Abstract
TAK1 (transforming growth factor (TGF)-β-activated kinase 1) is a serine/threonine kinase that is rapidly activated by TGF-β1 and plays a vital function in its signal transduction. Once TAK1 is activated, efficient down-regulation of TAK1 activity is important to prevent excessive TGF-β1 responses. The regulatory mechanism of TAK1 inactivation following TGF-β1 stimulation has not been elucidated. Here we demonstrate that protein phosphatase 2A (PP2A) plays a pivotal role as a negative regulator of TAK1 activation in response to TGF-β1 in mesangial cells. Treatment with okadaic acid (OA) induces autophosphorylation of Thr-187 in the activation loop of TAK1. In vitro dephosphorylation assay suggests that Thr-187 in TAK1 is a major dephosphorylation target of PP2A. TGF-β1 stimulation rapidly activates TAK1 in a biphasic manner, indicating that TGF-β1-induced TAK1 activation is tightly regulated. The association of PP2AC with TAK1 is enhanced in response to TGF-β1 stimulation and closely parallels TGF-β1-induced TAK1 activity. Attenuation of PP2A activity by OA treatment or targeted knockdown of PP2AC with small interfering RNA enhances TGF-β1-induced phosphorylation of TAK1 at Thr-187 and MKK3 (MAPK kinase 3). Endogenous TAK1 co-precipitates with PP2AC but not PP6C, another OA-sensitive protein phosphatase, and knockdown of PP6C by small interfering RNA does not affect TGF-β1-induced phosphorylation of TAK1 at Thr-187 and MKK3. Moreover, ectopic expression of phosphatase-deficient PP2AC enhances TAK1-mediated MKK3 phosphorylation by TGF-β1 stimulation, whereas the expression of wild-type PP2AC suppresses the MKK3 phosphorylation. Taken together, our data indicate that PP2A functions as a negative regulator in TGF-β1-induced TAK1 activation.
The transforming growth factor (TGF)3-β superfamily regulates a plethora of biological processes, such as cellular differentiation, proliferation, apoptosis, and wound healing (1, 2). Among TGF-β family members, TGF-β1, the prototype of the TGF-β family, is well established as a potent inducer of extracellular matrix synthesis and regarded as a key mediator in the pathogenesis of tissue fibrosis, including renal fibrosis (3, 4). During the past decade, important advances in our understanding of TGF-β1-induced signaling have been made, and the Smad signaling pathway is well documented as a canonical pathway induced by TGF-β1 (5, 6). However, a recently emerging body of evidence has demonstrated that TGF-β1 also induces various Smad-independent signaling pathways (7, 8) by which Rho small GTP-binding proteins (9, 10), Ras (11), phosphatidylinositol 3-kinase (12, 13), and TAK1 (TGF-β-activated kinase 1) (14, 15) can be activated. These Smad-independent signaling pathways subsequently trigger the activation of extracellular signal-regulated kinase 1/2 (16, 17), c-Jun N-terminal kinase (JNK) (18, 19), and p38 mitogen-activated protein kinases (MAPKs) (20-22).
TAK1, initially identified as a kinase activated by TGF-β and BMP family members, consists of ∼600 amino acids (14, 15). The N-terminal half, containing about 300 amino acids, bears the kinase domain and shares ∼30% identity with the catalytic domain of other mitogen-activated kinase kinase kinases (MKKKs) (e.g. Raf-1 and MEKK-1). TAK1 also mediates distinct signaling pathways induced by various stimuli, including environmental stress (23), inflammatory cytokines (24, 25), and lipopolysaccharide (26) as well as TGF-β1 (14). These observations suggest that TAK1 might be the point of convergence in various signaling pathways activated by a variety of stimuli. For its activation, phosphorylation of a threonine residue (Thr-187) and serine residue (Ser-192) in the activation loop of TAK1 protein is required (27-29). In addition, it has been shown that Ser-412 of TAK1 can be phosphorylated in a protein kinase A-dependent manner in RAW264.7 cells (30). Unlike other mitogen-activated kinase kinase kinases (MKKKs), however, TAK1 also requires a specific binding partner known as TAB1 (TAK1-binding protein 1) for its autophosphorylation, and coexpression of TAK1 and TAB1 strongly induces TAK1 activation through autophosphorylation (27-29). Besides TAB1, TAB2 and TAB3 interact with the C-terminal region of TAK1 and serve as adaptor molecules linking TAK1 to TNF-α receptor-associated factor 2 and 6 in TNF-α and IL-1 signaling pathways, respectively (31-35). However, it is not yet known whether TAB2 and TAB3 are required for TGF-β1-induced TAK1 activation. Once TAK1 is activated, it has the capability to trigger the activation of several downstream cell signaling cascades, including the MKK4/7 (MAPK kinase kinase 4/7)-JNK cascade, MKK3/6-p38 MAPK cascade, and nuclear factor-κB (NF-κB)-inducing kinase-IκB kinase cascade, which regulates NF-κB activation (24-26) in a cell type-specific and context-dependent manner.
TAK1 has been shown to mediate TGF-β-induced expression of type I and IV collagens and fibronectin in SV40 transformed mouse mesangial cells (36). We have previously reported that TGF-β1-induced activation of downstream MKK3-p38 MAPK cascade leads to type I collagen expression (22) and that TAK1 is a major upstream signaling molecule mediating TGF-β1-induced MKK3 activation in primary mouse mesangial cells (MMC) (37). In NIH3T3 fibroblasts, TGF-β-induced fibronectin expression is mediated by TAK1 through MKK4-JNK signaling cascade (38). Furthermore, increased expression and activation of TAK1 enhance p38 phosphorylation as well as interstitial fibrosis in myocardium from 9-day-old TAK1 transgenic mice (39), indicating that TAK1 plays a crucial role in tissue fibrosis and elaboration of extracellular matrix in vivo and in vitro. Additional functions of TAK1 in the TGF-β signaling pathway have also been described. For instance, TAK1 is implicated in TGF-β1-induced cell cycle regulation through the regulation of the expression of cyclin D1 and cyclin A in LLC-PK1 cells (40). TAK1 can also regulate TGF-β-induced activation of Smad signaling. The TAK1-p38 MAPK cascade induces Smad7 expression by regulation of the transcription factor ER81, a member of the ETS family (41). Furthermore, TAK1 directly interacts with the Mad homolog domain 2 of Smad family proteins, and the interaction of TAK1 with R-Smad results in interference with the nuclear localization and transactivation of R-smad in murine mesenchymal progenitors (42). Thus, TAK1 may function as an important regulator of the TGF-β-induced Smad-dependent and Smad-independent signaling pathway as well as an activator of JNK and p38 MAPK. However, the regulatory mechanism of TAK1 activation by TGF-β1 and its rapid inactivation remains incompletely understood.
In general, cyclic phosphorylation and dephosphorylation of kinases represent the major mechanism responsible for control of intracellular signaling cascades. TAK1 requires phosphorylation at Ser and Thr residues for its activation and is rapidly down-regulated. In this context, Ser/Thr protein phosphatases may play a crucial role in inactivation of TAK1. In mammalian cells, four major groups of Ser/Thr protein phosphatases (PP1, PP2A, PP2B, and PP2C) exist. Two isoforms of PP2C (PP2Cβ1 and PP2Cε) have been shown to be capable of binding with and dephosphorylating TAK1 in 293 cells under nonstimulated conditions (43, 44). More recently, it has been shown that a new Ser/Thr protein phosphatase family member, PP6, also interacts with and negatively regulates IL-1-induced TAK1 in 293 cells (45). In the present study, we sought to examine the role of another Ser/Thr protein phosphatase family member, PP2A, in TAK1 activation by TGF-β1. We reasoned that PP2A is a likely candidate, since it can associate with type I TGF-β receptor and Raf-1 whose kinase domain shares about 30% identity with TAK1 is consistently associated with PP2A (46, 47).
PP2A is an abundant and ubiquitous enzyme with pleiotropic functions (48). The predominant form of PP2A in cells contains a heterotrimeric subunit structure, consisting of a 36-kDa catalytic C subunit (PP2AC), a 65-kDa structural A subunit (PP2AA), and one of a variety of structurally distinct regulatory B subunits (PP2AB). In addition to the heterotrimeric form of PP2A, a core dimeric form of PP2A consisting of A and C subunits also exists in cells. The different B subunits are believed to change substrate specificity and subcellular localization of PP2A (49, 50). PP2AC undergoes at least three different post-translational modifications: phosphorylation of Tyr-307, carboxymethylation of Leu-309, and phosphorylation of unidentified threonine residues. By either mutation at Tyr-307 or Leu-309, PP2AAC core dimer loses the ability to interact with a regulatory B subunit (51, 52). Phosphorylation of Tyr-307 reduces its phosphatase activity (52, 53, 54), whereas the carboxymethylation at Leu-309 does not cause a large change in phosphatase activity (55). PP2ABα specifically interacts with activated TβRI through the WD-40 repeat domain and is phosphorylated by TGF-β stimulation (46). In addition, PP2A associates with and dephosphorylates p70S6 kinase in response to TGF-β stimulation, which in turn induces G1 arrest of polarized EpH4 mammary epithelial cells (56). Here, we demonstrate that PP2A associates with TAK1 and TAB1 and functions as a negative regulator in TGF-β1-induced TAK1 activation in mouse mesangial cells.
MATERIALS AND METHODS
Reagents—Recombinant human TGF-β1 was obtained from R & D Systems (Minneapolis, MN). Antibodies against HA (Y-11), TAK1, PP2AC, and horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Polyclonal antibodies against MKK3, p-MKK3/6, phospho-Thr-187-TAK1 (p187-TAK1), or phospho-Ser-412-TAK1 (p412-TAK1) were obtained from Cell Signaling Technologies (Beverly, MA). Anti-FLAG (M2), and anti-Myc (9E10) antibodies were from Sigma. Rabbit anti-PP6C antibody was obtained form Exalpha Biological Inc. (Watertown, MA). Anti-V5 antibody and Lipofectamine Plus™ reagent were purchased from Invitrogen. Okadaic acid (OA) was obtained from Calbiochem.
Cell Cultures and Transfection—Glomerular mesangial cells from male C57BL/6 mice were isolated and characterized as previously described (22). Primary MMC established in culture were maintained in RPMI 1640 medium supplemented with 15% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Transfection of expression vectors was performed using Lipofectamine Plus™ reagent according to the manufacturer's instruction. In brief, cells grown to ∼60% confluence on either a 100- or 60-mm dish were washed with phosphate-buffered saline and transfected with 1 μg (for a 100-mm dish) or 0.3 μg (for a 60-mm dish) of plasmid for 4 h under serum-free conditions. Total amount of DNA was adjusted with empty vector, pcDNA3.1. Following transfection, cells were washed with phosphate-buffered saline and incubated in medium supplemented with 15% FBS for 18 h prior to each experiment.
Expression Constructs—Mammalian expression constructs for wild-type TAK1 (HA-TAK1), kinase-deficient mutant of TAK1 (HA-TAK1DN, a K63W mutant of TAK1), FLAG-TAB1, Myc-TAB1, and bacterial expression vector for His-MKK6 (pQE-31-MKK6) were kindly provided by K. Matsumoto (Nagoya University) (24). First strand cDNA of mouse MKK3b was synthesized by Moloney murine leukemia virus reverse transcriptase with oligonucleotide dT12 according to the manufacturer's instructions and then amplified by PCR using a mouse MKK3b-specific PCR primer set (forward primer, 5′-GGGGTCCTGGGATCTGAATCCTCTCC-3′; reverse primer, 5′-GTCCATGGCTTTGGATCCGTCCCCAAGTAT-3′) synthesized according to the sequence from GenBank™ (accession number NM_008928). PCR products were then ligated with pcDNA3.1/TOPO cloning vector (Invitrogen). Correct clones were confirmed by DNA sequencing of the inserted MKK3b cDNA and its surrounding region and named pcDNA-V5-MKK3. Expression vectors for FLAG-tagged wild-type TAK1 (FLAG-TAK1) and C-terminal deleted TAK1 (FLAG-TAK1ΔC) harboring the N-terminal 300 amino acids of TAK1 were kindly provided by G. Gross (42). HA-tagged wild type PP2AC (HA-PP2AC) and its mutant variants, (HA-PP2AC (H118N), HA-PP2AC (Y307F), and HA-PP2AC (L309Q)) were gifts from D. L. Brautigan (University of Virginia) (52, 57). The His-118 mutant (H118N) is a phosphatase-deficient mutant of PP2AC, whereas the Tyr-307 mutant (Y307F) and Leu-309 mutant (L309Q) are the phosphorylation site mutant and methylation site mutant of PP2AC, respectively.
Small Interfering RNAs (siRNAs)—siGENOME SMARTpool targeted against mouse PP2AC (L-040657), PP6C (L-062583), and siCONTROL nontargeting pool (D-001206-13) were purchased from Dharmacon Inc. Transfection of siRNA using DharmaFECT™ reagent was carried out according to the manufacturer's instructions. Briefly, cells were grown to 50% confluence on a 60-mm dish in 3 ml of medium supplemented with 15% FBS. Two hundred microliters of siRNA transfection solution containing respective siRNA and 4 μl of DhamarFECT™ in serum-free medium were added to the cell culture, resulting in 100 nm siRNA for each transfection. Following transfection, cells were incubated in medium containing 15% FBS for 24 h and then rendered quiescent in medium supplemented with 0.5% FBS for 16 h prior to each experiment.
Immunoprecipitation and Western Blot Analysis—Cells were washed once with ice-cold phosphate-buffered saline and lysed in buffer containing 1% Nonidet P-40, 20 mm Tris (pH 8.0), 150 mm NaCl, 12.5 mm β-glycerophosphate, 1.5 mm MgCl2, 2 mm EGTA, 1 mm NaF, 2 mm dithiothreitol, 1 mm sodium orthovanadate (Na3VO4), 1 mm phenylmethylsulfonyl fluoride, and 20 μm aprotinin. Cell lysates were passed through 21-gauge needles several times and then centrifuged for 15 min at 14,000 × g at 4 °C to remove cellular debris. The protein concentration was determined by BCA protein assay reagent kit (Pierce). For Western blotting, protein samples (100 μg) were subjected to 10% SDS-PAGE and then transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% nonfat milk for 1 h and then incubated with primary antibodies overnight on a rocker at 4 °C. The membranes were washed three times (15 min/each) with TTBS buffer (10 mm Tris, pH 7.5, 50 mm NaCl, and 0.1% Tween 20) and then incubated with horseradish peroxidase-conjugated secondary antibodies for 30 min at room temperature. The target proteins were detected with LumiGLO (Cell Signaling Technologies). In the case of immunoprecipitation experiments, 200-500 μg of cell lysates were reacted with 1-2 μg of primary antibodies for 30 min at 4 °C, followed by precipitation with 20 μl of protein A/G-Sepharose for 2 h (for proteins expressed by transiently transfected cDNA) or overnight (for endogenous proteins) at 4 °C. The immunoprecipitates were then washed three times with the lysis buffer and subjected to Western blotting. Endogenous expression levels of phosphorylated Thr-187 in TAK1 activation loop (p187-TAK1) and phosphorylated MKK3 (p-MKK3) were quantitated using Image J software (Research Services Branch, National Institutes of Health). Statistical significance of the experimental data from three independent experiments was determined by Student's t test for paired data. p values of <0.05 were considered significant. Data are presented as mean ± S.E. of triplicate experiments.
In Vitro Dephosphorylation Assay—Phosphorylated TAK1 was obtained by coexpression of HA-TAK1 and FLAG-TAB1 in MMC followed by immunoprecipitation of TAK1 with anti-HA antibodies. An appropriate amount of phosphorylated TAK1 was incubated with varying amounts of purified PP2A (A and C complex) in the reaction buffer (20 mm Hepes, pH 7.2, 1 mm dithiothreitol, 1 mm MgCl2, 0.1 mg/ml bovine serum albumin, and 1 mm EDTA) for 20 min at 30 °C. Following the reaction, TAK1 dephosphorylation target sites of PP2A were evaluated by Western blotting with anti-p187-TAK1 and anti-p412-TAK1 antibodies.
Immune Complex Kinase Assay—At the indicated time points following TGF-β1 treatment, endogenous TAK1 protein was immunoprecipitated with anti-TAK1 antibody. Immunoprecipitates were washed with kinase buffer (20 mm Tris-HCl, pH 7.4, 1 mm dithiothreitol, 5 mm MgCl2) and resuspended in the same kinase buffer containing 1 μg of the specific substrate His-MKK6. The kinase reaction was initiated by the addition 5 μCi of [γ-32P]ATP (3000 Ci/mmol). After 2 min of incubation at 25 °C, the reactions were terminated by the addition of SDS sample buffer and boiled for 5 min. Samples were fractionated by 10% SDS-PAGE and visualized by autoradiography.
RESULTS
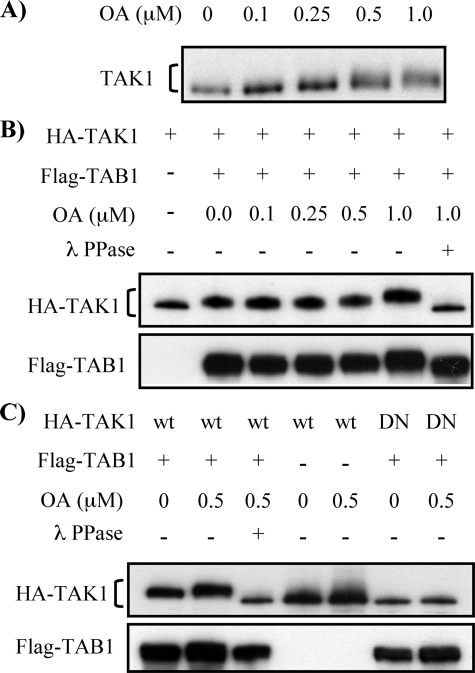
OA-sensitive Ser/Thr Protein Phosphatase(s) Negatively Regulates TAK1 Phosphorylation—TAK1 activation is induced by autophosphorylation and is rapidly inactivated. We hypothesized that a Ser/Thr protein phosphatase family member, PP2A, plays a crucial role in regulating TAK1 activation by TGF-β1 through dephosphorylation of TAK1. We initially examined the effects of OA, a specific inhibitor of PP2A, on endogenous TAK1 phosphorylation. OA treatment with levels up to 1 μm in various cell cultures has been shown to specifically inhibit PP2A without significantly affecting PP1 activity (58-60). MMC were treated with different concentrations of OA for 6 h, and mobility of TAK1 proteins obtained from cell lysates was examined on SDS-PAGE. As shown in Fig. 1A, OA treatment significantly reduced the migration of endogenous TAK1 in a dose-dependent manner. To confirm that the slower migration of TAK1 was due to its autophosphorylation, either wild-type TAK1 (HA-TAK1) or kinase-deficient mutant of TAK1 (HA-TAK1DN) was coexpressed with FLAG-TAB1 and subsequently treated with OA for 6 h. Coexpression of TAK1 and TAB1 induces TAK1 autophosphorylation and slower TAK1 migration on SDS-PAGE (Fig. 1B, top), as shown in previous reports (28, 29). OA treatment enhanced the retardation of TAK1 migration, consistent with our results of endogenous TAK1 in Fig. 1A. Conversely, treatment of cell lysates with λ protein phosphatase completely abolished the retardation of TAK1 migration induced by OA treatment and by coexpression of TAK1 with TAB1, indicating that the OA-induced slower migration of TAK1 was due to phosphorylation of TAK1. Moreover, TAK1DN, which possesses the same phosphorylation sites as wild-type TAK1 but lacks kinase activity, when coexpressed with TAB1 eliminated the slower migrating band induced by wild-type TAK1 expression (Fig. 1C). Taken together, these data suggest that OA-induced TAK1 phosphorylation is accomplished by its own kinase activity, and OA-sensitive Ser/Thr protein phosphatase(s) negatively regulates the autophosphorylation of TAK1.
FIGURE 1.
Inhibition of Ser/Thr protein phosphatases by OA induces TAK1 phosphorylation. A, OA treatment increases mobility shift in endogenous TAK1. MMC grown to subconfluence were treated with increasing concentrations of OA for 6 h as indicated. Cell lysates were subjected to Western blot analysis with anti-TAK1 antibody. B, OA treatment enhances TAK1 phosphorylation induced by coexpression of TAK1 and TAB1. MMC transfected with expression vector encoding HA-TAK1 alone or together with FLAG-TAB1 were treated with increasing concentrations of OA for 6 h, as indicated. Expression of HA-TAK1 and FLAG-TAB1 was determined by Western blot analysis with anti-HA and anti-FLAG antibodies, respectively. Four hundred units of λ protein phosphatase (λ PPase) were added to cell lysates and incubated for 30 min at 30 °C to dephosphorylate TAK1. C, TAK1 kinase activity is required for its phosphorylation. Wild-type HA-TAK1 (wt) or kinase-deficient mutant of HA-TAK1 (DN) was coexpressed with FLAG-TAB1 in MMC, as indicated, and treated with 0.5 μm OA for 6 h. Western blot analysis and treatment with λ PPase were performed as described in B.
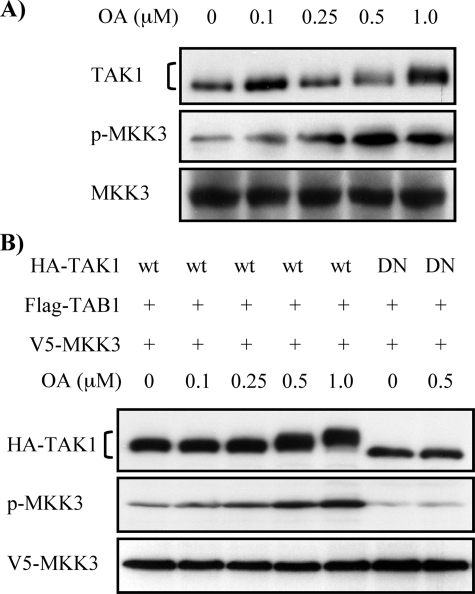
OA-induced TAK1 Phosphorylation Activates MKK3—We have previously reported that TAK1 is a major upstream signaling molecule activating MKK3 in MMC (37). Therefore, we next examined whether OA-enhanced TAK1 autophosphorylation is associated with downstream MKK3 activation in MMC. As shown in Fig. 2A, OA treatment increased phosphorylation of endogenous MKK3 in parallel with the OA-induced TAK1 phosphorylation in a dose-dependent manner. To determine whether the effect of OA on MKK3 phosphorylation was dependent on kinase activity of TAK1, either HA-TAK1 or HA-TAK1DN was coexpressed with FLAG-TAB1 and V5-MKK3 under different concentrations of OA, and changes in MKK3 phosphorylation and TAK1 migration were then determined by Western blot analysis. Fig. 2B demonstrates that coexpression of wild-type TAK1 and TAB1 increased phosphorylation of ectopically expressed V5-MKK3 in parallel with OA-induced TAK1 phosphorylation, similar to the endogenous MKK3 (Fig. 2A). However, no increases in phosphorylation of V5-MKK3 were observed after OA treatment when kinase-deficient mutant HA-TAK1DN was coexpressed (Fig. 2B). Thus, these data indicate that OA-induced MKK3 activation is not because of a direct effect of OA on MKK3 phosphorylation but because of OA-induced TAK1 phosphorylation and that MKK3 activation in MMC is predominantly dependent on TAK1 activity, as previously reported (37).
FIGURE 2.
OA-induced TAK1 phosphorylation mediates MKK3 phosphorylation. A, OA treatment increases endogenous MKK3 phosphorylation. MMC grown to subconfluence were treated with increasing concentrations of OA for 6 h as indicated. Cell lysates were subjected to Western blot analysis with anti-TAK1, anti-p-MKK3/6, and anti-MKK3 antibodies, respectively. B, OA-induced MKK3 phosphorylation is dependent on TAK1 activation. Wild-type HA-TAK1 (wt) or kinase-deficient mutant of HA-TAK1 (DN) was coexpressed with FLAG-TAB1 and V5-MKK3 in MMC and treated with increasing concentrations of OA for 6 h. Cell lysates were subjected to Western blot analysis with anti-HA, anti-p-MKK3/6, and anti-V5 antibodies, respectively.
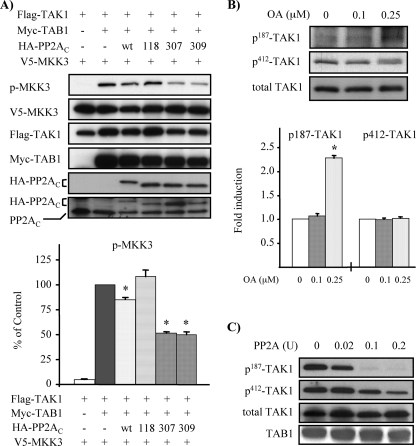
PP2A Inhibits TAK1 Activation Induced by Autophosphorylation—Given that OA can inhibit not only PP2A but also other protein phosphatases, such as PP6 (61, 62) and PP1 at high concentrations (58), we employed additional strategies to determine the role of PP2A in TAK1 activation. First, we examined the effects of ectopic expression of wild type and various mutants of PP2AC in MMC. TAK1 activation was evaluated by MKK3 phosphorylation, which was previously shown to be dependent on TAK1 activity (Fig. 2B). As shown in Fig. 3A, the ectopic expression of wild-type PP2AC reduced MKK3 phosphorylation induced by coexpression of TAK1 and TAB1. Moreover, the expression of either Y307F or L309Q, two nonphosphatase-deficient mutants of PP2AC, also resulted in reduced MKK3 phosphorylation. The expression levels of the transfected wild-type and mutant HA-PP2Ac, as compared with endogenous PP2Ac detected by Western blotting with anti-PP2Ac antibody (Fig. 3A, bottom), show that the seemingly modest changes in MKK3 phosphorylation may be because PP2Ac overexpression can be difficult, since this enzyme is subject to transcriptional autoregulation. Quantitative analysis show that the ectopic expression of wild-type PP2Ac and its mutants, Y307F and L309Q, reduced MKK3 phosphorylation by 20 ± 2, 51 ± 1, and 49 ± 2%, respectively. On the other hand, expression of H118N, a phosphatase-deficient mutant of PP2AC, exerted little additive effect on MKK3 activation, which suggests that the coexpression of TAK1 and TAB1 induced TAK1 activation almost to its maximal degree.
FIGURE 3.
PP2A down-regulates phosphorylation of TAK1 and MKK3. A, ectopic expression of wild type or mutants of PP2AC changes TAK1 activity. MMC were transfected as indicated with expression vectors for V5-MKK3, FLAG-TAK1, Myc-TAB1, and wild-type HA-PP2AC (wt) or mutants of PP2AC (H118N, Y307F, or L309Q). TAK1 activity was assessed by MKK3 phosphorylation. Expression of each transfected gene was confirmed by Western blot analysis with anti-V5, anti-FLAG, anti-Myc, and anti-HA antibodies, respectively. Expression levels of endogenous and exogenous PP2AC were evaluated by Western blot with anti-PP2AC antibody (bottom panel). Densitometry data are expressed as the percentage of p-MKK3 signals, quantified as the ratio to total V5-MKK3, compared with control transfection cells with FLAG-TAK1, Myc-TAB1, and V5-MKK3, but without HA-PP2AC (lane 2); the results represent mean ± S.E. of three independent experiments (*, p < 0.05 versus control transfection). B, OA treatment induces TAK1 phosphorylation at Thr-187. MMC grown to subconfluence were treated with OA for 30 min with the indicated concentrations. Cell lysates were subjected to Western blot analysis with anti-phospho-Thr-187-TAK1 (p187-TAK1), anti-phospho-Ser-412-TAK1 (p412-TAK1), and anti-TAK1 (total TAK1), respectively. Densitometry data are presented as -fold increase in p187-TAK1 or p412-TAK1 signals, quantified as the ratio to total TAK1, compared with control untreated cells; the results represent mean ± S.E. of three independent experiments (*, p < 0.05 versus untreated cells). C, dephosphorylation of TAK1 by PP2A primarily targets p-Thr-187 in the TAK1 activation loop. Phosphorylated HA-TAK1 was obtained by coexpression with FLAG-TAB1 in MMC followed by immunoprecipitation with anti-HA antibodies. The immunoprecipitates were incubated with 0.02, 0.1, and 0.2 units of purified PP2A for 20 min at 30 °C. Following the reaction, dephosphorylation of TAK1 was evaluated by Western blotting with anti-phospho-Thr-187-TAK1 and anti-phospho-Ser-412-TAK1 antibodies. A relative equivalent amount of HA-TAK1 in the immunoprecipitated samples was verified by Western blotting with anti-HA antibody. Expression of FLAG-TAB1 was confirmed by subjecting cell lysates to Western blot analysis with anti-FLAG antibody.
We next examined the effects of OA and PP2A on phosphorylation of TAK1 specifically at Thr-187 in the activation loop and Ser-412. To avoid the potential effect of OA on PP1 activity, we used low concentrations of OA (0.1 and 0.25 μm) and also reduced the treatment time of OA to 30 min. In MMC, treatment with 0.25 μm OA significantly induced endogenous TAK1 phosphorylation at Thr-187, but phosphorylation at Ser-412 was not affected (Fig. 3B). We next sought to determine whether PP2A may down-regulate TAK1 by directly dephosphorylating Thr-187 in the TAK1 activation loop required for its activation. An in vitro dephosphorylation assay with purified PP2A (AC heterodimeric form) was carried out, as shown in Fig. 3C, which demonstrates that TAK1 was phosphorylated at Thr-187 and Ser-412 by the coexpression of TAB1, and increasing concentrations of PP2A effectively dephosphorylated Thr-187 of TAK1, whereas dephosphorylation of Ser-412 was less effective by the same concentrations of PP2A. These data suggest that phosphorylated Thr-187 in the TAK1 activation loop may be the major target site of dephosphorylation by PP2A to negatively regulate TAK1.
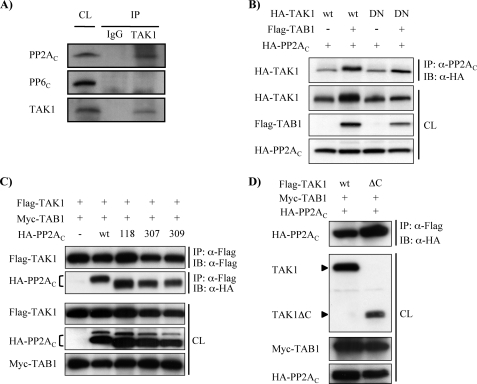
PP2A Associates with both TAK1 and TAB1—Given our findings indicating that PP2A dephosphorylates TAK1, we sought to determine whether PP2A might be associated with TAK1 to perform its inhibitory function even in the absence of stimuli. To verify the stable association of TAK1 with PP2A in MMC, cell lysates were subjected to immunoprecipitation with rabbit antibody against TAK1. Nonstimulated rabbit IgG was used as a control. Immunoprecipitates were then subjected to Western blotting with anti-PP2AC antibody. We also assessed for TAK1 interaction with PP6, another OA-sensitive phosphatase, by Western blotting with anti-PP6C antibody. Whole cell lysates not subjected to immunoprecipitation were also applied as positive controls for each protein. As shown in Fig. 4A, endogenous TAK1 was stably associated with PP2AC under nonstimulated conditions. However, TAK1-bound PP6C was not detectable in MMC, suggesting that PP2A rather then PP6 might regulate TAK1 activation in MMC.
FIGURE 4.
PP2A associates with TAK1. A, endogenous PP2AC is stably associated with TAK1. Cell lysates from MMC grown to subconfluence were immunoprecipitated with rabbit anti-TAK1 antibody. Nonimmunized rabbit IgG was used for control. Immunoprecipitates (IP) and cell lysates (CL) were subjected to immunoblotting with anti-PP2AC, anti-PP6C, and anti-TAK1 antibodies. B, phosphorylation status of TAK1 is not essential for the association of TAK1 with PP2AC. Wild-type HA-TAK1 (wt) or kinase-deficient mutant of HA-TAK1 (DN) was coexpressed with FLAG-TAB1 and HA-PP2AC, as indicated, in MMC. Cell lysates were subjected to immunoprecipitation with anti-PP2AC antibody, followed by immunoblotting (IB) with anti-HA antibody. Expression of the transfected genes was confirmed by subjecting cell lysates (CL) to Western blotting with corresponding anti-HA and anti-FLAG antibodies, as indicated. C, phosphatase activity of PP2AC does not affect the association of PP2AC with TAK1. Wild-type HA-PP2AC (wt) or mutants of PP2AC (H118N, Y307F, or L309Q) were coexpressed with FLAG-TAK1 and Myc-TAB1, as indicated, in MMC. Cell lysates were subjected to immunoprecipitation with anti-FLAG antibody for TAK1, followed by immunoblotting with either anti-FLAG or anti-HA antibodies. Expression of the transfected genes was confirmed by subjecting cell lysates to Western blotting with corresponding anti-FLAG, anti-HA, and anti-Myc antibodies. D, PP2AC interacts with the N-terminal region of TAK1. MMC were transfected with expression constructs encoding either full-length FLAG-TAK1 (wt) or C-terminally truncated TAK1 (ΔC) together with Myc-TAB1 and HA-PP2AC, as indicated. Cell lysates were subjected to immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with anti-HA antibody. Expression of the transfected genes was confirmed in cell lysates (CL) subjected to Western blotting with corresponding anti-TAK1, anti-Myc, and anti-HA antibodies.
To further determine whether either phosphorylation state or kinase activity of TAK1 is indispensable for the association with PP2AC, wild-type TAK1 (HA-TAK1) or kinase-deficient mutant of TAK1 (HA-TAK1DN) was coexpressed with HA-PP2AC and FLAG-TAB1, and then cell lysates were subjected to immunoprecipitation with anti-PP2AC antibody followed by Western blotting with anti-HA antibody. As shown in Fig. 4B, PP2AC coimmunoprecipitated with not only nonphosphorylated TAK1 (in the absence of coexpressed TAB1) but also kinase-deficient mutant of TAK1 (TAK1DN), indicating that neither phosphorylation state nor kinase activity of TAK1 is essential for its basal association with PP2AC. Conversely, we also examined if PP2AC activity affects the association with TAK1. Wild-type HA-PP2AC (wt in Fig. 4) or its mutant variants (HA-PP2AC (H118N), HA-PP2AC (Y307F), and HA-PP2AC (L309Q)) were coexpressed with FLAG-TAK1 and Myc-TAB1. As compared with wild-type PP2AC, however, all three mutants of PP2AC examined, including the phosphatase-deficient mutant H118N, did not show any significant differences in the association with TAK1 (Fig. 4C). Thus, the phosphatase activity of PP2AC may not affect the association with TAK1.
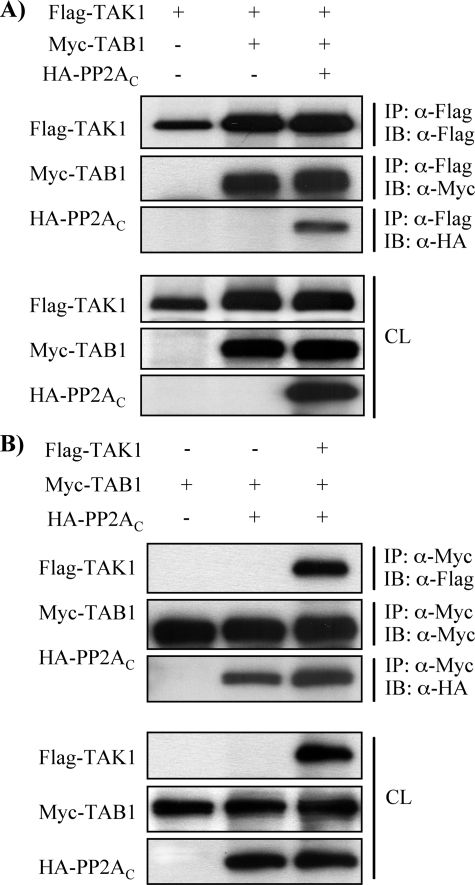
We next sought to determine which region of TAK1 is responsible for the association with PP2AC. Either full-length TAK1 (FLAG-TAK1) or C-terminal-truncated TAK1 (FLAG-TAK1ΔC) was coexpressed with Myc-TAB1 and HA-PP2AC. As shown in Fig. 4D, HA-PP2AC was associated with FLAG-TAK1ΔC as well as full-length FLAG-TAK1, indicating that the N-terminal region of TAK1 is responsible for its association with PP2AC. Intriguingly, it is known that the N-terminal half of TAK1 also bears a TAB1 binding domain (63) and a catalytic domain as well as phosphorylation sites. Therefore, it is plausible that PP2AC competes with TAB1 for the association with TAK1 to regulate TAK1 activation. To test this possibility, FLAG-TAK1 and Myc-TAB1 were coexpressed with or without HA-PP2AC. Unexpectedly, the expression of HA-PP2AC did not interfere with the interaction between FLAG-TAK1 and Myc-TAB1. In addition, HA-PP2AC was found to be a component of the TAK1-TAB1-PP2AC complex (Fig. 5A). However, it is possible that the formation of TAK1-TAB1-PP2AC complex was a consequence of the association of PP2AC with only TAK1 in the TAK1-TAB1 complex or with both TAK1 and TAB1 individually. To further analyze the association of PP2AC with TAK1 and TAB1, Myc-TAB1 and HA-PP2AC were coexpressed with or without FLAG-TAK1. As shown in Fig. 5B, HA-PP2AC coimmunoprecipitated with Myc-TAB1 in the absence of coexpression of FLAG-TAK1, whereas coexpression of FLAG-TAK1 and Myc-TAB1 enhanced the association of HA-PP2AC with Myc-TAB1. Thus, these data indicate that PP2AC is able to associate with TAK1 and TAB1 independently, and TAK1-TAB1 complex may facilitate the association with PP2AC.
FIGURE 5.
PP2AC associates with TAB1 independent of TAK1. A, FLAG-TAK1 was coexpressed with Myc-TAB1 and HA-PP2AC as indicated in MMC. Cell lysates were subjected to immunoprecipitation (IP) with anti-FLAG antibody, followed by immunoblotting (IB) with anti-FLAG, anti-Myc, and anti-HA antibodies, respectively. Expression of the transfected genes was confirmed in cell lysates (CL) subjected to Western blotting with corresponding anti-FLAG, anti-Myc, and anti-HA antibodies. B, Myc-TAB1 was coexpressed with FLAG-TAK1 and HA-PP2AC as indicated in MMC. Cell lysates were subjected to immunoprecipitation with anti-Myc antibody followed by immunoblotting with anti-FLAG, anti-Myc, and anti-HA antibodies, respectively. Expression of the transfected genes was confirmed in cell lysates subjected to Western blotting with corresponding anti-FLAG, anti-Myc, and anti-HA antibodies.
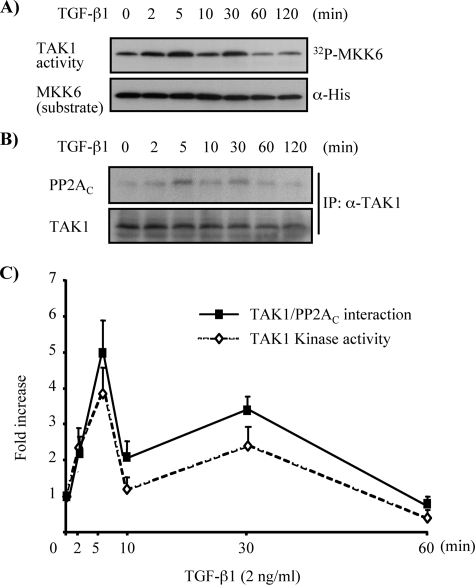
Association of PP2A with TAK1 Is Enhanced by TGF-β1 Stimulation—We next posed the question of whether PP2A also played a role in the regulation of TGF-β1-induced TAK1 activation in MMC. To address this, endogenous TAK1 protein was immunoprecipitated from cell lysates of MMC after stimulation with TGF-β1 for various time periods. One-half of the immunoprecipitate was subjected to an immune complex kinase assay using bacterial expressed His-MKK6 as a substrate of TAK1, and the other half was subjected to immunoblotting with anti-PP2AC antibody to evaluate potential correlation between TAK1 activation and PP2AC association with TAK1. As demonstrated in Fig. 6A, TGF-β1-induced endogenous TAK1 activation occurred in a biphasic manner. The initial activation was detectable within 2 min, the earliest time period examined, and peaked at 5 min after TGF-β1 stimulation. The primary activation declined rapidly thereafter to basal levels by 10 min after stimulation. A second wave of TAK1 activation was detected at 30 min after TGF-β1 treatment, which declined back to basal levels by 60 min after the treatment (Fig. 6A). Interestingly, the association of PP2AC with TAK1 (Fig. 6B) showed a close correlation with the time course of TAK1 activation by TGF-β1 (Fig. 6C).
FIGURE 6.
Association of PP2AC with TAK1 parallels activation and inactivation of endogenous TAK1 by TGF-β1 stimulation. MMC grown to subconfluence were rendered quiescent in medium supplemented with 0.5% FBS for 16 h and then stimulated with TGF-β1 (2 ng/ml) for the indicated times. Endogenous TAK1 in cell lysates was immunoprecipitated with anti-TAK1 antibody. One-half of the immunoprecipitate was subjected to immune complex kinase assay to assess endogenous TAK1 activity using His-MKK6 as a substrate and phosphorylation of MKK6 by TAK1 was visualized by autoradiography (A, top). The other half of the immunoprecipitate (IP) was subjected to immunoblotting with anti-PP2AC antibody to evaluate the association of endogenous TAK1 with PP2AC (B, top). Immunoblotting with anti-His and anti-TAK1 antibodies served as loading controls (bottom). C, densitometry data are presented as -fold increase in [32P]MKK6 and coprecipitated PP2AC, quantified as the ratio to total MKK6 substrate or total immunoprecipitated TAK1, compared with control untreated cells; the results represent mean ± S.E. of three independent experiments (⋄, 32P-labeled His-MKK6 versus His-MKK6; ▪, PP2AC versus TAK1 after immunoprecipitation).
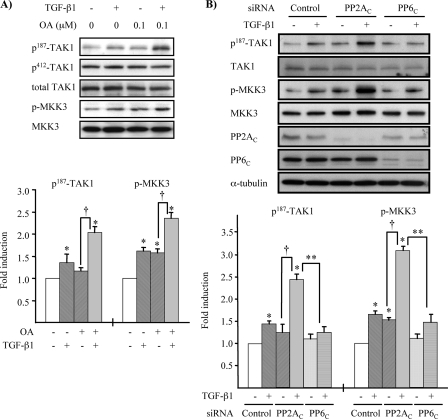
PP2A Negatively Regulates TGF-β1-induced TAK1 Activation—Given our findings indicating that the association of PP2AC with TAK1 showed a close correlation with the time course of TAK1 activation by TGF-β1, we sought to examine whether PP2A negatively regulates TGF-β1-induced TAK1 activation. To address this issue, PP2A activity was attenuated by OA treatment, and the activation of endogenous TAK1 by TGF-β1 stimulation was determined by phosphorylation of TAK1 and MKK3. MMC grown to subconfluence were pretreated with or without 0.1 μm OA for 30 min and then stimulated with TGF-β1, as indicated in Fig. 7A. Stimulation with TGF-β1 alone for 5 min or 0.1 μm OA for 30 min without TGF-β1 stimulation significantly induced phosphorylation of MKK3. The TGF-β1 induction of TAK1 phosphorylation at Thr-187 was to a lesser degree, but when combined with pretreatment with OA it significantly enhanced phosphorylation of TAK1 at Thr-187 as well as MKK3. In contrast, phosphorylation of Ser-412 did not respond to TGF-β1 stimulation even in the presence of OA pretreatment.
FIGURE 7.
Down-regulation of PP2A activity enhances TGF-β1-induced TAK1 and MKK3 phosphorylation. A, low dose OA increases TGF-β1-induced phosphorylation of Thr-187 in TAK1 and MKK3. MMC grown to subconfluence were rendered quiescent in medium supplemented with 0.5% FBS for 16 h and pretreated with 0.1 μm OA for 30 min, followed by treatment with TGF-β1 (2 ng/ml) for 5 min. Endogenous TAK1 and MKK3 phosphorylation were determined by Western blot analysis with anti-phospho-Thr-187-TAK1 (p187-TAK1), anti-phospho-Ser-412-TAK1 (p412-TAK1), and anti-phospho-MKK3/6 antibodies. Reblotting with anti-TAK1 (total TAK1) and anti-MKK3 antibodies was performed for loading controls. Quantitative analysis of levels of phospho-Thr-187-TAK1 and p-MKK3 was determined by densitometry, and data are presented as the mean ± S.E. band density quantified as the ratio to total TAK1 and MKK3 and normalized to nontreated conditions (*, p < 0.05 versus nontreated conditions; †, p < 0.05 versus OA pretreatment only). B, knockdown of PP2AC with siRNA enhances TGF-β1-induced phosphorylation of Thr-187 in TAK1 activation loop and MKK3. MMC transfected with siRNA targeted against PP2AC, PP6C, or control nontargeting siRNA (Control) were incubated in medium containing 15% FBS for 24 h and then rendered quiescent in medium supplemented with 0.5% FBS for 16 h prior to treatment with TGF-β1 (2 ng/ml) for 5 min. Endogenous TAK1 and MKK3 phosphorylation were determined by Western blot analysis with anti-phospho-Thr-187-TAK1 (p187-TAK1) and anti-p-MKK3/6 antibodies. Reblotting with anti-TAK1, anti-MKK3, and anti-α-tubulin antibodies served as loading controls. Reduction of PP2AC and PP6C protein expression was examined by Western blotting with anti-PP2AC and anti-PP6C antibodies. Densitometry data are presented as -fold increase in phospho-Thr-187-TAK1 or p-MKK3, quantified as the ratio to total TAK1 or MKK3, compared with control siRNA transfected cells without TGF-β1 treatment; the results represent mean ± S.E. of three independent experiments (*, p < 0.05 versus control siRNA-transfected cells without TGF-β1 treatment; **, p < 0.05 versus PP2AC siRNA-transfected cells with TGF-β1 treatment; †, p < 0.05 versus PP2AC siRNA-transfected cells without TGF-β1 treatment).
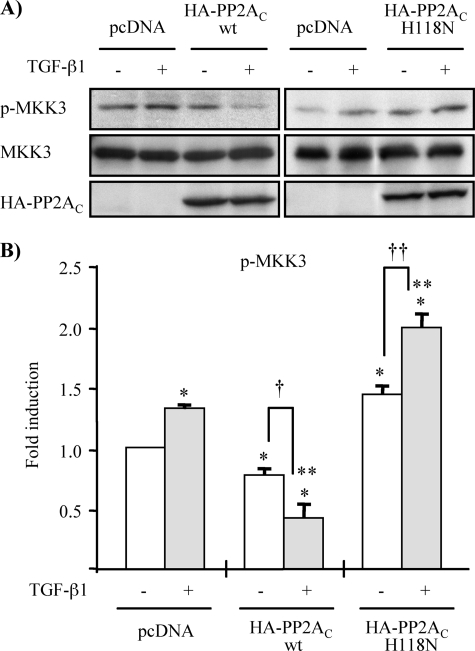
To further support the notion that PP2A and not PP6 is responsible for the negative regulation of TGF-β1-induced TAK1 activation, we determined the effects of targeted knockdown of PP2AC or PP6c by siRNA. As shown in Fig. 7B, TGF-β1-induced phosphorylation of TAK1 at Thr-187 and MKK3 was significantly enhanced by knockdown of PP2Ac but not by knockdown of PP6c. Moreover, overexpression of wild-type PP2AC severely hampered TGF-β1-induced endogenous MKK3 phosphorylation, whereas the ectopic expression of phosphatase-deficient mutant of PP2AC additively increased TGF-β1-induced endogenous MKK3 phosphorylation (Fig. 8). Collectively, our findings suggest that TGF-β1 stimulation activates TAK1, at least in part, through phosphorylation at Thr-187 in the activation loop that is negatively regulated by PP2A. Moreover, our results also suggest that monitoring MKK3 phosphorylation may be a more sensitive way to evaluate TGF-β1-induced endogenous TAK1 activation, since it is more readily detectable than immunoblotting with anti-phospho-Thr-187-TAK1 antibody in MMC.
FIGURE 8.
Ectopic expression of wild-type and phosphatase-deficient mutant of PP2AC affects TGF-β1-induced MKK3 phosphorylation. A, MMC were transiently transfected with expression vector encoding wild-type HA-PP2AC (wt) or phosphatase-deficient mutant of HA-PP2AC (H118N), as indicated. As controls, MMC were transfected with empty vector pcDNA3.1 (lanes 1 and 2 of each panel). Following transfection, cells were incubated in medium containing 15% FBS for 18 h and then rendered quiescent in medium supplemented with 0.5% FBS for 16 h, prior to treatment with TGF-β1 (2 ng/ml) for 5 min. Endogenous TAK1 activation was assessed by alteration in MKK3 phosphorylation determined by Western blot analysis with anti-p-MKK3/6 antibody. Immunoblotting with anti-MKK3 and anti-HA antibodies were performed for loading controls. B, densitometry data are presented as -fold increase in p-MKK3, quantified as the ratio to total MKK3, compared with empty vector pcDNA3.1 controls without TGF-β1 treatment; empty vector pcDNA3.1 controls without TGF-β1 treatment (*, p < 0.05 versus empty vector controls without TGF-β1 treatment; **, p < 0.05 versus empty vector controls with TGF-β1 treatment; †, p < 0.05 versus wild-type HA-PP2Ac-transfected cells without TGF-β1 treatment; ††, p < 0.05 versus mutant HA-PP2Ac (H118N)-transfected cells without TGF-β1 treatment).
DISCUSSION
TAK1 was initially identified as a kinase activated by members of the TGF-β and BMP family, and recent studies have demonstrated the role of TAK1 in TGF-β1-induced activation of MAPKs and extracellular matrix synthesis. We have previously demonstrated that the TAK1-MKK3-p38 MAPK signaling cascade is an indispensable mediator of TGF-β1-induced type I collagen expression in MMC (22, 37). However, specific mechanisms regulating TAK1 activation in response to TGF-β1 stimulation have not yet been fully elucidated. Our findings in MMC reveal that TAK1 is activated and deactivated very rapidly. This rapid activation and inactivation of TAK1 by TGF-β1 stimulation have been also observed in cardiac myocytes (39). TNF-α and IL-1 also induce rapid activation and inactivation of TAK1 (29, 45). Therefore, endogenous TAK1 activation and inactivation appear to be tightly regulated and may be controlled through rapid phosphorylation and dephosphorylation.
TAK1 requires phosphorylation at serine and threonine residues in the activation loop for its activation; thus, dephosphorylation by a Ser/Thr protein phosphatase would result in inactivation of TAK1. Instances of protein phosphatases functioning as negative regulators of TAK1 activation have been recently reported. PP6, a type 2A phosphatase family member, and two isoforms of PP2C (PP2Cβ-1 and PP2Cε) have been shown to dephosphorylate and inactivate TAK1 in 293 and COS7 cells (43-45). PP6 down-regulates TAK1 activation in response to IL-1 stimulation in 293 IL-1RI cells. In the case of PP2Cβ-1, inhibition of TAK1 activity occurs under nonstimulated conditions and does not respond to ligand stimulation, whereas PP2Cε is transiently dissociated from TAK1 by IL-1 stimulation, whereby TAK1 can be autophosphorylated and presumably activated.
Here, we demonstrate the first evidence that PP2A, another member of type 2A phosphatase family, also functions as a negative regulator of TAK1 activation in response to TGF-β1 stimulation. PP2A has the ability to associate with TAK1 and TAB1 individually and down-regulates autophosphorylation of TAK1. Interestingly, the mode of interaction of PP2A and PP2Cε with TAK1 in response to ligand stimulation is in the opposite direction. IL-1 stimulation results in transient dissociation of PP2Cε from TAK1 for its activation. Conversely, the association of PP2AC with TAK1 is increased in response to TGF-β1 stimulation. Moreover, IL-1-induced TAK1 activation is negatively regulated by PP6 and not by PP2A in 293 IL-1RI cells (45). However, our findings demonstrate that TAK1 stably interacts with PP2AC but not with PP6C in MMC, although PP6C is expressed. Therefore, control of TAK1 activation may be conducted in a cell type-specific or context-dependent manner. Thus, different protein phosphatases regulate TAK1 activity, and the opposite direction of the interaction of the phosphatases with TAK1 leads to inactivation or activation of TAK1.
Our findings show that the basal interaction of PP2AC with TAK1 is not dependent on either the phosphatase activity of PP2AC or phosphorylation state of TAK1. Interestingly, the presence of TAB1 enhanced the interaction of PP2AC with both wild-type and kinase-deficient TAK1, suggesting that the interaction not only occurs independently of TAK1 kinase activity (Fig. 4B) but also that PP2AC is able to interact with TAB1 independently, as shown in Fig. 5B, and/or the TAK1/TAB1 complex may facilitate the association with PP2AC.
The enhanced association of PP2AC with TAK1 by TGF-β1 stimulation in parallel with TAK1 activation suggests that TGF-β1-induced activation of TAK1 is controlled by binding of PP2AC. There are several plausible mechanisms by which TGF-β1 stimulation enhances the interaction of PP2AC with TAK1. First, TGF-β1 stimulation may enhance the formation of TAK1-TAB1 complex to activate TAK1 by autophosphorylation, which in turn enhances the interaction of PP2AC with TAK1 as shown in Figs. 4B and 5B. Second, TGF-β1 stimulation may induce dissociation of other phosphatases, such as PP2Cε, from TAK1, and that allows PP2A to associate with the phosphatase-free TAK1. Finally, TGF-β1 stimulation may specifically induce the association of PP2AC with TAK1. This is supported by previous reports that TGF-β1 stimulation induces the interaction of PP2A with certain proteins, such as p70S6 kinase and pRB (47, 64), although the precise mechanism remains to be defined. In addition, we observed that the ectopic expression of mutants of PP2AC (PP2AC (Y307F) and PP2AC (L309Q)) reduced the autophosphorylation of TAK1 more profoundly than wild-type PP2AC. This may be because these two mutants do not associate with the regulatory subunit Bα of PP2A (PP2ABα) (52). The H118N mutant of PP2AC harbors mutation in a region that is much farther upstream that would not affect interaction with PP2ABα. Upon TGF-β1 stimulation, PP2ABα associates with activated type I TGF-β receptor, which in turn, would decrease the availability of PP2ABα to interact with the PP2AAC core enzyme (46). Moreover, PP2ABα exhibits an inhibitory effect on PP2A activity in vitro (51). Therefore, given that the Y307F and L309Q mutants of PP2AC cannot associate with PP2ABα, the resultant decreased interaction of PP2ABα with the PP2AAC core enzyme would enhance the phosphatase activity of PP2A toward TAK1.
Phosphorylation of Thr-187 in the activation loop of TAK1 has been shown to be crucial for its activation not only by autophosphorylation but also by IL-1 and TNF-α stimulation. In addition, the Ser-412 residue is also phosphorylated in response to protein kinase A activation (30). However, our in vitro dephosphorylation assay and OA treatment results suggest that Thr-187 rather than Ser-412 may be the major target of PP2A in MMC. As others have reported, direct detection of phosphorylated endogenous TAK1 at Thr-187 by immunoblotting after ligand stimulation, such as TNF-α and IL-1, is difficult in the absence of a potent phosphatase inhibitor, such as OA (45). Detection of TGF-β1-induced endogenous TAK1 phosphorylation at Thr-187 by Western blotting with anti-phospho-Thr-187-TAK1 antibody was not efficient (Fig. 7, A and B). However, either OA treatment or knockdown of PP2AC with specific siRNA definitely enhanced TGF-β1-induced endogenous TAK1 phosphorylation at the Thr-187 residue, which in turn induced phosphorylation of MKK3, an immediate downstream effector of TAK1. These data indicate that the limited ability to detect TGF-β1-induced TAK1 phosphorylation might be due to tight regulation of TAK1 by PP2A.
TAK1 plays an essential function in TGF-β1 signaling through its ability to regulate at least two major signaling pathways, the MAPK and the Smad signaling pathways. Both the JNK and the p38 MAPK cascades are targets of TAK1 activation by TGF-β1. TAK1 can also regulate TGF-β-induced activation of Smad signaling by inducing Smad7 expression (41) and also interfering with transactivation of R-Smads by direct interaction with Mad homolog domain 2 of Smad proteins (42). Thus, activation of TAK1 may exert an inhibitory effect on the Smad signaling pathway. However, the inhibitory effect of TAK1 on Smad function is still controversial, since more recent studies have demonstrated that TAK1 induces the degradation of SnoN (Ski-related novel gene N), which in turn increases Smad-dependent transcriptional activation of TGF-β-responsible genes (65). In addition to the role of TAK1 in the regulation of Smad function, there is cross-talk between the Smad and downstream targets of TAK1, such as p38 MAPK and ATF2, in regulation of certain TGF-β1 target gene expression (20, 66, 67). Collectively, TAK1 plays a vital role in TGF-β1 signal transduction; therefore, tight regulation of endogenous TAK1 activation is imperative for fine tuning the multiple TGF-β1 actions in physiological state. Once TAK1 is rapidly activated, efficient down-regulation of TAK1 activity is important to prevent excessive TGF-β1 responses, since dysregulation and persistent TGF-β1 actions are thought to lead to pathological states. In the present study, we provide several lines of evidence indicating that PP2A plays a crucial role as a negative regulator in TGF-β1-induced TAK1 activation. Moreover, PP2A also participates in suppressing TAK1 basal activity under nonstimulated conditions.
This work was supported in part by NIDDK, National Institutes of Health, Grant R01 DK57661 and the M. James Scherbenske Grant from the American Society of Nephrology (to M. E. C.) and Beginning Grant-in-Aid 0665379U from the American Heart Association (to S. I. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TGF, transforming growth factor; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; OA, okadaic acid; MMC, primary mouse mesangial cells; IL, interleukin; p187-TAK1, phospho-Thr-187-TAK1; p412-TAK1, phospho-Ser-412-TAK1; p-MKK3, phospho-MKK3; FBS, fetal bovine serum; HA, hemagglutinin; siRNA, small interfering RNA.
References
- 1.Heldin, C. H., Miyazono, K., and ten Dijke, P. (1997) Nature 390 465-471 [DOI] [PubMed] [Google Scholar]
- 2.Massague, J. (1998) Annu. Rev. Biochem. 67 753-791 [DOI] [PubMed] [Google Scholar]
- 3.Border, W. A., and Noble, N. A. (1994) N. Engl. J. Med. 331 1286-1292 [DOI] [PubMed] [Google Scholar]
- 4.Cheng, J., and Grande, J. P. (2002) Exp. Biol. Med. 227 943-956 [DOI] [PubMed] [Google Scholar]
- 5.Massague, J., Blain, S. W., and Lo, R. S. (2000) Cell 103 295-309 [DOI] [PubMed] [Google Scholar]
- 6.de Caestecker, M. P., Piek, E., and Roberts, A. B. (2000) J. Natl. Cancer Inst. 92 1388-1402 [DOI] [PubMed] [Google Scholar]
- 7.Derynck, R., and Zhang, Y. E. (2003) Nature 425 577-584 [DOI] [PubMed] [Google Scholar]
- 8.Moustakas, A., and Heldin, C. H. (2005) J. Cell Sci. 118 3573-3584 [DOI] [PubMed] [Google Scholar]
- 9.Edlund, S., Landstrom, M., Heldin, C. H., and Aspenstrom, P. (2002) Mol. Biol. Cell 13 902-914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhowmick, N. A., Ghiassi, M., Bakin, A., Aakre, M., Lundquist, C. A., Engel, M. E., Arteaga, C. L., and Moses, H. L. (2001) Mol. Biol. Cell 12 27-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulder, K. M., and Morris, S. L. (1992) J. Biol. Chem. 267 5029-5031 [PubMed] [Google Scholar]
- 12.Bakin, A. V., Tomlinson, A. K., Bhowmick, N. A., Moses, H. L., and Arteaga, C. L. (2000) J. Biol. Chem. 275 36803-36810 [DOI] [PubMed] [Google Scholar]
- 13.Yi, J. Y., Shin, I., and Arteaga, C. L. (2005) J. Biol. Chem. 280 10870-10876 [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi, K., Shirakabe, K., Shibuya, H., Irie, K., Oishi, I., Ueno, N., Taniguchi, T., Nishida, E., and Matsumoto, K. (1995) Science, 270 2008-2011 [DOI] [PubMed] [Google Scholar]
- 15.Shibuya, H., Yamaguchi, K., Shirakabe, K., Tonegawa, A., Gotoh, Y., Ueno, N., Irie, K., Nishida, E., and Matsumoto, K. (1996) Science 272 1179-1182 [DOI] [PubMed] [Google Scholar]
- 16.Hartsough, M. T., and Mulder, K. M. (1995) J. Biol. Chem. 270 7117-7124 [DOI] [PubMed] [Google Scholar]
- 17.Mucsi, I., Skorecki, K. L., and Goldberg, H. J. (1996) J. Biol. Chem. 271 16567-16572 [DOI] [PubMed] [Google Scholar]
- 18.Atfi, A., Djelloul, S., Chastre, E., Davis, R., and Gespach, C. (1997) J. Biol. Chem. 272 1429-1432 [DOI] [PubMed] [Google Scholar]
- 19.Hocevar, B. A., Brown, T. L., and Howe, P. H. (1999) EMBO J. 18 1345-1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanafusa, H., Ninomiya-Tsuji, J., Masuyama, N., Nishita, M., Fujisawa, J., Shibuya, H., Matsumoto, K., and Nishida, E. (1999) J. Biol. Chem. 274 27161-27167 [DOI] [PubMed] [Google Scholar]
- 21.Chin, B. Y., Mohsenin, A., Li, S. X., Choi, A. M., and Choi, M. E. (2001) Am. J. Physiol. Renal. Physiol. 280 F495-F504 [DOI] [PubMed] [Google Scholar]
- 22.Wang, L., Ma, R., Flavell, R. A., and Choi, M. E. (2002) J. Biol. Chem. 277 47257-47262 [DOI] [PubMed] [Google Scholar]
- 23.Shirakabe, K., Yamaguchi, K., Shibuya, H., Irie, K., Matsuda, S., Moriguchi, T., Gotoh, Y., Matsumoto, K., and Nishida, E. (1997) J. Biol. Chem. 272 8141-8144 [DOI] [PubMed] [Google Scholar]
- 24.Ninomiya-Tsuji, J., Kishimoto, K., Hiyama, A., Inoue, J., Cao, Z., and Matsumoto, K. (1999) Nature 398 252-256 [DOI] [PubMed] [Google Scholar]
- 25.Sakurai, H., Suzuki, S., Kawasaki, N., Nakano, H., Okazaki, T., Chino, A., Doi, T., and Saiki, I. (2003) J. Biol. Chem. 278 36916-36923 [DOI] [PubMed] [Google Scholar]
- 26.Irie, T., Muta, T., and Takeshig, K. (2000) FEBS Lett. 467 160-164 [DOI] [PubMed] [Google Scholar]
- 27.Sakurai, H., Miyoshi, H., Mizukami, J., and Sugita, T. (2000) FEBS Lett. 474 141-145 [DOI] [PubMed] [Google Scholar]
- 28.Kishimoto, K., Matsumoto, K., and Ninomiya-Tsuji, J. (2000) J. Biol. Chem. 275 7359-7364 [DOI] [PubMed] [Google Scholar]
- 29.Singhirunnusorn, P., Suzuki, S., Kawasaki, N., Saiki, I., and Sakurai, H. (2005) J. Biol. Chem. 280 7359-7368 [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi, Y., Mizoguchi, T., Take, I., Kurihara, S., Udagawa, N., and Takahashi, N. (2005) J. Biol. Chem. 280 11395-11403 [DOI] [PubMed] [Google Scholar]
- 31.Takaesu, G., Kishida, S., Hiyama, A., Yamaguchi, K., Shibuya, H., Irie, K., Ninomiya-Tsuji, J., and Matsumoto, K. (2000) Mol. Cell. 5 649-658 [DOI] [PubMed] [Google Scholar]
- 32.Jiang, Z., Ninomiya-Tsuji, J., Qian, Y., Matsumoto, K., and Li, X. (2002) Mol. Cell. Biol. 22 7158-7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishitani, T., Takaesu, G., Ninomiya-Tsuji, J., Shibuya, H., Gaynor, R. B., and Matsumoto, K. (2003) EMBO J. 22 6277-6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin, G., Klika, A., Callahan, M., Faga, B., Danzig, J., Jiang, Z., Li, X., Stark, G. R., Harrington, J., and Sherf, B. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2028-2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung, P. C., Nebreda, A. R., and Cohen, P. (2004) Biochem. J. 378 27-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono, K., Ohtomo, T., Ninomiya-Tsuji, J., and Tsuchiya, M. (2003) Biochem. Biophys. Res. Commun. 307 332-337 [DOI] [PubMed] [Google Scholar]
- 37.Kim, S. I., Kwak, J. H., Zachariah, M., He, Y., Wang, L., and Choi, M. E. (2007) Am. J. Physiol. 292 F1471-F1478 [DOI] [PubMed] [Google Scholar]
- 38.Hocevar, B. A., Prunier, C., and Howe, P. H. (2005) J. Biol. Chem. 280 25920-25927 [DOI] [PubMed] [Google Scholar]
- 39.Zhang, D., Gaussin, V., Taffet, G. E., Belaguli, N. S., Yamada, M., Schwartz, R. J., Michael, L. H., Overbeek, P. A., and Schneider, M. D. (2000) Nat. Med. 6 556-563 [DOI] [PubMed] [Google Scholar]
- 40.Terada, Y., Nakashima, O., Inoshita, S., Kuwahara, M., Sasaki, S., and Marumo, F. (1999) Kidney Int. 56 1378-1390 [DOI] [PubMed] [Google Scholar]
- 41.Dowdy, S. C., Mariani, A., and Janknecht, R. (2003) J. Biol. Chem. 278 44377-44384 [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann, A., Preobrazhenska, O., Wodarczyk, C., Medler, Y., Winkel, A., Shahab, S., Huylebroeck, D., Gross, G., and Verschueren, K. (2005) J. Biol. Chem. 280 27271-27283 [DOI] [PubMed] [Google Scholar]
- 43.Hanada, M., Ninomiya-Tsuji, J., Komaki, K., Ohnishi, M., Katsura, K., Kanamaru, R., Matsumoto, K., and Tamura, S. (2001) J. Biol. Chem. 276 5753-5759 [DOI] [PubMed] [Google Scholar]
- 44.Li, M. G., Katsura, K., Nomiyama, H., Komaki, K., Ninomiya-Tsuji, J., Matsumoto, K., Kobayashi, T., and Tamura, S. (2003) J. Biol. Chem. 278 12013-12021 [DOI] [PubMed] [Google Scholar]
- 45.Kajino, T., Ren, H., Iemura, S., Natsume, T., Stefansson, B., Brautigan, D. L., Matsumoto, K., and Ninomiya-Tsuji, J. (2006) J. Biol. Chem. 281 39891-39896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Griswold-Prenner, I., Kamibayashi, C., Maruoka, E. M., Mumby, M. C., and Derynck, R. (1998) Mol. Cell. Biol. 18 6595-6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ory, S., Zhou, M., Conrads, T. P., Veenstra, T. D., and Morrison, D. K. (2003) Curr. Biol. 13 1356-1364 [DOI] [PubMed] [Google Scholar]
- 48.Virshup, D. M. (2000) Curr. Opin. Cell Biol. 12 180-185 [DOI] [PubMed] [Google Scholar]
- 49.Oliver, C. J., and Shenolikar, S. (1998) Front. Biosci. 3 D961-972 [DOI] [PubMed] [Google Scholar]
- 50.Hubbard, M. J., and Cohen, P. (1993) Trends Biochem. Sci. 18 172-177 [DOI] [PubMed] [Google Scholar]
- 51.Tolstykh, T., Lee, J., Vafai, S., and Stock, J. B. (2000) EMBO J. 19 5682-5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung, H., Nairn, A. C., Murata, K., and Brautigan, D. L. (1999) Biochemistry 38 10371-10376 [DOI] [PubMed] [Google Scholar]
- 53.Chen, J., Martin, B. L., and Brautigan, D. L. (1992) Science 257 1261-1264 [DOI] [PubMed] [Google Scholar]
- 54.Guo, H., and Damuni, Z. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 2500-2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Favre, B., Zolnierowicz, S., Turowski, P., and Hemmings, B. A. (1994) J. Biol. Chem. 269 16311-16317 [PubMed] [Google Scholar]
- 56.Petritsch, C., Beug, H., Balmain, A., and Oft, M. (2000) Genes Dev. 14 3093-3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chung, H., and Brautigan, D. L. (1999) Cell. Signal. 11 575-580 [DOI] [PubMed] [Google Scholar]
- 58.Favre, B., Turowski, P., and Hemmings, B. A. (1997) J. Biol. Chem. 272 13856-13863 [DOI] [PubMed] [Google Scholar]
- 59.Resjo, S., Oknianska, A., Zolnierowicz, S., Manganiello, V., and Degerman, E. (1999) Biochem. J. 341 839-845 [PMC free article] [PubMed] [Google Scholar]
- 60.Ugi, S., Imamura, T., Ricketts, W., and Olefsky, J. M. (2002) Mol. Cell. Biol. 22 2375-2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hastie, C. J., and Cohen, P. T. (1998) FEBS Lett. 431 357-361 [DOI] [PubMed] [Google Scholar]
- 62.Stefansson, B., and Brautigan, D. L. (2006) J. Biol. Chem. 281 22624-22634 [DOI] [PubMed] [Google Scholar]
- 63.Brown, K., Vial, S. C., Dedi, N., Long, J. M., Dunster, N. J., and Cheetham, G. M. (2005) J. Mol. Biol. 354 1013-1020 [DOI] [PubMed] [Google Scholar]
- 64.Van Berlo, J. H., Voncken, J. W., Kubben, N., Broers, J. L., Duisters, R., van Leeuwen, R. E., Crijns, H. J., Ramaekers, F. C., Hutchison, C. J., and Pinto, Y. M. (2005) Hum. Mol. Genet. 14 2839-2849 [DOI] [PubMed] [Google Scholar]
- 65.Kajino, T., Omori, E., Ishii, S., Matsumoto, K., and Ninomiya-Tsuji, J. (2007) J. Biol. Chem. 282 9475-9481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sano, Y., Harada, J., Tashiro, S., Gotoh-Mandeville, R., Maekawa, T., and Ishii, S. (1999) J. Biol. Chem. 274 8949-8957 [DOI] [PubMed] [Google Scholar]
- 67.Abecassis, L., Rogier, E., Vazquez, A., Atfi, A., and Bourgeade, M. F. (2004) J. Biol. Chem. 279 30474-30479 [DOI] [PubMed] [Google Scholar]