HpETE, 15-hydroperoxy-5Z,8Z,11Z,13E-eicosatetraenoic acid; 15-HETE, 15-hydroxy-5Z,8Z,11Z,13E-eicosatetraenoic acid; 5-OxoETE, 5-oxo-6E,8Z,11Z,14Z-eicosatetraenoic acid; hepoxilin B3, 10-hydroxy-11(S),12(S)-trans-epoxyeicosa-5Z,8Z,14Z-trienoic acid; 8-iso-PGE2, 9-oxo-11R,15S-dihydroxy-5Z-13E-(8β)-prostadienoic acid.
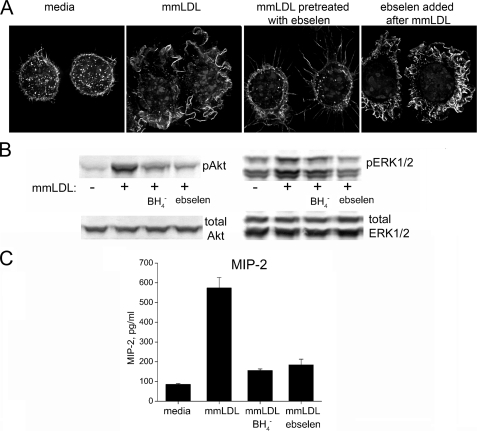
Ebselen Reduces Biological Effects of mmLDL—We previously reported that mmLDL induces macrophage spreading and membrane ruffling, phosphorylation of signaling kinases, Akt and ERK1/2, and secretion of proinflammatory cytokines (13, 17, 18). Pretreatment of mmLDL with ebselen abolished the mmLDL-induced membrane ruffling in J774 macrophages (Fig. 2A), and this was not due to a direct effect of ebselen on the cells because an addition of ebselen immediately after mmLDL did not affect membrane ruffling (far-right panel in Fig. 2A). Similarly, both ebselen and borohydride reduced the mmLDL-induced Akt and ERK1/2 phosphorylation, as well as the secretion of MIP-2 (Fig. 2, B and C). Thus, the sensitivity to ebselen implies that some of the biological activities of mmLDL are due to lipid hydroperoxides. We then focused on the mmLDL lipid components that were sensitive to reduction by ebselen.
FIGURE 2.
Reducing agents borohydride and ebselen diminish the biological effects of mmLDL. mmLDL was preincubated with or without ebselen (50 μm) or a borohydride resin for 1 h at 37 °C and then added to J774 macrophages. A, membrane ruffling. At the end of a 15-min incubation, the cells were fixed and stained for F-actin, and fluorescent images were captured as described under “Experimental Procedures.” To rule out a direct effect of ebselen on macrophages, ebselen was first added to the cells, followed by the addition of mmLDL (far-right panel). Extensive membrane ruffling in this sample suggests that the ebselen inhibitory effect was due to reducing hydroperoxide components of mmLDL and not due to any direct effect on the cells. B, Akt and ERK1/2 phosphorylation. At the end of a 15-min incubation, the cells were lysed, and the lysates were tested for the levels of phosphorylated and total Akt and ERK1/2. C, cytokine secretion. The cells were incubated with nonreduced or reduced mmLDL for 5 h, and the cell culture media were tested by enzyme-linked immunosorbent assay for the presence of MIP-2; presented as the means ± S.E.
Cholesteryl Ester Profile of mmLDL and Native LDL—Initially, we performed an HPLC-MS/MS survey of free fatty acids for the presence of oxidized FFA products but found no obvious differences between mmLDL and native LDL (not shown). We next focused on oxidized CEs, which we and others have reported to be generated in abundance in LDL modified with 12/15LO (10, 11, 21). We used a hexane-based method of lipid extraction (see “Experimental Procedures”), which favors the extraction of neutral lipids, CE and TG, whereas FFA and PL remain in the methanol/aqueous phase. The neutral lipid extract was then subjected to HPLC-MS/MS.
We analyzed these extracts by HPLC-MS/MS using MRM monitoring to look for expected CE oxidation products of cholesteryl arachidonate and linoleate, the two most common CEs found in LDL. As described under “Experimental Procedures,” fragmentation of the CE cations yielded only one fragment ion regardless of the CE moiety, the cholesterol portion of the molecule minus its C3-OH group having a m/z of 369. There was no evidence for the presence of oxysterols. Therefore, we limited our calculation of MRM pairs to predicted changes in arachidonate and linoleate but not in the sterol portion of CE. Possible oxidation products were predicted from an examination of the KEGG metabolic maps for AA and linoleic acid (Kyoto Encyclopedia of Genes and Genomes) and from a literature search as detailed under “Experimental Procedures.” Using the m/z value of 369 for the sterol moiety, we then calculated MRM pairs for theoretically expected oxidized arachidonate and linoyleate products based on the number of double bonds and the types of functional groups that might be generated as explained above (Table 1).
TABLE 1.
MRM pairs for oxygenated CEs used in this study
| CEs |
|---|
| Arachidonic acid |
| 707/369 |
| 4DB, 1 hydroxy (HETE) |
| 3DB, 1 epoxy (EpETrE) |
| 721/369 |
| 4DB, 1 hydroxy, 1 epoxy |
| 3DB, 1 keto, 1 epoxy |
| 3DB, 2 epoxys |
| 723/369 |
| 4DB, 1 hydroperoxy (HpETE) |
| 4DB, 2 hydroxy (DiHETE) |
| 737/369 |
| 2DB, cyclopentane, 1 epoxy, 1 ketone, 1 hydroxy |
| 4DB, 1 hydroperoxy, 1 epoxy |
| 739/369 |
| 4DB, 1 hydroperoxy, 1 hydroxy |
| 4DB, 3 hydroxys |
| 755/369 |
| 4DB, 1 hydroperoxy, 2 hydroxy |
| Linoleic acid |
| 683/369 |
| 2DB, 1 hydroxy (HODE) |
| 1DB, 1 epoxy (EpOME) |
| 697/369 |
| 2DB, 1 hydroxy, 1 epoxy |
| 1DB, 2 epoxys |
| 699/369 |
| 2DB, 1 hydroperoxy (HpODE) |
| 2DB, 2 hydroxy (DiHODE) |
| 1DB, 1 epoxy, 1 hydroxy (HEPO) |
| 713/369 |
| 2DB, 1 hydroperoxy, 1 epoxy |
| 2DB, 1 hydroperoxy, 1 keto |
| 715/369 |
| 2DB, 1 hydroperoxy, 1 hydroxy |
| 1DB, 1 hydroperoxy, 1 epoxy |
| 1DB, 1 hydroperoxy, 1 keto |
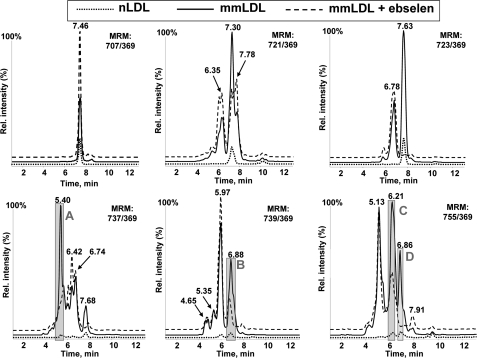
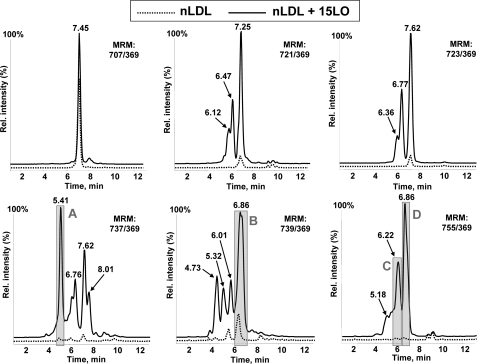
Analyzing the neutral lipid extracts for oxidized cholesteryl arachidonate (using six MRM pairs) and for oxidized cholesteryl linoleate (using five MRM pairs), we found that mmLDL was significantly enriched with oxygenated CE as compared with native LDL (Fig. 3 and supplemental Fig. S1). In addition to the parent mass indicated on each panel, the LC retention time (x axis in Fig. 3 and supplemental Fig. S1) informed us of the relative hydrophobicity of the molecules. In reverse phase LC, longer retention times indicate more hydrophobic molecules, and generally, hydroperoxides are slightly more hydrophobic than corresponding hydroxides. We found that mmLDL carried many hydrophilic CEs with increased masses, indicating the presence of polyoxygenated CEs.
FIGURE 3.
Oxygenated cholesteryl arachidonates in mmLDL and their reduction by ebselen. Native LDL, mmLDL, and mmLDL + ebselen (50 μm ebselen, incubated for 1 h at 37°C with mmLDL) were extracted with hexane:methanol as described under “Experimental Procedures” and analyzed by LC-MS using MRM pairs from Table 1. Base-line intensities of the MS signal are artificially altered to visually separate individual traces.
We also examined neutral lipid extracts of mmLDL incubated with ebselen. Remarkably, several of the peaks were reduced by the ebselen treatment, suggesting the presence of CE hydroperoxides. For example, the peak m/z 723/369, 7.63 min, corresponding to a mono-hydroperoxide CE (HpETE-CE), was completely reduced by ebselen (Fig. 3). Correspondingly, peak m/z 707/369, 7.46 min, corresponding to a mono-hydroxide CE (HETE-CE), was significantly increased, consistent with ebselen reduction of a hydroperoxide to its corresponding hydroxide. A similar trend was observed with the ebselen reduction of a HpODE-CE, yielding HODE-CE in supplemental Fig. S1. Ebselen also reduced several peaks with higher masses and higher hydrophilicity, both in cholesteryl arachidonate and linoleate products (see gray-shaded peaks A–E in Fig. 3 and supplemental Fig. S1). This result suggests the presence of polyoxygenated hydroperoxy-CE in mmLDL.
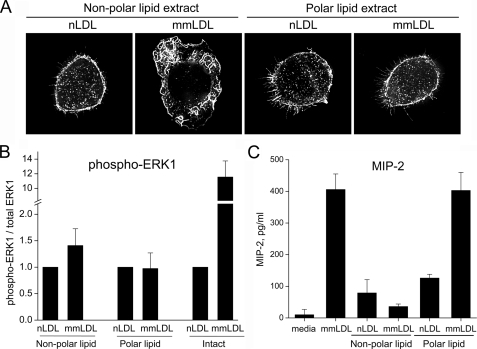
Biological Activity of Lipid Extracts of mmLDL—The nonpolar and polar lipid extracts (see “Experimental Procedures”) of mmLDL or native LDL were preincubated with intact native LDL and then added to J774 macrophage cells. We found that incorporating the lipid extracts into LDL provided an effective mode to present the oxidized lipids to cells and allow study of their biological effects. The nonpolar lipid extract of mmLDL, but not the polar lipid, induced macrophage membrane ruffling and spreading (Fig. 4A). In contrast, neither the polar or nonpolar extracts of native LDL had any effects. Phosphorylation of ERK1/2 was marginally stimulated by nonpolar lipid extracts of mmLDL (Fig. 4B). In contrast to the cytoskeletal changes, the polar lipids of mmLDL, but not the nonpolar lipid extracts, stimulated MIP-2 secretion (Fig. 4C).
FIGURE 4.
Biological activity of nonpolar lipid extracts of mmLDL. Native and mmLDL were extracted with hexane:methanol as described under “Experimental Procedures.” The hexane layer (nonpolar lipids) was collected, dried under argon, and reconstituted with serum-free DMEM containing 50 μg/ml native LDL. The remaining aqueous/methanol layer was re-extracted with chloroform. The chloroform layer (polar lipids) was dried under argon and reconstituted. mmLDL and native LDL (without the addition of lipid extracts) served as positive and negative controls, respectively. Membrane ruffling (A), ERK1 phosphorylation (B), and MIP-2 secretion (C) were assayed as in Fig. 2. The ERK1 phosphorylation (normalized to total ERK1) was quantified in three independent experiments and presented as the means ± S.E.
Biological Activity and HPLC-MS Characterization of LDL Modified by Immobilized 12/15LO—Next, we tested whether LDL oxygenation by isolated 12/15LO enzyme produced a modified LDL with an oxidized CE profile and biological properties similar to that of mmLDL. We have previously reported that intracellular 12/15LO plays an important role in the cytoskeleton regulation in macrophages and in phagocytosis of apoptotic cells (16, 17). Therefore, when testing the effect of 12/15LO-modified LDL on macrophages, to avoid direct effect of the 12/15LO itself on the cells, we needed to remove the enzyme from the modified LDL preparation. For this purpose, we immobilized soybean 15LO on magnetic beads and used the 15LO beads to modify LDL. We confirmed the biological activity of the 15LO beads by showing that the 15LO beads induced linoleic acid oxidation and diene formation as measured by UV absorption at 234 nm (not shown). In contrast to 15LO, 5LO does not directly oxidize LDL, and thus the 5LO beads (and bovine serum albumin-coated beads) were used as a negative control.
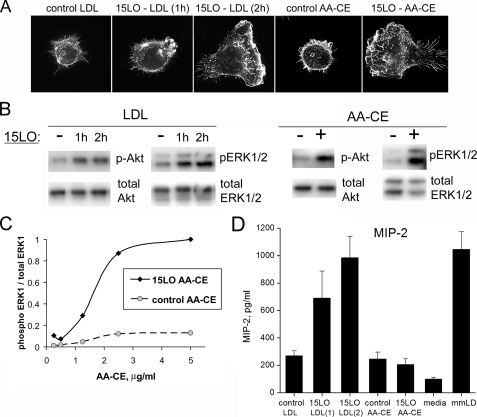
The 15LO-modified LDL stimulated macrophage membrane ruffling and cytoskeletal changes resembling lamelipodia formation at the leading edge of a migrating cell in a time-dependent manner. (Fig. 5A). After a 1-h incubation with 15LO, modified LDL produced a minimal degree of ruffling in macrophages, whereas a 2-h incubation was sufficient to produce a modified LDL with the biological activity comparable with that of mmLDL. This was accompanied by Akt and ERK1/2 phosphorylation (Fig. 5B). In addition, 15LO-modified LDL induced MIP-2 secretion by macrophage (Fig. 5C). The 5LO-modified LDL did not activate macrophages (not shown).
FIGURE 5.
Biological effects of native LDL and AA-CE modified by 15LO. Native LDL (200 μg/ml) was incubated with 15LO (soybean) immobilized on magnetic beads (see “Experimental Procedures”) for 1 and 2 h at room temperature. A control sample was incubated for 2 h with the beads coated with bovine serum albumin. The resulting modified LDL was separated from 15LO beads, diluted in DMEM to 50 μg/ml, and added to J774 cells. AA-CE (50 μg) was incubated in the presence or absence of 24,000 units 15LO (soybean) for 24 h at room temperature (see “Experimental Procedures” for detailed protocol), and then extracted with chloroform/methanol and 2.5 μg (A, B, and D) of control or 15LO-modified AA-CE was added in 1 ml of 10% fetal bovine serum/DMEM to J774 cells. A, membrane ruffling. B, ERK1/2 and Akt phosphorylation. C, cells were activated with control and 15LO-modified AA-CE at different concentrations (0.25–5.0 μg/ml), and the ratio of phospho-ERK1 to the total ERK1 protein in each sample was measured. D, MIP-2 secretion.
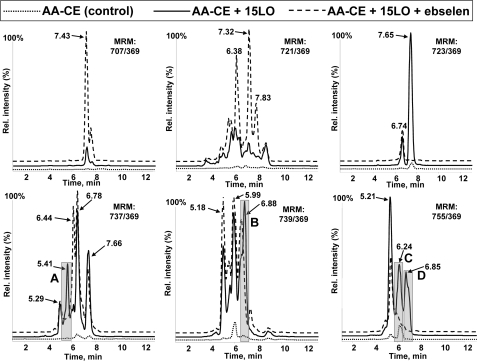
The LC-MS/MS analysis of 15LO-modified LDL demonstrated the presence of many polyoxygenated CE species found in mmLDL, with some differences in relative intensities between the peaks (compare gray-shaded peaks A–E in Fig. 6 and supplemental Fig. S2 with those in Fig. 3 and supplemental Fig. S1). The 5LO-modified LDL did not contain any detectable amounts of oxidized CE (not shown). These data confirm again that 15LO is capable of direct oxygenation of CE in LDL and also suggest that biologically active polyoxygenated CEs found in mmLDL can be directly generated by 15LO.
FIGURE 6.
Oxygenated cholesteryl arachidonate in 15LO-modified native LDL. Native LDL and 15LO-modified LDL were extracted with hexane/methanol as described under “Experimental Procedures” and analyzed by LC-MS using MRM pairs from Table 1. Base-line intensities of the MS signal are artificially altered to visually separate individual traces.
Biological Activity and HPLC-MS Characterization of 12/15LO-modified Cholesteryl Arachidonate—Because nonpolar lipid extracts from mmLDL were biologically active and a major component of these extracts was CE, we tested whether a direct CE oxidation by isolated 12/15LO produces biologically active molecules. AA-CE was incubated with or without soybean 15LO, and the lipid was extracted (purified from 15LO). The addition of 15LO-modified AA-CE induced robust J774 ruffling and spreading, as well as strong ERK1/2 phosphorylation in a dose-dependent manner (Fig. 5). The concentration of 2.5 μg/ml of 15LO-modified AA-CE was sufficient to maximally activate macrophages. The phosphorylation of Akt was less profound, and there was no MIP-2 secretion stimulated by 15LO-modified AA-CE, which is consistent with the lack of MIP-2 induction by nonpolar mmLDL extracts (Fig. 4B). The LC-MS/MS analysis of 15LO-modified AA-CE demonstrated the presence of many polyoxygenated CE species found in mmLDL and in 15LO-modified LDL, with some differences in relative intensities between the peaks (compare gray-shaded peaks A–D in Fig. 7 with those in Figs. 3 and 6). We calculated the intensities of peaks A–D in 50 μg of (protein) mmLDL and 2.5 μg of 15LO-modified AA-CE (supplemental Table S1) and found 1–4-fold differences in peaks A, B, and D. A complex lipid-protein organization of mmLDL may provide a more efficient presentation of oxidized CE and account for the concentration differences between the active component(s) in mmLDL and in 15LO-modified AA-CE that are sufficient to induce macrophage activation.
FIGURE 7.
Oxygenated cholesteryl arachidonate in 15LO-modified AA-CE. Control AA-CE and 15LO-modified AA-CE were extracted with chloroform/methanol as described under “Experimental Procedures” and analyzed by LC-MS using MRM pairs from Table 1. Base-line intensities of the MS signal are artificially altered to visually separate individual traces.
In an additional set of experiments, we tested the biological effect on macrophages of a variety of mono-oxygenated free fatty acids using HETEs, HpETEs, HODEs, HpODEs, as well as cholesterol-esterified HpODEs (all from Cayman Chemical) but did not observe noticeable macrophage activation (data not shown). These results suggest that the polyoxygenated CE species found in mmLDL and in 15LO-modified LDL are responsible for many of the biological activities induced by mmLDL.
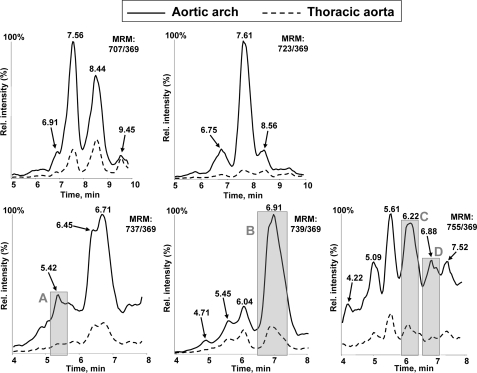
Active Components of mmLDL in Murine Atherosclerotic Lesions—Oxidized CE was previously detected in atherosclerotic lesions (22–24). We asked whether specific LC-MS/MS signatures of polyoxygenated CE hydroperoxides, characteristic for mmLDL and 15LO-modified LDL and AA-CE, could also be found in atherosclerotic lesions. In Fig. 8 and supplemental Fig. S3 we show representative LC-MS/MS analyses of the arch and the thoracic segments (see “Experimental Procedures”) of aorta from an apoE–/– mouse that was fed an atherogenic diet. The abdominal segment of the aorta from apoE–/– mice, which did not contain any visible lesions, and the arch and the thoracic segments of nonatherosclerotic C57BL6 mice did not give any MS signal in the region of the LC retention times of interest (data not shown). The aortic arch region, in which murine lesions first develop, contained many oxygenated CE species. Remarkably, we were able to detect in the murine lesions peaks that were close to or identical with peaks in mmLDL that were sensitive to reduction with ebselen, such as m/z 737/369, 5.42 min (peak A); m/z 739/369, 6.91 min (peak B); m/z 755/369, 6.22 min (peak C); and 6.88 min (peak D); and m/z 699/369, 7.76 min (peak E). These results suggest that CE hydroperoxides, the components of our model mmLDL with biological activity, are present in atherosclerotic lesions and may contribute to vascular inflammation.
FIGURE 8.
Oxygenated cholesteryl arachidonate in murine atherosclerotic lesions. Sections of aorta were isolated from high fat fed apoE–/– and chow-fed C57BL/6 mice and subjected to the lipid extraction procedure as detailed under “Experimental Procedures.” The extracts were analyzed by LC-MS using MRM pairs from Table 1. The extracts of atherosclerotic tissue contained many nonidentified peaks, and because the peaks of interest were of relatively low intensity, only the region of retention times from 4 to 8 min is shown. The extracts from chow fed C57BL/6 mice did not contain any detectable amounts of oxidized CE and are not shown in the figure.
DISCUSSION
Evidence has accumulated suggesting that 12/15LO-induced oxidation of LDL is important in the development of atherosclerosis. In this paper we used a model of biologically active minimally oxidized LDL, which was produced by the incubation of native LDL with cells overexpressing 12/15LO. In our original studies of 12/15LO cell-mediated generation of mmLDL, which were conducted in F-10 medium, we observed the progressive accumulation of CE hydroperoxides in mmLDL over time. In part, this may have reflected a degree of lipid peroxidation propagated by small amounts of transition metals present in the F-10, because EDTA partially inhibited this accumulation (11, 12). In our current mode of modifying LDL, in DMEM, such metal catalyzed propagation should not occur. However, we also observed the presence of oxidized CE in the mmLDL generated in DMEM, and many of these oxidized CE species were reduced by ebselen, suggesting that they were hydroperoxides (Fig. 3). Moreover, the fact that ebselen-treated mmLDL lost most of its biological activity (Fig. 2) led us to hypothesize that CE hydroperoxides are biologically active components of mmLDL.
To test this hypothesis we showed that when native LDL or isolated AA-CE were directly modified by 12/15-LO, they both developed many of the biological properties displayed by intact mmLDL (Figs. 4 and 5). Furthermore, nonpolar lipid extracts of mmLDL had similar activities, and HPLC-MS demonstrated similar m/z retention time peaks as found in the 12/15LO-modified LDL and CE profiles (Figs. 3, 6, and 7). Moreover, we showed a similar HPLC-MS profile of the same oxidized CE species in nonpolar extracts from murine atherosclerotic lesions as was found in the extracts of mmLDL (Fig. 8), supporting the in vivo occurrence of such oxidized CE. Indeed, accumulation of CE hydroperoxides, as a class of oxidized lipids, has been documented in human atherosclerotic lesions and in the lesions of apoE–/– mice fed a high fat diet (22–24). Our experiments demonstrated that oxidized CE are not the only biologically active lipid components of mmLDL. Polar lipid extracts of mmLDL stimulated MIP-2 secretion by macrophages (Fig. 4B), which is consistent with the results from Berliner's laboratory (32, 33) and our own, demonstrating that oxidized phospholipids stimulate human endothelial cells to produce interleukin-8, the analog of murine MIP-2.
Although considerable data were developed previously to support a role for cellular 12/15LO in mediating oxidation of extracellular LDL (11, 12, 34), there was little information to suggest how this could occur. We speculated that 12/15LO produced oxidized FFA in membrane PL, because PL are known to be substrates for 12/15-LO, and an activated phospholipase A2 would then release the oxidized FFA, which would then be “seeded” onto extracellular LDL and in some cases re-esterified into PL or CE (26). Recently, Yoshimoto and co-workers (35, 36) suggested an alternative and highly plausible mechanism. According to their hypothesis, supported by convincing experimental results, LDL binds to macrophage LDL receptor-related protein-1 (LRP1), which in turn induces 12/15LO translocation from the cytosol to the cell membrane, to the site of LDL-LRP1 binding. This would be compatible with our recent demonstration that mmLDL induced a similar translocation of 12/15LO in macrophages to the cell membrane, to sites where apoptotic cells were bound (16). At the site of the LDL ·LRP1 complex, LRP1 initiates an exchange of CE between LDL and the cell, leading to 12/15LO mediated oxygenation of the CE in an LRP1-dependent manner. Further, LRP1 contributes to the efflux of oxidized CE back to the LDL particle. This mechanism agrees well with the known preferential oxygenation of CE by 12/15LO-expressing cells (11). Selective affinity of another lipoprotein receptor, scavenger receptor class-B, type I, for oxidized CE has also been demonstrated in liver (37). Thus, a likely mechanism of the mmLDL formation in vivo involves a specific oxidation of CE by cellular 12/15LO and would be consistent with our model system in which oxidized CE appear to be prominent oxidized moieties of mmLDL generated by exposure to 15LO cells. In turn, this agrees with our data demonstrating that CE hydroperoxides constitute active components of mmLDL responsible in part for its biological effects.
Because a number of different possibilities of oxygenated CEs exist for each m/z noted (for example, as noted in Table 1, an m/z of 739/369 may correspond to molecules with two different sets of functional groups located at different carbon atoms, with different stereochemistry), definitive assignment of biological activities will require further analysis and synthesis of the individual oxidized CE moieties. However, our data to date suggest that CE should be polyoxygenated to induce the macrophage responses described in this paper. In our experiments, mono-oxygenated free fatty acids HETEs, HpETEs, HODEs, and HpODEs, as well as cholesterol-esterified HpODEs did not induce noticeable macrophage activation. This is in contrast to the reports that endothelial cells can be activated by 15-HpETE (38), 12-HETE (39), and 13-HODE (40). Similarly, it was reported that CE-HpODE activated endothelial cells and induced monocyte adhesion, although air-oxidized cholesteryl linoleate, which presumably contained a more diverse spectrum of oxidation products, was significantly more active than CE-HpODE (41). The different biological activity of oxygenated FFA and CE may be a consequence of their different orientation and active moiety presentation in LDL, as we discuss below. The difference between endothelial cell and macrophage responses to oxidized FFA and CE may be that endothelial cells have an enzymatic machinery to convert mono-hydroperoxide precursors into polyoxygenated products (42), which then activate the cells in an autocrine fashion. Numerous mechanisms have been described for transcellular biosynthesis of polyoxygenated eicosanoids when a precursor, made in one cell type, transfers into another cell type where it is transformed into a final bioactive molecule (43). For example, in experiments with several co-cultured cell types, neutrophil 5LO and platelet 12LO produced lipoxin A4 (44), and endothelial cell aspirin-modified COX-2 and neutrophil 5LO produced 15-epilipoxin A4 (45), both important anti-inflammatory in vivo regulators. Separately, these cells were unable to produce lipoxins. Similar mechanisms of formation of active oxygenated lipids in endothelial cells may explain the different responses by endothelial cells and macrophages to mono-oxygenated FFA. In addition, it has been reported that 12/15LO is capable of oxygenating several carbons on the same AA molecule, producing hepoxilin A3 (46). The 12/15LO specificity of AA oxygenation may also be affected by the AA esterification to cholesterol.
Our findings of biological activity of oxidized CE in mmLDL agree with previous reports that CE hydroperoxides in a moderately oxidized LDL were responsible for the increased CD36 expression in human macrophages (47) and that CE hydroperoxides induced ERK1/2-dependent monocyte adhesion to endothelial cells (41). The fact that oxidized CE in mmLDL initiates cell signaling may seem counterintuitive because mmLDL does not bind to scavenger receptors, and it is not clear how mmLDL would be internalized within the cell. Indeed, the hydrophobic CE are immersed in the central lipid core of LDL and do not have contact with the lipoprotein surface. However, oxygenation of the fatty acid moiety of CE would make it more polar and likely enable it to be inserted into the surface layer of the modified LDL. By analogy, we have shown that the PC moiety of oxidized PL is on the surface of oxidized LDL and becomes an epitope for the natural antibody EO6 (48), as well as being a ligand for CD36 (49). Thus, the PC of 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (PAPC), present in native LDL is not recognized by EO6 or by CD36. In contrast, when LDL is oxidized and POVPC (1-palmitoyl-2-oxovaleryl-sn-glycero-3-phosphocholine) and other similar OxPAPC moieties are generated, the PC headgroup must be expressed out of the plane of the lipid surface layer, where it can be recognized by both EO6 and CD36. In a recent paper, Li et al. (50) deduced the conformation of one of the OxPAPC molecular species within a bilayer membrane using nuclear Overhauser enhancement spectroscopy. They found that in OxPAPC the distal end of the oxidized sn-2 acyl chain protrudes into the aqueous phase, outside the bilayer, as was also the case for the PC headgroup, as we suggest above. Based on these findings, we speculate that an oxygenation of CE may also change the CE orientation in mmLDL so that an oxygenated acyl chain now projects outside the lipoprotein surface and the cholesteryl residue of the CE serves as an anchor holding the molecule in the lipoprotein. If this is the case, the oxidized acyl chain of the CE may become a ligand accessible to cellular receptors and/or may further participate in oxidation chemistry to modify cell surface proteins and lipids. Further studies will be needed to determine whether oxidized CE can bind to existing or novel receptors and in turn be internalized or, conceivably, directly activate a receptor to mediate signaling resulting in biological effects. Alternatively, the surface disposition of the oxidized CE in mmLDL may simply facilitate the transfer of the OxCE into the macrophage membrane, which in turn may lead to biological effects. It is important to point out that esterified eicosanoids are significantly more stable than corresponding FFA (51).
In summary, we demonstrate that polyoxygenated CE hydroperoxides, found in vivo in atherosclerotic tissue, are biologically active components of mmLDL generated by exposure to 12/15LO-containing cells. Studies are ongoing to identify and synthesize the specific oxidized moieties responsible for the different biological effects on macrophages noted, and the mechanisms by which this occurs.
Supplementary Material
Acknowledgments
We thank Dr. Robert Murphy for many helpful discussions and expert advice.
This work was supported by LIPID MAPS Large Scale Collaborative Grant GM069338 (to R. H., E. A. D., J. L. W., and Y. I. M.), Grant HL081862 (to Y. I. M.) from the NHLBI, National Institutes of Health, Grants 0530159N (to Y. I. M.) and 0630228N (to K. H.) from the American Heart Association, and a grant from the Leducq Fondation (to J. L. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
Footnotes
The abbreviations used are: LDL, low density lipoprotein; mmLDL, minimally oxidized LDL; LO, lipoxygenase; CE, cholesteryl ester; AA, arachidonic acid; FFA, free fatty acid; PL, phospholipid(s); PC, phosphocholine; ERK, extracellular signal regulated kinase; DMEM, Dulbecco's modified Eagle's medium; HPLC, high performance liquid chromatography; MRM, multiple reaction monitoring; TRITC, tetramethylrhodamine isothiocyanate; MS/MS, tandem mass spectrometry; LRP, LDL receptor-related protein; PAPC, 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine; OxPAPC, oxidized PAPC; LC, liquid chromatography.
References
- 1.Steinberg, D., Parthasarathy, S., Carew, T. E., Khoo, J. C., and Witztum, J. L. (1989) N. Engl. J. Med. 320 915–924 [DOI] [PubMed] [Google Scholar]
- 2.Glass, C. K., and Witztum, J. L. (2001) Cell 104 503–516 [DOI] [PubMed] [Google Scholar]
- 3.Cyrus, T., Witztum, J. L., Rader, D. J., Tangirala, R., Fazio, S., Linton, M. F., and Funk, C. D. (1999) J. Clin. Investig. 103 1597–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George, J., Afek, A., Shaish, A., Levkovitz, H., Bloom, N., Cyrus, T., Zhao, L., Funk, C. D., Sigal, E., and Harats, D. (2001) Circulation 104 1646–1650 [DOI] [PubMed] [Google Scholar]
- 5.Cyrus, T., Pratico, D., Zhao, L., Witztum, J. L., Rader, D. J., Rokach, J., FitzGerald, G. A., and Funk, C. D. (2001) Circulation 103 2277–2282 [DOI] [PubMed] [Google Scholar]
- 6.Reilly, K. B., Srinivasan, S., Hatley, M. E., Patricia, M. K., Lannigan, J., Bolick, D. T., Vandenhoff, G., Pei, H., Natarajan, R., Nadler, J. L., and Hedrick, C. C. (2004) J. Biol. Chem. 279 9440–9450 [DOI] [PubMed] [Google Scholar]
- 7.Huo, Y., Zhao, L., Hyman, M. C., Shashkin, P., Harry, B. L., Burcin, T., Forlow, S. B., Stark, M. A., Smith, D. F., Clarke, S., Srinivasan, S., Hedrick, C. C., Pratico, D., Witztum, J. L., Nadler, J. L., Funk, C. D., and Ley, K. (2004) Circulation 110 2024–2031 [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto, S. (1992) Biochim. Biophys. Acta 1128 117–131 [DOI] [PubMed] [Google Scholar]
- 9.Liavonchanka, A., and Feussner, I. (2006) J. Plant Physiol. 163 348–357 [DOI] [PubMed] [Google Scholar]
- 10.Belkner, J., Stender, H., and Kuhn, H. (1998) J. Biol. Chem. 273 23225–23232 [DOI] [PubMed] [Google Scholar]
- 11.Ezaki, M., Witztum, J. L., and Steinberg, D. (1995) J. Lipid Res. 36 1996–2004 [PubMed] [Google Scholar]
- 12.Benz, D. J., Mol, M., Ezaki, M., Mori-Ito, N., Zelaan, I., Miyanohara, A., Friedmann, T., Parthasarathy, S., Steinberg, D., and Witztum, J. L. (1995) J. Biol. Chem. 270 5191–5197 [DOI] [PubMed] [Google Scholar]
- 13.Miller, Y. I., Viriyakosol, S., Binder, C. J., Feramisco, J. R., Kirkland, T. N., and Witztum, J. L. (2003) J. Biol. Chem. 278 1561–1568 [DOI] [PubMed] [Google Scholar]
- 14.Sigari, F., Lee, C., Witztum, J. L., and Reaven, P. D. (1997) Arterioscler. Thromb. Vasc. Biol. 17 3639–3645 [DOI] [PubMed] [Google Scholar]
- 15.Michelsen, K. S., Wong, M. H., Shah, P. K., Zhang, W., Yano, J., Doherty, T. M., Akira, S., Rajavashisth, T. B., and Arditi, M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10679–10684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, Y. I., Chang, M. K., Funk, C. D., Feramisco, J. R., and Witztum, J. L. (2001) J. Biol. Chem. 276 19431–19439 [DOI] [PubMed] [Google Scholar]
- 17.Miller, Y. I., Worrall, D. S., Funk, C. D., Feramisco, J. R., and Witztum, J. L. (2003) Mol. Biol. Cell 14 4196–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, Y. I., Viriyakosol, S., Worrall, D. S., Boullier, A., Butler, S., and Witztum, J. L. (2005) Arterioscler. Thromb. Vasc. Biol. 25 1213–1219 [DOI] [PubMed] [Google Scholar]
- 19.Miller, Y. I. (2005) Future Cardiol. 1 785–792 [DOI] [PubMed] [Google Scholar]
- 20.Boullier, A., Li, Y., Quehenberger, O., Palinski, W., Tabas, I., Witztum, J. L., and Miller, Y. I. (2006) Arterioscler. Thromb. Vasc. Biol. 26 1169–1176 [DOI] [PubMed] [Google Scholar]
- 21.Upston, J. M., Witting, P. K., Alleva, R., and Stocker, R. (1997) J. Biol. Chem. 272 30067–30074 [DOI] [PubMed] [Google Scholar]
- 22.Letters, J. M., Witting, P. K., Christison, J. K., Eriksson, A. W., Pettersson, K., and Stocker, R. (1999) J. Lipid Res. 40 1104–1112 [PubMed] [Google Scholar]
- 23.Upston, J. M., Niu, X., Brown, A. J., Mashima, R., Wang, H., Senthilmohan, R., Kettle, A. J., Dean, R. T., and Stocker, R. (2002) Am. J. Pathol. 160 701–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leitinger, N. (2003) Mol. Aspects Med. 24 239–250 [DOI] [PubMed] [Google Scholar]
- 25.Havel, R. J., Bragdon, J. H., and Eder, H. A. (1955) J. Clin. Investig. 34 1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parthasarathy, S., Fong, L. G., Quinn, M. T., and Steinberg, D. (1990) Eur. Heart J. 11 (Suppl. E) 83–87 [DOI] [PubMed] [Google Scholar]
- 27.Sattler, W., Mohr, D., and Stocker, R. (1994) Methods Enzymol. 233 469–489 [DOI] [PubMed] [Google Scholar]
- 28.BLIGH, E. G., and DYER, W. J. (1959) Can. J. Biochem. Physiol. 37 911–917 [DOI] [PubMed] [Google Scholar]
- 29.Harkewicz, R., Fahy, E., Andreyev, A., and Dennis, E. A. (2007) J. Biol. Chem. 282 2899–2910 [DOI] [PubMed] [Google Scholar]
- 30.Tangirala, R. K., Rubin, E. M., and Palinski, W. (1995) J. Lipid Res. 36 2320–2328 [PubMed] [Google Scholar]
- 31.Muller, A., Cadenas, E., Graf, P., and Sies, H. (1984) Biochem. Pharmacol. 33 3235–3239 [DOI] [PubMed] [Google Scholar]
- 32.Walton, K. A., Hsieh, X., Gharavi, N., Wang, S., Wang, G., Yeh, M., Cole, A. L., and Berliner, J. A. (2003) J. Biol. Chem. 278 29661–29666 [DOI] [PubMed] [Google Scholar]
- 33.Chang, M. K., Binder, C. J., Miller, Y. I., Subbanagounder, G., Silverman, G. J., Berliner, J. A., and Witztum, J. L. (2004) J. Exp. Med. 200 1359–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheidegger, K. J., Butler, S., and Witztum, J. L. (1997) J. Biol. Chem. 272 21609–21615 [DOI] [PubMed] [Google Scholar]
- 35.Takahashi, Y., Zhu, H., Xu, W., Murakami, T., Iwasaki, T., Hattori, H., and Yoshimoto, T. (2005) Biochem. Biophys. Res. Commun. 338 128–135 [DOI] [PubMed] [Google Scholar]
- 36.Zhu, H., Takahashi, Y., Xu, W., Kawajiri, H., Murakami, T., Yamamoto, M., Iseki, S., Iwasaki, T., Hattori, H., and Yoshimoto, T. (2003) J. Biol. Chem. 278 13350–13355 [DOI] [PubMed] [Google Scholar]
- 37.Fluiter, K., Sattler, W., De Beer, M. C., Connell, P. M., van der Westhuyzen, D. R., and van Berkel, T. J. (1999) J. Biol. Chem. 274 8893–8899 [DOI] [PubMed] [Google Scholar]
- 38.Sultana, C., Shen, Y., Rattan, V., and Kalra, V. K. (1996) J. Cell. Physiol. 167 477–487 [DOI] [PubMed] [Google Scholar]
- 39.Bolick, D. T., Orr, A. W., Whetzel, A., Srinivasan, S., Hatley, M. E., Schwartz, M. A., and Hedrick, C. C. (2005) Arterioscler. Thromb. Vasc. Biol. 25 2301–2307 [DOI] [PubMed] [Google Scholar]
- 40.Chen, N. G., Sarabia, S. F., Malloy, P. J., Zhao, X. Y., Feldman, D., and Reaven, G. M. (1999) Biochem. Biophys. Res. Commun. 263 718–7225 [DOI] [PubMed] [Google Scholar]
- 41.Huber, J., Boechzelt, H., Karten, B., Surboeck, M., Bochkov, V. N., Binder, B. R., Sattler, W., and Leitinger, N. (2002) Arterioscler. Thromb. Vasc. Biol. 22 581–586 [DOI] [PubMed] [Google Scholar]
- 42.Pfister, S. L., Spitzbarth, N., Zeldin, D. C., Lafite, P., Mansuy, D., and Campbell, W. B. (2003) Arch. Biochem. Biophys. 420 142–152 [DOI] [PubMed] [Google Scholar]
- 43.Folco, G., and Murphy, R. C. (2006) Pharmacol. Rev. 58 375–388 [DOI] [PubMed] [Google Scholar]
- 44.Edenius, C., Stenke, L., and Lindgren, J. A. (1991) Eur. J. Biochem. 199 401–409 [DOI] [PubMed] [Google Scholar]
- 45.Claria, J., and Serhan, C. N. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 9475–9479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nigam, S., Patabhiraman, S., Ciccoli, R., Ishdorj, G., Schwarz, K., Petrucev, B., Kuhn, H., and Haeggstrom, J. Z. (2004) J. Biol. Chem. 279 29023–29030 [DOI] [PubMed] [Google Scholar]
- 47.Jedidi, I., Couturier, M., Therond, P., Gardes-Albert, M., Legrand, A., Barouki, R., Bonnefont-Rousselot, D., and Aggerbeck, M. (2006) Biochem. Biophys. Res. Commun. 351 733–738 [DOI] [PubMed] [Google Scholar]
- 48.Friedman, P., Hörkkö, S., Steinberg, D., Witztum, J. L., and Dennis, E. A. (2002) J. Biol. Chem. 277 7010–7020 [DOI] [PubMed] [Google Scholar]
- 49.Boullier, A., Friedman, P., Harkewicz, R., Hartvigsen, K., Green, S. R., Almazan, F., Dennis, E. A., Steinberg, D., Witztum, J. L., and Quehenberger, O. (2005) J. Lipid Res. 46 969–976 [DOI] [PubMed] [Google Scholar]
- 50.Li, X. M., Salomon, R. G., Qin, J., and Hazen, S. L. (2007) Biochemistry 46 5009–5017 [DOI] [PubMed] [Google Scholar]
- 51.Yin, H., Morrow, J. D., and Porter, N. A. (2004) J. Biol. Chem. 279 3766–3776 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.