Abstract
MEF2 (myocyte enhancer factor 2) proteins are a small family of transcription factors that play pivotal roles in striated muscle differentiation, development, and metabolism, in neuron survival and synaptic formation, and in lymphocyte selection and activation. Products of the four mammalian MEF2 genes, MEF2A, MEF2B, MEF2C, and MEF2D, are expressed with overlapping but distinct temporospatial patterns. Toward analysis of MEF2A functions and the determinants of its regulated expression, we have mapped and begun studies of the transcriptional control regions of this gene. Heterogeneous 5′-untranslated regions of MEF2A mRNAs result from use of alternative promoters and splicing patterns. The two closely approximated TATA-less promoters are ∼65 kb upstream of the exon containing the sole initiation codon. Ribonuclease protection and primer extension assays show that each promoter is active in various adult tissues. A canonical MEF2 site overlies the major promoter 1 transcription start site. This element specifically binds MEF2 factors, including endogenous nuclear MEF2A according to chromatin immunoprecipitation studies, and is critical to MEF2A transcription in myocytes. The site exerts reciprocal control of the alternative promoters, silencing promoter 1 and activating promoter 2 under some conditions. Erk5 and p38 MAPK signaling stimulate MEF2A expression by activating both promoters from the MEF2 element. MEF2A transcription is therefore subject to positive or negative regulation by its protein products, depending on signaling activities that influence MEF2 factor trans-activity. The sole MEF2 gene of the cephalochordate amphioxus has a similar regulatory region structure, suggesting that this mode of autoregulatory control is conserved among higher metazoan MEF2 genes.
MEF2 (myocyte enhancer factor 2) proteins are a subset of the minichromosome maintenance/agamous/deficiens/serum-response factor (MADS)5 box transcription factor superfamily (1). MEF2A was the first MEF2 to be identified (2, 3), and four splicing variants are encoded by the MEF2A gene of all vertebrates (4).6 Three additional mammalian MEF2 isotypes, MEF2B, MEF2C, and MEF2D, were subsequently described (5, 6). Other metazoans, from Porifera to Chordata, also have MEF2 genes. All MEF2 proteins have an N-terminal MADS box and an adjacent MEF2-specific signature domain that function together to specify MEF2 dimerization and sequence-specific dimer binding to CTA(T/A)4TAR (or YTA(T/A)4TAG) sequences in the regulatory regions of target genes (1, 5, 6).
Studies of MEF2 function initially focused on myogenesis and muscle-specific gene expression (6). It is now apparent that these factors play many roles in various tissues and cell types, both during development and after terminal differentiation. MEF2 proteins contribute to the orchestration of skeletal muscle differentiation (6, 7) and fiber type programming (8, 9), cardiac development and hypertrophy (10, 11), and vascular development and smooth muscle proliferation (12–14). These factors are also involved in excitation-dependent neuron survival (15, 16) and synapse formation (17, 18). Some MEF2 factors are expressed in immune cells where they control T cell selection and activation and cytokine expression (19, 20).
The vertebrate MEF2 gene products are differentially expressed spatially and temporally within tissues and tissue regions (21–23). In the mouse embryo, mef2a mRNA expression appears first in the myocardium at day 8.5 post-coitum, 1 day after mef2c mRNA first appears in mesodermal precursors of this tissue (21). Myocardial mef2a expression is maintained thereafter throughout development and after terminal differentiation. mef2a expression also lags slightly behind mef2c in the myotome, appearing first at day 9.5 post-coitum and within all muscle-forming regions of the embryo thereafter (21). Expression is also maintained in differentiated skeletal and smooth muscle cells. The order of MEF2 isotype gene expression in the embryo differs from that seen in differentiating cultured myoblasts (4, 24). Here, mef2d is an early marker, followed by mef2a after lineage commitment and mef2c late in differentiation. In the developing and adult brain, mef2a is widely expressed but is most abundant in the thalamus (23). Mechanisms controlling temporospatial MEF2A gene expression patterns are not known.
mef2a null mice survive to birth, but a large proportion die during the immediate postnatal period (25). These animals show right ventricular enlargement and reduced density and disorganization of myocardial mitochondria. The fraction of mutant mice that do survive to adulthood also have mitochondrial defects. Because a MEF2-responsive reporter gene was highly active in the myocardium of the mef2a–/– mice, and targeted disruption of mef2c does not give mitochondrial defects, MEF2A may serve a unique role among the MEF2 isotypes in mitochondrial biogenesis. The distinct phenotypes of mef2a–/– and mef2c–/– mice indicate that there are nonredundant functions of the MEF2 isotypes. Some are likely to be due to differential expression.
In contrast to extensive work on MEF2/MEF2 regulation at levels of modifications in response to signaling (5, 7, 18, 26–28) and transcription factor and co-regulator interactions (5, 6, 29), and some studies of translational (30) and pre-mRNA splicing control (4, 31), examination of MEF2 gene transcription has been limited to studies of Drosophila Mef2 (32, 33) and murine mef2c (34–36). As an initial step in the analysis of MEF2A transcriptional regulation, we report here the mapping and partial characterization of the highly conserved 5′-regulatory region of the mammalian MEF2A gene.
EXPERIMENTAL PROCEDURES
RNA Analyses—Human tissue total RNA was purchased from Clontech and Ambion, and murine tissue and C2C12 cell RNA was isolated by conventional procedures (37). Primer extension assays were performed as described (38) using primers hybridizing in human MEF2A exon A1 or exon A2. 5′-Rapid amplification of cDNA ends used human heart cDNA and reverse primers that hybridize in MEF2A exon 1 near the initiation codon. Ribonuclease protection assays (RPA) and radiolabeled cRNA probe syntheses were carried out as described (4, 31). MEF2A cRNA templates had fragments specific to A1/C and A2/C exon junctions. Templates for invariant segments of murine mef2a, mef2c, and mef2d were generated using RT-PCR on heart RNA with gene-specific primers. The human MEF2A cRNA probe template was a sequence at the 3′ end of the coding region.
Reporter Plasmid Construction—ptk-Luc was described (38). [MEF2MEF2A]3tk-Luc has three copies of the human MEF2A MEF2 element inserted into ptk-Luc upstream of the HSV tk promoter. The human JUN promoter (39) was obtained by PCR on genomic DNA template, and the amplicon was substituted for the tk promoter fragment of ptk-Luc to give pJUN-Luc. A subcloned RPCI-11C-231N21 BAC clone fragment containing ∼3.6 kb spanning the MEF2A promoter region was identified by hybridization screening. PCR on this template gave promoter 1 and 2 fragment amplicons that were substituted for tk sequence into ptk-Luc to give [L]-, [I]-, and [S]-MEF2Ap1-Luc and [L]-, [I]-, and [S]-MEF2Ap2-Luc, respectively. p1-[m1MEF2]-Luc and p2-[m1MEF2]-Luc were made using PCR mutagenesis.
Mammalian Expression Vector Construction—The pCDNA-MEF2A, -MEF2C, and -MEF2D splicing variant and -MEF2B7 constructs have been described (4, 31). A C-terminal FLAG epitope tag (DYKDDDDK) was appended to each using PCR to give pCDNA-MEF2(A/B/C/D)FLAG. MEF2A and MEF2D templates were FLAG-tagged constructs (gifts of J. Dilworth). Plasmids containing human MKK6 (40) mutants (MKK6EE = MKK6S207E/T211E; MKK6KA = MKK6K82A, gifts of J. Kyriakis) were PCR-amplified, and amplicons were subcloned into a derivatized pCDNA3 with sequence encoding the hemagglutinin (HA) epitope (YPYDVPDYA) to give pCDNA-HA-MKK6EE and -HA-MKK6KA. Coding sequences of a C-terminal truncation mutant of calcineurin A (41) and the protein kinases MEK5 and Erk5 (42) were obtained using RT-PCR on human heart RNA. Amplicons were subcloned into pCDNA3.1 hygro (Invitrogen) to give pCDNA-CnA+, pCDNA-MEK5, and pCDNA-Erk5, respectively. pCDNA-MEK5DD (=MEK5S311D,T315D) and pCDNA-Erk5AF (=Erk5T219A,Y221F) (42) were created using PCR mutagenesis.
Chromatin Immunoprecipitation—Procedures used were modified from the ChIP-IT kit using enzymatic DNA shearing (Active Motif). Methods are detailed in the supplemental material, as are human MEF2A and mouse mef2a genes and control primers. Results were taken only when validated with both positive and negative antibody and PCR controls. Processing of three independent cross-linked samples gave similar results.
Mobility Shift Assays—The various MEF2 α2/β coding region fragments (4, 31) were isolated from respective pCDNA3 plasmids and subcloned into pET28 to give pET-MEF2A, -MEF2C, and -MEF2D. Coding regions of murine mef2a, mef2c, and mef2d (4, 31) were obtained by RT-PCR using C2C12 myotube RNA, and each amplicon was subcloned into pET28 to give pET-mMEF2A, -mMEF2C, or -mMEF2D. In vitro transcription and translation reactions used these plasmid templates with the TnT system (Promega). Nuclear extracts were prepared from C2C12 myoblasts and myotubes as described (37, 38). Mobility shift assays were conducted as described (38), except that 0.25× TBE buffer, pH 7.5, was used.
Cultured Cell Transfection—C2C12 cells were maintained and differentiated as described (4, 31, 38). HeLa and HEK293 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum. Cells were plated in 12-well plates to achieve 25% confluence for transfection using Superfect (Qiagen) the following day. Triplicate wells received 0.5–1.0 μg of reporter plasmid, 0.3 μg of control reporter, and 0.05–0.5 μg of expression vector(s). Wells received identical amounts of reporter and expression vector by using plasmids lacking inserts as necessary. Cells were harvested 16–48 h after transfection for luciferase and β-galactosidase activity assays. For experiments involving modulation of p38 MAPK or calcineurin activities, Dulbecco's modified Eagle's medium without fetal bovine serum was used for the final 8–16 h before cell harvest. Luciferase readings were corrected for transfection efficiency using β-galactosidase activity from pSV40βGal, or Renilla luciferase activity from pRL-tk or pRL-SV40 (Promega). SB203580 was used at 1 μm, and cyclosporin A at 100 nm.
Amphioxus MEF2 cDNA and Gene Structure Determinations—GenBank™ expressed sequence tag partial cDNA sequences were compared with Joint Genome Institute amphioxus Branchiostoma floridae gene sequences to establish the intron/exon structure of the 5′ end of MEF2 (exons A → C and 1 → 4). Deduced MEF2 protein sequences of both Ciona intestinalis and Takifugu rubripes were then used as queries in TBLASTN to identify additional putative amphioxus MEF2 exons. Precise intron/exon boundaries were established by applying consensus splice site criteria (43). The predicted mRNAs, including splicing variants, extending from the complete 5′-UTR through the termination codon were then established.
Reagents and Miscellaneous Procedures—Plasmid segments derived from PCR were verified by dideoxy sequencing (37). Rabbit anti-MEF2 isotype-specific antibodies were generated using peptide antigens and conventional procedures (37). Other antibodies were from Santa Cruz Biotechnology (anti-MEF2, C-21), Upstate (anti-RNA polymerase II, 8WG16), and Sigma (anti-FLAG). PAGE-separated proteins were immunoblotted (IB) using chemiluminescence assays (ECL, Amersham Biosciences) with horseradish peroxidase-coupled secondary antibodies (Jackson ImmunoResearch). p38 assays used GST-ATF-2 as a phosphorylation substrate (40) (Cell Signaling Technology). Calcineurin activity in cell extracts was measured by quantitating release of free phosphate from RII phosphopeptide using a Malachite Green colorimetric assay. SB203580 and cyclosporin A were from Calbiochem.
RESULTS
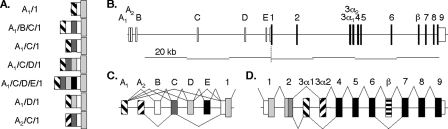
Heterogeneous MEF2A 5′-Untranslated Regions Result from Alternative Splicing—We used 5′-rapid amplification of cDNA ends, PCR, and data base cDNA sequence information to establish human MEF2A 5′-UTR sequences. Detected segments were compared with human chromosome 15q26 genomic sequence to complete the previously described partial MEF2A gene structure (44). Six exons (A1, A2, and B–E) exist in the ∼65-kb expanse upstream of the sole MEF2A initiation codon located in exon 1 (Fig. 1, A and B, and supplemental Table S1). Different 5′-UTRs result from various patterns of alternative splicing (Fig. 1C), with exon A1, C, and 1 sequence comprising the most common among them. The four MEF2 isotypes have highly similar gene structures and alternative splicing patterns among exons containing coding sequences (Fig. 1D) (4). This similarity does not extend to 5′-flanking regions, where the genes have different numbers and arrangements of alternative promoters, and different patterns of alternative splicing.6
FIGURE 1.
Seven exons contribute MEF2A 5′-UTR sequences. A, schematic of the various human MEF2A mRNA 5′-UTR sequences. Each shaded or patterned box represents a unique sequence segment. B, diagram of the entire ∼150-kb human MEF2A gene. Small open boxes denote UTR sequences and solid boxes denote coding sequences. C, alternative splicing patterns among MEF2A 5′ exons. A1 and A2 are alternative first exons. Exon 1 contains invariant 5′-UTR sequence and the sole initiation codon. D, alternative splicing patterns of vertebrate MEF2A among coding exons. MEF2C and MEF2D structures and splicing are similar to MEF2A, except that MEF2C has an alternative exon 9 splice acceptor (31).
Murine mef2a and other non-human mammalian MEF2A genes have structures that are similar to that of the human gene according to our in silico analyses. MEF2A cDNA data base entries from these species have 5′-UTR sequences consisting of exons A1, C, and 1 exclusively. However, sequences highly similar to human MEF2A exons A2, B, D, and E along with cognate splice sites are present at analogous locations in these other genes. It is therefore possible, if not likely, that murine mef2a and other mammalian MEF2A mRNAs have alternative 5′-UTRs that are comparable with those of human MEF2A.
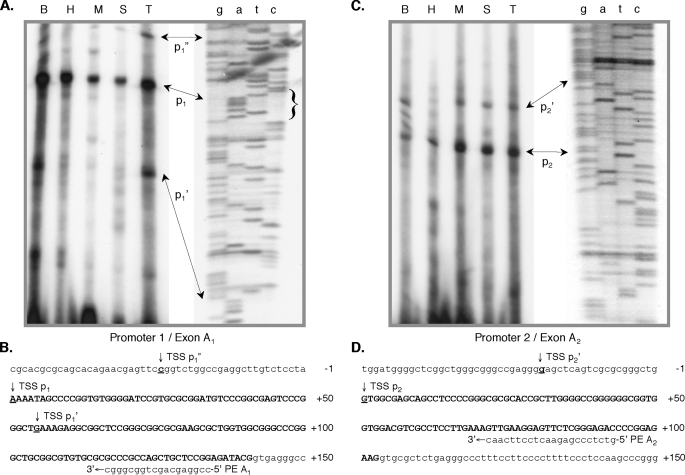
MEF2A Primary Transcripts Arise from Two Promoters—No human MEF2A cDNAs containing both A1 and A2 sequences were detected, and no obvious splice acceptor is present upstream of exon A2. We confirmed the implication that A1 and A2 are bona fide alternative first exons. RT-PCR with exon A1 forward and exon A2 reverse primers failed to produce an amplicon from various tissue RNA samples. We therefore designated the regions upstream of exons A1 and A2 as alternative promoters 1 (p1) and 2(p2), respectively. Primer extension and RPA were used to determine transcription start sites (TSS) and tissue distributions of MEF2A mRNA containing either exon A1 or A2. Using a primer hybridizing within exon A1, one major product predominated in all human tissue RNA samples examined (Fig. 2, A and B). The cognate TSS (TSS p1) corresponds to the 5′ end of several human expressed sequence tag clones, including some constructed using the “cap-trapper” strategy (45). Two additional extension products were seen with brain and testis RNA, the shorter of which (TSS p1′) corresponds to the 5′ end of multiple brain and epithelial cell data base cDNAs. Identification of this product in selected tissues suggests that this represents a true alternative TSS, although it may result from stalled RT as the sequence immediately upstream is G/C-rich.
FIGURE 2.
MEF2A has two TATA-less promoters. 2 μg of total RNA from murine brain (B), heart (H), skeletal muscle (M), spleen (S), or testis (T) was used in primer extension assays with antisense primers (PE A1 or PE A2) hybridizing within exon A1 (A) or A2 (C), respectively. The corresponding sequencing gel autoradiograms are vertically expanded for clarity. Brace in A delimits a canonical MEF2 element. B, sequence within and flanking exon A1. Numbers reference the major promoter 1 TSS (TSS p1). Alternative TSS (p1′ and p1″) are underlined, and the location of antisense primer PE A1 is shown. D, sequence within and flanking exon A2. Numbers reference the major promoter 2 TSS (TSS p2). The most 5′-alternative (TSS p2′) is underlined, and the location of antisense primer PE A2 is shown.
On similar analyses using a primer hybridizing within MEF2A exon A2, one major extension product was seen (Fig. 2, C and D). This corresponds to a site in MEF2A genomic sequence (TSS p2) immediately downstream of a group of GC boxes. Several minor alternative sites were apparent in brain, clustered upstream within 20 bases of this site, and one was also present in other tissues (TSS p2′). The total A2 extension product and the ratio of A2 to A1 product were consistently highest in skeletal muscle, suggesting that promoter 2 may be more active in this tissue. RPA were performed to further examine the tissue distribution of the alternative MEF2A transcripts. Complementary RNA probes derived from cDNAs containing either exons A1 and C or A2 and C were generated, but the A2/C probe was not useful because of extremely high G/C content and a lengthy near-palindromic sequence in exon A2. MEF2A A1/C mRNA was detected in all tissues (data not shown). We conclude from this and the primer extension results that the alternative TATA-less MEF2A promoters, each with a cluster of transcription initiation sites, are not selectively active among the tissues examined. We found no evidence for MEF2A promoters other than p1 and p2.
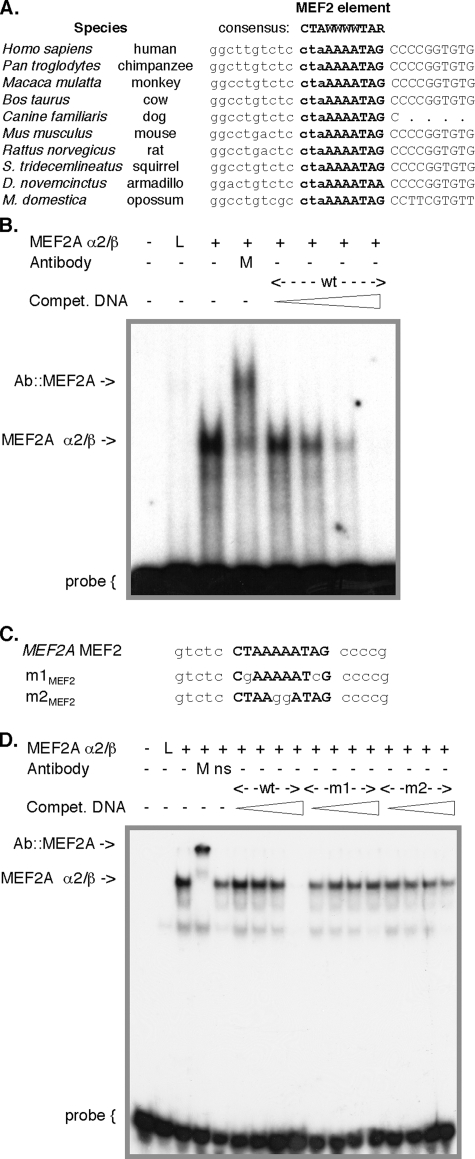
A Conserved MEF2 Element Overlies the MEF2A Promoter 1 Transcription Start Site—A canonical MEF2 element overlies the major MEF2A promoter 1 TSS (Fig. 2, A, brace, and B), and there is absolute conservation of this site among mammals (Fig. 3A). Promoter 1 also includes a functional canonical nuclear respiratory factor 1 (NRF1) (46) element at –53 relative to TSS p1,8 and potential NRF2 (46) (–273 and –307), NF-AT (47) (–250), and E box elements (48) (–398 and –782) within 800 bp of TSS p1. These elements are also entirely conserved in sequence and location in murine mef2a and other mammalian MEF2A genes.8 Promoter 2 has multiple GC boxes (49), including three within 80 bp of TSS p2. This regulatory region is otherwise unremarkable but is highly conserved among primates.6 We have begun to examine the control of MEF2A transcription by these elements, and we focus in this report on the MEF2 element.
FIGURE 3.
A canonical MEF2 element overlies the major MEF2A promoter 1 TSS. A, alignment of mammalian MEF2A gene sequences near the promoter 1 TSS shows evolutionary conservation of the MEF2 element (boldface letters). Transcribed sequences are in uppercase. Sequences are from GenBank™ entries AC013526 (human), AADA01045866 (chimpanzee), AANU01176656 (rhesus monkey), AC164694 (cow), CE302601 (dog), AC120123 (mouse), AC134737 (rat), AAQQ01634896 (squirrel), AAGV01302117 (armadillo), and AAFR03022775 (opossum). A high degree of sequence similarity also exists within 0.8 kb upstream of this site.8 B and D, autoradiograms of an EMSA using the MEF2A probe. Binding reactions included in vitro translated MEF2A α2/β+ or unprogrammed reticulocyte lysate (L), anti-MEF2 antibody (M), or antibody to the unrelated antigen (ns). Some reactions included competitor oligonucleotides in 3-, 10-, 30-, or 100-fold excess, as indicated by triangles. C, sense strand of the MEF2A promoter probe and competitor oligonucleotides used in EMSA. Residues that conform to a consensus MEF2 element are in boldface; mutated residues in the m1MEF2 and m2MEF2 oligonucleotides are in lowercase.
To verify binding of MEF2 factors to the MEF2A promoter element, we performed mobility shift assays with an oligonucleotide probe and in vitro translated MEF2A. For these studies, the MEF2A α2/β+ isoform was used, as this is the predominant MEF2A splicing form in differentiated muscle (4).7 A DNA-protein complex was identified that was supershifted by an antibody that recognizes all MEF2 proteins, confirming a MEF2A-MEF2A promoter probe interaction (Fig. 3B). This complex was competed by excess unlabeled probe, but not by otherwise analogous oligonucleotides containing mutations in either peripheral (m1MEF2) or A/T core (m2MEF2) residues within the site (Fig. 3, C and D). Specific retarded complexes were also seen in assays with this probe and other translated MEF2 proteins, as well as tissue and muscle cell nuclear extracts (see below). Thus, MEF2 factors can bind directly at the major site of MEF2A transcription initiation.
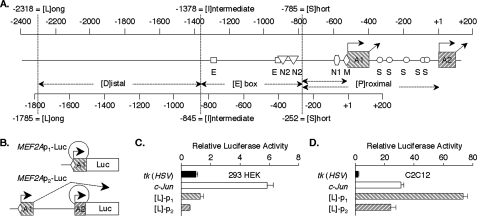
The MEF2 Element Regulates MEF2A Promoter Activity in Myocytes—We constructed reporters to evaluate MEF2A promoter activities (Fig. 4, A and B). Promoter 1 activity was assessed using p1-Luc in which the 3′ end of exon A1 is fused directly to luciferase. Promoter 2 was evaluated using p2-Luc. Here, the 3′ end of exon A2 is fused to luciferase such that transcription initiated at a TSS p2 generates luciferase mRNA, whereas transcripts from a TSS p1 are spliced to omit luciferase sequences. A deletion series of each construct included short (S), intermediate (I), and long (L) promoter segments. Proximal (P) promoter regions were present in each construct, and E box-containing (E) and distal (D) segments were variably included (Fig. 4A).
FIGURE 4.
MEF2A promoters are highly active in myocytes. A, diagram of the MEF2A transcriptional regulatory region. The upper index references distances from TSS p2; the lower references distances from TSS p1. Lengths of the short (S), intermediate (I), and long (L) promoter reporters are shown, and the regulatory region is divided into distal (D), E box-containing (E), and proximal (P) segments. Sites that are conserved among mammalian MEF2A promoters are shown, including the MEF2 site (M), an NRF1 (N1) and multiple NRF2 (N2), and E box (E) elements (Footnote 8). B, MEF2A promoter-reporters. The promoter 1 (p1) series has exon A1 fused directly to the luciferase coding region. The promoter 2 (p2) series has exon A2 fused to luciferase. C, indicated reporters (1.0 μg/well) were co-transfected with SV40-βgal (0.3 μg) into HEK293 cells in triplicate wells. After 18 h, cell extract luciferase activities were determined and normalized for transfection efficiency using β-galactosidase activity and then to the activity of ptk-Luc (=1.0). Data represent the average ± S.E. for triplicate wells in three independent experiments. D, C2C12 cells were transfected and analyzed as in C, except that cells were transfected at 50% confluence and allowed to begin differentiation (48–72 h) prior to harvesting.
The transcriptional activities of MEF2A promoters were compared with those of the herpes simplex virus thymidine kinase (ptk-Luc) (38) and human JUN (pJUN-Luc) genes. JUN has a single MEF2 element in the proximal promoter (39). In HEK293 cells, pJUN-Luc activity was substantially higher than L-p1-Luc (Fig. 4C). Qualitatively similar results were observed in transfected fibroblast cell lines (data not shown). By contrast, L-p1-Luc was much more active than both tk (∼70-fold) and JUN (∼2-fold) reporters in C2C12 cells (Fig. 4D). This promoter 1 reporter was ∼3-fold more active than L-p2-Luc in both cell types. The more robust activity of the MEF2A promoters in myocytes as compared with non-muscle cells is consistent with preferential expression of MEF2A mRNA in muscle (2, 4).
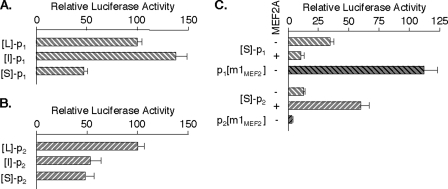
The two deletion series were used to determine regulatory sequences crucial to MEF2A transcription in C2C12 cells. Both S reporters were much more active than ptk-Luc, confirming that the P regions retain sequences critical to robust MEF2A promoter activity in myocytes. For promoter 1, activity of the I reporter was consistently higher than that of the L reporter (average ∼150%), whereas the S construct was approximately half as active (Fig. 5A). This indicates that promoter 1 is positively controlled from the E and P regions and that silencing element(s) may exist in the D segment. The upstream regulatory regions exert different effects on promoter 2. In this case, the I and S constructs each had approximately half the activity of the L-p2-Luc reporter (Fig. 5B). Therefore, the D and P regions have elements that function as enhancers for promoter 2.
FIGURE 5.
The MEF2A MEF2 element differentially controls promoter 1 and promoter 2 activities. C2C12 cells were transfected as described in Fig. 4D with indicated MEF2A promoter 1 (A) or promoter 2 (B) reporters. Values are normalized to the activity of the respective full-length (L) promoter reporter (=100). Averages from four independent experiments are shown. C, C2C12 cells were transfected with indicated reporters and analyzed as in A and B, except with or without pCDNA-MEF2A α2/β (0.5 μg/well), and activities are normalized to that of ptk-Luc. The m1MEF2 reporters have MEF2 element mutations identical to the m1MEF2 probes in Fig. 3B.
The function of the MEF2 element was evaluated by comparing activities of the S-Luc reporters with analogous constructs in which the MEF2 site was mutated as in the m1MEF2 competitor probe used in mobility shift assays. The mutation resulted in a marked reduction in promoter 2 activity (Fig. 5C). Coupled with the fact that forced expression of MEF2A α2/β stimulated the wild-type promoter, this shows that the MEF2 element can positively direct transcription from promoter 2. By contrast, p1-[m1MEF2]-Luc activity was consistently higher than that of p1-Luc in these and other cell types (Fig. 5C). Furthermore, forced expression of MEF2 factors inhibited p1-Luc activity to as low as ∼20% of control. MEF2 or associated proteins can therefore use this element to silence transcription from this alternative MEF2A promoter.
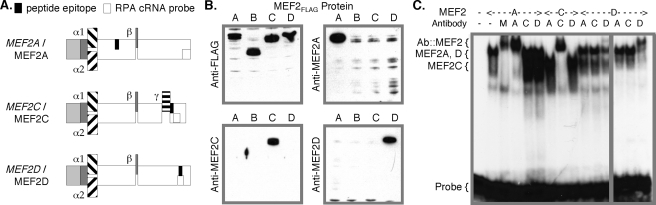
Endogenous Nuclear MEF2A Binds the MEF2A Promoter Element—We examined the composition of cellular MEF2 factor binding to MEF2A promoter using anti-peptide antibodies developed to discriminate MEF2A, MEF2C, and MEF2D. Epitopes were selected to permit immunodetection of all splicing isoforms of a particular MEF2 gene and to cross-react with known mammalian, avian, reptilian, amphibian, and actinopterygian MEF2 proteins where possible (Fig. 6A and supplemental Table S2). Antibody specificity was validated using IB (Fig. 6B) and IP (data not shown) with overexpressed FLAG epitope-tagged MEF2 proteins, as well as in mobility shift assays with the MEF2A MEF2 element probe and in vitro translated MEF2 proteins (Fig. 6C).
FIGURE 6.
MEF2 isotype-specific antibodies. A, MEF2A, MEF2C, and MEF2D splicing variant mRNAs and cognate proteins are depicted, along with the locations of antibody epitopes (solid boxes) and RPA cRNA probes (open boxes). The MADS box (light shading), MEF2S domain (dark shading) and the alternative α, β, and γ domains are indicated. B, HEK293T cells were transfected with vectors expressing FLAG epitope-tagged human MEF2A α2/β, MEF2B, MEF2C α2/β, or MEF2D α2/β. After 48 h, whole cell protein extracts were harvested, resolved, and (re)-immunoblotted using the four indicated antibodies. C, mobility shift assay using the MEF2A MEF2 element as in Fig. 3C. Binding reactions included in vitro translated mouse MEF2A (A), MEF2C (C), or MEF2D (D) α1/β isoforms, and either a commercially available anti-MEF2 antibody (M) (Santa Cruz Biotechnology, C-21) or anti-MEF2A (A), anti-MEF2C (C), or anti-MEF2D (D) antisera. Reactions containing two different titers of anti-MEF2D are shown.
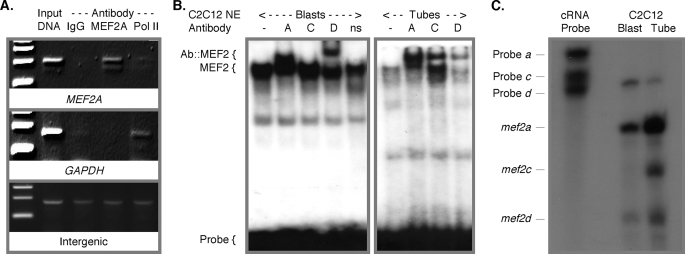
Having established efficacy of the anti-MEF2A antibody in IP and verified that the immunocomplex does not interfere with DNA binding, we used this reagent in ChIP assays to confirm endogenous nuclear MEF2A binding to the MEF2A promoter. In human HEK293T cells, which express a relatively low level of MEF2 proteins, ChIP detected MEF2A binding to the promoter 1/exon A1 region (Fig. 7A). Cellular MEF2 binding to the element was also confirmed using C2C12 myoblast and myotube nuclear extracts in mobility shift assays (Fig. 7B). According to supershifted bands in resolved reactions containing the MEF2 isotype-specific antibodies, the contributions of cellular MEF2A, MEF2C, and MEF2D to the DNA-protein complex correlated with the respective abundance in these samples. Thus, RPA using mef2a, mef2c, and mef2d cRNA probes (Fig. 6A) showed that mef2c is absent in early stages of differentiation, whereas differentiated myotubes express all three isotypes, and mef2a is most abundant (Fig. 7C) (24), and this was reflected in the supershifts. Taken together, these data show that MEF2A transcription is autoregulated, i.e. controlled by MEF2A, but suggest that the gene can also be controlled by other MEF2 factors.
FIGURE 7.
Endogenous nuclear MEF2A binds the MEF2A promoter element. A, ChIP using HEK293 cell nuclei and antibodies to MEF2A or RNA polymerase II (Pol II) or control IgG. The MEF2A proximal promoter 1 region, the GAPDH promoter, and an intergenic region were detected by PCR from co-precipitated chromatin. B, EMSA using the MEF2A MEF2 element probe as in Fig. 6C, except binding reactions included C2C12 myoblast or myotube nuclear extracts and the indicated anti-MEF2 or anti-Myc (ns) antibodies. C, RPA using RNA isolated from undifferentiated (Blast) and differentiated (Tube) C2C12 cells. The assay used pooled murine mef2-a, mef2-c, and mef2-d cRNA probes (Fig. 6A). n.b. correlation of MEF2 isotype expression with supershifted band intensities.
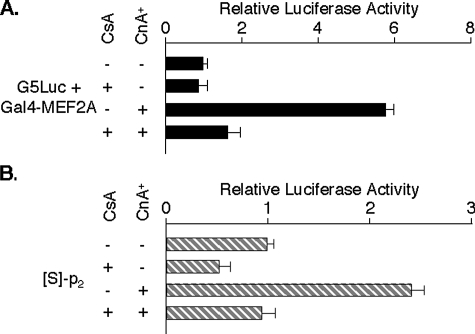
Calcineurin Activity Stimulates MEF2A Transcription—Contractile activity in striated muscle produces transient Ca2+ fluxes that activate the phosphoprotein phosphatase calcineurin (Cn) (9, 28). Because Cn activity is known to regulate the expression of a subset of MEF2-sensitive genes (18, 28, 41, 50), we examined whether MEF2A transcription is subject to this control. Forced expression of a constitutively active Cn catalytic subunit mutant (CnA+) and CsA were used to modulate Cn activity (41). In control experiments in C2C12 cells, the activity of Gal4-MEF2A α2/β on the Gal4-responsive reporter G5Luc was 6-fold higher in cells expressing CnA+ compared with controls (Fig. 8A). CsA exposure dampened the activity of the Gal4-MEF2A fusion and abolished the CnA+ effect. Expression of CnA+ led to a CsA-sensitive ∼4-fold stimulation of MEF2Ap2-Luc activity (Fig. 8B) but produced little effect on the promoter 1 reporters. This was dependent on an intact MEF2 element (data not shown), confirming that Cn influences MEF2A transcription from this site via direct or indirect activity of DNA-bound MEF2 factors. The net result of increased Cn activity was equilibration of the relative strengths of the alternative MEF2A promoters.
FIGURE 8.
Calcineurin activity activates MEF2A transcription from the promoter MEF2 element. C2C12 cells were transfected with G5Luc and Gal4-MEF2A α2/β (A) or with S-MEF2Ap2-Luc (B), each with or without CnA+, incubated in medium with or without 100 nm CsA for the final 8 h before cell harvesting and analyzed as in Fig. 4C. Values were normalized to basal activities of the respective reporters (=1.0).
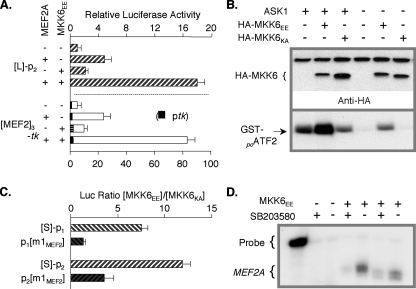
p38 MAPK and Erk5 Signaling Stimulate MEF2A Expression by Activating Its Promoters from the MEF2 Element—p38 MAPK phosphorylates two Thr residues on MEF2A, thereby stimulating its trans-activity (51). Coupled with MEF2A transcriptional autoregulation, this predicted sensitivity of MEF2A expression to p38 signaling. We tested this first by comparing MEF2A promoter reporter activities in cells in which p38 activity was modulated. Signaling was stimulated by forced expression of a constitutively active mutant of an upstream kinase, MKK6EE, and inhibited either by expression of an inactive kinase mutant (MKK6KA) or culture medium supplementation with SB203580 (40). In HeLa cells, which express little endogenous functional MEF2, MEF2A α2/β and MKK6EE synergistically stimulated L-p2-Luc (Fig. 9A). The isolated concatamerized MEF2A MEF2 element functioned similarly in a heterologous promoter context.
FIGURE 9.
p38 MAPK signaling stimulates MEF2A expression via the promoter MEF2 element. A, HeLa cells were transfected with indicated reporters (0.5 μg/well) with or without MEF2A α2/β (0.5 μg/well) and with or without MKK6EE (0.1 μg/well). Cells were treated and analyzed as in Fig. 4C, except values are normalized to the basal activity of L-p2MEF2A-Luc (upper) or ptk-Luc (lower)(=1.0). B, HEK293 cells were transfected with or without ASK1 and HA-tagged constitutively active (MKK6EE) or inactive (MKK6KA) MKK6 as indicated. p38 activity in cell extracts was determined by detecting modified GST-ATF-2 (lower). Resolved extracts were re-blotted with anti-HA antibody (upper). ASK1 activity activates MKK3/6 (83). C, HEK293 cells were transfected with indicated reporters (0.5 μg/well) and either MKK6EE or MKK6KA (0.5 μg/well). Ratios of the normalized luciferase activities are shown for each reporter. D, HEK293 cells were transfected with MKK6EE. Culture medium was replaced with serum-free medium with or without 1 μm SB203580 after 24 h. RNA harvested after an additional 18 h was used in RPA with a human MEF2A cRNA probe.
Efficacy of the MKK6 mutant constructs and SB203580 was confirmed in assays of p38 activity in HEK293 cells. These cells express endogenous MEF2 proteins at levels and isoform ratios that confer sensitivity of MEF2-responsive gene expression to p38 activity (51). These assays established that p38 signaling is low in these cells when grown in serum-free medium (Fig. 9B). In parallel reporter assays, S-p2-Luc was activated ∼9-fold in cells expressing MKK6EE, whereas MKK6KA expression both blunted this activation and modestly depressed basal activity of the reporter, as did SB203580 exposure (data not shown). The net ∼12-fold response of MEF2A promoter 2 activity to p38 signaling, determined by the ratio of reporter activity in cells expressing MKK6EE versus MKK6KA, was sharply dampened with mutation of the MEF2 element (Fig. 9C). Promoter 1 was similarly regulated by p38 activity. RNA was harvested from cells transfected with MKK6EE and incubated in the presence or absence of SB203580. RPA demonstrated an increase in MEF2A mRNA in p38 MAPK-activated versus in p38 MAPK-inhibited cell populations (Fig. 9D), confirming that p38 signaling stimulates MEF2A mRNA expression.
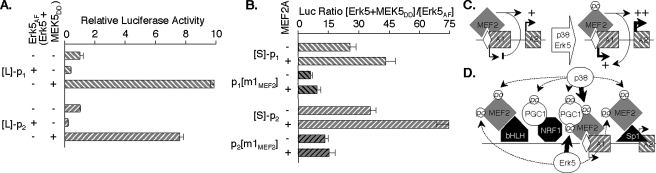
MEF2A factors are also substrates for Erk5 kinase, which phosphorylates two C-terminal Ser residues as well as the Thr that are substrates for p38 (52).7 We examined whether Erk5 signaling regulates the MEF2A promoters by modulating Erk5 activity using forced expression of a dominant interfering mutant (Erk5AF) or by co-expressing wild-type Erk5 with a constitutively active MEK5 (MEK5DD) (42). L-p1-Luc and L-p2-Luc activities were inhibited in C2C12 cells expressing Erk5AF (Fig. 10A), consistent with a modest Erk5 signaling activity in myocytes under control conditions, probably because of incubation medium serum (42). The MEF2A promoter 1 and 2 reporters were stimulated 10- and 8-fold, respectively, with co-expression of MEK5DD and Erk5. Because this response to Erk5 signaling activity was maintained in the S reporters, the contribution of the MEF2 element was explored by comparing responses of the wild-type and m1MEF2 S reporters. Mutation of the MEF2 element resulted in a marked reduction in promoter response to Erk5 signaling. Moreover, stimulation of the wild-type reporters by Erk5 activity was potentiated in cells in which MEF2A α2/β was overexpressed (Fig 10B).
FIGURE 10.
Erk5 signaling co-stimulates the MEF2A promoters via the MEF2 element. A, C2C12 cells were transfected with indicated reporters (0.5 μg/well) and either Erk5AF (0.5 μg/well) or MEK5DD and Erk5 (0.05 μg/well each). Cell extract luciferase activities are normalized to those in cells transfected with an equivalent amount of empty expression vector (=1.0). B, cells were transfected as in A, except with or without MEF2A α2.β, and analyzed as in Fig. 8C. C, schematic of the control of MEF2A alternative promoters from the MEF2 element by MAPK-modified MEF2 factors (see text). D, diagram of established (heavy solid arrows) and candidate (dashed arrows) pathways by which MAPK activity controls MEF2A transcription. MEF2 proteins bind the MEF2 element but also interact with DNA-bound myogenic and neurogenic bHLH factors (53, 61) and Sp1 (49). p38 phosphorylates MEF2A to stimulate trans-activity (51, 75) but also targets PGC-1α, a MEF2 co-activator (8, 55). MEF2A, MEF2C, and MEF2D can each be phosphorylated by Erk5, leading to potent increases in trans-activity (42, 52).
Our results show that p38 and Erk5 signaling positively coregulate the two MEF2A promoters, chiefly through MEF2 factors acting at the promoter MEF2 element. The net effect of this signaling is to amplify the positive control of promoter 2 and to convert the element from silencer to enhancer of promoter 1 activity (Fig. 10C). Because mutation of the MEF2 element did not entirely abolish responses of either promoter to signaling activity, additional factors binding elsewhere in the MEF2A promoters, or MEF2 proteins interacting with such DNA-bound factors, must also contribute to the MEF2A transcriptional response to signaling. The schematic of Fig. 10D suggests several candidate elements and transcription factors and cofactors (49, 53–55).
A MEF2 Element Overlies the Transcription Start Site of the Sole Cephalochordate Amphioxus B. floridae MEF2 Gene—All established mammalian genomes have four MEF2 gene isotypes. By contrast, metazoa ranging from Porifera (sponges, phylum 1) and Cnidaria (coral, hydra, and jellyfish, e.g. phylum 2) through Arthropoda (insects and crustaceans, e.g. phylum 17) and Echiniodermata (sea urchin, phylum 19) have a single MEF2 gene.6 The number of MEF2 genes in primitive vertebrates is not yet evident, but invertebrate chordates, including a Urochordate (Ciona sp., sea squirt) and a Cephalochordate (amphioxus B. floridae, lancelet), have but one.
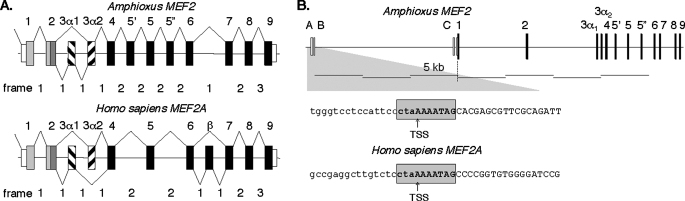
We determined the amphioxus MEF2 gene structure and found it to be analogous to vertebrate MEF2A, including evidence of a vertebrate-like alternative splicing feature6 (Fig. 11A and supplemental Table S1). Phylogenetic analyses showed that the deduced amphioxus MEF2 protein is most related to vertebrate MEF2A, and particularly the MEF2A α2 isoform. Taken together, these facts suggest that MEF2A most closely resembles an ancestral vertebrate MEF2 gene. We therefore determined the amphioxus MEF2 5′-UTR and the 5′-flanking region. The gene has three exons that contribute 5′-UTR sequences in addition to exon 1, which contains the initiation codon (Fig. 11B). A canonical MEF2 element overlies the MEF2 TSS. The element sequence and register with respect to the TSS are identical to those of vertebrate MEF2A (Fig. 11B).
FIGURE 11.
A MEF2 element overlies the amphioxus MEF2 gene TSS. A, cephalochordate amphioxus MEF2 gene structure is highly similar to mammalian MEF2A, and amphioxus has a mammalian-like alternative splicing option. The deduced amphioxus MEF2 protein most resembles the MEF2A α2 among all mammalian MEF2 isotypes and splicing variants (see Footnote 6). B, amphioxus MEF2 TSS (at exon A) is within a MEF2 element that is identical in sequence and register to that of MEF2A.
DISCUSSION
We have shown that primary transcripts of the MEF2A gene arise from two closely approximated promoters, both of which are active in all of the adult tissues examined. MEF2A transcription is regulated from a MEF2 element that directly overlies the promoter 1 TSS, and this site can function as either an enhancer or a silencer depending on MEF2 factor activity at the site. We propose that this provides for both positive and negative autoregulation of MEF2A transcription by MEF2A proteins, contributing to a wide dynamic range of MEF2A expression. Because all MEF2 factors share a common N-terminal MADS/MEF2 region with dimerization and sequence-specific DNA binding activities (5, 6), protein products of the other MEF2 isotype genes, i.e. MEF2B, MEF2C, MEF2D, are also predicted to regulate MEF2A transcription. The 5′-regulatory region of this gene is highly conserved,8 such that all mammalian MEF2A genes are likely to share this and other transcriptional control mechanisms. We are presently applying the results of these analyses to evaluations of MEF2A promoter and regulatory element functions in vivo using transgenic reporters.
Our reporter studies indicated that the MEF2A MEF2 element can function to silence promoter 1. Thus, mutation of the MEF2 site resulted in an increase in promoter activity, and forced expression of MEF2A gave repression. However, with signaling that stimulates MEF2 factor trans-activity, the element functioned as an enhancer. It is established that MEF2 factors can activate or repress transcription, depending on associations with its protein interactors (5, 8, 29, 53, 56–61) and on cell signaling activities (7, 15, 28, 41, 51, 52, 62). One major mechanism of repression involves class IIa histone deacetylases. These associate with MEF2 proteins and influence chromatin structure to inactivate target gene expression (29, 60). Transrepression is also produced by an independent but uncharacterized mechanism associated with the modification of a MEF2 γ domain Lys residue, γK, by small ubiquitin-like modifiers (SUMO) (18, 26, 27, 63). The mode by which the MEF2 element silences MEF2A promoter 1 may entail one or both of these processes. However, an independent means could be involved. For example, the binding of MEF2 factors at the TSS may block or constrain RNA polymerase II assembly. To our knowledge, there are no previous reports of a MEF2 element at a TSS. However, implementation of cap-trapper (45) and other improved cDNA library construction strategies has revealed the full 5′ extent of many mRNAs, and we have found several instances where MEF2 elements previously mapped to a proximal promoter actually do overlie a TSS.6 Mechanisms by which the MEF2A MEF2 element functions as silencer or enhancer are therefore likely to generalize to other MEF2-responsive genes.
In our promoter-reporter studies, the MEF2 element functioned as an enhancer of the downstream promoter 2 under all conditions. Here, mutation of the site gave a sharp reduction in promoter activity in cells expressing active endogenous MEF2 factors, and forced expression of MEF2 proteins stimulated this promoter in an element-dependent fashion. We had found a similar gene structure and MEF2 element activity on alternative CPT-IB promoters.9 In that case, a MEF2 element overlies an upstream TSS and silences the corresponding promoter but activates a muscle-specific downstream promoter. Although the activity of MEF2A promoter 2 is not strictly muscle-specific, its activity is highest in muscle. The similarity of CPT-IB and MEF2A regulatory region structure and alternative promoter behavior reinforces the likelihood that the gene architecture in this region is relevant to its complex regulation, and that a MEF2 element TSS locus is pivotal to silencing activity.
It is likely that the alternative MEF2A promoters allow for complexity in the temporal and spatial expression of this gene. However, it may also be that the corresponding alternative 5′-UTR sequences of exon A1 versus A2 exert control over MEF2A expression at some level, such as pre-mRNA splicing, mRNA stability, microRNA targeting or efficiency of translation. It is noteworthy that exon A2 is G/C-rich and contains an extended region of autocomplementarity. This may form a hairpin that could impact one or more of the aforementioned possibilities. This high G/C content may also account for relative under-representation of MEF2A cDNAs that contain this exon because of RT stalling during cDNA synthesis.
We found that MEF2A transcription was activated from the MEF2 element by Cn activity. There are two recognized mechanisms by which activity of this phosphoprotein phosphatase can control MEF2 activity to regulate the expression of target genes (9). The conventional mode involves the calcium-regulated forms of nuclear factor of activated T cells (NF-ATc), which are localized in the cytosol but translocate to the nucleus upon Cn-mediated dephosphorylation (9, 28, 41, 47, 64). The nuclear NF-ATc can interact with the MADS/MEF2S region of DNA-bound MEF2 factors, leading to transactivation of MEF2 targets. A second mode involves Cn-mediated dephosphorylation of several MEF2 poSer residues that in turn control γ domain Lys modifications (18, 26). This diminishes the transrepression function of γ, thereby increasing the net trans-activity of the MEF2 factor. Based on these paradigms, Cn activity could stimulate MEF2A transcription either via a NF-ATc-MEF2 interaction, or by the induction of γK acetylation or SUMOylation state(s) that increase MEF2 activity. Regardless of the mechanism, regulation of MEF2A transcription by Cn activity is likely to play a role in muscle differentiation, because that process requires both MEF2 and Cn activity (65). This Cn → MEF2A → MEF2A pathway may also account for the maintenance of MEF2A expression in differentiated neuromuscular tissues, where Cn is highly active (2, 9, 28, 65).
Using comparative global gene expression analyses, Tapscott and co-workers (66) determined that mef2a expression is responsive to MyoD activity. The conserved MEF2A gene MEF2 element described here indicates one mechanism by which this can occur, as myogenic bHLH factors, including MyoD, are understood to control transcription of target genes through direct interactions with DNA-bound MEF2 factors (6, 53). Although it remains for us to directly examine the function of the two conserved E boxes in the upstream “E” MEF2A regulatory region, these are also likely to play a role in the control of MEF2A expression by myogenic and neurogenic bHLH factors (48, 53, 61). It should be noted that there are multiple additional sequences in the human MEF2A regulatory region that conform to the E box consensus, but only the two sites described are conserved in sequence and location among mammalian MEF2A genes.8
MEF2 and myogenic or neurogenic bHLH factors are known to synergize in two fundamental ways. As described above, a member of one class can use a member of the other as DNA-bound scaffolding, allowing regulation of target genes of the scaffolding component (53, 61). A second mechanism for cooperative control involves cross-regulation of MEF2 and myogenic bHLH factor gene promoters. MRF4 (67, 68), myogenin (69), and MyoD (70) have MEF2 → bHLH cross-regulation. The existence of evolutionarily conserved E boxes in MEF2A suggest that MEF2A, like MEF2C (34, 35), is subject to bHLH → MEF2 cross-regulation.
The 5′-regulatory region of the mouse mef2c gene has been partially mapped and characterized (34–36). Transgenic promoter-reporter studies characterized the most 5′ of the promoters, and an E box and an adjacent MEF2 element of unusual structure were reported to be essential for transcription of the reporter in skeletal muscle (34). Although this claim of mef2c autoregulation has been disputed (35), we find that mammalian MEF2C genes have at least three different promoters that are separated by over 80 kb,6 and one has an evolutionarily conserved canonical MEF2 element.8 In conjunction with the findings reported here, this would allow for cross-talk at the transcriptional level between MEF2A and MEF2C. mef2a expression precedes that of mef2c in differentiating myoblasts (Fig. 7B) (4, 6, 31), but the converse is true in the developing embryo (10, 21, 22, 25, 35). Thus, cross-regulation may occur in either direction or both directions. Selective activity of MEF2 on the alternative gene promoters may represent a scheme to allow expression of these genes to escape positive auto- and cross-regulatory control.
MEF2A proteins are recognized targets of p38 MAPK α and β, and the resulting Thr modifications stimulate trans-activity of the factor through an unknown mechanism (51). Given MEF2A transcriptional autoregulation, it is not surprising that we observed sensitivity of MEF2A transcription and steady-state mRNA abundance to p38 signaling. Because p38 activity both stimulates and is required for myogenesis (7, 66, 71–74), MAPK-mediated stimulation of MEF2A autoregulation is likely to be involved. MEF2 factors can tether to DNA-bound bHLH factors at E boxes (53) and to Sp1 at GC boxes (49). The transcriptional co-activator PGC-1α that interacts with both NRF1 (54) and MEF2 (8) is also activated by p38 MAPK (55). The presence of conserved E boxes, GC boxes, and the NRF1 element in the MEF2A gene regulatory region may therefore allow for parallel transmission of stress signaling to expression of this gene (Fig. 10D). This pathway may also be relevant to macrophage activation (75), vascular smooth muscle proliferation (12), and excitation-dependent neuron survival (76), as each also involves gene regulation by p38 signaling through MEF2 factors. MEF2A is also a substrate for Erk5, and the resulting Ser/Thr phosphorylations stimulate factor trans-activity by SUMOylation-dependent and independent means.7 The Erk5 → MEF2 pathway is necessary for muscle differentiation (77), neuron survival (78), and smooth muscle proliferation (12). We propose that these processes also involve positive transcriptional autoregulation of MEF2A.
mef2a–/– mice exhibit diminished cardiac mitochondrial density and function and a neonatal sudden death syndrome (25). These animals have elevated cardiac expression of the other mef2 isotype mRNAs, and a MEF2-responsive transgene reporter is highly active. A loss of MEF2A transcriptional autoregulation may account for the lack of compensation by the up-regulated mef2 isotypes in these animals. This mef2a–/– phenotype, together with the fact that mef2c null embryos do not exhibit mitochondrial defects (10), indicates that MEF2A plays a specific role in mitochondrial biogenesis. The relevant mechanism remains to be elucidated. Based on the existence of conserved MEF2A promoter NRF1 and NRF2 elements, we have proposed that MEF2A acts as an intermediary in the communication of nuclear respiratory factor (46) activity in heart and muscle.8
MEF2A plays a second pivotal role in muscle metabolism. In specific, MEF2A DNA binding activity and mef2a mRNA expression are diminished in skeletal muscle of diabetic mice, and insulin administration corrects these deficits (79). This modulation of MEF2A abundance influences expression of glut4, a MEF2 target gene that encodes the insulin-responsive glucose transporter of muscle (80). It is not clear if this effect of insulin involves mef2a transcription, nor is it established that the effect is mediated directly by insulin-stimulated signaling. Our mapping of the MEF2A regulatory region establishes necessary substrate for transgenic promoter-reporter studies to address whether insulin-responsive MEF2A abundance is controlled at the transcriptional level in vivo.
Numerous antibodies exist that are purported to be specific for individual MEF2 isotypes (MEF2A, MEF2B, MEF2C, and MEF2D). We have not found these reagents to be satisfactory. This is a result of the following: 1) use of long partial protein or polypeptide antigens with segments that lack isotype specificity; 2) failure to account for alternative MEF2 splicing isoforms, such that antigens include alternative domains that are variably present and/or highly conserved across isotypes; and/or 3) use of antigens that correspond to domains that have regulated modification(s) that influence immunodetection. We have overcome this reagent limitation by developing three antibodies, and each specifically recognizes all splicing forms of MEF2A, MEF2C, or MEF2D. The antibodies identify human and mouse MEF2 proteins, and are predicted to recognize MEF2 factors of numerous mammalian, avian, reptilian, amphibian, and actinopterygian species (supplemental Table S2). In this study, we have used these reagents in ChIP and mobility shift assays to show that endogenous MEF2A protein binds the MEF2A MEF2 element and that other MEF2 factors occupy the element in proportion to their relative abundance. These antibodies, of utility in IB, IP, EMSA, and ChIP, as shown here, as well as in immunohistochemistry (81), should prove to be of significant value in studies of vertebrate MEF2 functions.
The Drosophila Mef2 (DMef2) transcriptional regulatory region consists of a complex array of elements that govern temporospatial gene expression (32). This region has several sites that bind DMEF2 according to ChIP (82), one of which is responsible for transcriptional autoregulation of this gene (33). Because true vertebrates probably originated from a cephalochordate-like ancestor, we mapped the sole MEF2 gene of a cephalochordate to investigate whether autoregulation generalizes to other species. We identified the 5′-regulatory region of the MEF2 gene of amphioxus B. floridae, a cephalochordate for which cDNA and genomic sequences are available. We found a MEF2 element whose sequence and register with respect to the TSS are identical to those of mammalian MEF2A. Although we did not perform functional studies, this striking similarity invites speculation that this gene is also subject to negative and positive control by its protein products, as for MEF2A. Thus it is likely that MEF2 transcriptional autoregulation is conserved among the higher metazoans, including arthropods, cephalochordates, and mammals.
Supplementary Material
Acknowledgments
We thank John Kyriakis (Tufts-New England Medical Center) for MKK6, Erk5, and MEK5 cDNAs; Jeffrey Dilworth (Ottawa Health Research Institute) for MEF2AFLAG and MEF2DFLAG constructs; Pan Yin and Chousheng Zou for technical assistance; and Tony Makkinje for p38 assays.
This work was supported in part by grants (to T. G.) from the American Heart Association, the Clinical Nutrition Research Center at Harvard Grant P30-DK40561, and National Institutes of Health Grants DK55875, DK02461, and HL72713. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Tables S1 and S2.
Footnotes
The abbreviations used are: MADS, minichromosome maintenance 1/agamous/deficiens/serum-response factor; bHLH, basic helix-loop-helix transcription factor; ChIP, chromatin immunoprecipitation; CnA(+), calcineurin A subunit (constitutively active); Erk, extracellular signal-regulated kinase; IB, immunoblot; IP, immunoprecipitation; MAP(K(K)), mitogen-activated protein (kinase (kinase)); NF-AT, nuclear factor of activated T cells; NRF, nuclear respiratory factor; RT, reverse transcriptase; SUMO, small ubiquitin-like modifier(s); tk, thymidine kinase; UTR, untranslated region; TSS, transcription start site; EMSA, electrophoretic mobility shift assay; HA, hemagglutinin; RPA, ribonuclease protection assays.
T. Gulick, unpublished observations.
B. Ramachandran and T. Gulick, submitted for publication.
G. Yu, B. Ramachandran, and T. Gulick, submitted for publication.
G. Yu and T. Gulick, unpublished observations.
References
- 1.Nurrish, S. J., and Treisman, R. (1995) Mol. Cell. Biol. 15 4076–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu, Y. T., Breitbart, R. E., Smoot, L. B., Lee, Y., Mahdavi, V., and Nadal-Ginard, B. (1992) Genes Dev. 6 1783–1798 [DOI] [PubMed] [Google Scholar]
- 3.Pollock, R., and Treisman, R. (1991) Genes Dev. 5 2327–2341 [DOI] [PubMed] [Google Scholar]
- 4.Zhu, B., Ramachandran, B., and Gulick, T. (2005) J. Biol. Chem. 280 28749–28760 [DOI] [PubMed] [Google Scholar]
- 5.Shalizi, A. K., and Bonni, A. (2005) Curr. Top Dev. Biol. 69 239–266 [DOI] [PubMed] [Google Scholar]
- 6.Black, B. L., and Olson, E. N. (1998) Annu. Rev. Cell Dev. Biol. 14 167–196 [DOI] [PubMed] [Google Scholar]
- 7.Naya, F. J., and Olson, E. (1999) Curr. Opin. Cell Biol. 11 683–688 [DOI] [PubMed] [Google Scholar]
- 8.Handschin, C., Rhee, J., Lin, J., Tarr, P. T., and Spiegelman, B. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7111–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, Y., Shen, T., Randall, W. R., and Schneider, M. F. (2005) J. Muscle Res. Cell Motil. 1–9 [DOI] [PubMed]
- 10.Lin, Q., Schwarz, J., Bucana, C., and Olson, E. N. (1997) Science 276 1404–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolodziejczyk, S. M., Wang, L., Balazsi, K., DeRepentigny, Y., Kothary, R., and Megeney, L. A. (1999) Curr. Biol. 9 1203–1206 [DOI] [PubMed] [Google Scholar]
- 12.Zhao, M., Liu, Y., Bao, M., Kato, Y., Han, J., and Eaton, J. W. (2002) Arch. Biochem. Biophys. 400 199–207 [DOI] [PubMed] [Google Scholar]
- 13.Lin, Q., Lu, J., Yanagisawa, H., Webb, R., Lyons, G. E., Richardson, J. A., and Olson, E. N. (1998) Development (Camb.) 125 4565–4574 [DOI] [PubMed] [Google Scholar]
- 14.Bi, W., Drake, C. J., and Schwarz, J. J. (1999) Dev. Biol. 211 255–267 [DOI] [PubMed] [Google Scholar]
- 15.Gong, X., Tang, X., Wiedmann, M., Wang, X., Peng, J., Zheng, D., Blair, L. A., Marshall, J., and Mao, Z. (2003) Neuron 38 33–46 [DOI] [PubMed] [Google Scholar]
- 16.Gaudilliere, B., Shi, Y., and Bonni, A. (2002) J. Biol. Chem. 277 46442–46446 [DOI] [PubMed] [Google Scholar]
- 17.Flavell, S. W., Cowan, C. W., Kim, T. K., Greer, P. L., Lin, Y., Paradis, S., Griffith, E. C., Hu, L. S., Chen, C., and Greenberg, M. E. (2006) Science 311 1008–1012 [DOI] [PubMed] [Google Scholar]
- 18.Shalizi, A., Gaudilliere, B., Yuan, Z., Stegmuller, J., Shirogane, T., Ge, Q., Tan, Y., Schulman, B., Harper, J. W., and Bonni, A. (2006) Science 311 1012–1017 [DOI] [PubMed] [Google Scholar]
- 19.Youn, H. D., Sun, L., Prywes, R., and Liu, J. O. (1999) Science 286 790–793 [DOI] [PubMed] [Google Scholar]
- 20.Pan, F., Ye, Z., Cheng, L., and Liu, J. O. (2004) J. Biol. Chem. 279 14477–14480 [DOI] [PubMed] [Google Scholar]
- 21.Edmondson, D. G., Lyons, G. E., Martin, J. F., and Olson, E. N. (1994) Development (Camb.) 120 1251–1263 [DOI] [PubMed] [Google Scholar]
- 22.Subramanian, S. V., and Nadal-Ginard, B. (1996) Mech. Dev. 57 103–112 [DOI] [PubMed] [Google Scholar]
- 23.Lyons, G. E., Micales, B. K., Schwarz, J., Martin, J. F., and Olson, E. N. (1995) J. Neurosci. 15 5727–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breitbart, R. E., Liang, C. S., Smoot, L. B., Laheru, D. A., Mahdavi, V., and Nadal-Ginard, B. (1993) Development (Camb.) 118 1095–1106 [DOI] [PubMed] [Google Scholar]
- 25.Naya, F. J., Black, B. L., Wu, H., Bassel-Duby, R., Richardson, J. A., Hill, J. A., and Olson, E. N. (2002) Nat. Med. 8 1303–1309 [DOI] [PubMed] [Google Scholar]
- 26.Gregoire, S., Tremblay, A. M., Xiao, L., Yang, Q., Ma, K., Nie, J., Mao, Z., Wu, Z., Giguere, V., and Yang, X. J. (2006) J. Biol. Chem. 281 4423–4433 [DOI] [PubMed] [Google Scholar]
- 27.Zhao, X., Sternsdorf, T., Bolger, T. A., Evans, R. M., and Yao, T. P. (2005) Mol. Cell. Biol. 25 8456–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2002) Trends Biochem. Sci. 27 40–47 [DOI] [PubMed] [Google Scholar]
- 29.McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2001) Curr. Opin. Genet. Dev. 11 497–504 [DOI] [PubMed] [Google Scholar]
- 30.Black, B. L., Lu, J., and Olson, E. N. (1997) Mol. Cell. Biol. 17 2756–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu, B., and Gulick, T. (2004) Mol. Cell. Biol. 24 8264–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen, H. T., and Xu, X. (1998) Dev. Biol. 204 550–566 [DOI] [PubMed] [Google Scholar]
- 33.Cripps, R. M., Lovato, T. L., and Olson, E. N. (2004) Dev. Biol. 267 536–547 [DOI] [PubMed] [Google Scholar]
- 34.Wang, D. Z., Valdez, M. R., McAnally, J., Richardson, J., and Olson, E. N. (2001) Development (Camb.) 128 4623–4633 [DOI] [PubMed] [Google Scholar]
- 35.Dodou, E., Xu, S. M., and Black, B. L. (2003) Mech. Dev. 120 1021–1032 [DOI] [PubMed] [Google Scholar]
- 36.De Val, S., Anderson, J. P., Heidt, A. B., Khiem, D., Xu, S. M., and Black, B. L. (2004) Dev. Biol. 275 424–434 [DOI] [PubMed] [Google Scholar]
- 37.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (eds) (1998) Current Protocols in Molecular Biology, John Wiley & Sons, Inc., New York
- 38.Yu, G. S., Lu, Y. C., and Gulick, T. (1998) J. Biol. Chem. 273 32901–32909 [DOI] [PubMed] [Google Scholar]
- 39.Han, T. H., and Prywes, R. (1995) Mol. Cell. Biol. 15 2907–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raingeaud, J., Whitmarsh, A. J., Barrett, T., Derijard, B., and Davis, R. J. (1996) Mol. Cell. Biol. 16 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, S., Liu, P., Borras, A., Chatila, T., and Speck, S. H. (1997) EMBO J. 16 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato, Y., Kravchenko, V. V., Tapping, R. I., Han, J., Ulevitch, R. J., and Lee, J. D. (1997) EMBO J. 16 7054–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharp, P. A. (2005) Trends Biochem. Sci. 30 279–281 [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, E., Lowry, J., Sonoda, G., Testa, J. R., and Walsh, K. (1996) Cytogenet. Cell Genet. 73 244–249 [DOI] [PubMed] [Google Scholar]
- 45.Sugahara, Y., Carninci, P., Itoh, M., Shibata, K., Konno, H., Endo, T., Muramatsu, M., and Hayashizaki, Y. (2001) Gene (Amst.) 263 93–102 [DOI] [PubMed] [Google Scholar]
- 46.Scarpulla, R. C. (2006) J. Cell. Biochem. 97 673–683 [DOI] [PubMed] [Google Scholar]
- 47.Hogan, P. G., Chen, L., Nardone, J., and Rao, A. (2003) Genes Dev. 17 2205–2232 [DOI] [PubMed] [Google Scholar]
- 48.Berkes, C. A., and Tapscott, S. J. (2005) Semin. Cell Dev. Biol. 16 585–595 [DOI] [PubMed] [Google Scholar]
- 49.Grayson, J., Bassel-Duby, R., and Williams, R. S. (1998) J. Cell. Biochem. 70 366–375 [PubMed] [Google Scholar]
- 50.Wu, H., Naya, F. J., McKinsey, T. A., Mercer, B., Shelton, J. M., Chin, E. R., Simard, A. R., Michel, R. N., Bassel-Duby, R., Olson, E. N., and Williams, R. S. (2000) EMBO J. 19 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao, M., New, L., Kravchenko, V. V., Kato, Y., Gram, H., di Padova, F., Olson, E. N., Ulevitch, R. J., and Han, J. (1999) Mol. Cell. Biol. 19 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kato, Y., Zhao, M., Morikawa, A., Sugiyama, T., Chakravortty, D., Koide, N., Yoshida, T., Tapping, R. I., Yang, Y., Yokochi, T., and Lee, J. D. (2000) J. Biol. Chem. 275 18534–18540 [DOI] [PubMed] [Google Scholar]
- 53.Molkentin, J. D., Black, B. L., Martin, J. F., and Olson, E. N. (1995) Cell 83 1125–1136 [DOI] [PubMed] [Google Scholar]
- 54.Wu, Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R. C., and Spiegelman, B. M. (1999) Cell 98 115–124 [DOI] [PubMed] [Google Scholar]
- 55.Puigserver, P., Rhee, J., Lin, J., Wu, Z., Yoon, J. C., Zhang, C. Y., Krauss, S., Mootha, V. K., Lowell, B. B., and Spiegelman, B. M. (2001) Mol. Cell 8 971–982 [DOI] [PubMed] [Google Scholar]
- 56.Youn, H. D., and Liu, J. O. (2000) Immunity 13 85–94 [DOI] [PubMed] [Google Scholar]
- 57.Sartorelli, V., Huang, J., Hamamori, Y., and Kedes, L. (1997) Mol. Cell. Biol. 17 1010–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morin, S., Charron, F., Robitaille, L., and Nemer, M. (2000) EMBO J. 19 2046–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han, A., Pan, F., Stroud, J. C., Youn, H. D., Liu, J. O., and Chen, L. (2003) Nature 422 730–734 [DOI] [PubMed] [Google Scholar]
- 60.Han, A., He, J., Wu, Y., Liu, J. O., and Chen, L. (2005) J. Mol. Biol. 345 91–102 [DOI] [PubMed] [Google Scholar]
- 61.Black, B. L., Ligon, K. L., Zhang, Y., and Olson, E. N. (1996) J. Biol. Chem. 271 26659–26663 [DOI] [PubMed] [Google Scholar]
- 62.Lazaro, J. B., Bailey, P. J., and Lassar, A. B. (2002) Genes Dev. 16 1792–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang, J., Gocke, C. B., and Yu, H. (2006) BMC Biochem. 7 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beals, C. R., Clipstone, N. A., Ho, S. N., and Crabtree, G. R. (1997) Genes Dev. 11 824–834 [DOI] [PubMed] [Google Scholar]
- 65.Friday, B. B., Horsley, V., and Pavlath, G. K. (2000) J. Cell Biol. 149 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bergstrom, D. A., Penn, B. H., Strand, A., Perry, R. L., Rudnicki, M. A., and Tapscott, S. J. (2002) Mol. Cell 9 587–600 [DOI] [PubMed] [Google Scholar]
- 67.Naidu, P. S., Ludolph, D. C., To, R. Q., Hinterberger, T. J., and Konieczny, S. F. (1995) Mol. Cell. Biol. 15 2707–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Black, B. L., Martin, J. F., and Olson, E. N. (1995) J. Biol. Chem. 270 2889–2892 [DOI] [PubMed] [Google Scholar]
- 69.Ridgeway, A. G., Wilton, S., and Skerjanc, I. S. (2000) J. Biol. Chem. 275 41–46 [DOI] [PubMed] [Google Scholar]
- 70.Wong, M. W., Pisegna, M., Lu, M. F., Leibham, D., and Perry, M. (1994) Dev. Biol. 166 683–695 [DOI] [PubMed] [Google Scholar]
- 71.Puri, P. L., Wu, Z., Zhang, P., Wood, L. D., Bhakta, K. S., Han, J., Feramisco, J. R., Karin, M., and Wang, J. Y. (2000) Genes Dev. 14 574–584 [PMC free article] [PubMed] [Google Scholar]
- 72.Zetser, A., Gredinger, E., and Bengal, E. (1999) J. Biol. Chem. 274 5193–5200 [DOI] [PubMed] [Google Scholar]
- 73.Lluis, F., Perdiguero, E., Nebreda, A. R., and Munoz-Canoves, P. (2006) Trends Cell Biol. 16 36–44 [DOI] [PubMed] [Google Scholar]
- 74.Penn, B. H., Bergstrom, D. A., Dilworth, F. J., Bengal, E., and Tapscott, S. J. (2004) Genes Dev. 18 2348–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han, J., Jiang, Y., Li, Z., Kravchenko, V. V., and Ulevitch, R. J. (1997) Nature 386 296–299 [DOI] [PubMed] [Google Scholar]
- 76.Okamoto, S., Krainc, D., Sherman, K., and Lipton, S. A. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 7561–7566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dinev, D., Jordan, B. W., Neufeld, B., Lee, J. D., Lindemann, D., Rapp, U. R., and Ludwig, S. (2001) EMBO Rep. 2 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cavanaugh, J. E. (2004) Eur. J. Biochem. 271 2056–2059 [DOI] [PubMed] [Google Scholar]
- 79.Mora, S., and Pessin, J. E. (2000) J. Biol. Chem. 275 16323–16328 [DOI] [PubMed] [Google Scholar]
- 80.Thai, M. V., Guruswamy, S., Cao, K. T., Pessin, J. E., and Olson, A. L. (1998) J. Biol. Chem. 273 14285–14292 [DOI] [PubMed] [Google Scholar]
- 81.Yu, G. S., Gulick, T., and Balos, L. (2007) Mod. Pathol. 20 310A17277765 [Google Scholar]
- 82.Junion, G., Jagla, T., Duplant, S., Tapin, R., Da Ponte, J. P., and Jagla, K. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 18479–18484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ichijo, H., Nishida, E., Irie, K., ten Dijke, P., Saitoh, M., Moriguchi, T., Takagi, M., Matsumoto, K., Miyazono, K., and Gotoh, Y. (1997) Science 275 90–94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.