Abstract
Transcriptional activation of many genes involved in peroxisome-related functions is regulated by the Oaf1p, Pip2p, and Adr1p transcription factors in Saccharomyces cerevisiae. We have analyzed the in vivo binding characteristics of Oaf1p-Pip2p and found that this complex is recruited to its target oleate-response element (ORE) under all growth conditions tested. In addition, this complex also binds to ORE-containing genes that do not appear to be regulated by these proteins, as well as to some genes lacking conventional OREs. The recruitment of the Oaf1p-Pip2p complex was greatly increased upon glucose derepression, possibly due to Oaf1p phosphorylation with only moderate increases upon oleate induction. Thus, this complex may receive a nutritional cue while it is already bound to DNA, suggesting that, in addition to the increase in Oaf1p-Pip2p binding, other mechanism(s) such as enhanced Adr1p association may drive the expression of highly inducible fatty acid-responsive genes. Adr1p binds to target genes in an oleate-dependent fashion and is involved in Oaf1p-Pip2p binding. In turn, the Oaf1p-Pip2p complex appears to be important for Adr1p binding to a subset of oleate-responsive genes. Adr1p is a positive regulator of ORE-containing genes, but it also acts as a negative factor in expression of some of these genes. Finally, we have also shown that Adr1p is directly involved in mediating oleate induction of Oaf1p-Pip2p target genes.
The yeast Saccharomyces cerevisiae is able to use a wide variety of carbon sources to maintain vegetative growth. When fermentable sugars, such as glucose, are supplied into culture media, genes involved in utilization of less favorable nonfermentable carbon sources are turned off via a mechanism called glucose repression (1). A switch from glucose to a nonfermentable carbon source triggers transcriptional activation of these genes through another pathway, termed glucose derepression (2). Some of these glucose-derepressible genes, however, are able to undergo further induction upon receiving additional nutritional signals. For example, the genes encoding β-oxidation enzymes become significantly induced when oleate is included in glycerol-containing growth media; the addition of oleate also leads to peroxisome proliferation (3). The promoters of these genes share a common sequence motif termed oleate-response element (ORE)4 (4). This sequence is sufficient to act as an in vitro binding target for a trans-acting factor(s) responsible for oleate induction (4). We and others (5–7) identified two highly homologous Zn2Cyc6 transcription factors, Oaf1p and Pip2p, which mediate oleate-dependent transcriptional activation of ORE-containing genes. Both of these proteins are required to trigger oleate-dependent induction and can be recruited by synthetic OREs in in vitro binding assays. Based on this experimental evidence, and by analogy with mammalian steroid hormone receptors (8–10), a model in which Oaf1p and Pip2p bind to OREs of target genes in the form of a heterodimer, presumably upon receiving an oleate nutritional signal, has been proposed (5–7).
Oaf1p and Pip2p play different roles in oleate induction. Although both of these proteins possess ligand binding domains, only Oaf1p binds free fatty acids (11) and is capable of sensing a fatty acid nutritional signal (12). The different regulation of genes encoding Oaf1p and Pip2p probably reflects different roles of these proteins in oleate response, whereas OAF1 is constantly expressed under all growth conditions, PIP2 is tightly controlled by an autoregulation mechanism (5, 6, 13, 14).
In an earlier study we conducted a global search for ORE-containing genes and identified 40 genes that have oleate response-like elements in their promoter regions (5). We demonstrated that more than 20 genes, encoding proteins with various subcellular locations, are regulated by the Oaf1p-Pip2p transcription factors. These include a number of highly inducible genes, as well as some genes that are only moderately induced by oleate. Subsequently, several genome-wide searches for ORE-containing and oleate-inducible genes were carried out, and most of the yeast genes that are significantly up-regulated by fatty acids have been identified (14–16). Interestingly, some of the Oaf1p-Pip2p-regulated genes lack a consensus ORE (5, 14). The minimal ORE is now defined as CGGN3TN(A/R)N8–12CCG (17).
In addition to the ORE, genes encoding peroxisomal proteins often contain a binding site for a sensor of less favored carbon sources, the C2H2 zinc-finger protein Adr1p (18–21). This sequence, referred to as type 1 upstream activation sequence (UAS1), is defined as CYCCR(A/T/GN4–36(T/A/C)YGGRG and is often found in close proximity to, or overlapping with, an ORE in the Oaf1p-Pip2p-regulated genes (17, 21, 22). Adr1p, originally identified as a regulator of the glucose-repressible gene ADH2 (23, 24), was later shown to be required for growth on oleic acid as a sole carbon source (18, 25). Adr1p regulates peroxisome-related target genes in two ways, directly via binding to the resident UAS1 sequences, and also through mediating expression of PIP2 (18–21). Adr1p target genes include the key peroxisome-proliferation peroxin gene PEX11, and thus peroxisome proliferation is also under Adr1p control (19). The binding of Adr1p to peroxisome-related genes has been demonstrated both in vitro with synthetic UAS1 targets under oleate induction (18–21) and in vivo using chromatin immunoprecipitation (ChIP) assays in the absence of glucose (26–28). Recently, Adr1p-regulated genes and Adr1p-binding sites have been identified using genome-wide global analyses; however, there have been no such analyses performed under oleate induction conditions (26–31).
In recent years, chromatin immunoprecipitation in combination with quantitative PCR has become a powerful and widely used tool for detecting the associations of regulatory proteins with specific DNA sequences in vivo (32, 33). Although a number of genome-wide location analyses using ChIP followed by DNA microarray (ChIP on chip) assays have been completed to date (27, 29, 30, 34), none of them focused specifically on Oaf1p-Pip2p interactions with their target sequences, and there have been no such studies addressing regulation under fatty acid-inducing conditions. In this study, we have examined the binding characteristics of Oaf1p and Pip2p to their target sequences under three different growth conditions, including oleate induction, in vivo. We confirmed that Oaf1p requires Pip2p for binding and that these proteins act as a heterodimer on their binding sites under all growth conditions tested. We also demonstrated that, in contrast to the current view, the amount of Oaf1p-Pip2p complex bound to the ORE is significantly increased upon glucose derepression and increases only moderately upon oleate induction. Furthermore, we provide evidence that Oaf1p undergoes phosphorylation when cells are grown on glycerol- or oleate-containing media, and this post-translational modification may cause an increased Oaf1p-Pip2p binding under these conditions. Thus, Oaf1p most likely receives a fatty acid nutritional signal while it is already bound to DNA, therefore receiving such a signal does not cause the initial binding of Oaf1p-Pip2p to the ORE, as was previously thought. These data also suggest that the increase in Oaf1p-Pip2p binding upon a shift from glycerol to oleate is not sufficient to achieve maximal induction of highly inducible oleate-responsive genes, and therefore there must be an additional mechanism(s) involved in up-regulation of these genes. We have also demonstrated that Adr1p binds to its target sequences under derepression and induction growth conditions, and this binding appears to be oleate-dependent. Our findings indicate that Adr1p is involved in Oaf1p-Pip2p binding, and this complex, in turn, stimulates Adr1p recruitment to the promoters of a subset of genes. In addition, we determined that Adr1p is not only required for achieving maximal expression levels, but it also contributes to oleate induction of the Oaf1p-Pip2p target genes. Finally, we have shown that Adr1p, being a positive regulator of oleate induction of peroxisome-related genes, plays an additional role in maintaining low expression levels of some oleate-responsive genes under glucose derepression conditions (negative regulation).
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Conditions
The yeast strains used in this study are listed in Table 1. Yeast strains were grown in YPD, YPG, or YPGO media as described previously (5, 6, 35).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Ref. |
|---|---|---|
| W3031A | MATaleu2 ura3 trp1 ade2 his3 | 51 |
| OAF1-HA3 | MATaleu2 ura3 [pRS306-URA3-OAF1-HA3] trp1 ade2 his3 oaf1::HIS3 | This study |
| PIP2-HA3 | MATaleu2 pip2::LEU2 ura3 trp1 [pRS304-TRP1-PIP2-HA3] his3 | This study |
| PIP2-HA3/OAF1-myc13 | MATaleu2 pip2::LEU2 ura3 trp1 [pRS304-TRP1-PIP2-HA3] his3 oaf1:: [OAF1-13Myc-TADH1-HIS5] | This study |
| ΔΔOAF1-HA3 | MATaleu2 pip2::LEU2 ura3 [pRS306-URA3-OAF1-HA3] trp1 his3 oaf1::HIS3 | This study |
| ΔΔPIP2-HA3 | MATaleu2 pip2::LEU2 ura3 trp1 [pRS304-TRP1-PIP2-HA3] his3 oaf1::HIS | This study |
| ADR1-myc13 | MATaleu2 ura3 trp1 ade2 his3 adr1:: [ADR1-13Myc-TADH1-HIS5] | This study |
| ΔADR1 | MATaleu2 ura3 trp1 adr1::13Myc-TADH1-TRP1 ade2 his3 | This study |
| PIP2-HA3/ΔADR1 | MATaleu2 pip2::LEU2 ura3 trp1 [pRS304-TRP1-PIP2-HA3] his3 adr1::13Myc-TADH1-HIS5 | This study |
| ΔPIP2/ADR1-myc13 | MATaleu2 pip2::LEU2 ura3 trp1 adr1:: [ADR1-13Myc-TADH1-TRP1] his3 | This study |
| ΔO1ΔP2/ADR1-myc13 | MATaleu2 pip2::LEU2 ura3 trp1 adr1:: [ADR1-13Myc-TADH1-TRP1] his3 oaf1::HIS3 | This study |
| OAF1-HA3/ADR1-myc13 | MATaleu2 ura3 [pRS306-URA3-OAF1-HA3] trp1 adr1:: [ADR1-13Myc-TADH1-TRP1] ade2 his3 oaf1::HIS3 | This study |
| OAF1-HA3/ΔADR1 | MATaleu2 ura3 [pRS306-URA3-OAF1-HA3] trp1 adr1::13Myc-TADH1-TRP1 ade2 his3 oaf1::HIS3 | This study |
| ΔOAF1/ADR1-myc13 | MATaleu2 ura3 trp1 oaf1::13Myc-TADH1-TRP1 ade2 his3 adr1:: [ADR1-13Myc-TADH1-HIS5] | This study |
| PIP2-HA3/OAF1-myc13/ΔADR1 | MATaleu2 pip2::LEU2 ura3 adr1Δ::URA3 trp1 [pRS304-TRP1-PIP2-HA3] his3 oaf1:: [OAF1-13Myc-TADH1-HIS5] | This study |
| MCY2692OAF1-HA3 | MATaleu2 ura3 [pRS306-URA3-OAF1-HA3] snf1-K84R | This study |
| ΔO1ΔP2 | MATaleu2 pip2::LEU2 ura3 trp1 his3 oaf1::HIS3 | 5 |
Epitope Tagging
OAF1-HA3—The nine-amino acid epitope of influenza virus hemagglutinin (HA) was used for C-terminal tagging the OAF1 gene product in our previous studies (5). To create a plasmid that contains both Oaf1p-HA3 and the URA3 selectable marker, a DNA fragment encoding the OAF1-HA3 allele was subcloned into pRS306 (36) to create pRS306OAF-1HA3. This plasmid was then linearized and integrated into the URA3 locus of the appropriate recipient yeast cells.
PIP2-HA3—Three tandem copies of the HA epitope were added to the C terminus of the PIP2 gene product directly upstream from the stop codon, using a combination of PCR and subcloning techniques. The resulting DNA fragment encoding PIP2-HA3 was subcloned into pRS304 to create pRS304PIP-2HA3. This plasmid was then linearized and integrated into the TRP1 locus of our pip2Δ recipient strain.
OAF1-myc13 and ADR1-myc13—Thirteen copies of the 10-amino acid human c-Myc epitope were integrated into the OAF1 or ADR1 chromosomal loci using pF6a-13Myc-TRP1 or pF6a-13MycHis3-MX6 heterologous modules for PCR-based gene targeting (37).
Gene Disruption by PCR-based Gene Targeting
PCR-based gene targeting approach was also used to disrupt the OAF1 gene in the ADR1-myc13 strain using pF6a-13Myc-TRP1 as a template. Similarly, the ADR1 gene was disrupted in our wild-type strain (see Table 1). To disrupt the ADR1 gene in the PIP2-HA3/OAF1-myc13 strain, a URA3 template was used instead of the pF6-based cassettes. The correct integration of the cassettes was verified by diagnostic PCR.
ChIP
The procedures described below for single and sequential ChIPs are modifications of the methods of Aparicio et al. (32) and Geisberg et al. (38), respectively. All buffers and stock solution recipes were taken from original protocols, except where described below.
Lysate Preparation—Yeast cultures were grown in 200 ml of YPD, YPG, or YPGO as was described previously (5). Cells were subsequently cross-linked in the media, harvested, and washed as described (32). The fixed cells were resuspended in 6–10 ml of FAPI buffer (FA buffer supplemented with complete EDTA-free protease inhibitor mixture (Roche Applied Science) and 1 mm phenylmethylsulfonyl fluoride). 1-ml aliquots of this suspension were placed in 2-ml Eppendorf tubes, and ∼1 ml of glass beads (diameter ∼ 0.5 mm) was added to every sample. The suspensions were incubated, with shaking, for 1.5–2 h at 4 °C. The cell lysates were collected together and centrifuged as described (32). The resultant pellets, containing cross-linked chromatin, were resuspended in 4 ml of FAPI buffer, and the volume was brought to 5 ml if required. The suspension was sonicated on ice 20 times for 30 s using continuous pulse with 1-min intervals at a power output of ∼15% using a W-385 sonicator (Heat Systems-Ultrasonics, Farmingdale, NY). Following sonication, the lysate was split into 2-ml Eppendorf tubes and centrifuged in a refrigerated microcentrifuge at maximum speed for 30 min. The recovered supernatants were pooled and passed through a 0.22-μm syringe filter. A 100-μl sample was then taken to confirm DNA fragmentation (32), and the volume of the remaining lysate was adjusted to 5 ml with FAPI buffer. The lysate was divided into 500-μl aliquots placed in Eppendorf tubes, frozen in liquid nitrogen, and stored at –80 °C. Before using the lysate in ChIP assays, it was titrated to determine the amount of respective antibody required to precipitate between 50 and 90% of the antigen in question as described (32).
Single ChIP Assay—For an individual single ChIP, 250 –500 μl of the lysate was taken into an Eppendorf tube, and the volume was brought to 1 ml with FAPI buffer. A 50-μl sample was taken to determine the amount of DNA present in the assay (input sample), and a 30-μl sample was taken to determine the amount of antigen in the assay (T-sample). These samples were processed as described (32). Upon addition of antibody, the tube was incubated at 4 °C for 2 h on an Orbitron Rotator. 50 μl of protein G-agarose beads (Roche Applied Science) were then added, and the incubation was continued overnight. The sample was washed, eluted with 100 μl of the appropriate peptide elution buffer, and reverse cross-linked as described (32). DNA was then purified using phenol/chloroform extraction followed by ethanol precipitation with GlycoBlue carrier (Ambion, Austin, TX) as described (32) and was then dissolved in 40–100 μl of TE as required. A 30-μl supernatant sample (S-sample) was taken before washing and run along with the T-sample onto an 8% SDS-PAGE followed by ECL Western blotting to determine the percentage of the antigen precipitated by ChIP. The input samples were reverse cross-linked as described (32), purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA) according to the manufacturer's instructions, and eluted in 200 μl. Both ChIP and input samples were stored at –20 °C.
Sequential ChIP Assay—Sequential ChIPs were performed in both the forward and reverse direction. For this purpose, 1–2 ml of cross-linked lysates was taken for each direction. 50-μl input samples were processed as described above. The first ChIP samples were set up and then carried out as described above for single chips. After elution with the corresponding peptide, 10 μl of the first ChIP eluates was taken into a PCR tube containing 90 μl of TE, 100 μl of ChIP elution buffer, and 20 μl of Pronase. The samples were subjected to reverse cross-linking and then purified as described (38). The samples were resuspended in 20–100 μl of TE and stored at –20 °C as “first ChIP.” To set up second ChIPs, the remaining 90 μl of the first ChIP eluate were mixed with 600 μl of FABPR buffer (FA buffer containing 5 mg/ml bovine serum albumin, 25 μg/ml λ phage DNA (New England Biolabs, Beverly, MA)) and 50 μg/ml E. coli tRNA (Roche Applied Science), and second antibody (50% of the amount used above) was added to the samples. The second ChIP samples were incubated for 2 h and then 50 μl of protein G-agarose beads were added, and incubation was continued overnight. The samples were washed, eluted, and purified as was described above for the single ChIPs. The purified samples, “second ChIP,” were stored at –20 °C.
RNA Purification
Total yeast RNA was purified according to a hot phenol extraction procedure as described (5). mRNA was prepared from total yeast RNA using Oligotex milk as specified by the manufacturer (Qiagen, Valencia, CA). Before use in the real time RT-PCR assays, both yeast total and mRNA were liberated from DNA using the DNA-free™ kit (Ambion Inc., Austin, TX) according to the manufacturer's instructions.
Quantitatative Real Time PCR
Real time PCR was performed using the LightCycler 2.0 Instrument (Roche Applied Science) and LightCycler software version 4.0. The primers were designed using LightCycler Probe Design 2 software based on the sequences of the specific genes downloaded from the yeast genome data base. All oligonucleotide sequences are available upon request. The amplification programs were designed according to manufacturer's recommendations. 10-μl reactions were set up in 20-μl LightCycler capillaries (Roche Applied Science). DNA (ChIPs and inputs) samples were amplified using either LightCycler Fast-Start DNA MasterPLUS SYBR Green I kit (Roche Applied Science) or homemade 2× SYBR Green Taq mix (32). The single ChIP data were processed using Microsoft Office Excel software and the following formula: Chip% = (2Cp input–Cp chip) × Fd × 100, where Chip% is the ChIP efficiency expressed in percent when compared with total DNA; Cp chip and Cp input are crossing points (threshold cycles) for ChIP and input samples, respectively; and Fd is the dilution factor. The relative occupancy was calculated as the fold enrichment of the gene-specific DNA over a reference gene DNA (GPD1 or POX1). For sequential ChIPs, C values, or efficiencies of co-occupancy, in percent, were calculated according to the following formula: C = 100(AB-A)/(A × B – A), where A, B, and AB represent the fold enrichments for the first, second, and sequential ChIP, respectively (38). For real time RT-PCR, the one-step Quanti-Test Multiplex RT-PCR NR kit (Qiagen) was used. The one-step reactions were set up in 10 μl, and the RT-PCR programs were designed according to the manufacturer's recommendations. The relative mRNA expression (R) for every gene was calculated using the formula R = 2Cp actin –Cp gene taking the ACT1 gene as a reference.
Phosphatase Treatment of Oaf1p-HA3
Yeast strains were grown on YPD, YPG, or YPGO media, and cell extracts were prepared as described previously (35). For phosphatase treatment of Oaf1p-HA3 expressed in wild-type cells (strain OAF1-HA3), cell extracts containing 10 μg of total protein were incubated at 37 °C for 1 h in the presence of 20 units of calf intestinal phosphatase (CIP, New England Biolabs) in a total volume of 10–15 μl. Phosphatase inhibitor mixture (5 mm sodium phosphate, pH 7.5, 10 mm sodium pyrophosphate, and 5 mm EDTA) and/or 0.2% SDS was added to the samples if required. Control sample was treated identically, but CIP and phosphatase inhibitors were not included in the reaction mixture. The treated samples were then separated on a 7% SDS-PAGE followed by immunoblotting with 12CA5 anti-HA antibody. To analyze the migration pattern of Oaf1p-HA3 expressed in snf1 mutant cells under glucose, glycerol, and oleate growth conditions, cell lysates containing 50 μg of total protein were separated on a 7% SDS-PAGE followed by immunoblotting with 12CA5 anti-HA antibody. Densitometric analysis was carried out to identify the exact position of each protein band using ImageQuant software (GE Healthcare).
Antibodies
Rabbit polyclonal affinity purified HA and c-Myc antibodies were obtained from Bethyl Laboratories (Montgomery, TX); mouse monoclonal anti-HA antibody (clone 12CA5), rat monoclonal anti-HA antibody (clone 3F10), anti-HA-peroxidase high affinity and mouse monoclonal anti-c-Myc antibody (clone 9E10), and anti-(c-Myc)-peroxidase were obtained from Roche Applied Science; anti-rabbit IgG (whole molecule) peroxidase conjugate was obtained from Sigma.
RESULTS
Choosing a System to Study Oaf1p and Pip2p Binding in Vivo—The binding of the Oaf1p-Pip2p complex to a synthetic POX1 ORE in vitro is strongly regulated by the carbon source supplied in the growth media. In our previous studies we observed a weak band shift using protein extracts prepared from glucose-grown cells (39). The signal was increased upon derepression (glycerol media) and reached its highest levels under oleate-growth conditions. One interpretation of these results is that the induction of the fatty acid-responsive genes in the presence of oleate is because of an increase in the amount of Oaf1p-Pip2p complex binding to the target OREs in vivo. As a first step toward testing this hypothesis, we measured the binding of Oaf1p and/or Pip2p to the target promoters in vivo, using chromatin immunoprecipitation followed by real time PCR analysis of the immunoprecipitated DNA. Using a single ChIP assay, Pip2p-HA3 binding was highest when cells were grown on oleate for most of the genes analyzed. However, Oaf1p-HA3 binding was highest under glycerol growth conditions when compared with oleate (data not shown). It seems likely that this difference in Oaf1p-HA3 and Pip2p-HA3 binding levels in these experiments is an artifact caused by HA-tagging Oaf1p. To overcome this potential problem, we created an additional yeast strain that co-expresses a functional Pip2p-HA3 allele and a Myc-tagged Oaf1p allele (Oaf1p-myc13). Using Western blot analysis with a polyclonal antibody to the acyl-CoA oxidase protein, we found that these co-expressing cells are able to induce POX1 expression to a level similar to that seen for our wild-type cells grown on oleate medium, indicating that the double-tagged (Oaf1p-myc13)-(Pip2p-HA3) complex is functional. We tested lysates prepared from this co-expressing strain in single ChIP pilot experiments and found that the binding patterns for both Oaf1p and Pip2p were similar to that seen for Pip2p in previous experiments with a Pip2p-HA3 single-expressing strain (see below).
In Vivo Binding of the Oaf1p and Pip2p Transcription Factors to the Promoters of the Target Genes—Table 2 lists 14 genes that are potential targets for Oaf1p and Pip2p. These genes can be separated into four different categories according to the mode of regulation by fatty acids. Type I genes are highly induced by unsaturated fatty acids such as oleate. These genes contain a consensus ORE and are regulated by the Oaf1p and Pip2p transcription factors. Type II genes are also induced by oleate under the regulation of Oaf1p and Pip2p, but to a lesser extent (2–4-fold). Type III genes contain a consensus ORE; however, these genes are repressed by unsaturated fatty acids while they are induced by saturated fatty acids such as myristate. Finally, type IV genes contain an ORE, but it appears to be nonfunctional because they are not regulated by fatty acids. We also note that there are three genes (PXA1, YLR287c, and YMR018w) that lack an obvious ORE in their promoters but nevertheless are up-regulated by oleate. Previously, we have shown that PXA1 is induced 2-fold by oleate (5), whereas two other genes (YLR287c and YMR018w) were selected as candidates in a global microarray expression search for oleate-inducible genes (14).
TABLE 2.
ORE-containing genes studied for Oaf1p, Pip2p, and Adr1p in vivo binding
| Gene name | Protein | Presence of ORE | Approximate fold induction on YPGO versus YPG (5) |
Binding of a transcription factor to the promoter on YPG versus
YPGO
|
||
|---|---|---|---|---|---|---|
| Oaf1p | Pip2p | |||||
| -fold | % | |||||
| Type I | ||||||
| POX1 | Peroxisomal acyl-CoA oxidase | Functional | 10 | 58 ± 24 | 49 ± 5 | |
| FOX3 | Peroxisomal thiolase | Functional | 10 | 62 ± 19 | 63 ± 13 | |
| ECI1 | Peroxisomal enoyl-CoA isomerase | Functional | 10 | 63 ± 20 | 58 ± 11 | |
| PIP2 | Transcription factor involved in peroxisome proliferation | Functional | 10 | 53 ± 18 | 58 ± 12 | |
| PEX11 | Peroxisomal membrane protein | Functional | 10 | 48 ± 16 | 54 ± 11 | |
| Type II | ||||||
| PEX5 | Peroxin | Functional | 2 | 73 ± 21 | 68 ± 9 | |
| CRC1 | Mitochondrial inner membrane carnitine transporter | Functional | 2 | 65 ± 22 | 58 ± 7 | |
| CTA1 | Peroxisomal catalase | Functional | 4 | 65 ± 20 | 53 ± 12 | |
| QDR1 | Multidrug transporter | Functional | 2 | 101 ± 21 | 53 ± 11 | |
| Type III | ||||||
| IZH2/ORE20 | Osmotin receptor | Functional | Repressed | 82 ± 27 | 73 ± 5 | |
| IZH1 | Membrane protein involved in zinc metabolism | Functional | Repressed | 106 ± 34 | 49 ± 6 | |
| IZH4 | Membrane protein involved in zinc metabolism | Functional | Repressed | 142 ± 31 | 70 ± 1 | |
| Type IV | ||||||
| DUS3 | Dihydrouridine synthase | Nonfunctional | None | 69 ± 14 | 61 ± 10 | |
| SER1 | 3-phosphoserine aminotransferase | Nonfunctional | None | 71 ± 10 | 44 ± 4 | |
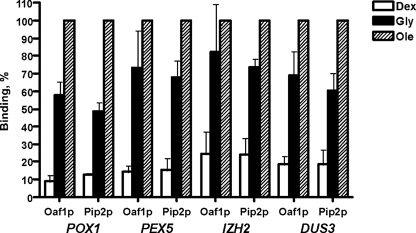
To confirm that Oaf1p and Pip2p bind to the promoter of the genes described above, we performed a series of single ChIP experiments. Lysates prepared from cells co-expressing Pip2p-HA3 and Oaf1p-myc13 grown on YPD, YPG, and YPGO media were used in single ChIP assays with either HA or c-Myc antibodies. This series of experiments demonstrated that both Oaf1p and Pip2p bind to all of the target genes listed in Table 2 under all growth conditions. The association of these transcription factors with OREs depended on the target gene and on the growth conditions (Fig. 1 and Tables 2 and 3). With the exception of three genes (QDR1, IZH1, and IZH4), the association of both transcription factors was low on glucose and significantly greater on glycerol (3–10-fold), and the highest binding was detected when cells were grown in the presence of oleate (Fig. 1 and Table 2). The association of both transcription factors, however, was only moderately increased (∼2-fold or less) on oleate compare with glycerol. We and others previously demonstrated that transcription of the highly inducible oleate-responsive genes (type I) is increased at least 10-fold in cells that have been shifted from glycerol to oleate media (3, 5, 39, 40), whereas others (type II) are only moderately induced by oleate (Table 2). Because Oaf1p and Pip2p binding patterns were similar for all of these genes, there must be an additional mechanism(s) implicated in oleate induction of highly inducible genes. Surprisingly, we also found that the Oaf1p-Pip2p complex binds to the promoters of two oleate-responsive genes, PXA1 and YMR018w, that lack a conventional ORE (Fig. 2 and data not shown). This finding suggests that perhaps the widely accepted definition of the ORE as a simple DNA sequence may be too narrow. It is possible that Oaf1p-Pip2p binding to a target sequence also requires an interaction of this complex with chromatin proteins. Furthermore, Oaf1p and Pip2p also bind to two ORE-containing genes, DUS3 and SER1, that are not induced by fatty acids (Fig. 1 and Tables 2 and 3). The role of Oaf1p-Pip2p transcription factors in regulating these genes remains to be investigated.
FIGURE 1.
Association of Oaf1p and Pip2p with the promoters of genes representing type I–IV (as listed in Table 2). The lysates prepared from PIP2-HA3/OAF1-myc13 double-expressing cells grown on YPD, YPG, and YPGO media were used in single ChIP experiments with either anti-HA or anti-c-Myc polyclonal antibody as described (see “Experimental Procedures”). Both ChIP and input samples were assayed in a real time PCR as described under “Experimental Procedures.” The binding (immunoprecipitation efficiency) was calculated as a percentage of gene-specific DNA pulled down by an antibody from the lysate in a single ChIP experiment. Association of the transcription factor under oleate growth conditions was taken to be 100%. Numbers are the mean of three experiments. Dex, glucose; Gly, glycerol; Ole, oleate.
TABLE 3.
Relative occupancies of Oaf1p and Pip2p on target genes
The relative occupancies were calculated as the fold enrichments of a given genomic region over the GPD1 background according to the formula RO = IPgene/IPGPD1, where IPgene and IPGPD1 are the immunoprecipitation efficiencies for a given target gene and the GPD1 gene, and RO is the relative occupancy expressed in GPD1 units, respectively.
|
Gene
|
Transcription factor
|
Media
|
||
|---|---|---|---|---|
| Glucose | Glycerol | Oleate | ||
| POX1 | Oaf1p | 2.24 ± 0.35 | 7.75 ± 1.76 | 12.25 ± 4.51 |
| Pip2p | 4.38 ± 1.96 | 12.13 ± 3.83 | 14.57 ± 2.34 | |
| PEX5 | Oaf1p | 11.82 ± 2.02 | 25.79 ± 1.65 | 37.59 ± 7.90 |
| Pip2p | 14.99 ± 12.06 | 43.33 ± 14.61 | 36.88 ± 4.77 | |
| IZH2 | Oaf1p | 5.09 ± 0.92 | 7.46 ± 0.10 | 9.57 ± 0.92 |
| Pip2p | 6.75 ± 4.95 | 13.61 ± 3.83 | 10.67 ± 2.87 | |
| DUS3 | Oaf1p | 14.52 ± 5.51 | 22.31 ± 3.19 | 34.58 ± 8.89 |
| Pip2p | 16.59 ± 13.79 | 35.79 ± 12.25 | 33.69 ± 1.57 | |
| GPD1 | Oaf1p | 1.00 | 1.00 | 1.00 |
| Pip2p | 1.00 | 1.00 | 1.00 | |
FIGURE 2.
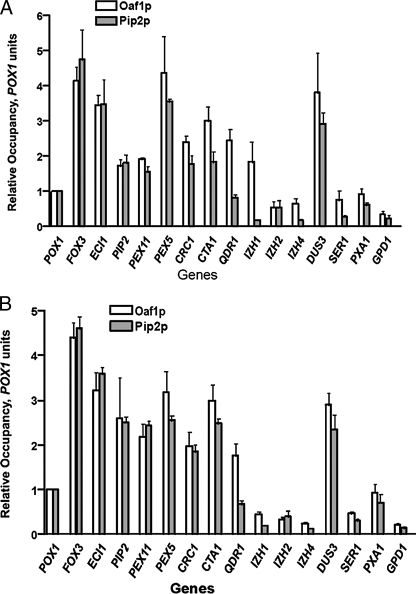
Comparison of the relative occupancies of the Oaf1p and Pip2p transcription factors on the promoters of target genes under glycerol (A) and oleate (B) growth conditions. Relative occupancies were calculated as described under “Experimental Procedures” and Table 3 using the POX1 gene as a reference. Numbers are the mean of three experiments.
For Oaf1p, the relative occupancy of binding (see “Experimental Procedures” and Table 3) determined using GPD1 as a reference gene mimicked the pattern observed for its IP efficiency under all growth conditiGons. The increases in relative occupancy when moving from glucose to glycerol media range from 1.5- to 3.5-fold and are statistically significant. However, although moving from glycerol to oleate media also leads to moderate increases in relative occupancy of Oaf1p in all cases, the changes for Pip2p are variable. Furthermore, these latter fluctuations are not statistically significant. Thus, for Pip2p, the relative occupancy does not appear to be increased upon oleate induction (Table 3) and therefore does not follow its IP efficiency pattern. Because Oaf1p and Pip2p IP efficiency patterns appear to be similar for an individual gene, the observed difference in relative occupancy patterns of these proteins is most likely caused by variations in the experimental background when a nonbinding GPD1 locus is used as a reference gene (32).
To reduce the potential for experimental error, we determined the relative occupancy of Oaf1p and Pip2p using an Oaf1p-Pip2p-binding gene (POX1) as a reference gene, rather than genes that do not recruit these transcription factors. For most of the genes studied, the relative occupancy to an individual gene was similar for both transcription factors on a given growth condition (glycerol or oleate), when expressed in POX1 units (Fig. 2). These findings indicate that Oaf1p and Pip2p bind to target genes in an equimolar ratio, and strongly support a model in which Oaf1p and Pip2p act as a heterodimer to trigger oleate-inducible transcription. Thus, it appears that the difference in patterns for Oaf1p and Pip2p relative occupancy seen previously (Table 3) is most likely because of the background variations for these two proteins when GPD1 is used as a reference gene. In contrast, we also observed that the relative occupancy of Oaf1p was greater than that of Pip2p for QDR1, CTA1, and IZH genes in glycerol growth conditions (Fig. 2B). This suggests that Oaf1p alone, most likely in the form of a homodimer, may be recruited to the promoter of a subset of target genes.
The binding of both Oaf1p and Pip2p under a given growth condition was variable among the genes investigated (Fig. 2). We have found no correlation between the groups of genes and the relative occupancies of both proteins under a specific growth condition. For example, the DUS3 gene is not regulated by the Oaf1p-Pip2p complex; however, both transcription factors bind to this gene, and their association with the DUS3 ORE was almost three times higher than the association with ORE of the highly inducible POX1 promoter (Fig. 2).
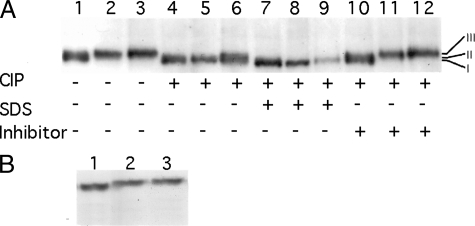
Regulated Phosphorylation of Oaf1p—Phosphorylation has been shown to be required for activation of yeast transcription factors, such as Gal4p (41). Because expression of OAF1 is constitutive, irrespective of the growth conditions, we hypothesized that Oaf1p may be phosphorylated, and this modification may increase Oaf1p-Pip2p binding upon glucose derepression and oleate induction. Immunoblot analysis showed that Oaf1p-HA3 expressed in glycerol-grown cells appeared to migrate slightly slower than when this protein was expressed in glucose-grown cells (Fig. 3A). Upon switching from glycerol to oleate, an additional increase in Oaf1p-HA3 mobility was seen (Fig. 3A). Decreased mobility of Oaf1p-HA3 in glycerol- and oleategrown cells compared with that seen in glucose-grown cells suggests that the protein undergoes a post-translational modification(s) upon switching from glucose to glycerol or oleate media. To test whether Oaf1p gets phosphorylated upon a shift from glucose to glycerol- and oleate-containing media, cell extracts were treated with CIP. The results showed that the addition of CIP to the cell extracts from glycerol- and oleate-grown cells gave rise to a single Oaf1p band, whereas the presence of phosphatase inhibitors in the reaction mixture prevented this mobility shift (Fig. 3A). Therefore it appears that phosphorylation accounts for the reduced mobility of Oaf1p in cells grown in glycerol- and oleate-containing media. We next investigated whether Snf1p, a protein kinase that is essential for glucose derepression as well as for utilizing of fatty acids (17), is involved in phosphorylation of Oaf1p. For this purpose we expressed Oaf1p-HA3 in a strain that carries a mutation in the SNF1 gene (42). The electrophoretic migration pattern of Oaf1p-HA3 in the snf1 mutant was similar to that in wild-type cells (Fig. 3B), suggesting that SNF1 is not essential for the phosphorylation of Oaf1p. However, the possibility that Snf1p plays some role in phosphorylation of Oaf1p, but does not cause a detectable change in its migration in SDS-PAGE, cannot be ruled out.
FIGURE 3.
Oaf1p is phosphorylated upon glucose derepression and oleate induction. A, phosphatase treatment of Oaf1p-HA3. Reactions were carried out with cell extracts from glucose-grown cells (lanes 1, 4, 7, and 10), glycerol-grown cells (lanes 2, 5, 8, and 11), and oleate-grown cells (lanes 3, 6, 9, and 12). The addition of CIP, phosphatase inhibitor mixture and 0.2% SDS is indicated below each lane. Numbers I–III show exact positions of each form of Oaf1p-HA3. B, migration pattern of Oaf1p-HA3 expressed in the snf1 mutant cells grown on glucose-containing media (lane 1), glycerol-containing media (lane 2), and oleate-containing media (lane 3).
Oaf1p and Pip2p Co-occupy Target Sequences and Pip2p Is Required for Oaf1p Binding—We have previously shown, using in vitro binding assays, that Oaf1p and Pip2p predominantly act as a heterodimer to activate transcription of many fatty acid-responsive genes (5, 6). We have also demonstrated here that these proteins bind to DNA in an equimolar ratio in vivo. Asa next step to confirm that these proteins bind simultaneously in vivo, we analyzed the co-occupancy of Oaf1p and Pip2p on the promoter of each of the target genes by performing quantitative sequential ChIP using our Pip2p-HA3/Oaf1p-myc13 co-expressing strain. For every gene positively tested, we found a partial co-occupancy of Oaf1p and Pip2p on the respective promoter under all growth conditions, irrespective of the sequential ChIP direction (Table 4).5 These include QDR1 and CTA1, which are genes that do not recruit Oaf1p and Pip2p at an equimolar ratio under glycerol and oleate growth conditions (Fig. 2). For the genes that do not significantly recruit these proteins, such as IZH1, the results were inconclusive (data not shown), probably because of the limitations of the sequential ChIP procedure. Thus, Oaf1p and Pip2p (at least partially) co-occupy OREs of target genes in vivo. These data, together with our previous findings (6, 39) and additional data presented here, provide strong evidence for the presence of Oaf1p-Pip2p heterodimers on the promoters of oleate-responsive genes. To determine whether Pip2p is required for Oaf1p binding to a target sequence, we measured Oaf1p binding in the absence of Pip2p. Using lysates prepared from our wild-type and pip2Δ strains expressing Oaf1p-HA3 in a single ChIP real time PCR assay, we demonstrated that Oaf1p binding to the target genes in the pip2Δ strain was significantly lower than our wild-type strain under all growth conditions (Table 5, see IPΔpip2/Ipwt rows). Thus, to efficiently bind to target sequences, Oaf1p depends on Pip2p. We also noted that GPD1 background levels on glycerol and oleate were drastically reduced in PIP2 mutant cells, suggesting that the nonspecific association of Oaf1p to chromatin was lower in the absence of Pip2p. As a result, Oaf1p relative occupancy values in the pip2Δ strain were significantly above 1 GDP1 unit under both glycerol and oleate growth conditions for all genes except IZH2 (Table 5, see RO rows). Therefore, we can conclude that Oaf1p is recruited to target genes even in the absence of Pip2p, most likely in a form of a homodimer, although at a much lower efficiency than the Oaf1p-Pip2p heterodimer. Unfortunately, we were not able to measure the binding of Pip2p in the absence of Oaf1p because the PIP2 gene is expressed at extremely low levels in oaf1Δ cells (5, 6).
TABLE 4.
Oaf1p-Pip2p sequential ChIP
C values, or efficiencies of co-occupancy, in percentages, were calculated according to the following formula: C = 100(AB-A)/(A × B — A), where A, B, and AB represent the fold enrichments for the first, second, and sequential ChIP, respectively (37). The threshold point for the AB/A ratio was taken to be 2.0 to call for an ensured co-occupancy. However, if the AB/A ratio was found to be in a range from 1.5 to 2.0, a possible co-occupancy was called if the corresponding C values were found to be in a range from 1.0 to 100.0.
|
Direction of IP
|
Samples
|
POX1
|
PEX5
|
IZH2
|
QDR1
|
DUS3
|
YMR018w
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | AB/A | C | AB/A | C | AB/A | C | AB/A | C | AB/A | C | AB/A | ||
| % | % | % | % | % | % | ||||||||
| Oaf1p | Glucose | 58.1 | 2.11 | 58.9 | 17.51 | 420.7 | 23.75 | 23.47 | 1.34 | 107.5 | 25.63 | 166.4 | 9.78 |
| Then | Glycerol | 57.4 | 3.80 | 24.2 | 16.45 | 139.6 | 18.00 | 32.75 | 2.13 | 40.5 | 19.84 | 83.4 | 11.31 |
| Pip2p | Oleate | 22.5 | 3.04 | 11.9 | 8.63 | 56.6 | 7.16 | 9.23 | 1.42 | 17.9 | 10.70 | 23.1 | 7.52 |
| Pip2p | Glucose | 83.5 | 2.71 | 16.0 | 3.76 | 168.8 | 3.41 | 18.71 | 1.59 | 25.0 | 4.26 | 38.1 | 2.04 |
| Then | Glycerol | 29.9 | 1.79 | 5.7 | 2.91 | 47.3 | 3.20 | 26.33 | 2.38 | 9.0 | 3.01 | 25.2 | 2.79 |
| Oaf1p | Oleate | 16.3 | 1.69 | 4.1 | 2.22 | 22.9 | 2.31 | 14.94 | 2.21 | 6.2 | 2.50 | 11.3 | 3.27 |
TABLE 5.
Binding of Oaf1p to the promoters of target genes in the absence of Pip2p
Binding in Δpip2 strain is expressed as percentage of the corresponding IP efficiency values in W3031A wild-type strain (IPΔpip2/IPwt), relative occupancies (RO) in Δpip2 strain are expressed in GPD1 units using GPD1 IP enrichment values found in this strain.
|
Gene
|
Oaf1p binding in Δpip2
|
Media
|
||
|---|---|---|---|---|
| Glucose | Glycerol | Oleate | ||
| POX1 | IPΔpip2/IPwt | 22.63 ± 19.65 | 3.64 ± 0.58 | 12.58 ± 0.24 |
| RO | 0.70 ± 0.41 | 4.72 ± 2.33 | 5.42 ± 2.74 | |
| PIP2 | IPΔpip2/IPwt | 6.54 ± 4.49 | 1.48 ± 0.43 | 4.06 ± 1.74 |
| RO | 1.26 ± 0.11 | 5.94 ± 3.48 | 6.36 ± 4.94 | |
| PEX5 | IPΔpip2/IPwt | 33.17 ± 31.55 | 4.19 ± 1.77 | 10.92 ± 0.58 |
| RO | 6.75 ± 4.95 | 13.61 ± 3.83 | 10.67 ± 2.87 | |
| CTA1 | IPΔpip2/IPwt | 49.69 ± 47.81 | 11.54 ± 9.79 | 20.78 ± 1.88 |
| RO | 2.4 ± 0.25 | 35 ± 30 | 28 ± 11 | |
| IZH2 | IPΔpip2/IPwt | 17.67 ± 12.48 | 1.47 ± 0.30 | 6.28 ± 0.57 |
| RO | 0.63 ± 0.34 | 1.67 ± 0.39 | 2.23 ± 1.38 | |
| DUS3 | IPΔpip2/IPwt | 49.35 ± 33.48 | 9.35 ± 3.67 | 19.89 ± 4.82 |
| RO | 3.18 ± 0.92 | 27.92 ± 17.01 | 19.89 ± 13.51 | |
| GPD1 | IPΔpip2/IPwt | 100.65 ± 69.30 | 7.28 ± 0.78 | 14.84 ± 11.56 |
| RO | 1.00 | 1.00 | 1.00 | |
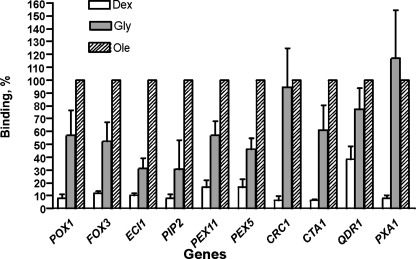
Adr1p Binds to the Promoters of Oaf1p and Pip2p Target Genes under Derepression and Induction Conditions—The pleiotropic activator Adr1p is required for oleate-dependent induction of a number of genes involved in peroxisome function (18–20, 26). To confirm that this transcription factor binds to Oaf1p-Pip2p target genes in vivo, we created a strain that expresses a c-Myc-tagged allele of Adr1p (Adr1p-myc13). We performed a series of single ChIP experiments with lysates prepared from these cells and found that Adr1p binds to the type I and type II genes listed in Table 2, with the exception of QDR1 and PEX5 (Fig. 4 and Table 6). For QDR1 and PEX5, the relative occupancy ranged up to 2 GPD1 units, suggesting that weak Adr1p binding to the promoters of these genes might have taken place under oleate growth conditions. We did not detect binding to genes belonging to the IZH family, and we only found weak association (relative occupancy 2.5 GPD1 units or less) with genes harboring a nonfunctional ORE (DUS3 and SER1) (Table 6). Interestingly, Adr1p binding appears to be oleate-responsive for the majority of genes tested in this series of experiments.
FIGURE 4.
Association of Adr1p with promoters of Oaf1p-Pip2p target genes. Lysates were prepared from ADR1-myc13 cells grown on YPD, YPG, and YPGO media and used in single ChIP experiments with anti-c-Myc polyclonal antibody as described (see “Experimental Procedures”). Adr1p binding (IP efficiency) was calculated as a percentage of gene-specific DNA pulled down by c-Myc antibody from the lysate in a single-ChIP experiment. Binding of Adr1p under oleate growth conditions was taken to be 100%. Numbers are the mean of three experiments. Dex, glucose; Gly, glycerol; Ole, oleate.
TABLE 6.
Relative occupancy of Adr1p on the promoters of Oaf1p-Pip2p target genes
See Table 3 for definition of GPD1 units.
|
Gene
|
Media
|
||
|---|---|---|---|
| Glucose | Glycerol | Oleate | |
| POX1 | 1.04 ± 0.16 | 3.49 ± 2.14 | 5.64 ± 2.64 |
| FOX3 | 1.36 ± 0.40 | 2.74 ± 1.48 | 4.80 ± 1.57 |
| ECI1 | 1.03 ± 0.34 | 1.36 ± 0.61 | 4.06 ± 0.60 |
| PIP2 | 1.55 ± 1.31 | 2.66 ± 2.60 | 7.45 ± 3.75 |
| PEX11 | 0.90 ± 0.16 | 1.43 ± 0.64 | 2.31 ± 0.62 |
| PEX5 | 0.95 ± 0.18 | 1.04 ± 0.58 | 1.87 ± 0.52 |
| CRC1 | 0.92 ± 0.78 | 5.02 ± 2.00 | 5.01 ± 0.87 |
| CTA1 | 1.05 ± 0.32 | 4.68 ± 2.41 | 7.10 ± 1.34 |
| QDR1 | 1.15 ± 0.26 | 1.08 ± 0.44 | 1.32 ± 0.67 |
| PXA1 | 1.27 ± 0.15 | 8.98 ± 5.08 | 7.05 ± 1.20 |
| IZH1 | 1.00 ± 0.16 | 0.62 ± 0.11 | 0.70 ± 0.30 |
| IZH2 | 0.95 ± 0.46 | 0.42 ± 0.24 | 0.61 ± 0.24 |
| IZH4 | 0.95 ± 0.16 | 0.83 ± 0.19 | 0.73 ± 0.21 |
| DUS3 | 1.49 ± 0.18 | 1.30 ± 0.32 | 1.60 ± 0.37 |
| SER1 | 1.59 ± 0.32 | 1.71 ± 0.64 | 1.65 ± 0.90 |
| GPD1 | 1.00 | 1.00 | 1.00 |
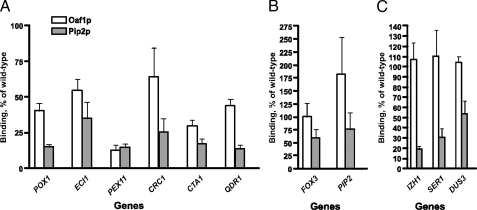
Is the Binding of Adr1p and the Oaf1p-Pip2p Complex Dependent on One Another?—Adr1p binds to a sequence, referred to as UAS1, in target genes (14, 22). The UAS is often found in close proximity or overlaps with the ORE of Oaf1p-Pip2p target genes (17, 19–21, 26). To investigate the relationship between Oaf1p, Pip2p, and Adr1p on the promoters of target genes, we first analyzed the binding of Oaf1p and Pip2p in an ADR1 deletion strain. Single ChIP assays were carried out with lysates prepared from Oaf1p-myc13/Pip2p-HA3 co-expressing strain that lacked Adr1p (Table 1). We found that there was a decrease in Oaf1p binding to the majority of type I and type II genes listed in Table 2, in the ADR1 deletion strain (Fig. 5A). However, for other genes, Oaf1p binding was either unaffected or enhanced (i.e. for PIP2) (Fig. 5B). For genes that weakly bind Adr1p, or do not bind this transcription factor at all, Oaf1p binding remained unchanged (Fig. 5C). We also noted that binding of Pip2p was decreased for each gene analyzed. For the PEX11 gene, the binding of both Oaf1p and Pip2p was greatly decreased (∼5-fold). These results indicate that the Oaf1p-Pip2p complex is dependent on Adr1p for binding to target genes.
FIGURE 5.
The effect of deleting ADR1 on Oaf1p and Pip2p recruitment by target genes. Lysates prepared from the ΔADR1(PIP2-HA3/OAF1-myc13) strain grown on oleate were used in single ChIP experiments as described in Fig. 1. The binding of Oaf1p and Pip2p to each individual gene was calculated as described in Fig. 1 and expressed as a percentage of the wild-type value (PIP2-HA3/OAF1-myc13 co-expressing strain). A, reduced recruitment of Oaf1p and Pip2p. B, unchanged or increased recruitment of Oaf1p and Pip2p. C, unchanged recruitment of Oaf1p and reduced recruitment of Pip2p. Numbers are the mean of three experiments.
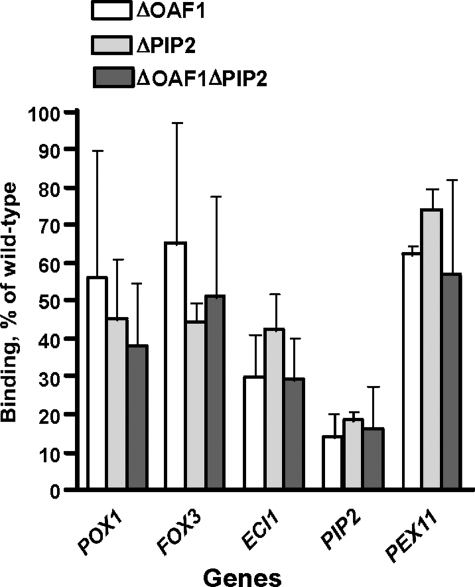
We next investigated whether the Oaf1p-Pip2p complex is important for Adr1p binding using the same approach. Three additional Adr1p-myc13-expressing strains that lack either Oaf1p or Pip2p or both of these proteins were created for this purpose (Table 1). These experiments showed that Adr1p binding to target genes was moderately reduced compared with our “wild-type” Adr1p-myc13-expressing strain under both glycerol and oleate growth conditions for all genes with the exception of PIP2 (Fig. 6 and data not shown). Adr1p binding to the PIP2 promoter, however, was significantly reduced in the absence of Oaf1p and Pip2p. These results indicate that, for the majority of the genes studied, both Oaf1p and Pip2p are important but not essential for Adr1p binding to its target sequences.
FIGURE 6.
The effect of deleting OAF1, PIP2, or both of these genes on Adr1p recruitment by ORE-containing genes. Lysates prepared from ΔOAF1/ADR1-myc13, ΔPIP2/ADR1-myc13, and ΔO1ΔP2/ADR1-myc13 cells grown on YPGO media were subjected to a single ChIP assay with anti-c-Myc polyclonal antibody as described (see “Experimental Procedures”). The binding to every gene was calculated and expressed as a percentage of the wild-type values. Adr1p binding to promoters of Oaf1p-Pip2p target genes is decreased in oaf1Δ, pip2Δ, and oaf1Δpip2Δ strains. Numbers are the mean of three experiments.
It was previously shown that Oaf1p, but not Pip2p, is able to receive a fatty acid nutritional signal (12). Our own single ChIP experiments (see above) revealed that Adr1p binding to Oaf1p-Pip2p target genes is oleate-responsive (Fig. 4). To investigate whether Adr1p itself is able to receive an oleate nutritional signal, we compared the ratios of binding in oleate versus glycerol for Adr1p in our wild-type and oaf1Δpip2Δ mutant cells (Table 7 and data not shown). Although these ratios ranged up to 4-fold in wild-type cells, the ratio was approximately 1 or unity in mutant cells for all genes tested. This finding suggests that, unlike Oaf1p in pip2Δ cells, Adr1p binding is not responsive to oleate in oaf1Δpip2Δ mutant cells. Therefore, it appears that Adr1p itself is not able to function as an acceptor of the oleate nutritional signal.
TABLE 7.
Adr1p binding ratios (oleate versus glycerol) in wild type and an oaf1Δ pip2Δ strain
|
Gene
|
Strain
|
|
|---|---|---|
| Wild type | oaf1Δpip2Δ | |
| POX1 | 1.85 ± 0.76 | 1.29 ± 0.46 |
| FOX3 | 2.10 ± 1.18 | 1.00 ± 0.24 |
| ECI1 | 3.43 ± 1.39 | 0.96 ± 0.29 |
| PIP2 | 4.19 ± 3.02 | 0.63 ± 0.15 |
| PEX11 | 1.87 ± 0.92 | 1.16 ± 0.35 |
Expression of the Oaf1p-Pip2p Target Genes in the Absence of Adr1p—Previously, Adr1p was implicated in the regulation of a number of ORE-containing genes by its involvement in relieving glucose repression, as well as achieving fatty acid-dependent induction of these genes (19–22, 43, 44). We analyzed and compared the expression of the Oaf1p-Pip2p target genes listed in Table 2 in our wild-type and adr1Δ cells using real time RT-PCR analysis. Our data indicate that deletion of ADR1 drastically changes the expression of Oaf1p-Pip2p target genes under glycerol and oleate growth conditions (Table 8). Some genes, such as POX1 and CTA1, show a dramatic decrease in their expression levels under glycerol and oleate growth conditions. These results are in accordance with previously published findings indicating that a deletion of ADR1 leads to a decrease in the expression of a number of peroxisome-related genes upon oleate induction (18–21, 26). However, the expression of other genes, such as PEX5, was increased in glycerol growth conditions when compared with the wild-type grown under the same conditions. Interestingly, Oaf1p-Pip2p target genes that do not recruit Adr1p to their promoters (e.g. IZH1, IZH4) also followed this pattern. Thus, Adr1p appears to act on Oaf1p-Pip2p target genes in two different ways; it is required to maintain high levels of expression of a subset of oleate-responsive genes under glycerol and oleate growth conditions (positive regulation), and it also controls the low glycerol derepression levels of another subset of the genes (negative regulation). In addition, we have shown that a lack of Adr1p leads to a decrease in the oleate response of some oleate-inducible genes (Table 8).
TABLE 8.
Behavior of Oaf1p-Pip2p target genes in wild-type and adr1Δ strains
The mRNA levels in adr1Δ strain are expressed as percentages of wild-type levels. Oleate induction is expressed as oleate versus glycerol mRNA ratios for both wild-type and adr1Δ strains.
|
Genes
|
Expression in adr1Δ
|
Fold induction (oleate versus glycerol)
|
|||
|---|---|---|---|---|---|
| Glucose | Glycerol | Oleate | Wild type | adr1Δ | |
| POX1 | 125 ± 3 | 6.2 ± 1.6 | 5.5 ± 1.7 | 42 ± 6 | 36 ± 3 |
| FOX3 | 91 ± 1 | 20 ± 8 | 18 ± 5 | 27 ± 1.5 | 24 ± 1.5 |
| ECI1 | 84 ± 10 | 54 ± 7 | 13 ± 3 | 62 ± 14 | 15 ± 1.5 |
| PIP2 | 119 ± 40 | 71 ± 24 | 29 ± 14 | 11 ± 2 | 4.6 ± 1.4 |
| EX11 | 89 ± 8 | 35 ± 11 | 21 ± 2 | 5.8 ± 1.6 | 3.6 ± 0.2 |
| PEX5 | 120 ± 21 | 332 ± 99 | 42 ± 21 | 23 ± 5 | 2.8 ± 0.8 |
| CRC1 | 84 ± 18 | 39 ± 17 | 39 ± 13 | 7.9 ± 3.4 | 7.8 ± 1.5 |
| CTA1 | 123.5 ± 7 | 9.3 ± 4.2 | 9.3 ± 5.4 | 16 ± 3 | 15 ± 3 |
| QDR1 | 124 ± 13 | 48 ± 13 | 16 ± 3 | 6.3 ± 1.3 | 2.1 ± 0.2 |
| IZH1 | 83 ± 10 | 268 ± 48 | 46 ± 9 | 1.5 ± 0.5 | 0.25 ± 0.04 |
| IZH2 | 112 ± 30 | 174 ± 19 | 101 ± 52 | 2.5 ± 1.2 | 1.23 ± 0.22 |
| IZH4 | 114 ± 30 | 561 ± 187 | 56 ± 27 | 0.14 ± 0.05 | 0.012 ± 0.01 |
| SER1 | 117 ± 15 | 175 ± 40 | 46 ± 15 | 7.1 ± 1.1 | 1.8 ± 0.3 |
| PXA1 | 128 ± 8 | 35 ± 8 | 5.8 ± 1.9 | 9.0 ± 1.4 | 1.5 ± 0.2 |
| PGK1 | 114 ± 20 | 92 ± 20 | 66 ± 8 | 0.78 ± 0.16 | 0.56 ± 0.07 |
Finally, we should note that by using the precise and sensitive technique of real time RT-PCR, we were able to re-evaluate the levels of expression and induction of the genes under study. Our previous studies, using Northern blot analyses, indicated an ∼10-fold induction of POX1 (5, 39), whereas the real time RT-PCR analysis indicated an ∼42-fold induction. A detailed analysis of the Oaf1p-Pip2p target genes expression using real time RT-PCR will be described elsewhere.
DISCUSSION
In this study we analyzed the binding of the Oaf1p-Pip2p transcription factors to promoters of target genes in vivo. Using quantitative single ChIP assays on a single gene basis, we have shown that both Oaf1p and Pip2p bind to their target sequences under glucose, glycerol, and oleate growth conditions in vivo, although this binding was much lower on glucose when compared with glycerol or oleate. This should not come as a surprise, because Oaf1p and Pip2p are constantly localized to the nucleus (17), and the OREs to which they bind usually reside in nucleosome-free regions and remain accessible to them irrespective of the growth conditions (45).
The patterns of binding for both Oaf1p and Pip2p were similar for the majority of the genes tested (with only three exceptions, see “Results”). The pattern of binding mimicked that of mRNA expression for genes that are only moderately induced by oleate, but not for highly inducible genes (5). Many genes that are under control of the Oaf1p-Pip2p complex are regulated by two distinct mechanisms, glucose repression and oleate induction. We have shown here that a shift from glucose to glycerol media causes a large increase in binding of the Oaf1p-Pip2p complex to its response element, with only a moderate further increase in the amount of binding when cells are shifted to inducing conditions (Fig. 1). What could trigger this increase in binding upon glycerol derepression? We have demonstrated that Oaf1p undergoes phosphorylation in cells grown on glycerol or oleate media and that Snf1p kinase does not appear to be essential for this phosphorylation. Based on these findings, and by analogy with other yeast transcription factors (41), we hypothesize that this post-translational modification causes the increase in the Oaf1p-Pip2p binding under glucose derepression. Upon receiving a fatty acid nutritional cue, Oaf1p appears to undergo an additional phosphorylation and that, in turn, might further increase Oaf1p-Pip2p binding to an ORE. It is also possible that this additional phosphorylation causes a conformational change in the Oaf1p-Pip2p complex that leads to unmasking its transcriptional activity. It remains unclear whether Pip2p, the Oaf1p counterpart, is similarly phosphorylated and what role this post-translational modification may play in regulating transcriptional activity of the Oaf1p-Pip2p complex upon glucose derepression and oleate induction. A possible role of Adr1p phosphorylation is discussed below. The nature of a putative protein kinase(s) involved in the phosphorylation of both Oaf1p and Pip2p remains to be determined.
In addition to the genes that contain obvious OREs in their promoters, we were able to detect Oaf1p-Pip2p binding to two genes that lack a clearly defined ORE. The minimal consensus ORE is defined as CGGN3TN(A/R)N8–12CCG (17), but perhaps this definition is too restrictive. A genome-wide search for all Oaf1p-Pip2p-binding sites is required to define a consensus ORE that is physiologically meaningful. Furthermore, it is possible that interaction of the Oaf1p-Pip2p transcription factors with a target DNA sequence is more complex than simple protein-DNA binding, and it may also involve interaction of this complex with proteins associated with nucleosome-free OREs (45). In this study we also found that some OREs previously defined to be nonfunctional (5) appear to recruit the Oaf1p-Pip2p complex. In this case it is possible that Pip2p, rather than Oaf1p, plays a role as an acceptor of a nutritional signal because both of these proteins have ligand binding domains (11), but only Oaf1p binds free fatty acids. The nature of any additional signal, as well as the role of Oaf1p-Pip2p in the regulation of genes harboring “nonfunctional” OREs, has yet to be determined.
We and others previously postulated that the Oaf1p-Pip2p complex acts in a form of a heterodimer to regulate target genes (6, 7). The mammalian transcription factors, peroxisome proliferator-activated receptor α and retinoic acid receptor α, dimerize and subsequently bind to peroxisome proliferator-response elements of genes encoding peroxisomal proteins only upon ligand binding (8–10). By analogy, it has also been proposed that the Oaf1p-Pip2p complex binds to OREs upon receiving a nutritional signal (5–7), although the possibility of Oaf1p and Pip2p being permanently bound to OREs was also raised (17, 41). We show here that Oaf1p and Pip2p co-occupy the promoter regions of target genes in vivo, that they bind to most of the target sequences at an equimolar ratio, and that the binding of one protein appears to be dependent on the other. In addition, our results indicate that target genes preferably recruit the Oaf1p-Pip2p heterodimer rather than an Oaf1p-Oaf1p homodimer. These findings strongly support the current view that the Oaf1p-Pip2p transcription factors act predominantly as a heterodimer on target genes. It appears, however, that homodimers of Oaf1p may play an additional role in regulating certain genes such as QDR1, CTA1, and the IZH genes. This is in accordance with our previous finding that Oaf1p is capable of mediating partial oleate-dependent induction of CTA1 and QDR1 as well as maintaining sub-wild-type levels of IZH2 expression under glucose repression in the absence of Pip2p (5). It is possible that both Oaf1p-Oaf1p homodimers and Oaf1p-Pip2p heterodimers control the expression of these genes.
Our findings that the Oaf1p-Pip2p complex is able to bind to DNA without receiving an oleate nutritional signal raises the question as to whether both forms (DNA-bound and DNA-free) of this complex are capable of receiving this signal. Other transcription activators, such as Put3p and Leu3p, although always bound to a promoter are controlled by direct interaction with low molecular weight effectors (41, 46–49). In light of the recent identification of the Oaf1p ligand-binding domain that is capable of binding free fatty acids (11), it now seems likely that transcriptional activity of the Oaf1p-Pip2p complex is controlled by a similar mechanism, rather than by the peroxisome proliferator-activated receptor α-retinoic acid receptor α model of regulation.
In addition to the Oaf1p-Pip2p complex, Adr1p is involved in the regulation of many peroxisome-related genes via binding to its resident target UAS1 sequences in their promoters (18–21). This binding was previously demonstrated in vivo using ChIP assays (26–28); however, none of these studies were performed under oleate induction conditions, and there have been no such studies focused on Adr1p interactions with the Oaf1p-Pip2p complex. We show here that Adr1p binds in vivo to the promoters of Oaf1p-Pip2p target genes (with the exception of the IZH genes) under glucose derepression and oleate induction conditions. Because the expression of peroxisome-related genes appears to be impaired in ADR1 mutant cells (17), we also asked whether Adr1p acts on these genes by mediating the Oaf1p-Pip2p binding to their OREs. Indeed, the association of Oaf1p and Pip2p with OREs was lower in the adr1Δ mutant cells then in our wild-type cells for most of the genes studied. It is note-worthy that binding of these proteins to PEX11 was most reduced in adr1Δ cells. Because Pex11p is a key regulator of peroxisome proliferation, this supports the involvement of Adr1p in controlling this process (19).
As discussed above, Adr1p generally acts as a positive regulator of Oaf1p-Pip2p binding. However, the association of Oaf1p to the PIP2 promoter was increased in ADR1 mutant cells, suggesting that Adr1p regulates Oaf1p-Pip2p binding to this promoter in a negative fashion. Because PIP2 plays a unique regulatory role among fatty acid-responsive genes, it is perhaps not surprising that Oaf1p-Pip2p binding to the PIP2 promoter is also regulated in a distinctive fashion.
We also analyzed the importance of the Oaf1p-Pip2p complex for Adr1p binding and found that this complex is important but not essential. Our results also show that Adr1p binding to the PIP2 ORE appears to be under the stringent control of the Oaf1p-Pip2p complex. It is interesting to note that Adr1p binds to the majority of Oaf1p-Pip2p target genes in an oleate-dependent fashion in wild-type cells, but it loses its ability to respond to oleate in oaf1Δpip2Δ mutant cells. Thus, it appears that this protein is not capable of receiving an oleate nutritional signal independently, but rather it receives such a signal through the Oaf1p-Pip2p complex. The issue of whether Adr1p receives an oleate nutritional signal directly (i.e. by physically interacting with the Oaf1p-Pip2p complex) or indirectly (i.e. by interacting with another intermediate factor(s)) remains to be addressed.
Adr1p positively regulates the expression of Oaf1p-Pip2p target genes, although the level of regulation is variable (17). Our findings indicate that Adr1p is required for achieving the maximum expression of these genes when oleate is supplied in the growth media. This raises the possibility that, in addition to catabolite derepression, Adr1p might also be involved in the oleate induction of Oaf1p-Pip2p target genes. Interestingly, when ADR1 is disrupted, the response to oleate is reduced in a number of these genes, whereas others respond normally. Together, these results suggest that Adr1p is directly involved in oleate induction of at least some target genes, including highly inducible genes such as ECI1 and PIP2. Because an increase in Adr1p binding occurs under oleate growth conditions, it is reasonable to speculate that such an increase contributes to the up-regulation of Oaf1p-Pip2p target genes seen upon induction. However, the oleate induction is not completely abolished in the adr1Δ strain; therefore, there must be additional mechanism(s) involved in this up-regulation. One possibility might be the recruitment of chromatin remodeling factors by the Oaf1p-Pip2p complex and/or by Adr1p upon receiving a nutritional cue.
The mechanism controlling Adr1p binding upon switching from glycerol- to oleate-containing media remains unclear. Based on the finding that the Snf1p kinase is required for Adr1p to bind to DNA, it has been suggested that initial binding of Adr1p to UAS1 under derepression conditions is because of its phosphorylation by the Snf1p kinase (see Ref. 17 and references therein). It is possible that Adr1p undergoes additional phosphorylation under oleate induction, and such phosphorylation is accountable for the increase in its binding. It is also clear that the regulatory role of Adr1p is complex and variable for different genes. For example, the expression of PEX5 is increased in cells lacking Adr1p when cells are grown in the presence of glycerol, but it decreased when the cells are switched to media containing oleate. Thus, Adr1p appears to act as a positive or negative regulator, depending on the environmental conditions. It is likely that the positive regulation includes the stimulation of Oaf1p-Pip2p binding to a target sequence by Adr1p under both glucose repression and oleate induction. However, the mechanism by which Adr1p negatively regulates Oaf1p-Pip2p target genes remains to be investigated.
The results obtained during the course of this study, together with previous findings from our group and others, lead us to propose a model of Oaf1p-Pip2p and Adr1p action that induces the expression of oleate-responsive genes. When cells are grown in the presence of glucose, Pip2p is expressed at extremely low levels, whereas Oaf1p is constitutively expressed at higher levels. Under these conditions, Pip2p is most likely present exclusively in the form of heterodimers with Oaf1p, and these heterodimers bind only weakly to the ORE. Under the same glucose repression conditions, Adr1p is not able to bind to its target sequence UAS1. Together this results in a very low level of expression of oleate-responsive genes in this nutritional environment. Upon derepression, Oaf1p and perhaps Pip2p undergo phosphorylation, and this post-translational modification greatly increases Oaf1p-Pip2p binding to the ORE and initiates a higher expression of PIP2 by an autoregulatory mechanism. Adr1p also binds to UAS1 under derepression conditions, probably because of its phosphorylation. This binding increases Oaf1p-Pip2p association with the ORE whereas this complex, in turn, positively influences Adr1p recruitment to UAS1. The presence of both Oaf1p-Pip2p and Adr1p on the promoters of oleate-responsive genes turns on the transcriptional machinery and allows it to drive the expression of target genes to the moderate derepression levels. When oleate is supplied into glycerol-containing media, Oaf1p receives a fatty acid nutritional signal and appears to undergo additional phosphorylation. This causes a further moderate increase in Oaf1p-Pip2p binding to OREs. The oleate nutritional cue turns the DNA-bound Oaf1p-Pip2p heterodimer to the active transcriptional state, probably via a conformational change because of its phosphorylation. It is possible that Adr1p also undergoes additional phosphorylation upon receiving a fatty acid nutritional signal from the Oaf1p-Pip2p complex, and this phosphorylation causes an increase in its binding. All of these events increase the level of expression of the target genes, including PIP2, and trigger the autoregulatory mechanism to achieve maximal transcription under oleate induction conditions.
While this manuscript was under review, Smith et al. (50) published a report describing a systematic analysis of the response to fatty acids in yeast, using a combination of genome-wide chromatin localization and expression analyses. In their study, the authors investigated a regulatory network of four transcription factors, Oaf1p, Pip2p, Adr1p, and an additional negative regulator of the oleate response Ykr064wp, named Oaf3p (50). The authors demonstrated that this regulatory network expanded, and there was more combinatorial control, in the presence of oleate. These results are in accordance with our data that demonstrate an increase in Oaf1p-Pip2p association with target genes, as well as new interactions of Adr1p with target sequences, upon switching from glucose to oleate growth conditions. We should note, however, that the systematic analysis of Smith et al. (50) did not detect some well established interactions such as the association of the Oaf1p-Pip2p complex with the POX1 ORE or Adr1p with the POX1 UAS1 (5, 6, 19, 39, 42). Clearly, additional studies are necessary to characterize transcriptional regulatory networks of yeast response to fatty acids and the details of the precise mechanism by which a fatty acid nutritional signal causes transcriptional activation of target genes.
Acknowledgments
We gratefully acknowledge Alexandra Obregon, Vitaly Zyhadlo, and Juncheng Li for excellent technical assistance. We also thank Drs. Avrom Caplan and Gintaras Deikus for reading and helpfully discussing the manuscript.
This work was supported by American Heart Association Grant 0350364N, NIGMS Grant SO6 GM-08168 from the National Institutes of Health, and National Institutes of Health Grant RCMI RR 03060. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ORE, oleate-response element; ChIP, chromatin immunoprecipitation; RT, reverse transcription; CIP, calf intestinal phosphatase; UAS, upstream activation sequence; HA, hemagglutinin; IP, immunoprecipitation.
I. V. Karpichev, J. M. Durand-Heredia, and G. M. Small, unpublished results.
References
- 1.Gancedo, J. M. (1998) Microbiol. Mol. Biol. Rev. 62 334–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuller, H. J. (2003) Curr. Genet. 43 139–160 [DOI] [PubMed] [Google Scholar]
- 3.Veenhuis, M., Mateblowski, M., Kunau, W. H., and Harder, W. (1987) Yeast 3 77–84 [DOI] [PubMed] [Google Scholar]
- 4.Einerhand, A. W. C., Kos, W. T., Distel, B., and Tabak, H. F. (1993) Eur. J. Biochem. 314 323–331 [DOI] [PubMed] [Google Scholar]
- 5.Karpichev, I. V., and Small, G. M. (1998) Mol. Cell. Biol. 18 6560–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karpichev, I. V., Luo, Y., Marians, R. C., and Small, G. M. (1997) Mol. Cell. Biol. 17 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rottensteiner, H., Kal, A. J., Hamilton, B., and Tabak, H. F. (1997) Eur. J. Biochem. 247 776–783 [DOI] [PubMed] [Google Scholar]
- 8.Issemann, I., and Green, S. (1990) Nature 347 645–650 [DOI] [PubMed] [Google Scholar]
- 9.Issemann, I., Prince, R. A., Tugwood, J. D., and Green, S. (1993) J. Mol. Endocrinol. 11 37–47 [DOI] [PubMed] [Google Scholar]
- 10.Reddy, J. K., and Hashimoto, T. (2001) Annu. Rev. Nutr. 21 193–230 [DOI] [PubMed] [Google Scholar]
- 11.Phelps, C., Gburcik, V., Suslova, E., Dudek, P., Forafonov, F., Bot, N., MacLean, M., Fagan, R. J., and Picard, D. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 7077–7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumgartner, U., Hamilton, B., Piskacek, M., Ruis, H., and Rottensteiner, H. (1999) J. Biol. Chem. 274 22208–22216 [DOI] [PubMed] [Google Scholar]
- 13.Rottensteiner, H., Kal, A. J., Filpits, M., Binder, M., Hamilton, B., Tabak, H. F., and Ruis, H. (1996) EMBO J. 15 2924–2934 [PMC free article] [PubMed] [Google Scholar]
- 14.Smith, J. J., Marelli, M., Christmas, R. H., Vizeacoumar, F. J., Dilworth, D. J., Ideker, T., Galitski, T., Dimitrov, K., Rachubinski, R. A., and Aitchison, J. (2002) J. Cell Biol. 158 259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kal, A. J., van Zonneveld, A. J., Benes, V., van den Berg, M., Koerkamp, M. G., Albermann, K., Strack, N., Ruijter, J. M., Richter, A., Dujon, B., Ansorge, W., and Tabak, H. F. (1999) Mol. Biol. Cell 10 1859–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koerkamp, M. G., Rep, M., Bussemaker, H. J., Hardy, G. P., Mul, A., Piekarska, K., Szigyarto, C. A., De Mattos, J. M., and Tabak, H. F. (2002) Mol. Biol. Cell 13 2783–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurvitz, A., and Rottensteiner, H. (2006) Biochim. Biophys. Acta 1763 1392–1402 [DOI] [PubMed] [Google Scholar]
- 18.Simon, M., Adam, G., Rapatz, W., Spevak, W., and Ruis, H. (1991) Mol. Cell. Biol. 11 699–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurvitz, A., Hiltunenm, J. K., Erdmann, R., Hamilton, B., Hartig, A., Ruis, H., and Rottensteiner, H. (2001) J. Biol. Chem. 276 31825–31830 [DOI] [PubMed] [Google Scholar]
- 20.Gurvitz, A., Wabnegger, L., Rottensteiner, H., Dawes, I. W., Hartig, A., Ruis, H., and Hamilton, B. (2000) Mol. Cell. Biol. Res. Commun. 4 81–89 [DOI] [PubMed] [Google Scholar]
- 21.Rottensteiner, H., Wabnegger, L., Erdmann, R., Hamilton, B., Ruis, H., Hartig, A., and Gurvitz, A. (2003) J. Biol. Chem. 278 27605–27611 [DOI] [PubMed] [Google Scholar]
- 22.Cheng, C., Kacherovsky, N., Dombek, K. M., Camier, S., Thukral, S. K., Rhim, E., and Young, E. T. (1994) Mol. Cell. Biol. 14 3842–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciriacy, M. (1975) Mol. Gen. Genet. 138 157–164 [DOI] [PubMed] [Google Scholar]
- 24.Denis, C. L., and Young, E. T. (1983) Mol. Cell. Biol. 3 360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon, M., Binder, M., Adam, G., Hartig, A., and Ruis, H. (1992) Yeast 8 303–309 [DOI] [PubMed] [Google Scholar]
- 26.Young, E. T., Dombek, K. M., Tachibana, C., and Ideker, T. (2003) J. Biol. Chem. 278 26146–26158 [DOI] [PubMed] [Google Scholar]
- 27.Harbison, C. T., Gordon, D. B., Lee, T. I., Rinaldi, N. J., Macisaac, K. D., Danford, T. W., Hannett, N. M., Tagne, J. B., Reynolds, D. B., Yoo, J., Jennings, E. G., Zeitlinger, J., Pokholok, D. K., Kellis, M., Rolfe, P. A., Takusagawa, K. T., Lander, E. S., Gifford, D. K., Fraenkel, E., and Young, R. (2004) Nature 431 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tachibana, C., Yoo, J. Y., Tagne, J. B., Kacherovsky, N., Lee, T. I., and Young, E. (2005) Mol. Cell. Biol. 25 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., Zeitlinger, J., Jennings, E. G., Murray, H. L., Gordon, D. B., Ren, B., Wyrick, J. J., Tagne, J. B., Volkert, T. L., Fraenkel, E., Gifford, D. K., and Young, R. A. (2002) Science 298 763–764 [DOI] [PubMed] [Google Scholar]
- 30.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., Volkert, T. L., Wilson, C. J., Bell, S. P., and Young, R. A. (2000) Science 290 2306–2309 [DOI] [PubMed] [Google Scholar]
- 31.Gunji, W., Kai, T., Takahashi, Y., Maki, Y., Kurihara, W., Utsugi, T., Fujimori, F., and Murakami, Y. (2004) DNA Res. 11 163–177 [DOI] [PubMed] [Google Scholar]
- 32.Aparicio, O., Geisberg, J. V., Sekinger, E., Yang, A., Moqtaderi, Z., and Struhl, K. (2005) in Current Protocols in Molecular Biology (Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K., eds) pp. 21.3.1–21.3.33, John Wiley & Sons, Inc., New York [DOI] [PubMed]
- 33.Orlando, V. (2000) Trends Biochem. Sci. 25 99–104 [DOI] [PubMed] [Google Scholar]
- 34.Robert, F., Pokholok, D. K., Hannett, N. M., Rinaldi, N. J., Chandy, M., Rolfe, A., Workman, J. L., Gifford, D. K., and Young, R. (2004) Mol. Cell 16 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karpichev, I. V., Cornivelli, L., and Small, G. M. (2002) J. Biol. Chem. 277 19609–19617 [DOI] [PubMed] [Google Scholar]
- 36.Sikorski, R. S., and Hieter, P. (1989) Genetics 122 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bahler, J., Wu, J.-Q., Longtine, M., Shan, N. G., Amos, M., Steever, A. B., Wach, A., Philippsen, P., and Pringle, J. R. (1998) Yeast 14 943–951 [DOI] [PubMed] [Google Scholar]
- 38.Geisberg, J. V., and Struhl, K. (2005) in Current Protocols in Molecular Biology (Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K., eds) pp. 21.8.1–21.8.7, John Wiley & Sons, Inc., New York
- 39.Luo, Y., Karpichev, I. V., Kohanski, R. A., and Small, G. M. (1996) J. Biol. Chem. 271 12068–12075 [DOI] [PubMed] [Google Scholar]
- 40.Skoneczny, M., Chelstowska, A., and Rytka, J. (1988) Eur. J. Biochem. 174 297–302 [DOI] [PubMed] [Google Scholar]
- 41.MacPherson, S., Larochelle, M., and Turcotte, B. (2006) Microbiol. Mol. Biol. Rev. 70 583–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo, Y. (1997) Molecular Mechanisms Regulating Transcription of the Gene Encoding Peroxisomal Acyl-CoA Oxidase in Saccharomyces cerevisiae. Ph.D. dissertation, Mount Sinai School of Medicine, New York
- 43.Filipits, M., Simon, M. M., Rapatz, W., Hamilton, B., and Ruis, H. (1993) Gene (Amst.) 132 49–55 [DOI] [PubMed] [Google Scholar]
- 44.Simon, M. M., Pavlik, P., Hartig, A., Binder, M., Ruis, H., Cook, W. J., Denis, C. L., and Schanz, B. (1995) Mol. Gen. Genet. 249 289–296 [DOI] [PubMed] [Google Scholar]
- 45.Agricola, E., Verdone, L., Xella, B., Di Mauro, E., and Caserta, M. (2004) Biochemistry 43 8878–8884 [DOI] [PubMed] [Google Scholar]
- 46.Sellick, C. A., and Reece, R. (2005) Trends Biochem. Sci. 30 405–412 [DOI] [PubMed] [Google Scholar]
- 47.Sellick, C. A., and Reece, R. (2003) EMBO J. 22 5147–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirkpatrick, C. R., and Schimmel, P. (1995) Mol. Cell. Biol. 15 4021–4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sze, J. Y., Woontner, M., Jaehning, J. A., and Kohlhaw, G. (1992) Science 258 1143–1145 [DOI] [PubMed] [Google Scholar]
- 50.Smith, J. J., Ramsey, S. A., Marelli, M., Marzolf, B., Hwang, D., Saleem, R. A., Rachubinski, R. A., and Aitchison, J. D. (2007) Mol. Systems Biol. 3 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas, B. J., and Rothstein, R. (1989) Cell 56 619–630 [DOI] [PubMed] [Google Scholar]