Abstract
Human DNA ligase III contains an N-terminal zinc finger domain that binds to nicks and gaps in DNA. This small domain has been described as a DNA nick sensor, but it is not required for DNA nick joining activity in vitro. In light of new structural information for mammalian ligases, we measured the DNA binding affinity and specificity of each domain of DNA ligase III. These studies identified two separate, independent DNA-binding modules in DNA ligase III that each bind specifically to nicked DNA over intact duplex DNA. One of these modules comprises the zinc finger domain and DNA-binding domain, which function together as a single DNA binding unit. The catalytic core of ligase III is the second DNA nick-binding module. Both binding modules are required for ligation of blunt ended DNA substrates. Although the zinc finger increases the catalytic efficiency of nick ligation, it appears to occupy the same binding site as the DNA ligase III catalytic core. We present a jackknife model for ligase III that posits conformational changes during nick sensing and ligation to extend the versatility of the enzyme.
DNA ligase III is one of three mammalian DNA ligases (LigI, LigIII, and LigIV)2 that function in different DNA replication and repair pathways. LigI is the major replicative ligase with limited functions in some DNA repair pathways (1-4). LigIV joins DNA double-stranded breaks in the nonhomologous end joining and during V(D)J recombination in the developing immune system (5). LigIII functions in a variety of DNA repair pathways, including single-stranded break repair, base excision repair, and nucleotide excision repair. A recent report indicates that LigIII also functions in a back-up pathway for nonhomologous end joining when the major NHEJ pathway is compromised (6-8). LigIII is the only DNA ligase present in mammalian mitochondria and probably participates in all DNA repair and replication activities in that organelle (9-11). Inactivation of the LIGIII gene causes early embryonic lethality (12), highlighting the essential function(s) of this enzyme.
LigIII and its partner protein XRCC1 interact with aprataxin, a phosphodiesterase that removes the 5′-adenylate group from abortive ligation intermediates arising from unsuccessful attempts at joining DNA strands with damaged termini (13). Left unrepaired, these abortive ligation intermediates are cytotoxic and especially dangerous in irreplaceable, post-mitotic cells such as neurons. Mutation of aprataxin causes ataxia oculomotor apraxia-1, a neurodegenerative disease (14, 15). Thus, promiscuous substrate selection by LigIII poses a risk for the development of some neurodegenerative diseases.
Despite their distinct biological roles, the three mammalian ligases utilize the same reaction mechanism (Fig. 1) (16, 17) and have similar core structures, including a highly divergent DNA-binding domain (DBD) and a well conserved catalytic core (CC) made up of the nucleotidyltransferase (NTase) and oligonucleotide/oligosaccharide-binding domains (OBD) (18, 19). Together, the DBD and CC enable mammalian ligases to fully encircle a DNA substrate, stabilizing a highly distorted, partially underwound structure that positions the DNA termini within the active site (18). The DBD of LigI was shown to be essential for DNA nick joining activity, and it makes the greatest contribution of any domain to DNA binding affinity (18). DNA ligases of bacteria and viruses also appear to be capable of wrapping around DNA (18, 20). Escherichia coli LigA and related NAD-dependent ligases have a C-terminal helix-hairpin-helix domain that functions similarly to the N-terminal DBD of LigI (20, 21). Even Chlorella virus ligase, which is the smallest known eukaryotic DNA ligase and lacks a DBD, has a small latch structure that allows the enzyme to fully encircle the DNA (22).
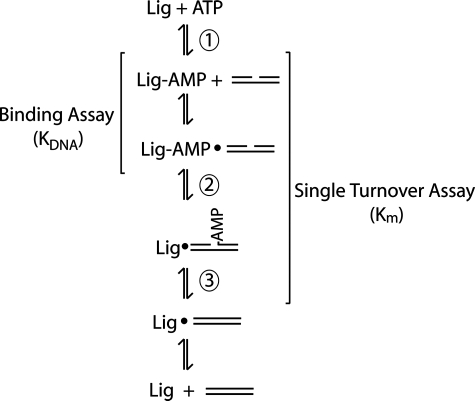
FIGURE 1.
DNA ligation reaction. The reaction mechanism shared by ATP-dependent ligases has three catalytic steps. First, the ligase adenylates itself on an active site lysine to form an enzyme-AMP intermediate. The AMP group is then transferred to the 5′-phosphate of the nicked DNA (step 2) before a phosphodiester bond is formed during step 3 to yield the ligated product. All three catalytic steps require magnesium or another divalent metal. The binding assay, performed in the absence of magnesium, measures the dissociation constant of adenylated ligase binding to DNA. DNA end joining activity measured under single turnover conditions with adenylated DNA ligase III reflects the rate of steps 2 and 3, without the contribution of step 1 or dissociation of the ligated DNA product.
The CC of DNA ligases contains five conserved sequence motifs that include the catalytic lysine and residues surrounding the ATP-binding pocket (18, 23-25). The mammalian ligases contain additional domains that mediate protein-protein interactions targeting these enzymes to specific pathways (26, 27). For example, LigIIIα contains a C-terminal BRCA1 C-terminal domain that interacts with XRCC1 (27-30). LigIIIβ, an isoform of LigIII expressed in the testis during the latter stages of meiotic prophase, lacks the C-terminal BRCA1 C-terminal domain and does not interact with XRCC1 (30, 31).
LigIII is distinguished from other DNA ligases by the presence of a unique zinc finger domain that improves ligation efficiency by an unknown mechanism (32-35). The sequence and structure of the LigIII zinc finger (ZnF) identify it as a member of the poly(ADP-ribose) polymerase (PARP) family of zinc fingers (32). The two N-terminal zinc fingers of PARP1 bind to DNA breaks, particularly single-stranded breaks (36, 37). PARP-like zinc fingers also recognize flexible joints within double-stranded DNA, typically favoring gaps of any size over nicks (38), and two enzymes with PARP-like zinc fingers have been shown to bend DNA 73-102° upon binding (39, 40).
The LigIII ZnF reportedly binds specifically to nicks, and deletion of the ZnF reduces affinity for DNA secondary structures, including nicks, flaps, gaps, and Holliday junctions (32, 33, 35). However, the ZnF is not required for enzymatic activity in vitro or to complement an E. coli ligA mutant (33). Although a recent NMR structure of the LigIII ZnF identified several residues with resonance peaks that shift in the presence of DNA, the mode of nick recognition by the zinc finger and its biological significance have not been established (32). In LigIII, the ZnF is immediately adjacent to the DBD of the catalytic core. Notably, the potential influence of the DBD was not considered during the initial studies characterizing the LigIII ZnF.
Here we report a quantitative analysis of the DNA binding and nick recognition functions of the constituent domains of LigIII. We demonstrate that LigIII has two independent DNA-binding modules participating in two different modes of nick recognition. The ZnF alone fails to bind tightly or specifically to nicked DNA. Instead, the ZnF functions in a cooperative manner with the DBD to promote DNA end joining by LigIII. Furthermore, we show that the ZnF is essential for double-stranded break ligation activity in vitro, consistent with the reported role of LigIII in a backup pathway of NHEJ (8, 41, 42).
EXPERIMENTAL PROCEDURES
Expression and Purification of LigIIIβ—The pQE-32 vector containing full-length LigIIIβ (herein referred to as LigIII; Fig. 2) with an N-terminal histidine tag has been described previously (33). Expression of full-length LigIII followed the published protocol, with two modifications. At the time of induction, 10 μm ZnCl2 was added to the LB medium, and following induction, the cultures were incubated at 16 °C for 16 h before bacterial cells were harvested by centrifugation.
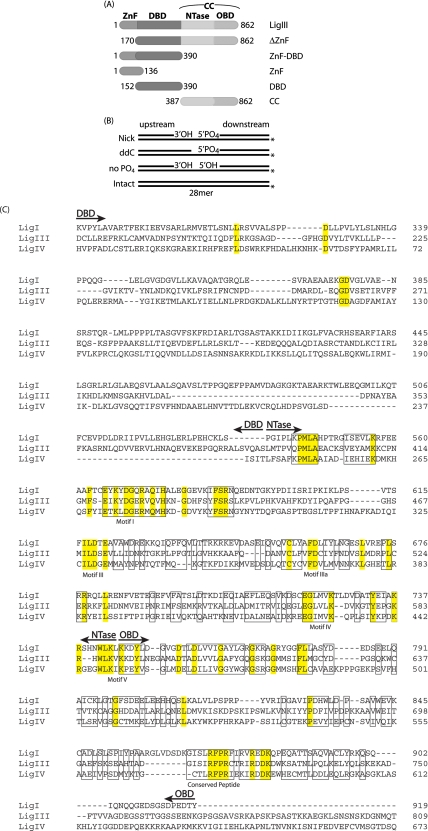
FIGURE 2.
Constructs and substrates used in this study. A, DNA encoding deletions of LigIIIβ were cloned into pET-28a with an N-terminal hexahistidine tag. The N- and C-terminal amino acids are listed for each construct that span the following domains: ZnF, DBD, NTase, and OBD. B, DNA substrates used in the fluorescence anisotropy binding experiments and end joining assays are shown. For DNA binding experiments, the template strand was 5′ fluorescein-labeled, as denoted by the asterisk. DNA end joining was assayed using substrates radiolabeled with 32P on the 5′ terminus of the downstream strand. C, multiple sequence alignment of human DNA ligases showing only the conserved DBD, NTase, and OBD domains. The flanking N- and C-terminal sequences of the full-length proteins are not shown. Identical residues are highlighted, and conserved residues are boxed (18).
Full-length LigIII was purified using a protocol similar to that previously reported (33). Bacterial cell pellets were resuspended in 50 mm Tris, pH 7.5, 50 mm NaCl, 2% Nonidet P-40 substitute, 1 mm phenylmethanesulfonyl fluoride (PMSF), 1 mm benzamidine-HCl, 10 mm β-mercaptoethanol, and 0.1 mm EDTA. The cells were lysed by two passages through a high pressure cell disrupter, and the lysate was clarified by centrifugation at 30,000 × g for 1 h at 4 °C. The clarified lysate was passed through a 0.45-μm filter prior to loading on a P-11 phosphocellulose column. The P11 column was blocked with bovine serum albumin and then pre-equilibrated with Buffer A (50 mm Tris-HCl, pH 7.5, 10% glycerol, 50 mm NaCl, 1 mm PMSF, 1 mm benzamidine-HCl, 5 mm β-mercaptoethanol) prior to loading the cell lysate. After washing with Buffer A, LigIII was eluted with a gradient from 50 mm to 2 m NaCl. Fractions containing LigIII were pooled and loaded onto a 5-ml HiTrap nickel Chelating column (GE Healthcare) equilibrated with Buffer B (50 mm Tris-HCl, pH 7.5, 10% glycerol, 10 mm imidazole, 1 mm PMSF, 1 mm benzamidine-HCl, and 100 mm NaCl). LigIII was eluted stepwise with Buffer B containing 100, 300, and 500 mm imidazole. Fractions containing LigIII were pooled, concentrated, and loaded onto a Sephadex S200 (GE Healthcare) gel filtration column equilibrated with Buffer C (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2 mm dithiothreitol (DTT), 0.1 mm EDTA). Purified protein was concentrated to 30-40 mg/ml, flash-frozen in liquid nitrogen, and stored at -80 °C. The protein concentrations were determined by absorbance at 280 nm in 6 m guanidine HCl using the calculated extinction coefficients.
Cloning, Expression, and Purification of LigIII Domains—LigIII domains (Fig. 2) were cloned into pET-28a with an N-terminal hexahistidine tag and expressed in E. coli BL21(DE3) cells. The cells were grown in LB medium containing kanamycin at 37 °C. The cultures were induced with 1 mm isopropyl thiogalactoside and supplemented with 1 μm ZnCl2 if the LigIII domain being expressed contained the zinc finger domain. The cultures were then incubated at 37 °C for 2 h, and the cells were harvested by centrifugation. Bacterial cell pellets were stored at -20 °C.
Thawed bacterial cells were resuspended in Lysis Buffer (50 mm Tris-HCl, pH 8, 500 mm NaCl, 2% Nonidet P-40, 0.5 mm PMSF, 5 mm β-mercaptoethanol, 1 mm benzamidine-HCl, 1 μg/ml aprotinin, 5 μg/ml leupeptin, 0.7 μg/ml pepstatin A) and lysed by two passages through a high pressure cell disruptor. The lysate was clarified by centrifugation at 30,000 × g for 1 h at 4 °C and passed through a DE52 column. The clarified lysate was loaded onto a 5-ml HiTrap nickel chelating column (GE Healthcare) equilibrated with Buffer A (50 mm Tris-HCl, pH 8, 500 mm NaCl, 0.5 mm PMSF). Following a wash with Buffer A, the LigIII domains were eluted stepwise with Buffer A plus 100, 300, and 500 mm imidazole. Fractions containing the LigIII domain were pooled, and the salt was diluted to 80 mm NaCl by the addition of buffer B (50 mm Tris-HCl, pH 7.5, 0.5 mm PMSF, 0.1 mm EDTA, 2 mm DTT). This was loaded onto a HiTrap S-Sepharose column (GE Healthcare) equilibrated with Buffer B plus 100 mm NaCl and eluted with a gradient up to 1 m NaCl. Fractions containing the LigIII domain were pooled, concentrated, and loaded onto a Sephadex S200 gel filtration column equilibrated with Buffer C (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2 mm DTT, 0.1 mm EDTA). Purified protein was concentrated to 30-40 mg/ml, flash-frozen in liquid nitrogen, and stored at -80 °C. Protein concentration was determined by absorbance at 280 nm in 6 m guanidine HCl using the calculated extinction coefficients.
DNA Binding Assays—The change in fluorescence polarization of a DNA duplex containing a 5′-labeled strand was measured to determine the DNA binding affinity of LigIII and its domains. A 28-nucleotide oligomer (GTGCTGATGCGTCGTCGGACTGATTCGG) with a 5′ 6-carboxyfluorescein label was annealed at equimolar concentrations to a complementary 28-mer to make an intact duplex DNA or to two complementary strands, 13-mer (GTGCTGATGCGTC), and a 15-mer (GTCGGACTGATTCGG) to make a nicked DNA substrate for ligation (Fig. 2B). Duplex DNA oligomers (25 nm) were mixed with protein at concentrations ranging from 50 nm to 30 μm, reaction buffer (50 mm MOPS-KOH, pH 7.5, 1 mm DTT, 50 μg/ml bovine serum albumin, 10 mm EDTA), and NaCl at the listed concentration in a total reaction volume of 150 μl. ATP and divalent cations were omitted to prevent ligation of the substrates. A 2-mm cuvette was used. The data were collected at 22 °C with a C-60 spectrofluorometer (Photon Technology International), using excitation and emission wavelengths of 495 and 520 nm, respectively. Apparent Kd values for the DNA (KDNA) were calculated by fitting the data to the equation (18): r = r0 + (rmax - r0)*f where r is the fluorescence anisotropy determined experimentally and f = p*d/([DNA] + Kd) with p = [Protein]/(1 + d/Kd), and d = (-[Protein]/Kd + [DNA]/Kd - 1 + sqrt[([Protein]/Kd - [DNA]/Kd + 1)2 + (4 * [DNA]/Kd)])/(2/Kd).
DNA Ligation Activity of LigIII and ΔZnF—The single-stranded DNA nick ligation activities of LigIII and ΔZnF were measured in a single turnover reaction at 4 °C to retard the reaction to a measurable rate. The nicked DNA substrate was identical to the one used in the DNA binding experiments, but without the 5′ 6-carboxyfluorescein label. The 15-mer was 5′-radiolabeled with polynucleotide kinase. 15-mer (0.5 μm), [γ-32P]ATP (0.5 μm), 1× polynucleotide kinase buffer (70 mm Tris-HCl, pH 7.6, 10 mm MgCl2, 5 mm DTT), and 0.5 unit/μl polynucleotide kinase were mixed in a 20-μl reaction and incubated for 25 min at 37 °C. The reaction was arrested by incubation at 65 °C for 10 min and 95 °C for 5 min.
Labeled DNA was annealed in equimolar concentrations to the template 28-mer and the upstream 13-mer. 0.25 nm DNA duplex was mixed with an excess of protein in reaction buffer (50 mm HEPES, pH 7.5, 120 mm NaCl, 50 μg/ml bovine serum albumin, 5 mm MgCl2, 5 mm DTT, 1 mm ATP) in a 55-μl reaction. To evaluate the extent of ligation as a function of time, the aliquots were removed and quenched periodically over the course of the reaction. The reaction was quenched by the addition of EDTA-formamide and incubation at 95 °C for 5-10 min. The 15-mer substrate was separated from the 28-mer ligated product by denaturing PAGE. Both substrate and product bands were quantified by a phosphorimaging device (Fuji BAS1000).
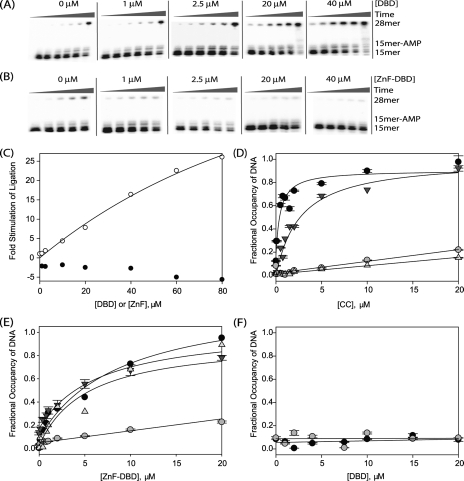
Catalytic Core Complementation with DBD or ZnF-DBD—Ligation activity of the CC was measured in a single turnover reaction at 25 °C. A higher temperature was used for the CC than for LigIII and ΔZnF to increase its ligation rate. The nicked DNA substrate and labeling reaction were identical to the one described above. 4 nm DNA duplex was mixed with 100 nm CC and various concentrations of DBD or ZnF-DBD in reaction buffer (50 mm HEPES, pH 7.5, 120 mm NaCl, 50 μg/ml bovine serum albumin, 5 mm MgCl2, 5 mm DTT, 1 mm ATP) in a 46-μl reaction. As described above, the aliquots were removed and quenched periodically over the course of the reaction. The separation and quantitation of substrates and products are also described above.
Ligation of Substrates with Blunt Ends or Short Overhangs—Ligation activity on DNA substrates with blunt or sticky ends (complementary 2-bp 5′ overhangs) was measured in a multiple turnover reaction at 30 °C. As shown in Fig. 7, the substrates contained only one 5′-phosphate and one 3′-hydroxyl to allow ligation in only one orientation. The top strand of the blunt substrate had the sequence 5′-GTCGGACTGATTCGGGCTGGAGGAC; the bottom strand had the sequence (5′-GTCCTCCAGCCCGAATCAGTCCGAC. The sticky end substrate was constructed by annealing the top strand of the blunt substrate to a bottom strand with a two-nucleotide overhang on the 3′-terminus (5′-GTCCTCCAGCCCGAATCAGTCCGACAT). A diagram of the substrates with blunt ends or short overhangs is shown in Fig. 7A. 4 nm blunt or sticky end substrate was mixed with 100-1000 nm LigIII or ΔZnF in reaction buffer (50 mm HEPES, pH 7.5, 50 μg/ml bovine serum albumin, 5 mm MgCl2, 5 mm DTT, 50 mm NaCl, 1 mm ATP) in a 20-μl reaction. Quenching of the reactions and separation of substrate and products are described above.
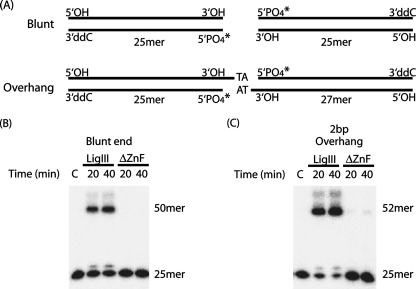
FIGURE 7.
Blunt end joining by LigIII requires two independent DNA-binding modules. A, DNA substrates with blunt ends or complementary TA 3′-overhangs were designed such that they could be ligated in only one orientation. B and C, DNA end joining of blunt-ended substrates (B) or DNAs with short, complementary overhangs (C) was measured for LigIII and ΔZnF as described under “Experimental Procedures.” Control reactions lacked enzyme (lanes C).
RESULTS
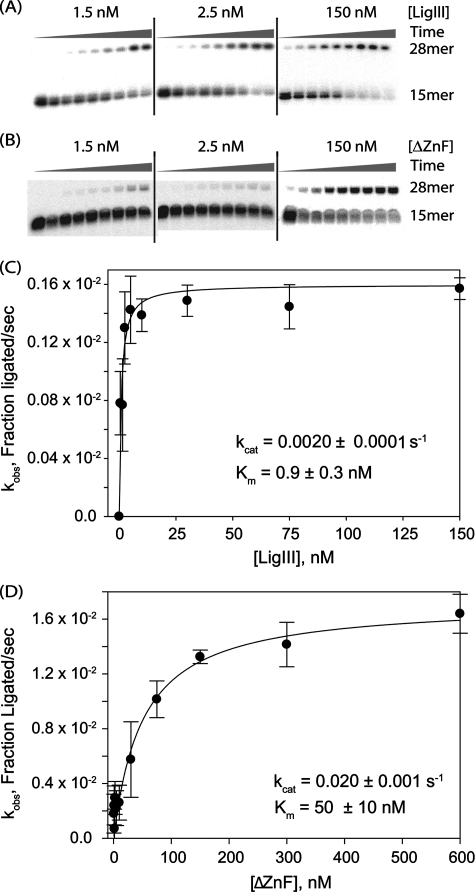
Comparison of DNA Ligation Activity of LigIII and ΔZnF—Although the ZnF of LigIII is not required for enzymatic activity in vitro (33, 34) or to complement E. coli cells lacking the endogenous LigA protein (33), it does broaden the DNA substrate specificity of LigIII (34, 35). To determine how the ZnF contributes to the mechanism of ligation, we examined the kinetics of DNA nick joining in vitro using a single-turnover ligation assay. This assay reports on the kinetics of the DNA-dependent steps of the end joining process (i.e. DNA binding, adenylation, and nick joining) independent of enzyme adenylation or product release. As previously reported, the ZnF is not required for ligation of a single-stranded nick (Fig. 3, A and B). Deletion of the ZnF actually increases the kcat 10-fold, from 0.0020 ± 0.0001 s-1 for LigIII to 0.020 ± 0.001 s-1 for ΔZnF (Fig. 3, C and D). However, the Km for DNA is enhanced 50-fold for LigIII (Km = 0.9 ± 0.3 nm) in comparison with the ΔZnF protein (Km = 50 ± 10 nm). These differences are reflected in a 5-fold higher catalytic efficiency of LigIII (kcat/Km = 2 × 106 m-1 s-1) compared with ΔZnF (kcat/Km = 4 × 105 m-1 s-1).
FIGURE 3.
Deletion of the ZnF domain lowers the catalytic efficiency of end joining. A and B, purified LigIII (A) or ΔZnF (B) was assayed for DNA end joining activity in a single turnover assay. The rate of ligation of nicked DNA (0.25 nm) was measured at different enzyme concentrations (1.5, 2.5, 5, 10, 30, 75, 150, 300, or 600 nm). The extent of ligation over time was monitored by periodically removing and quenching aliquots of the reaction (15 s, 30 s, 1 min, 2 min, 4 min, 10 min, 1 h, 2 h, or 3 h) for reactions performed at 0 °C to slow ligation to a measurable rate. C and D, observed rate constants (kobs) for each protein concentration were plotted to determine the rate constant (kcat) and Michaelis constant (Km) for LigIII (C) or ΔZnF (D).
The 50-fold increase in Km for DNA caused by deletion of the LigIII ZnF suggested that the ZnF may help stabilize the DNA in the active site of LigIII. We therefore measured the DNA binding affinities of LigIII and ΔZnF for nicked and intact DNA duplexes. Fluorescence anisotropy measurements with labeled DNA duplexes were performed in the absence of divalent cations to prevent ligation. We confirmed that Step 2 transfer of AMP to the DNA 5′PO4 does not occur under these conditions (data not shown). In 80 mm NaCl, LigIII binds nonspecifically to DNA, showing equal affinity for nicked and intact DNA (KDNA = 0.23 ± 0.06 μm and 0.24 ± 0.05 μm, respectively; Table 1). Because 80 mm NaCl is well below the optimal salt concentration for ligation by LigIII (33), binding measurements were repeated at 250 mm NaCl. Under these conditions, LigIII binds specifically to nicked DNA (KDNA = 0.3 ± 0.08 μm) and does not bind measurably to duplex DNA (>20 μm; Fig. 4 and Table 2). This is in contrast to LigI, which is strongly inhibited by elevated salt concentrations (18).
TABLE 1.
DNA binding affinities (KDNA, μm) measured in 80 mm NaCl
| Nicked duplex | Intact duplex | |
|---|---|---|
| LigIII | 0.23 ± 0.06 | 0.24 ± 0.05 |
| ZnF | 2.2 ± 0.5 | 1.4 ± 0.2 |
| DBD | >20 | >30 |
| ZnF-DBD | 0.36 ± 0.06 | 0.4 ± 0.03 |
| ΔZnF | 0.22 ± 0.05 | 0.84 ± 0.2 |
| CC | 0.75 ± 0.07 | 0.68 ± 0.07 |
FIGURE 4.
The zinc finger of LigIII contributes DNA binding affinity and specificity. Binding of LigIII (A), ΔZnF (B), and ZnF (C) to DNA was measured by fluorescence anisotropy in buffer containing 250 mm NaCl. The DNA substrate was an intact duplex (○) or contained a single-stranded nick (•). Nicked DNA lacking the 3′-hydroxyl (▾) or the 5′-phosphate (▵) was also assayed.
TABLE 2.
DNA binding affinities (KDNA, μm) measured in 250 mm NaCl
| Nicked duplex | Intact duplex | |
|---|---|---|
| LigIII | 0.30 ± 0.08 | >20 |
| ZnF | >20 | >20 |
| DBD | >30 | >30 |
| ZnF-DBD | 2.5 ± 1.8 | >30 |
| ΔZnF | 0.63 ± 0.24 | >30 |
| CC | 0.46 ± 0.15 | >20 |
Deletion of the ZnF did not significantly change LigIII DNA binding affinity, nor did it abrogate the specificity of the enzyme for a nicked substrate (Fig. 4 and Table 2). The ΔZnF binds to nicked DNA with an affinity of 0.63 ± 0.24 μm and to intact duplex with an affinity of >30 μm in 250 mm NaCl. This indicates that the nick binding activity of LigIII is not solely dependent upon the ZnF domain. In contrast, LigI, which lacks a ZnF, fails to bind selectively to nicked DNAs (18), highlighting functional differences between these structurally related enzymes.
The dissociation constants measured for DNA binding by LigIII and ΔZnF are higher than the Michaelis constants measured in enzymatic activity assays (Table 2). The divalent cations that are present in enzymatic activity assays but absent from DNA binding assays may increase the affinity for nicked DNA. Alternatively, if adenylation of the DNA substrate is not a rate-limiting step for the single turnover reaction (Fig. 1), then the Km value measured in our single turnover assay could reflect contributions from both DNA binding and adenylation.
The isolated ZnF domain binds weakly to DNA, with about 10-fold less affinity for nicked DNA than the LigIII or ΔZnF enzymes in low salt conditions (80 mm NaCl). In high salt conditions (250 mm NaCl), the ZnF does not bind measurably to DNA. Although the ZnF significantly enhances the catalytic efficiency of LigIII, it is neither necessary nor sufficient for the DNA binding activity and nick specificity of LigIII. We therefore explored the contributions of the other LigIII domains to these activities.
A Second DNA Nick Sensor in LigIII—The DBD of human LigI and Vaccinia DNA ligase accounts for most of the DNA binding affinity of these enzymes (18, 43). In contrast, the DBD is not required for LigIII to bind tightly to DNA, and the isolated DBD does not bind tightly to DNA (Fig. 5D and Tables 1 and 2). In low salt concentrations, the isolated DBD has an affinity of 21 μm for nicked DNA and 33 μm for intact DNA (Table 1), whereas the DBD shows no detectable binding in the high salt conditions used for most of our activity assays (Fig. 5F and Table 2).
FIGURE 5.
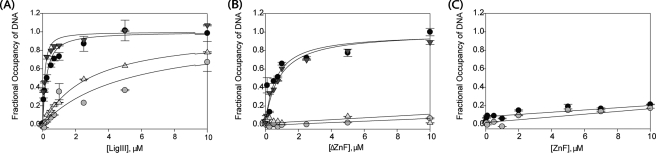
DNA end joining activity is stimulated by the DNA-binding domain of LigIII. A and B, to measure the stimulation of CC ligation activity by DBD and ZnF-DBD, purified CC (100 nm) was mixed with nicked, radiolabeled DNA (4 nm) and increasing concentrations (1, 2.5, 5, 7.5, 10, 20, 30, 40, 60, and 80 μm) of the DBD (A) or ZnF-DBD (B). The extent of ligation at various times (30 min, 1, 2, 3, 6, or 9 h) was determined by periodically removing and quenching aliquots of the reaction and measuring the extent of conversion of a 15-mer substrate to 28-mer ligated product. C, ligation activity of the CC is stimulated when the DBD is added in trans (open circles) but inhibited when the ZnF-DBD is added in trans (closed circles). D-F, DNA binding by the CC (D), ZnF-DBD (E), and DBD (F) was measured using fluorescence anisotropy. The affinity for nicked DNA (•), intact DNA (○), nicked DNA lacking a 3′hydroxyl (▾), and nicked DNA lacking a 5′ phosphate (▵) was measured.
In contrast to the DBD, the CC of LigIII binds nearly as tightly to nicked DNA as the full-length enzyme (KDNA = 0.46 ± 0.15 μm versus 0.30 ± 0.08 μm; Table 2) and with slightly higher affinity than the ΔZnF enzyme (KDNA = 0.63 ± 0.24 μm). The CC exhibits greater than 40-fold specificity for nicked DNA versus an intact duplex (Table 2) and therefore constitutes an independent DNA nick-binding module of LigIII. In contrast, the CC of LigI lacks measurable DNA binding activity (18).
Stimulation of CC Ligation Activity by Addition of the DBD in Trans—Although the DBD of LigIII is dispensable for DNA nick binding activity, it strongly augments the enzymatic activity of the CC. The DNA nick ligation of the CC was barely detectable, even when assayed at 25 °C (Fig. 5, A and B;0 μm). At this elevated temperature, the rate of DNA nick ligation by the CC was about 700-fold slower than that of intact LigIII measured at 0 °C. The addition of the DBD in trans stimulates DNA nick joining by the CC (Fig. 5A), perhaps by helping to correctly position DNA within the active site of the CC. Nonetheless, the DBD makes little apparent contribution to the overall affinity of LigIII for DNA (Fig. 5, D and F).
The ZnF and DBD of LigIII Function Cooperatively—Whereas the conserved DBDs of LigI and LigIII stimulate the DNA ligation activity of their cognate CC domain in trans (Ref. 18 and Fig. 5), the LigIII DBD lacks robust DNA binding activity (Table 1) in contrast to the DBD of LigI (18). The ZnF of LigIII has been predicted to function as a DNA nick sensor (32, 33), yet the ZnF shows weak DNA binding activity (Table 1). Nonetheless, the ZnF contributes significantly to the catalytic efficiency of LigIII (Fig. 3). To reconcile these observations, we considered the possibility that the ZnF functions in concert with the adjoining DBD of LigIII.
To test this hypothesis, the DNA binding affinity of the tandem ZnF-DBD construct was directly measured. Surprisingly, the tandem combination of ZnF and DBD binds DNA with much higher affinity than either domain alone (Fig. 5), rivaling the affinity of full-length LigIII (Table 2). The cooperative DNA binding behavior of the ZnF and DBD implies that these domains together constitute a functional unit for DNA nick sensing. In 250 mm NaCl, the ZnF-DBD fragment bound with high specificity to nicked DNA, exhibiting more than 10-fold higher affinity for nicked DNA (KDNA = 2.5 ± 1.8 μm; Fig. 5E and Table 2) over intact duplex DNA (KDNA > 30 μm). Thus, the ZnF-DBD comprises a second, independent DNA nick-binding module of LigIII.
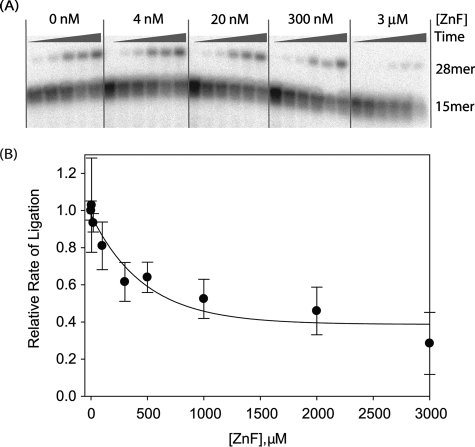
We then tested whether the ZnF-DBD can stimulate DNA ligation by the CC when added in trans. The ZnF-DBD actually inhibits the weak enzymatic activity of the CC (Fig. 5, B and C), suggesting that this fragment competes for binding to the nicked DNA substrate. The addition of the ZnF domain also inhibits the nick ligation activity of the ΔZnF enzyme (Fig. 6). Thus, the two independent DNA nick-binding modules of LigIII, the ZnF-DBD and the CC, appear to have at least partially overlapping DNA-binding sites.
FIGURE 6.
The ZnF domain added in trans inhibits DNA ligation by ΔZnF LigIII. A, increasing concentrations of the ZnF were added to reactions containing ΔZnF (20 nm) and a nicked DNA substrate (4 nm). Ligation of the 15- and 13-mer strands of the substrate generates a 28-mer product that was detected during the time course of the reaction (5 s, 30 s, 1 min, 5 min, 10 min, and 50 min). B, the observed rate constants (kobs) for ligation were determined in the presence of the different ZnF concentrations shown. Increasing concentrations of ZnF inhibit end joining activity by the ΔZnF enzyme.
DNA Structure-Activity Relationship and Nick Sensing—A previous study indicated that the ZnF diversifies the substrate specificity of LigIII (35). This led us to hypothesize that the CC and ZnF-DBD DNA-binding modules may bind in different ways or to different parts of the nicked DNA. In an attempt to parse the roles of the different domains in nick binding and catalysis, we tested whether the 3′-hydroxyl or 5′-phosphate of a nicked DNA substrate are selectively recognized by the CC and the ZnF-DBD fragments of LigIII. In other ligases, the 5′-phosphate group strongly contributes to nick sensing activity (44), whereas removal of the 3′-hydroxyl has less effect (45). This is also the case for LigIII because removal of the 3′-hydroxyl had no effect on nick sensing by LigIII, whereas removal of the 5′-phosphate reduced binding affinity by an order of magnitude (Table 3). The ZnF-DBD fragment is relatively tolerant of modifications at the nick termini; its affinity was reduced 4-fold by removal of the 3′-hydroxyl and 2-fold by removal of the 5′-phosphate (Table 3). In contrast, the CC is more dramatically affected by the fine structure of the nick, losing 10-fold binding affinity upon removal of the 3′-hydroxyl and showing no DNA binding activity for a DNA nick lacking a 5′-phosphate. We confirmed that AMP is not transferred to the 5′-phosphate end of the DNA in these conditions (not shown).
TABLE 3.
Effect of nick structure on DNA binding: DNA binding affinities (KDNA, μm) measured in 250 mm NaCl
| Ligatable nick | ddC | No 5′ PO4 | |
|---|---|---|---|
| LigIII | 0.30 ± 0.08 | 0.11 ± 0.02 | 2.8 ± 0.27 |
| ZnF-DBD | 2.5 ± 1.8 | 9.8 ± 2.3 | 5.1 ± 3.1 |
| ΔZnF | 0.63 ± 0.24 | 0.87 ± 0.15 | >30 |
| CC | 0.46 ± 0.15 | 2.4 ± 0.7 | >30 |
With the DBD present, the nick sensing activity of the ΔZnF protein is more tolerant of removal of the 3′hydroxyl, indicating that the DBD contributes to the intrinsic nick sensing activity of the CC in addition to being a component of the ZnF-DBD DNA-binding module. Thus, intact LigIII has two nick sensing activities: the relatively nonspecific nick binding activity of the ZnF-DBD and the CC preference for a ligatable nick with both a3′ hydroxyl and a 5′ phosphate.
Two Binding Modules of LigIII Stimulate Intermolecular Ligation—The presence of two independent DNA-binding modules in LigIII may confer an advantage in the intermolecular ligation of two DNA molecules. Indeed, LigIII efficiently catalyzes the ligation of two DNAs with blunt or short complementary ends, whereas the ΔZnF protein is notably deficient in this activity under assay conditions that support robust nick ligation activity by ΔZnF (Fig. 7). In fact, LigIII is significantly more effective at blunt end ligation than T4 DNA ligase, an enzyme routinely used for the shotgun cloning of blunt-ended inserts into recombinant vectors (not shown). Thus, the ZnF of the ZnF-DBD DNA-binding module plays a major role in intermolecular ligation, whereas it is relatively dispensable for single-stranded nick joining in vitro.
DISCUSSION
The presence of an N-terminal ZnF distinguishes LigIII from the other mammalian DNA ligases. Initial characterizations of the LigIII ZnF failed to take into account the DBD, which had not yet been identified and was variably present or absent from LigIII deletions used to assess nick recognition by the ZnF (33, 35). Here, with the benefit of the crystal structure of the homologous LigI protein, we have prepared a more precise collection of LigIII deletion constructs (Fig. 2) and shown that the ZnF and DBD together form a DNA-binding module that interacts specifically with nicked DNA (Fig. 4) and is relatively insensitive to the fine structure of the nick termini (Table 3). The CC of LigIII also binds selectively to nicked DNA but exhibits a marked preference for ligatable nicks with 3′OH and 5′PO4 termini (Tables 2 and 3). The CC supports a low level of nick joining activity that is greatly accelerated by the ZnF-DBD module in full-length LigIII (Fig. 5), indicating functional interplay between the two DNA-binding modules within LigIII during catalysis.
Unlike the DBD alone, the ZnF-DBD added in trans does not stimulate the ligation activity of the CC and instead acts as an inhibitor of the CC. Our data suggest that the ZnF-DBD and CC function as separate DNA-binding modules that bind to overlapping sites on a nicked DNA. By analogy to the continuous ring structure of LigI that envelopes a DNA nick (18), we predict that the additional ZnF of LigIII would be unable to access the nick when the enzyme is catalyzing step 3 of the ligation reaction (Fig. 1). Chemical footprinting data indicate that LigIII has a 15-20-nucleotide footprint on DNA, which is similar to the DNA footprint of LigI (18, 33, 46). These data suggest that the ZnF does not expand the footprint of LigIII and perhaps is unlikely to bind to the flanking DNA while the CC ligates the nick.
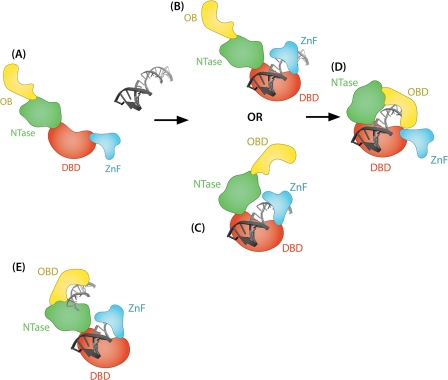
To explain the stimulation of DNA end joining by the ZnF (Figs. 3 and 7), we propose that the ZnF and CC of LigIII engage the ends of a nicked DNA substrate in a sequential manner, like the blades of a jackknife that can open and close (Fig. 8). Our jackknife model posits that the two DNA-binding modules of LigIII engage the ends of the DNA substrate at different times during the end joining reaction. The ZnF-DBD binds to the nick first then subsequently must disengage from the nick to allow access by the CC for catalysis. The DBD also enhances the nick binding and catalytic activities of the CC. The DBD may act as a pivot, linking nick recognition with the transition to the catalytically active conformation in which the ligase encircles the DNA nick.
FIGURE 8.
Jackknife model of DNA recognition by LigIII. In the proposed jackknife model, unliganded DNA ligase III is in an extended conformation that enables loading onto DNA (A), similar to the homologous DNA ligase from Sulfolobus solfataricus (49). B, upon binding to DNA, the ZnF-DBD initially engages an interruption in the DNA backbone, functioning as a nick sensor. At this step, unligatable DNA ends may be further processed by repair enzymes before the ligation reaction can be completed. C and D, the NTase domain of LigIII may also bind to DNA (C) before the enzyme transitions to a catalytically active conformation with the catalytic core (NTase and OBD) engaging the nick (D). With the DBD, NTase and OBD fully encircling the DNA substrate, the ZnF is effectively excluded from binding to the nick. When joining double-stranded breaks, two independent DNA-binding modules of LigIII (the ZnF-DBD and the CC) might allow the enzyme to bind the two DNAs simultaneously (E).
Although the ZnF-DBD and CC DNA-binding modules bind to ligatable DNA nicks with similar affinity, the ZnF-DBD is more tolerant of modifications at the nick termini. During the repair of DNA damage, LigIII is recruited to DNA single-stranded breaks through its interaction with ADP-ribosylated PARP1 (46). One implication of the jackknife model is that the initial nick binding activity of the ZnF-DBD could either displace ADP-ribosylated PARP1 from the strand break or enable LigIII to recognize the strand break despite the presence of the negatively charged ADP-ribose polymers generated by PARP1. The ability of the ZnF-DBD to recognize nicks with different structures is well suited to the initial recognition step because the majority of single-stranded breaks generated by ionizing radiation and reactive oxygen species will not be substrates for ligation. The initial engagement of the strand break by the ZnF-DBD presumably leaves the ends of the DNA accessible to other repair enzymes such as polynucleotide kinase. Once a ligatable nick has been generated by the end processing, the CC will bind to the nick in a catalytically active conformation.
LigIII also has robust intermolecular ligation activity, joining together duplex DNAs with either cohesive or blunt ends. Strikingly, the blunt end DNA joining activity is absolutely dependent upon the ZnF domain (Fig. 7). The other mammalian DNA ligases, LigI and LigIV, fail to show this activity in the absence of accessory proteins (Refs. 47 and 48 and data not shown). We attribute the pronounced blunt end joining activity of LigIII to two independent DNA-binding modules, the ZnF-DBD and the CC, which may simultaneously engage two DNAs for intermolecular ligation (Fig. 8).
In summary, ZnF and DBD combine to form a novel intrinsic DNA nick-sensing module that facilitates LigIII functions in a variety of pathways, including nuclear DNA repair, meiotic recombination, and mitochondrial DNA replication and repair (48). There is functional interplay between the ZnF-DBD and CC-binding modules during nick ligation. Moreover, the ZnF-DBD module expands the catalytic repertoire of LigIII to include intermolecular ligation, an activity that presumably underlies the participation of LigIII in a back-up pathway of nonhomologous end joining that functions when the major NHEJ pathway is compromised (8, 42).
This work was supported by Structural Biology of DNA Repair Program Project Grant P01 CA092584 and National Institutes of Health Grants GM52504 (to T. E.) and ES12512 (to A. E. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Lig, ligase; DBD, DNA-binding domain; CC, catalytic core; NTase, nucleotidyltransferase; OBD, oligonucleotide/oligosaccharide-binding domain; ZnF, zinc finger; PARP, poly(ADP-ribose) polymerase; PMSF, phenylmethanesulfonyl fluoride; DTT, dithiothreitol.
References
- 1.Waga, S., Bauer, G., and Stillman, B. (1994) J. Biol. Chem. 269 10923-10934 [PubMed] [Google Scholar]
- 2.Barnes, D. E., Tomkinson, A. E., Lehmann, A. R., Webster, A. D., and Lindahl, T. (1992) Cell 69 495-503 [DOI] [PubMed] [Google Scholar]
- 3.Mackenney, V. J., Barnes, D. E., and Lindahl, T. (1997) J. Biol. Chem. 272 11550-11556 [DOI] [PubMed] [Google Scholar]
- 4.Sleeth, K. M., Robson, R. L., and Dianov, G. L. (2004) Biochemistry 43 12924-12930 [DOI] [PubMed] [Google Scholar]
- 5.Li, Z., Otevrel, T., Gao, Y., Cheng, H. L., Seed, B., Stamato, T. D., Taccioli, G. E., and Alt, F. W. (1995) Cell 83 1079-1089 [DOI] [PubMed] [Google Scholar]
- 6.Moser, J., Kool, H., Giakzidis, I., Caldecott, K., Mullenders, L. H., and Fousteri, M. I. (2007) Mol. Cell 27 311-323 [DOI] [PubMed] [Google Scholar]
- 7.Wang, M., Wu, W., Wu, W., Rosidi, B., Zhang, L., Wang, H., and Iliakis, G. (2006) Nucleic Acids Res. 34 6170-6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang, H., Rosidi, B., Perrault, R., Wang, M., Zhang, L., Windhofer, F., and Iliakis, G. (2005) Cancer Res. 65 4020-4030 [DOI] [PubMed] [Google Scholar]
- 9.Lakshmipathy, U., and Campbell, C. (1999) Mol. Cell. Biol. 19 3869-3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogenhagen, D. F., Pinz, K. G., and Perez-Jannotti, R. M. (2001) Prog. Nucleic Acids Res. Mol. Biol. 68 257-271 [DOI] [PubMed] [Google Scholar]
- 11.Lakshmipathy, U., and Campbell, C. (2001) Nucleic Acids Res. 29 668-676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puebla-Osorio, N., Lacey, D. B., Alt, F. W., and Zhu, C. (2006) Mol. Cell. Biol. 26 3935-3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahel, I., Rass, U., El-Khamisy, S. F., Katyal, S., Clements, P. M., McKinnon, P. J., Caldecott, K. W., and West, S. C. (2006) Nature 443 713-716 [DOI] [PubMed] [Google Scholar]
- 14.Date, H., Onodera, O., Tanaka, H., Iwabuchi, K., Uekawa, K., Igarashi, S., Koike, R., Hiroi, T., Yuasa, T., Awaya, Y., Sakai, T., Takahashi, T., Nagatomo, H., Sekijima, Y., Kawachi, I., Takiyama, Y., Nishizawa, M., Fukuhara, N., Saito, K., Sugano, S., and Tsuji, S. (2001) Nat. Genet. 29 184-188 [DOI] [PubMed] [Google Scholar]
- 15.Moreira, M. C., Barbot, C., Tachi, N., Kozuka, N., Uchida, E., Gibson, T., Mendonca, P., Costa, M., Barros, J., Yanagisawa, T., Watanabe, M., Ikeda, Y., Aoki, M., Nagata, T., Coutinho, P., Sequeiros, J., and Koenig, M. (2001) Nat. Genet. 29 189-193 [DOI] [PubMed] [Google Scholar]
- 16.Tomkinson, A. E., and Mackey, Z. B. (1998) Mutat. Res. 407 1-9 [DOI] [PubMed] [Google Scholar]
- 17.Tomkinson, A. E., and Levin, D. S. (1997) Bioessays 19 893-901 [DOI] [PubMed] [Google Scholar]
- 18.Pascal, J. M., O'Brien, P. J., Tomkinson, A. E., and Ellenberger, T. (2004) Nature 432 473-478 [DOI] [PubMed] [Google Scholar]
- 19.Martin, I. V., and MacNeill, S. A. (2002) Genome Biol. 3 Reviews 3005.1-3005.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nandakumar, J., Nair, P. A., and Shuman, S. (2007) Mol. Cell 26 257-271 [DOI] [PubMed] [Google Scholar]
- 21.Lee, J. Y., Chang, C., Song, H. K., Moon, J., Yang, J. K., Kim, H. K., Kwon, S. T., and Suh, S. W. (2000) EMBO J. 19 1119-1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nair, P. A., Nandakumar, J., Smith, P., Odell, M., Lima, C. D., and Shuman, S. (2008) Nat. Struct. Mol. Biol. 14 770-778 [DOI] [PubMed] [Google Scholar]
- 23.Subramanya, H. S., Doherty, A. J., Ashford, S. R., and Wigley, D. B. (1996) Cell 85 607-615 [DOI] [PubMed] [Google Scholar]
- 24.Hakansson, K., Doherty, A. J., Shuman, S., and Wigley, D. B. (1997) Cell 89 545-553 [DOI] [PubMed] [Google Scholar]
- 25.Odell, M., Sriskanda, V., Shuman, S., and Nikolov, D. B. (2000) Mol. Cell 6 1183-1193 [DOI] [PubMed] [Google Scholar]
- 26.Dulic, A., Bates, P. A., Zhang, X., Martin, S. R., Freemont, P. S., Lindahl, T., and Barnes, D. E. (2001) Biochemistry 40 5906-5913 [DOI] [PubMed] [Google Scholar]
- 27.Taylor, R. M., Wickstead, B., Cronin, S., and Caldecott, K. W. (1998) Curr. Biol. 8 877-880 [DOI] [PubMed] [Google Scholar]
- 28.Thornton, K. H., Krishnan, V. V., West, M. G., Popham, J., Ramirez, M., Thelen, M. P., and Cosman, M. (2001) Protein Expression Purif. 21 401-411 [DOI] [PubMed] [Google Scholar]
- 29.Krishnan, V. V., Thornton, K. H., Thelen, M. P., and Cosman, M. (2001) Biochemistry 40 13158-13166 [DOI] [PubMed] [Google Scholar]
- 30.Nash, R. A., Caldecott, K. W., Barnes, D. E., and Lindahl, T. (1997) Biochemistry 36 5207-5211 [DOI] [PubMed] [Google Scholar]
- 31.Mackey, Z. B., Ramos, W., Levin, D. S., Walter, C. A., McCarrey, J. R., and Tomkinson, A. E. (1997) Mol. Cell. Biol. 17 989-998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulczyk, A. W., Yang, J. C., and Neuhaus, D. (2004) J. Mol. Biol. 341 723-738 [DOI] [PubMed] [Google Scholar]
- 33.Mackey, Z. B., Niedergang, C., Murcia, J. M., Leppard, J., Au, K., Chen, J., de Murcia, G., and Tomkinson, A. E. (1999) J. Biol. Chem. 274 21679-21687 [DOI] [PubMed] [Google Scholar]
- 34.Taylor, R. M., Whitehouse, J., Cappelli, E., Frosina, G., and Caldecott, K. W. (1998) Nucleic Acids Res. 26 4804-4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor, R. M., Whitehouse, C. J., and Caldecott, K. W. (2000) Nucleic Acids Res. 28 3558-3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menissier-de Murcia, J., Molinete, M., Gradwohl, G., Simonin, F., and de Murcia, G. (1989) J. Mol. Biol. 210 229-233 [DOI] [PubMed] [Google Scholar]
- 37.Gradwohl, G., Menissier de Murcia, J. M., Molinete, M., Simonin, F., Koken, M., Hoeijmakers, J. H., and de Murcia, G. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 2990-2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrucco, S. (2003) Nucleic Acids Res. 31 6689-6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Cam, E., Fack, F., Menissier-de Murcia, J., Cognet, J. A., Barbin, A., Sarantoglou, V., Revet, B., Delain, E., and de Murcia, G. (1994) J. Mol. Biol. 235 1062-1071 [DOI] [PubMed] [Google Scholar]
- 40.Petrucco, S., Volpi, G., Bolchi, A., Rivetti, C., and Ottonello, S. (2002) J. Biol. Chem. 277 23675-23683 [DOI] [PubMed] [Google Scholar]
- 41.Windhofer, F., Wu, W., and Iliakis, G. (2008) J. Cell. Physiol. 213 475-483 [DOI] [PubMed] [Google Scholar]
- 42.Audebert, M., Salles, B., and Calsou, P. (2004) J. Biol. Chem. 279 55117-55126 [DOI] [PubMed] [Google Scholar]
- 43.Sekiguchi, J., and Shuman, S. (1997) Nucleic Acids Res. 25 727-734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sekiguchi, J., and Shuman, S. (1997) J. Virol. 71 9679-9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Odell, M., and Shuman, S. (1999) J. Biol. Chem. 274 14032-14039 [DOI] [PubMed] [Google Scholar]
- 46.Leppard, J. B., Dong, Z., Mackey, Z. B., and Tomkinson, A. E. (2003) Mol. Cell. Biol. 23 5919-5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen, L., Trujillo, K., Sung, P., and Tomkinson, A. E. (2000) J. Biol. Chem. 275 26196-26205 [DOI] [PubMed] [Google Scholar]
- 48.Tomkinson, A. E., Vijayakumar, S., Pascal, J. M., and Ellenberger, T. (2006) Chem. Rev. 106 687-699 [DOI] [PubMed] [Google Scholar]
- 49.Pascal, J. M., Tsodikov, O. V., Hura, G. L., Song, W., Cotner, E. A., Classen, S., Tomkinson, A. E., Tainer, J. A., and Ellenberger, T. (2006) Mol. Cell 24 279-291 [DOI] [PubMed] [Google Scholar]