Abstract
Ubiquitination of proteins modifies protein function by either altering their activities, promoting their degradation, or altering their subcellular localization. Deubiquitinating enzymes are proteases that reverse this ubiquitination. Previous studies demonstrate that proteins that contain an ovarian tumor (OTU) domain possess deubiquitinating activity. This domain of ∼130 amino acids is weakly similar to the papain family of proteases and is highly conserved from yeast to mammals. Here we report structural and functional studies on the OTU domain-containing protein from yeast, Otu1. We show that Otu1 binds polyubiquitin chain analogs more tightly than monoubiquitin and preferentially hydrolyzes longer polyubiquitin chains with Lys48 linkages, having little or no activity on Lys63- and Lys29-linked chains. We also show that Otu1 interacts with Cdc48, a regulator of the ER-associated degradation pathway. We also report the x-ray crystal structure of the OTU domain of Otu1 covalently complexed with ubiquitin and carry out structure-guided mutagenesis revealing a novel mode of ubiquitin recognition and a variation on the papain protease catalytic site configuration that appears to be conserved within the OTU family of ubiquitin hydrolases. Together, these studies provide new insights into ubiquitin binding and hydrolysis by yeast Otu1 and other OTU domain-containing proteins.
The covalent addition or removal of ubiquitin on target proteins controls numerous cellular processes, including cell cycle progression, the immune and inflammatory response, differentiation and development, and intracellular trafficking (1, 2). Ubiquitin, a 76-amino acid protein, can be conjugated by the sequential action of E1,5 E2, and E3 enzymes forming an isopeptide bond between the COOH terminus of ubiquitin and lysine residues of target proteins. Ubiquitin is attached either singly or as a polyubiquitin chain where the COOH terminus of one ubiquitin moiety is covalently attached to one of seven lysine residues of an adjacent ubiquitin (3-5). Lys48-linked polyubiquitination serves to target and deliver specific proteins to the proteasome for degradation, whereas monoubiquitination and Lys63-linked polyubiquitination signal nondegradative events (6, 7).
Deubiquitination involves the hydrolysis of these isopeptide bonds and requires the activity of a deubiquitinating enzyme (DUB) (8). DUBs can be divided into six distinct families: the ubiquitin-specific processing proteases (UBPs), the ubiquitin carboxyl-terminal hydrolases (UCHs), the Ataxin-3/Josephin domains, the ovarian tumor domain-containing proteases (OTUs) (9, 10), the viral processing proteases (11, 12), and the JAMM proteases. The JAMM metalloproteases are unique among the DUBs in that zinc is required to catalyze the reaction (13), whereas the other DUB families use an active site cysteine to hydrolyze the isopeptide bond between ubiquitin and the target. Although much is known, both structurally and biochemically, about UBPs and UCHs, relatively little is known about OTU domains.
The ovarian tumor (Otu) gene was originally identified as a gene required for Drosophila oogenesis (14). The Otu gene product is involved in the control of cell division and differentiation of the cystoblast into an oocyte and 15 nurse cells (15, 16). Located within the Drosophila Otu gene is an ∼130-residue domain that is highly conserved from human to yeast (17). This region, termed the OTU domain, displays weak similarity to the papain-like family of proteases (18). The OTU domain is found in a number of conserved genes from all eukaryotes and a number of viruses that infect eukaryotes. For example, the OTU domain is found in the tumor necrosis factor α-induced protein, A20. A20 protects cells from apoptosis and blocks activation of lymphoid cells by negatively regulating NF-κB, presumably by altering the ubiquitination state of proteins involved in the signaling pathway (19, 20). In the yeast Saccharomyces cerevisiae, two open reading frames contain the OTU domain (Otu1 and Otu2).
Recently, Rumpf and Jentsch (21) have implicated yeast Otu1 in the ER-associated degradation (ERAD) pathway. Otu1 and numerous other proteins interact with Cdc48, an AAA ATPase that plays a central role in the ERAD pathway by chaperoning proteins to the proteasome for destruction. Ufd3, a protein of unknown function, and Ufd2, a so-called E4 ubiquitin ligase, also interact with Cdc48 and promote the degradation of ubiquitin fusion proteins. Both Otu1 and Ufd3 can independently counteract the activity of Ufd2 by separate mechanisms. In the case of Otu1, the deubiquitination activity of Otu1 seems to act to remove ubiquitin molecules from Ufd2 substrates. Interestingly, overexpression of Otu1 is able to suppress the Δufd3 mutant.
The structure of an OTU domain, from human otubain 2, was determined by Balakirev et al. (9). The structure reveals a five-stranded β-sheet flanked by two α-helical domains that is novel for DUB enzymes. Despite the novel DUB fold and low sequence homology with other DUB families, otubain 2 shows a similar active site configuration with UCHs and UBP deubiquitinating enzymes.
In this report, we examine the substrate preference and enzymatic activity of Otu1 and show that it preferentially hydrolyzes polyubiquitin chains with Lys48 linkages and that an NH2-terminal ubiquitin fold domain in Otu1 mediates interaction with Cdc48. The Otu1/Cdc48 oligomer preferentially binds polyubiquitin analogs, suggesting a role in polyubiquitin metabolism. We also report the x-ray crystal structure of the OTU domain of Otu1 in covalent complex with ubiquitin and carry out structure-guided mutagenesis revealing a novel mode of ubiquitin recognition and a variation on the papain protease catalytic site configuration that appears to be conserved within the OTU family of ubiquitin hydrolases. Implications of these studies for the molecular mechanism of Otu1 recognition of polyubiquitin and for the biochemical consequence of its association with Cdc48 are discussed.
EXPERIMENTAL PROCEDURES
Preparation of the Otu1-Ubiquitin Complex—The genes encoding amino acids 87-301 of Otu1 and full-length ubiquitin were PCR-amplified and cloned into the pTYB2 vector. Bacteria containing either the pTYB2-Otu1ΔUBX (amino acids 87-301) or pTYB2-ubiquitin-(1-75) plasmids were grown in LB medium at 37 °C to a density of A600 of 0.6-0.8 and induced with isopropyl 1-thio-β-d-galactopyranoside overnight at 18 °C. Cells were harvested and lysed by sonication. After centrifugation, the cell lysates were bound to a pre-equilibrated chitin column (New England Biolabs) in a buffer containing 50 mm Tris, pH 8.5, and 1 m NaCl. Chemical cleavage of the Otu1-intein-chitin binding domain fusion was performed using a solution containing 50 mm DTT overnight at 25 °C. The sample was then run over a Superdex-200 gel filtration column (Amersham Biosciences) and frozen at -70 °C until further use. Selenomethionine-substituted Otu1 was prepared by transforming the pTYB2-Otu1-(87-301) construct into the methionine auxotroph strain, B834 (Novagen), and growing and inducing the cells in a minimal medium (Molecular Dimension, Ltd.) containing 50 μg/liter selenomethionine (Sigma).
To create a covalent Otu1 complex with ubiquitin, an irreversible ubiquitin inhibitor was created, ubiquitin-Br3, in which glycine 76 was substituted with a bromopropylamine group as described in Ref. 22. Briefly, after attaching the ubiquitin-intein-chitin binding domain fusion protein to the chitin column, cleavage was carried out overnight using 50 mm β-mercaptoethanesulfonate sodium salt. After elution and concentration, 0.2 mmol of 3-bromopropylamine hydrobromide (Sigma) and 100 μl of 2 m sodium hydroxide were added to 500 μl of ∼2 mg/ml ubiquitin-β-mercaptoethanesulfonate sodium salt. The tubes were vortexed and incubated at room temperature for 20 min, after which 100 μl of HCl was added. The solution was then dialyzed against 50 mm sodium acetate, pH 4.5, for 2 h and then dialyzed with phosphate-buffered saline, pH 7.4, and 1 mm Tris(2-carboxyethyl)phosphine overnight. The solution of Ub-Br3 was then concentrated to ∼1.5 mg/ml. To create the Otu1-ubiquitin complex, concentrated Otu1 was mixed with Ub-Br3 in 1:1 stoichiometric amounts in a solution containing 100 mm HEPES, pH 7.4, and incubated for 2 h at 37 °C. The complex was enriched using a HiTrap Q column (Amersham Biosciences) with a NaCl gradient at pH 8.5 and further purified on a Superdex-200 gel filtration column equilibrated with phosphate-buffered saline, pH 7.4, and 1 mm Tris(2-carboxyethyl)phosphine. Fractions containing the Otu1-Ub-Br3 complex were concentrated to ∼15 mg/ml for crystallization.
Crystallization and Structure Determination—All crystals were grown using the hanging drop vapor diffusion method. Orthorhombic crystals were obtained by mixing 2 μl of 15 mg/ml protein complex with 2 μl of reservoir solution and equilibrating over 0.5 ml of reservoir solution containing 100 mm Bis-Tris, pH 6.5, 100-250 mm magnesium chloride, 16-21% polyethylene glycol 3350. The crystals belong to space group P212121 with unit cells dimensions of a = 46.3, b = 73.1, and c = 89.3 Å and contain 1 complex per asymmetric unit cell. A hexagonal crystal form of the protein complex was prepared by mixing 2 μl of 15 mg/ml protein complex with 2 μl of reservoir and equilibration solution containing 100 mm MES, pH 6.5, 50-200 mm ammonium acetate, 17-22% polyethylene glycol 3350. The hexagonal crystals belong to the space group P64 and have unit cell dimension of a = 107.3, b = 107.3, and c = 98.9 Å and contain two molecules in the asymmetric unit cell.
To determine phases, orthorhombic crystals were soaked for 4 h in a 5 mm potassium tetracyanoplatinate, K2Pt(CN)4, solution (Hampton Research). For the hexagonal crystals, selenomethionine-substituted Otu1 protein was used for crystallization. Heavy atom-derivatized crystals were cryoprotected for data collection by transferring the crystals to a solution containing reservoir solution supplemented with 25% glycerol prior to freezing in liquid nitrogen for data collection. Data for the Otu1-ubiquitin complex crystals were collected at the X29 beamline at the National Synchrotron Light Source at Brookhaven National Laboratory. Data processing was carried out with the HKL2000 (23) implementation of the programs DENZO and SCALEPACK. Three-wavelength multiwavelength anomalous dispersion experiments were carried out for both the platinum-soaked orthorhombic crystals and the selenomethionine-substituted hexagonal crystals. Data for both complex crystal forms were treated and solved independently from each other. Data collection statistics are described in Table 1. Shelx (24) was used to identify the heavy atom sites, and SHARP (25) was used to refine their position, occupancy, and B-factors. Using the heavy atom sites, the initial experimental phases were calculated and improved by solvent flattening using DM (26). From the resultant electron density map, the program ARP/wARP (27) was able to unambiguously build a model of the OTU complex. Alternating rounds of model building and refinement were performed to get the R-factor to 18.9% and R-free to 21.1% using Phenix. There are 143 waters. Of all of the residues, 97.5% (241 residues) are in the most favored regions of the Ramachandran plot, 2.5% (6 residues) are in the allowed regions, and none are in disallowed regions.
TABLE 1.
Data collection and refinement statistics for the OTU-ubiquitin complexes
|
OTU-ubiquitin complex (orthorhombic form,
P212121 platinum-soaked)
|
OTU-ubiquitin complex (hexagonal form, P64
Se-Met-substituted)
|
||||||
|---|---|---|---|---|---|---|---|
| 0.9795 Å wavelength (native) | 1.0722 Å wavelength (peak) | 1.0725 Å wavelength (inflection) | 1.0450 Å wavelength (remote) | 0.9793 Å wavelength (peak) | 0.9795 Å wavelength (inflection) | 0.9566 Å wavelength (remote) | |
| Resolution (Å) | 50-1.50 (1.55-1.50) | 20-2.0 (2.07-2.00) | 20-2.0 (2.07-2.00) | 20-2.0 (2.07-2.00) | 50-2.31 (2.39-2.31) | 50-2.31 (2.39-2.31) | 50-2.45 (2.56-2.45) |
| Observations | 314,652 | 295,383 | 295,672 | 296,114 | 517,121 | 360,539 | 263,582 |
| Unique reflections | 48,644 (4677) | 39,220 (3893) | 39,562 (3941) | 39,392 (3931) | 28,554 (2785) | 28,173 (2553) | 23,627 (2740) |
| Completeness (%) | 98.6 (96.4) | 99.9 (100) | 99.9 (99.9) | 99.9 (100) | 99.8 (98.2) | 98.3 (89.4) | 98.1 (91.4) |
| I/σ | 46.8 (3.2) | 11.5 (13.8) | 20.0 (5.08) | 25.8 (9.11) | 30.2 (7.12) | 26.6 (3.29) | 23.7 (2.75) |
| Rsym | 4.6 (38.0) | 9.5 (16.4) | 9.5 (38.1) | 7.3 (22.8) | 8.0 (35.7) | 7.2 (52.9) | 6.7 (62.9) |
| Rworking | 18.9% | 19.6% | |||||
| Rfree | 21.1% | 24.3% | |||||
| Root mean square deviation, bond length (Å) | 0.005 | 0.010 | |||||
| Root mean square deviation, angle (degrees) | 0.85 | 1.23 | |||||
Preparation of Full-length Otu1—Full-length Otu1 was PCR-amplified from yeast genomic DNA and cloned into the pRSET vector. Escherichia coli cells harboring pRSET-Otu1 were inoculated into LB medium at 37 °C and grown to A600 of 0.6. Cultures were induced with isopropyl 1-thio-β-d-galactopyranoside, and the temperature was shifted to 18 °C overnight. Cells were harvested and resuspended in a lysis buffer containing 50 mm Tris, pH 8.0, 10 mm DTT, 50 μm phenylmethylsulfonyl fluoride, 1 mm EDTA, and 10 mm MgCl2. Lysozyme and sonication were used to lyse the cells, and after centrifugation the cell lysate was loaded on a Fast Flow Q (Amersham Biosciences) column. Fractions that had high amounts of DUB activity using ubiquitin 7-amidomethylcourmarin (Ub-AMC) as a substrate were analyzed by SDS-PAGE and determined to be >95% pure. Active Otu1 fractions were pooled and dialyzed against sodium acetate, pH 7.0, 10 mm DTT and frozen at -80 °C for later use.
Otu1 Binding to Ub4 Analog Resins—Purified Otu1 (100 ng) was diluted to 500 μl in binding buffer (50 mm Tris-HCl, pH 7.2, 150 mm NaCl, 2 mm DTT) and incubated with 100 μl of the appropriate Ub4 resin for 90 min at 4 °C with gentle shaking. Identical experiments using ethanolamine blocked NHS-activated Sepharose (i.e. no conjugated ubiquitin) or a monoubiquitin resin were performed as negative controls. After incubation, reactions were briefly centrifuged, and the supernatants were removed and saved for SDS-PAGE analysis. Resin-bound Otu1 was batch-eluted by boiling in 500 μl of SDS-sample buffer. Bound and unbound fractions were analyzed by SDS-PAGE and Sypro Ruby Red (Invitrogen) or Imperial (Pierce) staining to estimate the amounts of Otu1 present (28).
Otu1 Deubiquitination Assay—300 ng of Otu1 and 250 ng of the appropriate Ub4 chain (gift of C. Pickart) were diluted to 30 μl in reaction buffer (Tris-HCl, pH 7.5, MgCl2, and DTT). A control reaction lacking enzyme was also prepared for each polyubiquitin chain analyzed. Reactions were incubated for 2 h at 37 °C. The reactions were then quenched with 3× SDS sample buffer and boiled for 3 min. Five μl of each reaction was then loaded onto a 10-20% gradient Tricine gel (Bio-Rad). For Western analysis, the protein was transferred to nitrocellulose in 20 mm Tris-HCl, 150 mm glycine, 20% methanol, pH 8.0, for 1 h at 100 V. After blocking with a 5% milk solution for 1 h, the blot was washed and incubated with a 1:500 dilution of a monoclonal anti-ubiquitin antibody, Ub (P4D1), from Santa Cruz Biotechnology for 1 h. Primary antibody was detected with a horseradish peroxidase-conjugated anti-rabbit immunoglobulin G secondary antibody (Amersham Biosciences) followed by enhanced chemiluminescence detection.
Otu1 Ubiquitin-AMC Kinetics—All reactions were performed in a 200-μl cuvette at 37 °C using an Aminco-Bowman Series 2 Luminescence Spectrometer to collect data. Settings for the analysis were as follows: excitation wavelength of 340 nm, emission wavelength of 440 nm, a sensitivity of 680 V, and an analysis time of 300 s. Kinetic parameters were determined from the linear phase of each reaction. 124 μl of assay buffer (50 mm Tris-HCl, pH 7.5, 2 mm DTT, and 10 μg/ml ovalbumin) with varying amounts of Ub-AMC were mixed in the cuvette and allowed to thermoequilibrate in the spectrometer for 2 min. Thirty s after the start of a run, Otu1 was added to the cuvette to 0.5 nm and mixed, and the cuvette was returned to the spectrometer for luminescence measurements.
Identification of Otu1 Interactors—Lysates were prepared from ΔOtu1 cells (strain gift of A. Corbett). The appropriate Ub4 analog or control resins (100 μl) were incubated separately with each of the following: 1) 500 μl of ΔOtu1 lysate, 2) 2 μg of full-length Otu1 or NH2-terminally truncated Otu1 diluted to 500 μl in lysate buffer, and 3) 500 μl of ΔOtu1 lysate supplemented with 2 μg of the appropriate Otu1. Binding reactions were incubated overnight at 4 °C with gentle agitation. After binding, the resin was pelleted by a brief centrifugation at 13,000 × g, and the supernatant was removed. Resins were washed with 1 ml oflysate buffer followed by two 1-ml washes of lysate buffer without Triton X-100. After washing, resins were eluted by adding 500 μl of SDS sample buffer, mixing, and boiling for 3 min. Twenty μl of each elution fraction was loaded and run on a 10% SDS-polyacrylamide gel that was then stained with Sypro Ruby Red. Potential interactors (bands present in incubation 3 elution fractions but not in incubation 1 or 2) were identified by MALDI-TOF analysis as previously described (28).
Demonstration of Direct Cdc48-Otu1 Interaction—An Otu1 affinity resin was synthesized by conjugating full-length Otu1 to Sulfolink (Pierce) resin. Otu1 was prepared for conjugation, and conjugation was carried out according to the manufacturer's instructions, except that β-mercaptoethanol was used to block unused active sites after Otu1 conjugation rather than the suggested l-cysteine.
All binding experiments were performed in duplicate with or without the presence of 1 mm ATP in the binding buffer. Control or Otu1-Sulfolink resin (50 μl) was equilibrated in binding buffer (50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 2 mm DTT, 0.1% Triton X-100). After equilibration, 17 μg of His6Cdc48 in binding buffer plus a protease inhibitor mixture (100 μm phenylmethylsulfonyl fluoride, pepstatin, leupeptin, aprotinin, chymostatin at final concentrations of 3 μg/ml) was added to the resins, mixed, and incubated overnight at 4 °C with gentle agitation. After binding, the mixtures were briefly centrifuged, and the supernatant was removed. Resins were then washed twice in 500 μl with binding buffer followed by two 500-μl washes using binding buffer without Triton. Resins were then eluted with 1 ml of 8 m urea plus 1 mm HCl buffer. Five μl of each elution was run on a 10% SDS-polyacrylamide gel. Protein was then transferred to nitrocellulose and prepared for Western analysis by standard procedures. The blot was incubated with anti-His6 antibody at a 1:2000 dilution for 60 min. The blot was then incubated with an anti-rabbit horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences) for 60 min at a 1:4000 dilution. Bands were visualized by enhanced chemiluminescence detection.
RESULTS
Otu1 Substrate Specificity—Rumpf and Jentsch (21) showed that yeast Otu1 is a deubiquitinating enzyme. To determine the functionality of our recombinant protein, we labeled recombinant full-length Otu1 with ubiquitin-vinyl sulfone (UbVS), an active site-directed irreversible inhibitor that reacts specifically with the active site cysteine of DUBs. Titration of 100 ng of Otu1 with increasing amounts of UbVS clearly shows the formation of a covalent adduct migrating about 10 kDa higher than Otu1 alone on SDS-PAGE (Fig. 1A). Significantly, the quantitative UbVS labeling of Otu1 indicates that almost all of the recombinant Otu1 is enzymatically competent. UbVS also completely inhibits Otu1 cleavage of Ub-AMC in fluorometric assays, indicating that the UbVS binds to the Otu1 active site (data not shown).
FIGURE 1.
Kinetic analysis of Otu1. A, Otu1 labeling by ubiquitin vinyl sulfone. The quantitative labeling of Otu1 by ubiquitin vinyl sulfone was analyzed by SDS-polyacrylamide gel and stained with Sypro Ruby Red. B, deubiquitinase activity of Otu1. Lineweaver-Burk plot of activity data obtained for recombinant full-length Otu1 (blue diamonds) and Otu1ΔUBX (magenta boxes) using a ubiquitin-AMC substrate. The release of AMC as a function of added protein is monitored by fluorescence. C, Otu1 catalytic activity in the presence of Cdc48. Activity of Otu1 is measured in the presence or absence of a 2-fold molar excess of the Cdc48 hexamer. D, Otu1 cleavage of native polyubiquitin chains. Otu1-mediated cleavage of Lys29-, Lys48-, or Lys63-linked Ub4 chains is analyzed by SDS-polyacrylamide gel and Western blotting with a monoclonal ubiquitin antibody and visualized by ECL.
We next examined the specific activity of Otu1. Previous data suggested that Otu1 was a very sluggish DUB, with an estimated velocity of ∼0.01 s-1 at low substrate concentration (21). We speculated that this slow activity might be due to the NH2-terminal ubiquitin foldlike (UBX) domain acting as an inhibitor by occupying the ubiquitin binding site of the enzyme. To test this hypothesis, recombinant full-length Otu1 and a truncated version missing an 86-residue NH2-terminal UBX domain, herein called Otu1ΔUBX, were incubated with increasing concentrations of the generic DUB substrate, Ub-AMC. Cleavage of Ub-AMC was monitored by the release of the fluorescent AMC molecule (Fig. 1B). These experiments reveal comparable kinetic behavior for both proteins with no evidence of saturation below 2 μm Ub-AMC and an apparent kcat/Km of 1 × 103m-1 s-1. This result shows that the UBX fold does not participate in catalysis by Otu1. We next asked whether Cdc48 association with Otu1 has an effect on the deubiquitination activity of Otu1. The ability of the Cdc48-Otu1 complex to cleave Ub-AMC was analyzed and found to be only slightly slower than that catalyzed by Otu1 alone (Fig. 1C). This result shows that the association of Cdc48 with Otu1 has little effect on hydrolyzing ubiquitin-AMC, although this experiment does not eliminate the possibility that other proteins that interact with Otu1 or Cdc48 might alter the deubiquitination activity of Otu1 in a Cdc48-dependent or independent way. Together, these results show that Otu1 contains modest DUB activity but little affinity for monoubiquitin.
It has been previously shown that Otu1 also cleaves polyubiquitin and that a 4-fold molar excess of enzyme cleaved Lys48-linked chains somewhat more efficiently than Lys63-linked chains (21). Chains of three or fewer ubiquitins were not cleaved at all. We extended these studies using stoichiometric amounts of Otu1 incubated with Lys29, Lys48, or Lys63-linked Ub4, and the reaction was analyzed for deubiquitination activity with anti-ubiquitin immunoblotting. This analysis demonstrates that Otu1 has a clear preference for Lys48-linked Ub4, although it has some activity on K63-linked Ub4 as well (Fig. 1D). Lys29-linked chains were not cleaved at all. It is notable that the extent of chain cleavage is very limited. It appears that single cleavage events are detected, with the majority of cleavages occurring in the middle of the tetraubiquitin chain. This suggests that the enzyme may be unable to efficiently release product and brings up the possibility that the action of the Cdc48 chaperone is necessary to release bound products. Alternatively, the enzyme may prefer to cleave in the middle of a chain releasing diubiquitin.
Substrate Binding by Otu1—To determine if Otu1 preferentially bound polyubiquitin chains over monoubiquitin and if Cdc48 or other cellular cofactors modulated this ubiquitin binding activity, we carried out binding studies of recombinant Otu1 to mono-Ub and Ub4 analog resins (28),6 in the presence or absence of ΔOtu1 yeast lysate. Each resin was incubated with ΔOtu1 lysate, recombinant Otu1 protein (∼20-fold excess to endogenous Otu1 levels), or both. These experiments show that the Otu1 protein does not bind to control or monoubiquitin resins under any experimental conditions (Fig. 2A, lanes 2-7). However, Otu1 protein binds efficiently to 11-, 29-, 48-, and 63-Ub4 analog resins, whether ΔOtu1 lysate is present or not (see arrow labeled Otu1 in Fig. 2A, lanes 8-19). These results demonstrate that Otu1 binds directly to polyubiquitin chains and that this binding can occur in the absence of additional cellular cofactors.
FIGURE 2.
Protein-protein interactions of Otu1. A, ubiquitin binding activity of Otu1. Otu1 binding to ubiquitin and Ub4 resins in the presence or absence of ΔOtu1 yeast lysate is analyzed by SDS-polyacrylamide gel stained with Sypro Ruby Red. The properties of the resin are indicated above the gel, and the presence or absence of Otu1 protein and ΔOtu1 yeast lysate is indicated below the gel. B, requirement of the Otu1 NH2 terminus for Cdc48 association. Cdc48 association with resins containing monoubiquitin or Lys48-linked Ub4 in the presence or absence of Otu1 and Otu1ΔUBX is analyzed by a SDS-polyacrylamide gel and stained with Sypro Ruby Red. The Otu1 load lanes are 4% of total Otu1 input, and the remaining lanes are all elution fractions from the resins. The arrows indicate the Otu1 interactor, Cdc48. C, requirement of the NH2 terminus of Otu1 for Cdc48 association. His6Cdc48 was incubated with control or OTU1 containing resins in the presence or absence of 1 mm ATP. Cdc48 in the flow-through and elution fractions was detected with anti-His6 antibody.
Despite the lack of a requirement for other cellular proteins to facilitate the binding of Otu1 protein to Ub4 analog resins, we noticed that a lysate protein band (migrating at an apparent molecular mass of 110 kDa) was strongly enriched in the bound fractions containing Otu1 (see arrow in Fig. 2A). Significantly, this band did not appear in bound fractions when Otu1 protein was not present, suggesting that this band represents a protein recruited to the Otu1-Ub4 complex. To establish the identity of this protein band, it was excised from the gel, trypsin-digested, and subjected to MALDI-TOF analysis. Of 56 peptides detected, 18 were a match to Cdc48 using Profound (Genomic Solutions, Ann Arbor, MI). Based on the 22% sequence coverage and a high Z-score of 2.38 (confidence level of >95%), the protein band was confidently identified as Cdc48, an essential AAA ATPase involved in numerous cellular functions ranging from chaperoning polyubiquitinated proteins to the proteasome to regulating membrane fusion and previously reported to bind Otu1 (21).
Otu1 Simultaneously Interacts with Polyubiquitin and with Cdc48—To understand how Otu1 interacts with Cdc48, we focused on UBX, the NH2-terminal 86 amino acid portion of Otu1. UBX is an ∼80-amino acid domain where the secondary structure of the domain closely mimics the ubiquitin fold, but the primary sequence has minimal homology to ubiquitin. Other yeast proteins, such as Npl4 and Shp1, are known to interact with Cdc48 through UBX domains. To test if the UBX domain of Otu1 bound to polyubiquitin is involved in binding to Cdc48, we repeated ubiquitin-analog binding experiments using ΔOtu1 yeast lysate with recombinant full-length Otu1 and a truncated Otu1 construct lacking the NH2-terminal 100-residue UBX region (Otu1ΔUBX). These studies reveal that although Otu1ΔUBX bound to the Ub4 analog resins indistinguishably from the intact Otu1 protein, the protein band corresponding to the Cdc48 protein (Fig. 2B, arrows) was noticeably absent in the presence of the Otu1ΔUBX construct. Based on this observation, we conclude that the UBX region of Otu1 plays an important role in Cdc48 interaction.
To determine if the interaction between Otu1 and Cdc48 was direct, we immobilized purified Otu1 on an affinity support and incubated it with His6-tagged Cdc48 purified to ∼80% homogeneity from E. coli lysates and analyzed by SDS-PAGE and immunoblotting. Analysis demonstrates that recombinant Cdc48 protein binds to the immobilized Otu1 in an ATP-independent fashion (Fig. 2C) but not to the control resin. Note that Cdc48 is not bound to resins containing Ub4 analogs but not Otu1 (Fig. 2A). These results confirm that Otu1 and Cdc48 associate through a direct interaction and that both free Otu1 and the Otu1-Cdc48 oligomeric complex bind polyubiquitin chains.
Structure of the Otu1 Catalytic Domain Bound to Ubiquitin—To understand the molecular basis for ubiquitin recognition and cleavage by the OTU domain, we determined the structures of Otu1 bound in a covalent complex with ubiquitin. In our crystallization studies, we used an Otu1 construct lacking the UBX domain but containing the zinc domain, Otu1ΔUBX, amino acids 87-301. To make the covalent complex, we employed a published protocol (22) to create a ubiquitin derivative that is modified at the COOH terminus with an electrophile that will react with the catalytically active cysteine. The COOH-terminal glycine, Gly76, was replaced with 3-bromopropylamine to produce Ub75-NH-CH2-CH2-CH2-Br, which was mixed with the OTU domain of Otu1 to form the thiol ether-linked covalent complex that was purified to homogeneity for crystallization.
Two different crystal forms of the covalent Otu1-Ub complex were obtained. An orthorhombic form crystallized in the P212121 space group diffracted to 2.0 Å resolution and contained one covalent complex per asymmetric unit cell. A hexagonal form crystallized in space group P64 and diffracted to 2.4 Å resolution with two covalent complexes in the asymmetric unit. Both crystal forms were solved independently by multiwavelength anomalous dispersion, using platinum and selenium for the orthorhombic and hexagonal crystal forms, respectively. The structures from the two crystal forms are essentially superimposable, with a root mean square deviation for Cα atoms of 0.68 and 0.58 Å, comparing each of the two molecules in the hexagonal crystal form with the orthorhombiccrystal form. In further discussions of the structure, we will concentrate on the orthorhombic form due to its higher resolution.
The catalytic domain of Otu1 is made up of two globular lobes separated by a large, relatively shallow cleft (Fig. 3A). The larger lobe is composed of a six-β sheet core in a β1↑ β2↓ β6↑ β5↓ β3↓ β4↑ configuration surrounded by three α-helices, α1, α5, and α6. The smaller lobe is made up of two scaffolding α-helices, α2 and α4, that support a short third α-helix, α3. The two Otu1 lobes come together to form a clamp around a cleft that forms the binding site for ubiquitin (Fig. 3B). The ubiquitin binding cleft narrows into a channel where the very COOH terminus of ubiquitin is proximal to an active site cysteine at the bottom of a channel. Although contained in the crystals, we did not see electron density for the zinc-binding domain. We also did not see any evidence of degradation of the Otu1 protein in the crystals (data not shown).
FIGURE 3.
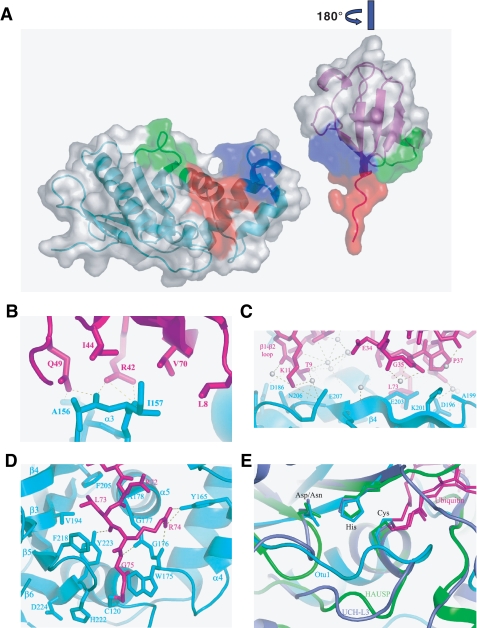
Overall structure of the Otu1-ubiquitin complex. A, overall structure of the Otu1-Ub complex. A schematic representation of the complex is shown with Otu1 and ubiquitin color-coded cyan and purple, respectively. Secondary structure elements are as labeled. B, the ubiquitin binding pocket on Otu1 (a view orthogonal to A looking down on the ubiquitin binding site Otu1). Otu1 is shown in a surface representation with a mapping of OTU domain-conserved residues mapped onto the protein and ubiquitin shown in a schematic representation. The color-coding for conservation is as follows: yellow, 50% conserved; orange, 75% conserved; red, 95% conserved. C, sequence alignment of OTU domains. A subset of OTU domains from different organisms is shown. The degree of conservation is color-coded according to B. The secondary structure of the Otu1 catalytic domain is shown above the sequence alignment, and residues that participate in ubiquitin contact are indicated with a star above the sequence alignment. Mutated residues (Fig. 5A) are underlined.
The Otu1 and ubiquitin interface is quite extensive, burying a total of about 1900 Å2 of surface area. The protein-protein interface also harbors the greatest degree of sequence conservation among the OTU domains (Fig. 3, B and C), further highlighting the importance of this pocket of Otu1 for ubiquitin recognition and suggesting that the complex observed here is representative of ubiquitin complexes with other OTU domains from other proteins and from other species (Fig. 3C).
The Interface between Otu1 and Ubiquitin—The Otu1-ubiquitin interface can be divided into three different regions of ubiquitin that participate in hydrolase interaction (Fig. 4A). Regions 1 (blue) and 2 (green) are on opposite sides of the surface of ubiquitin that contact the small and large Otu1 domains, respectively, and flank the COOH-terminal extension of ubiquitin that forms region 3 (red) and that makes the most extensive contacts to the Otu1 cleft.
FIGURE 4.
The Otu1-ubiquitin interface. A, three regions of interaction between Otu1 and ubiquitin. The figure is rendered in a surface representation and shown in “open book” format highlighting contact regions 1 (blue), 2 (green), and 3 (red). B, close-up of Otu1-ubiquitin interactions in region 1. Side chains that mediate hydrogen bonds (dotted line) and van der Waals interactions are shown. C, close-up of Otu1-ubiquitin interactions in region 2. Water molecules are indicated as white spheres. D, close-up of Otu1-ubiquitin interactions in region 3. E, superimposed active sites of deubiquitinating enzymes. The catalytic triad cysteine, histidine, and aspartate as well as ubiquitin (magenta) are shown as a stick models. Otu1 is shown as cyan, UCH-L3 (Protein Data Bank code 1xd3) is shown in violet, and HAUSP/USP7 (Protein Data Bank code 1nbf) is shown in green.
Region 1 centers around residue Ile44 of ubiquitin and the small lobe of Otu1 (Fig. 4B). Two scaffolding α-helices of the small Otu1 lobe, α2 and α4, emanate from the central region of the protein to support an ubiquitin recognition helix, α3, also in the Otu1 small lobe. Helix α3 makes both van der Waals and hydrogen bonding contacts to ubiquitin (Fig. 4B). In particular, Ile157 of Otu1 abuts the hydrophobic patch of Leu8, Ile44, and Val70 of ubiquitin. In addition, a hydrogen bonding network exists between the main chain carbonyl groups of Ala156 and Ile157, the carboxyl group of Asp159 of Otu1, and the guanidinium group of Arg40 and amide group of Gln49 of ubiquitin.
On the other side of the ubiquitin molecule, region 2 involves interactions between β4 of the Otu1 large lobe with the β1-β2 loop of ubiquitin (Fig. 4C). Eight of the 11 residues in this region are within 4 Å of the ubiquitin core. Among the direct interactions are the hydrogen bonding contacts between Thr9 and Lys11 of ubiquitin and Asn206 and Glu207 of Otu1 just COOH-terminal to β4. In the β4 region, a number of residues from ubiquitin and Otu1 are with in 4 Å of each other, including Asp196 of Otu1 and Gln40 of ubiquitin, which interact with the same water molecule. Most of the interactions in region 2 are indirect and mediated through hydrogen bonding with ordered water molecules. Indeed, 12 water molecules appear to be closely associated with the interface in this region.
The most extensive interaction between Otu1 and ubiquitin occurs in ubiquitin region 3, where Otu1 flanks the COOH terminus of ubiquitin (Fig. 4D). Otu1 utilizes both hydrophobic interactions and hydrogen bonding to orient the COOH-terminal 4 amino acids of ubiquitin into the active site (Fig. 4D). The clamp that flanks the ubiquitin COOH terminus is composed of two loop regions: the WGGA loop between α4 and α5 and Phe205, Phe218, and Tyr223 from different secondary structural elements of Otu1. Specifically, the main chain atoms of the glycine-glycine sequence in the WGGA loop of Otu1 make a number of hydrogen bonds with the main chain atoms of Arg72 and Gly75 of ubiquitin to form one side of the clamp. Trp175 of this loop also makes van der Waals interactions with the COOH-terminal ubiquitin glycine residue and appears to help wedge it toward the active site cysteine of Otu1. The hydroxyl group of Tyr165 of Otu1 makes a critical contact with the guanidinium group of ubiquitin Arg74. The other side of the clamp is composed of Phe205, Val194, Phe218, and Tyr223. Phe205 and Val194 of Otu1 create a hydrophobic sandwich of the Leu73 side chain of ubiquitin, whereas the edges of the aromatic rings of Phe218 and Tyr223 of Otu1 interact with the main chain atoms of Leu73, Arg74, and Gly75. Taken together, the Otu1-ubiquitin complex forms an extensive interface, and the observation that nearly all of the residues that make ubiquitin contacts by Otu1 are highly conserved within the OTU family (Fig. 3C) suggests that the observed interface is characteristic of the OTU family of hydrolases.
The primary sequence of Otu1 shows weak similarity to the papain family of cysteine proteases. In cysteine proteases, the cysteine thiol is deprotonated by a histidine, which is polarized by an aspartic acid residue. Although the catalytic triad in Otu1 shows a similar configuration of the corresponding catalytic triad residues (Cys120 and His222 on Otu1), the corresponding aspartic acid (Asp224 in Otu1) emanates from a different position in the primary sequence (Fig. 4E). In contrast to other cysteine proteases, the loop immediately preceding the helix containing the active site cysteine is also shifted closer to the active site, resulting in a more spatially restricted active site (Fig. 4E). Conservation of the catalytic triad residues among the OTU domains (Fig. 3C) and the spatial overlap of these residues and the active site loop with the OTU domain of otubain 2 (Fig. 4E) suggest that the observed active site is characteristic of the OTU family of hydrolases.
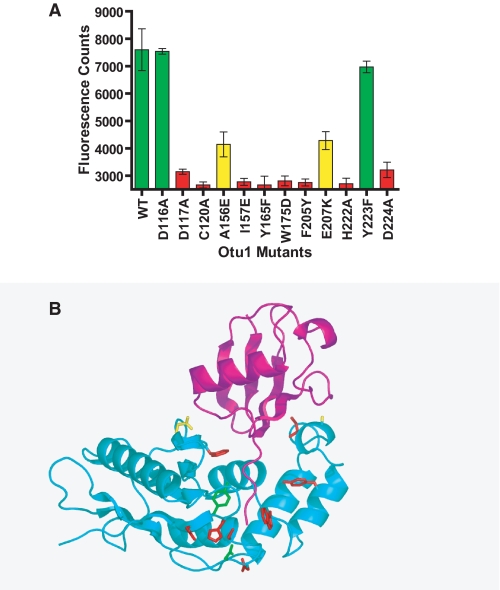
Mutational Analysis of the Otu1-Ubiquitin Interface—To establish the functional importance of the observed Otu1-ubiquitin interactions, we carried out a mutational analysis. For the mutagenesis, we targeted selected residues within contact regions 1-3 of Otu1 as well as the active site residues. Each mutant was prepared by site-directed mutagenesis in the context of the recombinant Otu1 domain used for structural analysis, purified to homogeneity, and assayed for deubiquitinating enzyme activity using the fluorescence assay described earlier with Ub-AMC as the substrate (Fig. 1B). The results of this assay are summarized in Fig. 6A. As a base line for these experiments, we mutated the catalytic triad residues (C120A, H222A, and D224A), and as expected, the C120A and H222A residues showed background levels of catalytic activity. However, the D224A mutant still showed about 30% of wild-type activity, suggesting that it plays a less critical role in catalysis consistent with its poorer conservation relative to the cysteine and histidine residues of the catalytic triad (Fig. 3C).
FIGURE 6.
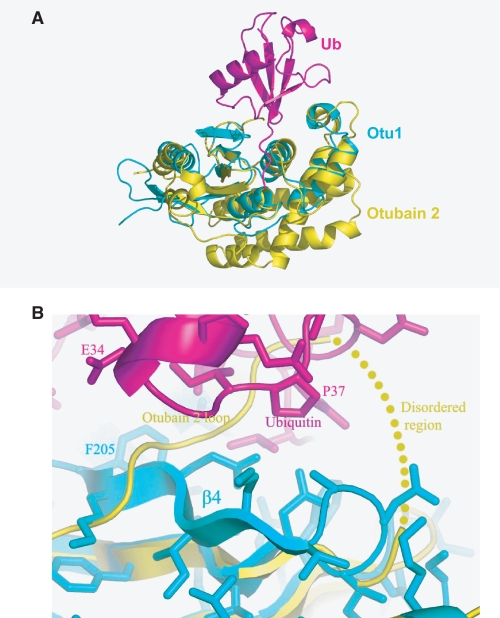
Comparison of the Otu1-ubiquitin complex with otubain 2. A, superposition of the Otu1-ubiquitin complex (cyan and magenta, respectively) with nascent otubain 2 (yellow). B, close-up view of the β4-loop region of Otu1 (cyan) with bound ubiquitin (magenta) with the corresponding region of nascent otubain 2 (yellow).
Within region 1 of the Otu1-ubiquitin interface, we mutated alanine 156 and isoleucine 157 to glutamate residues. Although the A156E mutation showed 50% of wild-type activity, the I157E mutation showed less than 10% of wild-type activity (Fig. 6A). Both residues mediate van der Waals contacts to a hydrophobic patch on ubiquitin that is centered around Ile44, a residue that appears to be widely recognized by ubiquitin binding domains, such as UIM, CUE, and GAT domains among many others (30). The conservation of residues Ala156 and Ile157 also points to their functional importance (Fig. 3C).
Within region 2 of the Otu1-ubiquitin interface, we mutated glutamate 207, a residue that appeared from the structure to mediate important direct and water-mediated interactions with Lys11 and Thr9 of ubiquitin. Surprisingly, the E207K mutant showed 50% of wild-type activity, thus arguing against the relative importance of region 2 for Otu1-ubiquitin complex formation.
The structure of the Otu1-ubiquitin complex suggests that region 3 plays a particularly important role in complex formation, and this is supported by the mutational analysis. Specifically, three of the four mutations that we prepared in this region (Y165F, W175D, and F205Y) were significantly compromised in deubiquitination activity. The Y223F mutant, however, showed wild-type levels of activity (Fig. 5A). The three mutationally sensitive residues are also the most highly conserved among the OTU domains, further supporting their functional importance for ubiquitin recognition. A mapping of the mutational results, color-coded by the severity of the effect of the mutation on Otu1 in vitro activity, is shown in Fig. 5B and highlights the importance of Otu1 interaction with the COOH-terminal ubiquitin tail for substrate recognition by Otu1 and probably other OTU domain proteins.
FIGURE 5.
Mutational analysis of the Otu1-ubiquitin interface. A, deubiquitinase activity of recombinant Otu1 and single site mutants. Each of the reactions was performed in triplicate and in the linear range of time and enzyme and substrate concentration. Activities are color-coded according to severity (green, wild type activity; yellow, about 50% of wild-type activity; red, less than 30% of wild-type activity). B, mapping of mutational studies onto the Otu1-ubiquitin complex. Side chains that were mutated in A are color-coded according to their severity on enzymatic activity as described in A.
Comparison of the Otu1-Ubiquitin Complex with Otubain 2—The structure of a nascent OTU domain from otubain 2 was previously reported (9), giving us an opportunity to assess the structural variability of the OTU domain fold and to investigate any structural rearrangement that might accompany ubiquitin substrate binding by OTU domains. A superposition of the Otu1 and Otubain 2 structures reveals that, not surprisingly, the proteins have similar overall folds, however with a higher root mean square deviation than expected of 2.46 Å over 110 structurally aligned Cα positions (Fig. 6A). The greatest similarity maps to the central core region composed of the small and large Otu1 lobes that recognize ubiquitin and participate in catalysis, suggesting that this region represents a conserved feature among the OTU family of deubiquitinating enzymes. There are, however, three notable differences. First, the NH2 termini of the two proteins are dissimilar. Otu1 contains one β sheet, β1, whereas otubain 2 has an α-helix and a β strand, β1 and α1 (9). The COOH-terminal portion of otubain 2, α1, however, structurally aligns with the last helix, α6, of Otu1. Second, otubain 2 also has an additional three α-helices, α4, α5, and α6, which buttress the structurally conserved α7, α8, and α9 helices (α2, α3, and α4 in the small lobe of Otu1). Together, these two structural differences between Otu1 and Otubain 2 create a divergent scaffold that appears to buttress the structurally conserved core of the OTU fold that participates in ubiquitin recognition and catalysis (9).
The third region of structural divergence between Otu1 and otubain 2 appears to be directly correlated with ubiquitin recognition (Fig. 6B) (9). Specifically, otubain 2 has a disordered loop region from residue 198 to 204, and 6 residues COOH-terminal to this disordered region (residues 205-210) are in position to sterically clash with the bound ubiquitin molecule. In contrast, Otu1 forms a β strand (β4)-turn in the corresponding region, and residues within this segment form specific stabilizing contacts to ubiquitin as described earlier. In particular, residues Phe205 in the β4 strand and Glu207 in the succeeding loop are mutationally sensitive for ubiquitin binding (Fig. 5, A and B). Taking these results together suggests that this region of the OTU domain proteins may adopt an inhibitory conformation similar to that observed for the nascent otubain 2 in the absence of ubiquitin but undergoes a structural transition to an active form for ubiquitin binding that is similar to that seen for the Otu1-ubiquitin complex.
DISCUSSION
The functional studies that we present here demonstrate that Otu1 prefers to bind polyubiquitin chains and interacts with monoubiquitin very weakly. In vitro studies show that full-length Otu1 has a preference for Lys48 linked tetraubiquitin over Lys29- or Lys63-linked tetraubiquitin. Kinetic studies (Fig. 1C) suggest the possibility that it acts by an endo mechanism releasing diubiquitin from polyubiquitin chains. Interestingly, the OTU protein, A20, seems to prefer Lys63-linked substrates (19, 31), although it is not known if other domains outside the OTU domain are responsible for this selectivity.
In this study, we have confirmed that Otu1 interacts via its UBX with Cdc48, a component of the ERAD pathway and the ubiquitin-fusion protein degradation pathway. Interestingly, the biological consequences of the Otu1/Cdc48 interaction are unclear. Cdc48 plays an important role in recognizing and chaperoning polyubiquitinated proteins, especially ERAD substrates. Both membrane-bound and luminal ERAD substrates require the activity of Cdc48, and it delivers the substrates to the proteasome or to proteasome adapters, such as Rad23. Otu1 binds to the N-domain of Cdc48 and/or the Cdc48-Ufd1-Npl4 complex (32). It is thought that binding of the Ufd1-Npl4 heterodimer results in a conformational change in the other N-domains of the hexameric Cdc48, both preventing the binding of additional Ufd1-Npl4 heterodimers and recruiting UBX-containing adapters, such as Otu1, Shp1, and Ubx-1 to -7. These adapters then mediate the diverse activities of Cdc48 in ERAD, ubiquitin-fusion protein degradation pathway, and heterotypic membrane fusion. Additionally, these complexes can contain other proteins bound to the D1 and D2 ATPase domains, such as components of the ubiquitin-fusion protein degradation pathway, Ufd2 (an ubiquitin E4 ligase) or Ufd3 (a regulator of ubiquitin levels). Thus, this complicated machine synthesizes, chaperones, remodels, and binds polyubiquitinated substrates involved in numerous cellular functions.
The structure that we present of the Otu1-ubiquitin complex clearly shows how the first ubiquitin is recognized but gives no direct insights into the recognition of the second ubiquitin. Nonetheless, a number of the structural and functional observations presented here suggest how Otu1 might bind a second ubiquitin. Specifically, although the structure shows that the ubiquitin COOH-terminal tail enters the Otu1 clamp from the top and is surrounded on three sides by the small and large lobes and the α5 helix of Otu1, the COOH-terminal tail of ubiquitin is accessible from one side of Otu1 (opposite the α5 helix) and the bottom that is blocked by only the β1-α2 loop (Fig. 4A). It therefore seems likely that the P1′ (leaving group) ubiquitin that is attached to the COOH-terminal glycine of the bound ubiquitin exits from this accessible region of Otu1. Consistent with this idea, we find that mutation of a strictly conserved aspartic acid to alanine, D117A, within the β1-α2 loop causes a significant decrease in Otu1 activity in vitro (Fig. 6A).
If the second ubiquitin molecule interacts through the area of the β1-α2 loop, what contacts it? It is possible that the second ubiquitin molecule is contacted by a cysteine-rich and putative zinc binding domain that is COOH-terminal to the catalytic domain of Otu1 described here. Interestingly, this C2H2 domain is present in the protein construct that was crystallized for these studies, but neither the domain nor a bound zinc atom is visible in the electron density map, presumably due to disorder. We propose that the putative zinc binding domain of Otu1 may become ordered only in the presence of a second ubiquitin molecule. Consistent with this idea, there are now several examples of zinc binding domains that bind ubiquitin, including isopeptidase T/USP5 (33), Rabex-5 (34, 35), and Npl4 (36). Interestingly and perhaps most relevant to Otu1, the C2H2 domain of the OTU-containing protein A20 recognizes Asp58 of ubiquitin. It remains to be seen whether the C2H2 domain of Otu1 recognizes ubiquitin in a similar way despite the lack of sequence homology. An alternative hypothesis is suggested by the fact that diubiquitin is apparently not a good substrate for this enzyme and may even be tightly bound. In this model, the C2H2 domain may in fact form an S2 pocket, resulting in tight binding of diubiquitin.
It is interesting to see how different classes of deubiquitinating enzymes, which share little primary sequence homology, recognize and cleave the same ubiquitin moiety. There are currently seven structures of deubiquitinating enzyme-ubiquitin complexes in the Protein Data Bank, all but one of which belong to the UBP and UCH classes of deubiquitinating enzymes. The other structure complex, a viral processing protease from herpes virus, M48USP, has been determined recently (12). Van der Waals interactions via Ile44 of ubiquitin appear to play an important role in hydrolase recognition in each of these complexes. Another common theme is that deubiquitinating enzymes recognize ubiquitin through an extensive network of ordered water molecules. USP2 uses 25 waters, UCH-L3 uses 18 waters, and Otu1 uses 23 ordered water molecules to mediate recognition. The most extensive interactions between deubiquitinating enzymes and ubiquitin are contained in the region surrounding COOH-terminal 6 residues of ubiquitin. Deubiquitinating enzymes create a specific hydrogen bonding network with the guanidinium groups of Arg72 and Arg74 as well as with main chain atoms of the COOH terminus ubiquitin. These interactions are direct and are rarely mediated by water molecules.
There are subtle differences in the active sites between the UCH/UBP classes and OTU family of deubiquitinating enzymes. When we compare the catalytic residues from Otu1 with those of HAUSP (37) (Protein Data Bank code 1nbf) and UCH-L3 (38) (Protein Data Bank code 1xd3), we see a root mean square deviation for the 24 atoms of 1.61 and 1.68 Å, respectively (Fig. 4E). UBPs and UCHs have active sites where the cysteine, histidine, and aspartate side chains emerge from different secondary structural elements. Indeed, in the case of HAUSP, the substantial rearrangement of the active site residues upon ubiquitin binding would be difficult to achieve if they were linked more closely. However, in the case of the Otu domain, the active site residues His222 and Asp224 are just 2 residues apart. In the Otu1-ubiquitin structure, Asp224 is poised to polarize the His222, which in turn deprotonates active site cysteine. Interestingly, Asp224 is not conserved among all OTU domain members. In an alignment of OTU domains from Cezanne, A20, and VCIP135, this residue is serine, valine, and isoleucine, respectively (8). A20 has been shown to possess both a deubiquitinating enzyme and an E3 ligase activity (31). Perhaps for A20, the absence of aspartate or asparagine is important in allowing the zinc E3 ligase domain to cooperate with the deubiquitinating enzyme domain in facilitating the “editing” of Lys63-linked ubiquitin molecules to Lys48-linked polyubiquitin chains.
It is generally thought that the activity of deubiquitinating enzymes must be tightly regulated. When we compare the structure of the OTU domain alone (otubain 2) with the Otu1-ubiquitin complex (9), we notice that otubain 2 may be in a self-inhibited form. Residues 198-210 of otubain 2 are in a position to sterically clash with a bound ubiquitin. These residues may form an ordered β-strand upon ubiquitin binding as in the Otu1-ubiquitin structure. This mode of self-inhibition by loops is not unprecedented. In the structures of USP14 alone and in complex with ubiquitin, one clearly sees that two loops, termed BL1 and BL2, adopt different conformations (29). In the unbound form, BL1 and BL2 seem to block access of ubiquitin to the ubiquitin binding pocket, and in the bound form, BL1 and BL2 adopt an open conformation.
Broadly speaking, a comparison of the OTU DUB enzymes with other DUB families suggests that deubiquitinating enzymes have evolved subtle differences around a structurally conserved scaffold to mediate different substrate specificities with different biological consequences. Further, Otu1 appears to acquire its substrate by interacting with Cdc48 and/or Cdc48-associated adapters. This is a common theme among deubiquitinating enzymes, and several DUBs have been shown to have weak affinity for ubiquitin and to form macromolecular complexes with proteins that may act as adapters or scaffolds (8). The mechanistic link to the biology of Otu1 will be an important area for future investigation.
Acknowledgments
We thank Santosh Hodawadekar, Michael Brent, and Brandi Sanders for useful discussions and the administration and staff at the National Synchrotron Light Source at Brookhaven National Laboratory for access to and assistance with the use of beamline X29.
The atomic coordinates and structure factors (codes 3C0R and 3BY4) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by Department of Defense Grant W81XWH-04-1-0539 (to T. M.), and National Institutes of Health Grant GM30308 (to K. D. W.), and a grant from the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health (to the Wistar Institute). Use of the X29 beamline at the National Synchrotron Light Source at Brookhaven National Laboratory was supported by the Offices of Biological and Environmental Research and of Basic Energy Sciences of the United States Department of Energy and by the National Center for Research Resources of the National Institutes of Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; MALDI-TOF, matrix-assisted laser desorption ionization-time-of-flight; DUB, deubiquitinating enzyme; UBP, ubiquitin-specific processing protease; UCH, ubiquitin carboxyl-terminal hydrolase; OTU, ovarian tumor; ER, endoplasmic reticulum; ERAD, ER-associated degradation; DTT, dithiothreitol; Ub, ubiquitin; MES, 4-morpholineethanesulfonic acid; AMC, 7-amidomethylcourmarin; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; UbVS, ubiquitin-vinyl sulfone; UBX, ubiquitin foldlike.
N. S. Russell and K. D. Wilkinson, manuscript in preparation.
References
- 1.Haglund, K., and Dikic, I. (2005) EMBO J. 24 3353-3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reinstein, E., and Ciechanover, A. (2006) Ann. Intern. Med. 145 676-684 [DOI] [PubMed] [Google Scholar]
- 3.Hochstrasser, M. (2000) Nat. Cell Biol. 2 153-157 [DOI] [PubMed] [Google Scholar]
- 4.Pickart, C. M. (2001) Annu. Rev. Biochem. 70 503-533 [DOI] [PubMed] [Google Scholar]
- 5.Fang, S., and Weissman, A. M. (2004) Cell Mol. Life Sci. 61 1546-1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerscher, O., Felberbaum, R., and Hochstrasser, M. (2006) Ann. Rev. Cell Dev. Biol. 22 159-180 [DOI] [PubMed] [Google Scholar]
- 7.Babst, M. (2005) Traffic 6 2-9 [DOI] [PubMed] [Google Scholar]
- 8.Amerik, A. Y., and Hochstrasser, M. (2004) Biochim. Biophys. Acta 1695 189-207 [DOI] [PubMed] [Google Scholar]
- 9.Nanao, M. H., Tcherniuk, S. O., Chroboczek, J., Dideberg, O., Dessen, A., and Balakirev, M. Y. (2004) EMBO Rep. 5 783-788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, P. C., Smith, T. S., Lai, M. J., Williams, M. G., Burke, D. F., Heyninck, K., Kreike, M. M., Beyaert, R., Blundell, T. L., and Kilshaw, P. J. (2003) J. Biol. Chem. 278 23180-23186 [DOI] [PubMed] [Google Scholar]
- 11.Kattenhorn, L. M., Korbel, G. A., Kessler, B. M., Spooner, E., and Ploegh, H. L. (2005) Mol. Cell 19 547-557 [DOI] [PubMed] [Google Scholar]
- 12.Schlieker, C., Weihofen, W. A., Frijns, E., Kattenhorn, L. M., Gaudet, R., and Ploegh, H. L. (2007) Mol. Cell 25 677-687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verma, R., Aravind, L., Oania, R., McDonald, W. H., Yates, J. R., III, Koonin, E. V., and Deshaies, R. J. (2002) Science 298 611-615 [DOI] [PubMed] [Google Scholar]
- 14.Steinhauer, W. R., Walsh, R. C., and Kalfayan, L. J. (1989) Mol. Cell. Biol. 9 5726-5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodesch, C., Geyer, P. K., Patton, J. S., Bae, E., and Nagoshi, R. N. (1995) Genetics 141 191-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sass, G. L., Comer, A. R., and Searles, L. L. (1995) Dev. Biol. 167 201-212 [DOI] [PubMed] [Google Scholar]
- 17.Glenn, L. E., and Searles, L. L. (2001) Mech. Dev. 102 181-191 [DOI] [PubMed] [Google Scholar]
- 18.Makarova, K. S., Aravind, L., and Koonin, E. V. (2000) Trends Biochem. Sci. 25 50-52 [DOI] [PubMed] [Google Scholar]
- 19.Evans, P. C., Ovaa, H., Hamon, M., Kilshaw, P. J., Hamm, S., Bauer, S., Ploegh, H. L., and Smith, T. S. (2004) Biochem. J. 378 727-734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, E. G., Boone, D. L., Chai, S., Libby, S. L., Chien, M., Lodolce, J. P., and Ma, A. (2000) Science 289 2350-2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rumpf, S., and Jentsch, S. (2006) Mol. Cell 21 261-269 [DOI] [PubMed] [Google Scholar]
- 22.Borodovsky, A., Ovaa, H., Kolli, N., Gan-Erdene, T., Wilkinson, K. D., Ploegh, H. L., and Kessler, B. M. (2002) Chem. Biol. 9 1149-1159 [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 24.Sheldrick, G. M., and Schneider, T. R. (1997) Methods Enzymol. 227 319-343 [PubMed] [Google Scholar]
- 25.Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M., and Paciorek, W. (2003) Acta Crystallogr. 59 2023-2030 [DOI] [PubMed] [Google Scholar]
- 26.Cowtan, K. D., and Main, P. (1996) Acta Crystallogr. 52 43-48 [DOI] [PubMed] [Google Scholar]
- 27.Morris, R. J., Perrakis, A., and Lamzin, V. S. (2003) Methods Enzymol. 374 229-244 [DOI] [PubMed] [Google Scholar]
- 28.Russell, N. S. (2005) Identification and Characterization of Polyubiquitin-binding Proteins in Saccharomyces cerevisiae Using Polyubiquitin Analogs, Ph.D. dissertation, Department of Biochemistry, Emory University, Atlanta, GA
- 29.Hu, M., Li, P., Song, L., Jeffrey, P. D., Chenova, T. A., Wilkinson, K. D., Cohen, R. E., and Shi, Y. (2005) EMBO J. 24 3747-3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hicke, L., Schubert, H. L., and Hill, C. P. (2005) Nat. Rev. 6 610-621 [DOI] [PubMed] [Google Scholar]
- 31.Wertz, I. E., O'Rourke, K. M., Zhou, H., Eby, M., Aravind, L., Seshagiri, S., Wu, P., Wiesmann, C., Baker, R., Boone, D. L., Ma, A., Koonin, E. V., and Dixit, V. M. (2004) Nature 430 694-699 [DOI] [PubMed] [Google Scholar]
- 32.Shcherbik, N., and Haines, D. S. (2007) Mol. Cell 25 385-397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reyes-Turcu, F. E., Horton, J. R., Mullally, J. E., Heroux, A., Cheng, X., and Wilkinson, K. D. (2006) Cell 124 1197-1208 [DOI] [PubMed] [Google Scholar]
- 34.Penengo, L., Mapelli, M., Murachelli, A. G., Confalonieri, S., Magri, L., Musacchio, A., Di Fiore, P. P., Polo, S., and Schneider, T. R. (2006) Cell 124 1183-1195 [DOI] [PubMed] [Google Scholar]
- 35.Lee, S., Tsai, Y. C., Mattera, R., Smith, W. J., Kostelansky, M. S., Weissman, A. M., Bonifacino, J. S., and Hurley, J. H. (2006) Nat. Struct. Mol. Biol. 13 264-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alam, S. L., Sun, J., Payne, M., Welch, B. D., Blake, B. K., Davis, D. R., Meyer, H. H., Emr, S. D., and Sundquist, W. I. (2004) EMBO J. 23 1411-1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu, M., Li, P., Li, M., Li, W., Yao, T., Wu, J. W., Gu, W., Cohen, R. E., and Shi, Y. (2002) Cell 111 1041-1054 [DOI] [PubMed] [Google Scholar]
- 38.Misaghi, S., Galardy, P. J., Meester, W. J., Ovaa, H., Ploegh, H. L., and Gaudet, R. (2005) J. Biol. Chem. 280 1512-1520 [DOI] [PubMed] [Google Scholar]