Abstract
Targeted deletion of acyl-CoA:cholesterol acyltransferase 2 (ACAT2) (A2), especially in the liver, protects hyperlipidemic mice from diet-induced hypercholesterolemia and atherosclerosis, whereas the deletion of ACAT1 (A1) is not as effective, suggesting ACAT2 may be the more appropriate target for treatment of atherosclerosis. Among the numerous ACAT inhibitors known, pyripyropene A (PPPA) is the only compound that has high selectivity (>2000-fold) for inhibition of ACAT2 compared with ACAT1. In the present study we sought to determine the PPPA interaction site of ACAT2. To achieve this goal we made several chimeric proteins where parts of ACAT2 were replaced by the analogous region of ACAT1. Differences in the amino acid sequence and the membrane topology were utilized to design the chimeras. Among chimeras, A2:1–428/A1:444–550 had 50% reduced PPPA selectivity, whereas C-terminal-truncated ACAT2 mutant A2:1–504 (C-terminal last 22 amino acids were deleted) remained selectively inhibited, indicating the PPPA-sensitive site is located within a region between amino acids 440 and 504. Three additional chimeras within this region helped narrow down the PPPA-sensitive site to a region containing amino acids 480–504, representing the fifth putative transmembrane domain of ACAT2. Subsequently, for this region we made single amino acid mutants where each amino acid in ACAT2 was individually changed to its ACAT1 counterpart. Mutation of Q492L, V493L, S494A resulted in only 30, 50, and 70% inhibition of the activity by PPPA, respectively (as opposed to greater than 95% with the wild type enzyme), suggesting these three residues are responsible for the selective inhibition by PPPA of ACAT2. Additionally, we found that PPPA non-covalently interacts with ACAT2 apparently without altering the oligomeric structure of the protein. The present study provides the first evidence for a unique motif in ACAT2 that can be utilized for making an ACAT2-specific drug.
In mammals, intracellular cholesterol esterification is performed by two enzymes, ACAT12 and ACAT2, that use two lipophilic substrates, cholesterol and acyl-CoA. Both the enzymes are polytopic integral membrane proteins localized in the endoplasmic reticulum (ER) (1, 2). Expression of ACAT1 occurs in a wide variety of cell types, whereas ACAT2 is localized only to the enterocytes of the intestine and the hepatocytes of the liver (3). We have demonstrated that ACAT2 is an important cholesterol-esterifying enzyme in the human liver specifically within the hepatocytes (4). ACAT enzymes have long been associated with the pathogenesis of atherosclerosis. In particular, it has been shown in nonhuman primates that hepatic ACAT activity is associated with cholesteryl oleate enrichment of low density lipoprotein and increased coronary artery atherosclerosis (5–7).
To elucidate the role of ACAT enzymes in atherosclerosis, gene knock-out studies were performed in hyperlipidemic mouse models. ACAT1 knock-out (KO) mice were not well protected from atherosclerosis, whereas both apoE KO mice and low density lipoprotein receptor-deficient mice were dramatically protected by ACAT2 gene deletion (8–13). Furthermore, liver-specific knockdown of ACAT2 using antisense oligonucleotides significantly reduced hepatic cholesterol concentration, plasma low density lipoprotein cholesterol oleate, and aortic atherosclerosis (14). Together, the data of these studies suggest that inhibition of ACAT2 may be preferable to inhibition of ACAT1 for providing protection against atherosclerosis.
Many studies have been performed to identify the inhibitors for ACAT enzymes (15). Although many potent inhibitors of ACAT are available, most have been found to inhibit both isoforms with similar efficiencies (16).3 One exception is pyripyropene A (PPPA), that has more than a 2000-fold higher selectivity for inhibition of ACAT2 (16). PPPA-mediated ACAT2 inhibition can be achieved in a cell-free system as well as in intact cells (16), suggesting the possibility that PPPA may directly interact with ACAT2 and inhibit its activity. In this report we sought to identify the PPPA-sensitive site of ACAT2. As PPPA is specific for ACAT2, we hypothesized that if we replace the PPPA-sensitive site of ACAT2 with the analogous region of ACAT1, the chimeric protein would loose PPPA sensitivity. With this approach we succeeded in identifying the PPPA-sensitive residues of ACAT2.
EXPERIMENTAL PROCEDURES
Generation of Chimeras and Mutants—All the chimeras and single amino acid mutants were generated by an overlap PCR method. African green monkey (AGM) ACAT1 and ACAT2 sequences were used as the template for making all the chimeras and the mutants. For construction of chimeras, each of the fragments of the chimeras was amplified separately with suitable primers (obtained from IDT DNA Technologies). All the internal primers were designed with a forward primer with a 5′ overhang sequence to its preceding fragment and a reverse primer with a 5′ overhang sequence to a fragment immediately after it. In the first step, each of the fragments of a chimera was amplified using proofstart DNA polymerase (Qiagen). In the final step, all the fragments were mixed to perform an overlap PCR using two external primers having 5′ Kpn1 and 3′ Not1 restriction sites. The full-length DNA construct was gel-extracted (Qiagen gel extraction kit) followed by Kpn1 and Not1 digestion and ligation (Fast Link DNA ligase, Epicenter Biotechnologies) into a pre-digested pCDNA3 vector (Invitrogen). For making a C-terminal-truncated ACAT2 construct termed CT504, a 3′ reverse primer was designed that introduced a stop codon after residue 504 followed by Not1 restriction site. All single amino acid mutants were made by site-directed mutagenesis approach using suitable primers that introduce the desired mutation into the full-length DNA sequence. Full-length DNA sequences containing the point mutations were then digested and ligated into pCDNA3 in similar ways as for the chimeras. All the PCR reactions were run with the following conditions: 95 °C for 5 min, 1 cycle; 94 °C for 30 s, 55 °C for 30 s, 72 °C for 90 s for 25 cycles; followed by 1 cycle at 72 °C for 10 min. All modified sequences were confirmed by DNA sequencing. DNA with confirmed sequences was then further purified using Endo free Maxi kit (Qiagen) to get transfection quality cDNA.
Cell Culture—AC29 cells (a Chinese hamster ovary cell-derived cell line, which was a gift from T. Y. Chang) lack any endogenous ACAT activity, mRNA, or protein and were used for all of these experiments. Cells were maintained in monolayer at 37 °C in 5% CO2 in Ham's F-12 medium supplemented with 1% Eagle's vitamins, penicillin (100 units/ml), streptomycin (100 μg/ml), and 10% heat-inactivated fetal bovine serum, and cells were typically grown to 70–90% confluence for all experiments.
Cell-based ACAT Assay—3 × 106 AC29 cells were transiently transfected with 6 μg of various constructs of ACAT cDNA using nucleofection technology (Amaxa Biosystems) according to the manufacturer's instructions. Suspended cells were divided into two aliquots after transfection; one aliquot was used to seed four 35-mm dishes to be used for kinetic assay, and the remaining aliquot of cells was plated into a 60-mm dish to use for subsequent immunoblotting. 72 h post-transfection the cells in 35-mm dishes were incubated for 30 min with either Me2SO (2 dishes) as control or 5 μm PPPA (2 dishes). Thereafter cells were pulsed-labeled with 1μCi of [9,10-3H]oleic acid (PerkinElmer Life Sciences, NET-289) for 2 h to stay within a linear response range. Cells were then removed by scraping and added directly to a glass tube containing 3.75 ml of 2:1 (v/v) chloroform:methanol. Extraction of total cellular lipids was completed by the addition of 1.25 ml of chloroform and 1.25 ml of water, respectively, to the tube followed by vigorous shaking. Finally, 3 ml of the lipid layer was isolated, and CE standards were added to samples. Samples were dried down under nitrogen, and lipid classes were separated by thin layer chromatography (TLC) using Silica Gel 60 plates in a solvent system containing hexane:diethyl ether:acetic acid (70:30:1). CE bands were visualized by exposure to iodine vapor and scraped, and radioactivity was determined using a liquid scintillation counter.
Preparation of Post-nuclear Supernatant (PNS)—72 h post-transfection cells from 60-mm dishes were washed twice with ice-cold balance salt solution. Cells were solubilized in 60 μl of RIPA buffer (0.1% SDS, 0.5% sodium deoxycholate and 1% Nonidet P-40 in phosphate-buffered saline) in the presence of 3 μl of protease inhibitor cocktails (Sigma). This was followed by sonication and removal of the nucleus and the cell debris by centrifugation at 14,000 rpm at 4 °C for 15 min. Supernatant was isolated, and 3-μl protease inhibitor cocktails were added to it and saved at –80 °C before use. Protein concentration of the PNS was measured by BCA assay (Pierce).
Preparation of Microsomes—Cells were washed twice with ice-cold balanced salt solution and were scraped from the dish. Excess balanced salt solution was removed from the cells by centrifugation, and cells were suspended in homogenization buffer (0.25 m sucrose, 0.1 m K2HPO4, 1 mm EDTA, pH 7.4). Protease inhibitor mixture (Sigma) was added to the cells, and the cells were lysed by sonication. The nucleus and cellular debris were removed and discarded after centrifugation at 14,000 rpm at 4 °C. PNS was then subjected to ultracentrifugation at 100,000 rpm for 30 min. The pellet containing the microsomes was collected and subsequently suspended in ice-cold 0.1 m K2HPO4 buffer at pH 7.4. Protein concentration was measured by BCA assay (Pierce). Microsomes in suspension were stored at –80 °C until assays were done.
Microsomal ACAT Assay—Microsomes were thawed, and about 25–100 μg of proteins were mixed with 1 mg of bovine serum albumin and 20 μl of a cholesterol-saturated solution of β-cyclodextrin, and the final volume was brought to 300 μl. The samples were equilibrated in a 37 °C water bath for 30 min, and then [14C]oleyl-CoA (Amersham Biosciences) was added to the tubes and incubated for 20 min. To stop the reaction 6 ml of chloroform:methanol, 2:1, was added to the samples. 1.2 ml of KCl was then added, and the mixture was allowed to sit overnight at room temperature. An aliquot (3 ml) of the organic phase (containing lipids) was removed and evaporated to dryness under nitrogen. The residue was resuspended in 100 μl of chloroform containing CE standard and then applied to a Silica Gel 60 TLC plate with subsequent separation in hexane:ethyl ether:acetic acid 70:30:1. The portion of the TLC plate containing the CE was scraped and suspended in scintillation fluid, and radioactivity was determined.
Western Blotting—Proteins from microsomes or PNS were suspended in an equal volume of protein solubilization buffer, (120 mm Tris, pH 6.8, 20%(v/v) glycerol, 4%(w/v) SDS, 0.01% (w/v) bromphenol blue) 100 mm DTT, and incubated at room temperature for 30 min. 50 mm iodoacetamide was added to samples and incubated at room temperature for another 30 min. Proteins were electrophoretically separated using a 4–12% NuPAGE® Novex® Bis-Tris Mini Gels (Invitrogen) and were transferred to a nitrocellulose membrane for 1 h at 115 V using a Western blot apparatus (Bio-Rad). The membrane was blocked overnight in 5% nonfat dry milk in TBST buffer (0.1 m Tris, pH 7.5, 0.15 m NaCl, 0.1% (v/v) Tween 20) at 4 °C which was followed by its incubation with the purified ACAT primary antibody (1 μg/ml) for 2 h at room temperature. The primary antibody was then removed, and the membrane was washed 3 times (10 min each) with TBST. Thereafter, the membrane was incubated with a goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (Sigma) at 1:20,000 dilution at room temperature for 1 h. After removing the secondary antibody, the membrane was washed 3 times with TBST (10 min each). The peroxidase signal was detected using Western illuminating reagents (PerkinElmer Life Sciences) and was captured on film (Eastman Kodak Co. BioMax light film). Affinity-purified ACAT antibodies were made as described before (3). Specificity of the antibodies was determined by running the PNS of empty vector-transfected cells on the gel as the negative control. We did not get any band on our negative control lane (data not shown).
Cross-linking Experiment—Microsomes were incubated with either Me2SO or 5 μm PPPA at room temperature for 30 min. Disuccinimidyl glutarate (solubilized in Me2SO) was then added to the samples, and mixtures were incubated for 30 min at room temperature. The cross-linking reaction was stopped by adding 1 m Tris at pH 7.5. All the samples were then subjected to Western blotting.
RESULTS
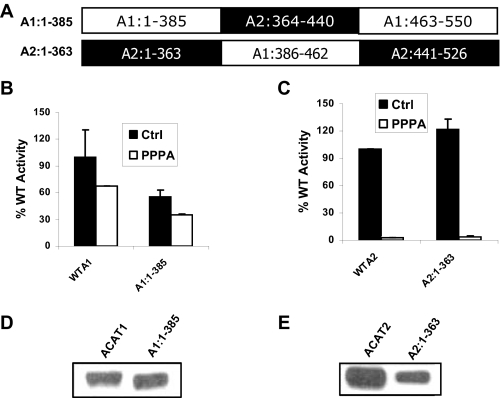
A PPPA-sensitive Site Is Located Outside the Putative Active Site Domain of ACAT2—ACAT enzymes share more than 50% sequence similarity to each other after the first 100 amino acid residues, where the sequence similarity is only 2%. The area of highest sequence similarity among these proteins is toward the C-terminal end, including amino acid residues 386–462 of ACAT1 and amino acid residues 364–440 of ACAT2. Interestingly, this region contains two conserved motifs, FYXDWWN and HEY. Because the sequence in this region is so highly conserved from yeast to human (supplemental Fig. C), we hypothesized that this region contains the active site domain of ACAT isozymes, and thus, both enzymes would have the same active site. Because PPPA is a highly selective inhibitor of ACAT2, we hypothesized that the PPPA-sensitive site of ACAT2 would be located outside of the putative active site domain of the enzyme. To test this hypothesis we made two chimeric proteins, A1:1–385 and A2:1–363, where the putative active site was swapped between the two enzymes (Fig. 1A). Kinetic assay result showed upon PPPA treatment ACAT1 activity was decreased 30% compared with the control cells, whereas this reduction was >95% for ACAT2-transfected cells, indicating PPPA is selective for ACAT2 (compare wild type enzymes of Fig. 1, B and C). Among the chimeras, A1:1–385 was inherently less catalytically efficient compared with its wild type (WT) counterpart since it had only 55% of the WT activity. With PPPA treatment, activity of this chimera in cells was lower by about the same 30% as seen when WT ACAT1 cells were treated with PPPA (Fig. 1B). This stands in direct contrast to the much greater decrease (>95%) induced by PPPA in activity of A2:1–363, a decrease that was comparable with WT ACAT2 enzyme (Fig. 1C). In each case the expression levels of the chimeras were substantial and comparable with the respective WT proteins (Fig. 1, D and E). Taken together, these data support our hypothesis and suggest that the PPPA-sensitive site is located outside the highly conserved putative active site domain of ACAT2 (A2:364–440).
FIGURE 1.
PPPA-sensitive site of ACAT2 is located outside the putative active site domain of the enzyme. A, primary structure of the chimeric proteins termed A1:1–385 and A2:1–363 are indicated where sequences from ACAT1 are in open boxes and sequences from ACAT2 are in filled boxes. B, kinetic assay data for WTA1 and A1:1–385. AC29 cells were transfected with the cDNA encoding WTA1 and A1:1–385 proteins. 72 h post-transfection cells were incubated with either vehicle (Me2SO) or 5μm PPPA for 30 min at 37 °C. There after cells were pulse-labeled with 1 μCi of [3H]oleic acid for 2 h. The incorporation of [3H]oleic acid into cellular CE pool was measured as the determinant of the enzymatic activity of the respective proteins. Background activity was obtained by a parallel kinetic assay where cells were transfected with an empty vector. All activities were corrected by background subtraction and were normalized against the control (Ctrl) WT activity. Data represent the mean ± S.E. for n = 2. This experiment was repeated three times with similar results. C, kinetic assay data for of WTA2 and A2:1–363. The assay was performed essentially as described above, and data are presented as above. D, PNS made from cells transfected with WTA1 and A1:1–385 cDNAs were subjected to immunoblot using affinity-purified ACAT1 antibody as described under “Experimental Procedures.” E, PNS, made from the cells transfected with WTA2 and A2:1–363 cDNAs, were subjected to Western blot analysis as described above.
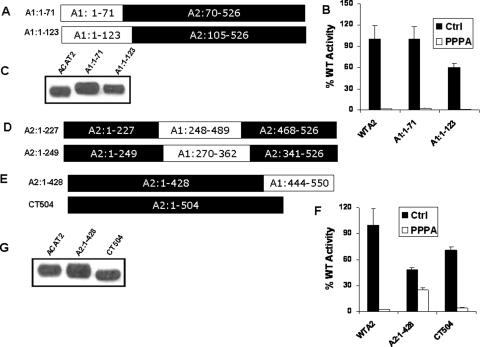
PPPA-sensitive Site Is Located between a Region Located within Amino Acids 428–504—The sequence similarity among the first 100 amino acids of the ACAT enzymes is only 2%, suggesting this region might be a good candidate region for the PPPA-sensitive site. We made two chimeras, A1:1–71 and A1:1–123, to replace the entire N terminus of ACAT2 with that of ACAT1 (Fig. 2A). Both of these chimeras retained cholesterol esterification activity with normal expression levels when transfected into AC-29 cells (Fig. 2, B and C). However, in the presence of PPPA, activity of both the chimeras was fully inhibited, indicating the PPPA-sensitive site is not located within the N-terminal sequences of ACAT2.
FIGURE 2.
PPPA-sensitive site of ACAT2 is located near the C-terminal end of the enzyme. A, amino acid sequences of the chimeras A1:1–71 and A1:1–123 are represented as in Fig. 1A. B, kinetic assay data for WTA2, A1:1–71, and A1:1–123 proteins. Assays were done as described in Fig. 1B. Data represent the mean ± S.E. for n = 2, and the experiment was repeated twice with similar results. C, PNS obtained from the cells transfected with WTA2, A1:1–71, and A1:1–123 cDNAs were subject to immunoblotting as described under “Experimental Procedures.” Primary antibodies used here were as follows. Affinity-purified ACAT2 antibody was used to detect WTA2 band, whereas affinity-purified ACAT1 antibody was used for A1:1–71 and A1:1–123 proteins. D, primary sequences of the chimeras A2:1–227 and A2:1–249 are indicated as Fig. 1A. E, amino acid sequence of the chimera A2:1–428- and ACAT2-C-terminal truncated mutant CT504. F, kinetic activity of WTA2, A2:1–428, and CT504. The enzymatic assay was performed as described in Fig. 1B. Data represent mean ± S.E. for n = 2. This experiment was repeated three times with similar outcomes. Ctrl, control. G, PNS obtained from the cells transiently transfected with WTA2, A2:1–428, and CT504 cDNAs were used for Western blotting.
Our experimentally obtained topology model of ACAT enzymes shows both have Ncyto Cexo orientation with five potential transmembrane domains (19). The computer-predicted transmembrane domain D was not utilized, whereas domain F was utilized in ACAT2; the opposite was true for ACAT1 (supplemental Fig. A). Transmembrane domains with the intervening letters were likewise not utilized in either enzyme (19). Because of this difference in membrane topology, a large part of the ACAT1 sequence (amino acids 265–502) is predicted to be located on the cytoplasmic side of the membrane where the analogous sequence from amino acids 222–344 of ACAT2 is predicted to be on the luminal side of ER. To test whether the PPPA-sensitive site was located within this region, we made two different chimeras. For A2:1–227 we swapped the entire sequence of this region of ACAT2 with the analogous region of ACAT1, and for A2:1–249 we swapped only the segment of this region whose subcellular localization was to be opposite among the enzymes (Fig. 2D). However, when we transfected these chimeras into AC-29 cells, they were not expressed perhaps due to the more gross structural changes in this region of the proteins.
Next, we made chimera A2:1–428 and CT 504, a C-terminal truncated version of ACAT2 where last 22 amino acids were removed from ACAT2 sequence (Fig. 2E). Transfection of the cDNAs encoding each of these two modified proteins shows both have cholesterol esterification activity (Fig. 2F), and their expression levels were comparable with the WT ACAT2 (Fig. 2G). Surprisingly, A2:1–428 showed only 50% inhibition of ACAT activity under PPPA treatment, suggesting the PPPA-sensitive site is located after amino acid 428. However, the activity of CT504 was nearly completely inhibited by PPPA in similar fashion to WT ACAT2. Thus, these two versions of modified ACAT2 helped narrow down the PPPA interaction site to be within the region located between amino acids 428 and 504.
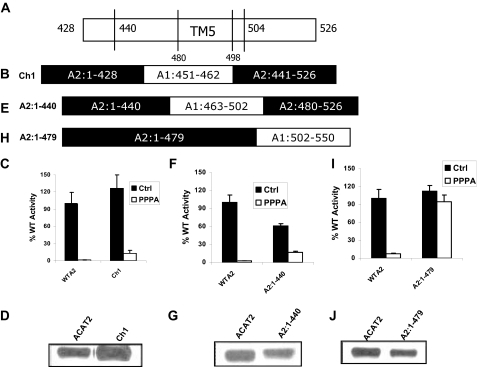
The PPPA-sensitive Site Is Located within the Fifth Transmembrane Domain of ACAT2—The region between amino acids 428 to 504 of ACAT2 contains part of the putative active site domain and the fifth transmembrane domain of the enzyme. We can divide this region into three segments as follows; 1) amino acids 428–440, the C-terminal boundary of the putative active site domain of the enzyme; 2) amino acids 480–504, which contain the fifth transmembrane domain; 3) amino acids 440–480, a region located in between the putative active site domain and the fifth transmembrane domain of the enzyme (Fig. 3A). In chimera Ch1 we replaced the 428–440 segment of ACAT2 with its ACAT1 counterpart (Fig. 3B). Ch1 was expressed as an active enzyme (Fig. 3C) with normal levels of expression (Fig. 3D); however, its response to PPPA was similar with that of the WT protein, suggesting the PPPA-sensitive site is not located within the sequence 428–440 of ACAT2. We then swapped the sequence between amino acids 440 and 480 of ACAT2 with the analogous sequence of ACAT1 in the chimera A2:1–440 (Fig. 3E). This chimera was active (Fig. 3F) and had normal protein expression level (Fig. 3G). PPPA also inhibits activity of this chimera, although the inhibition was 80% as oppose to 95% in the WT enzyme. Finally, we made chimera A2:1–479, where after residue 479 the remainder of the ACAT2 sequence was replaced by ACAT1 sequence (Fig. 3H). This chimera was enzymatically active (Fig. 3I), and its expression level was comparable with the WT protein (Fig. 3J). However, this chimera was completely insensitive to PPPA inhibition. Because we have already demonstrated that the sequence after residue 504 is not important for PPPA selective inhibition of ACAT2 (Fig. 2F), we concluded that the PPPA-sensitive site was located within a region between amino acid residues 480 and 504, which includes the putative fifth transmembrane domain.
FIGURE 3.
PPPA-sensitive site is located within the fifth transmembrane domain of ACAT2. A, amino acid sequence representation for ACAT2 within the region 428–526 indicating the 5th transmembrane domain with amino acids 480–498 (TM5) near the C-terminal end of the enzyme. B, amino acid sequence of the chimera Ch1 represented as Fig. 1A. C, the kinetic assay data WTA2 and Ch1. The assay was performed as described in Fig. 1A. Data represent the mean ± S.E. for n = 2. This experiment was repeated twice with similar results. D, PNS obtained from WTA2- and Ch1-transfected cells were used for Western blotting using the protocol as described under “Experimental Procedures.” E, amino acid sequence of the chimera A2:1–440 indicated as Fig. 1A. F, the kinetic activity data for WTA2 and the chimera A2:1–440. The assay was performed as described for Fig. 1B. The assay was repeated twice with identical outcome. Data represent the mean ± S.E. for n = 2. G, Western blot analysis was performed using PNS obtained from WTA2- and A2:1–440-transfected AC29 cells. H, amino acid sequence representation of chimera A2:1–479 indicated as Fig. 1A. I, kinetic activity assay data for WTA2 and A2:1–479. The assay was performed as described in Fig. 1B. Data represent the mean ± S.E. for n = 4 of two independent experiments. J, PNS obtained from WTA2 and A2:1–479-transfected cells Western blotted as described under “Experimental Procedures.”
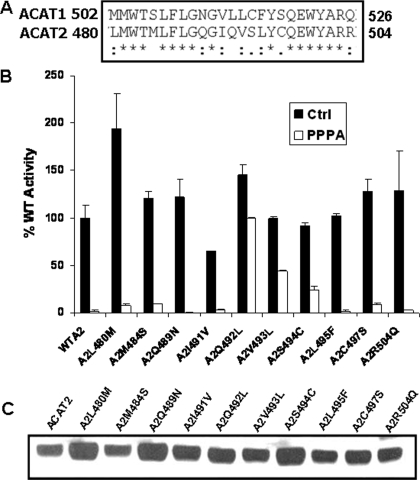
Residues 492, 493, and 494 Constitute the PPPA Interactive Site of ACAT2—To further narrow down the PPPA-sensitive site of ACAT2, we performed sequence alignment of ACAT2 481–504 sequence with the analogous sequence of ACAT1 (Fig. 4A). Ten amino acids within this region were different between two enzymes. We speculated that because PPPA is an ACAT2-specific inhibitor, the PPPA-sensitive site must correspond to 1 or more of these 10 amino acids. Thus, we made the 10 single amino acid mutants of ACAT2 where each amino acid was mutated to its ACAT1 counterpart. All the mutants were active (Fig. 4B) and had comparable expression levels to the WT ACAT2 protein (Fig. 4C). Interestingly, among the 10 mutant proteins, A2Q492L, A2V493L, and A2S494C had only 30, 50 and 70% of the inhibition in enzymatic activity due to PPPA, respectively. More than 90% inhibition the activity was observed with PPPA for all the other mutants along with the WT enzyme. These results clearly suggest Gln-492 is required for PPPA selective inhibition of ACAT2. In addition, Val-493 and Ser-494 may also be required for PPPA-selective inhibition of ACAT2 perhaps by providing steric interactions around Gln-492 residue.
FIGURE 4.
Identification of the residues responsible for PPPA-mediated inhibition of ACAT2. A, amino acid sequence alignment of the PPPA-sensitive region of ACAT2 with its ACAT1 counterpart. This region represents the fifth transmembrane domain of both enzymes. The asterisk represents sequence identity, the colon represents conserved substitutions, the dot represents semi-conserved substitutions, and no symbol indicates no match. The alignment was performed using ClustalW program from EMBL web site. B, enzymatic assay data of each of the single amino acid mutants where the amino acid in ACAT2 is replaced with the analogous amino acid in ACAT1 as indicated for the x axis. AC29 cells were transfected with the cDNA encoding either WTA2 or the various single amino acid mutants of ACAT2. Assays were done as indicated in Fig. 1B. Data represent the mean ± S.E. for n = 2. This experiment was repeated twice with similar results. C, Western blot analysis of the WT and each of various mutant ACAT2 proteins as indicated, performed as described under “Experimental Procedures.”
Kinetic Properties of PPPA-insensitive Recombinants—It is possible that the mutant and chimeric enzymes might have altered kinetic properties compared with the WT enzyme that contribute to the loss of PPPA sensitivity. To test this possibility, we determine the apparent Vmax and Km values for PPPA-insensitive recombinants and WT enzyme using the microsomal ACAT assay (Table 1). Our results indicate the Vmax value was highest for WT enzyme, whereas the various recombinants had lower but comparable maximum reaction velocities. Likewise, the apparent substrate affinity (Km) of A2:1–428, A2:1–479, and A2S494C was comparable with that for the WT enzyme. Although the apparent Km of A2Q492L averaged about 2-fold higher than WT ACAT2 and the value for A2V493L was about 50% higher, these were not statistically significant differences, so that the substrate concentration requirement for each of the enzyme constructs remained similar. Furthermore, to determine whether CE synthesis rates in whole cells of these recombinants may have been altered due to PPPA treatment, we performed whole cell-based ACAT assays that were terminated at different time intervals (1, 2, 4, and 8 h). For each recombinant, we then compare the plot of CE synthesis rate (y axis CE synthesis versus x axis time) in the presence of either Me2SO vehicle or PPPA. Our result showed that the shape of the curves remained similar for up to 4 h regardless of vehicle or inhibitor treatment (data not shown) so that the 2-h time point used for most comparisons was part of an apparent linear first order response. All together these results suggest that the kinetic characteristics of the recombinant enzymes were quite similar to those of the wild type enzyme and should not confound the interpretation of PPPA-induced inhibition.
TABLE 1.
Kinetic properties of PPPA-insensitive chimeras and mutants
Microsomes were prepared from AC-29 cells that had been transiently transfected with either WT or various mutant and chimeric constructs of ACAT2 protein as indicated below. For the assay, 50 μg of microsomal protein was used for measurement of ACAT activity by the method described under “Experimental Procedures,” which was run at four different [14C]oleyl-CoA (5,15,30, and 50 nmol) concentrations. Using Western blotting, expression levels of the proteins were determined. Specific activity was calculated as the dpm of cholesteryl esters formed divided by the protein mass obtained from densitometric analysis of the Western blots. Substrate saturation curves of each of the recombinant constructs were made by plotting specific activity against its corresponding [14C]oleyl-CoA concentrations. Kinetics parameters were calculated using Hyperbolic Regression Analysis software. This program employs nonlinear regression to calculate kinetic parameters as shown earlier (17, 24). Each assay was run in duplicate.
| Clone | Vmax | Km |
|---|---|---|
| Arbitrary units | nm | |
| ACAT2 | 4237 ± 800 | 5.53 ± 4.44 |
| A2:1-428 | 970.8 ± 198.3 | 3.33 ± 3.71 |
| A2:1-479 | 707.1 ± 75.39 | 2.66 ± 1.75 |
| A2Q492L | 1268 ± 178.3 | 12.18 ± 5.14 |
| A2V493L | 1400 ± 198.2 | 8.094 ± 4.11 |
| A2S494C | 1371 ± 386.6 | 5.87 ± 6.85 |
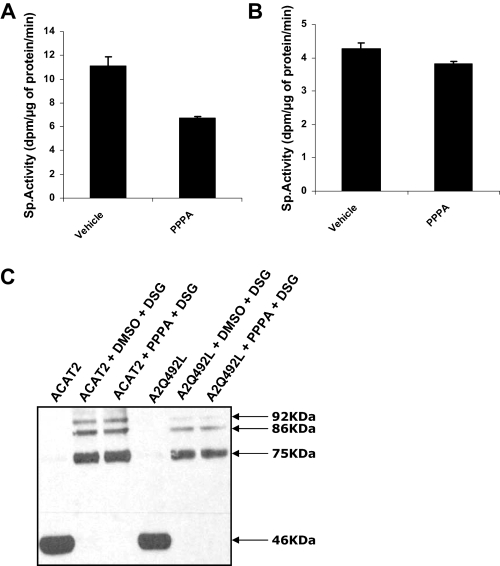
Nature of Interaction of PPPA with ACAT2—Although PPPA inhibits ACAT2 activity, the mechanism of this was not known. Because PPPA can inhibit the activity of the enzyme in a cell-free system (microsomal assay), it is possible that it directly interacts with the enzyme to inhibit its activity. Because of the lack of a purified protein, we were not able to directly test this hypothesis; however, we performed two experiments to assess the nature of the protein-inhibitor interaction. In the first experiment, ACAT2 stable cells were incubated with the vehicle (Me2SO) or 5 μm of PPPA in a 150-mm cell culture dish, and microsomes were then prepared from these cells. ACAT assay was performed thereafter with the microsomes, and only 35% inhibition was achieved with microsomes isolated from PPPA-treated cells (Fig. 5A). In another experiment, microsomes were prepared from ACAT2 stably transfected cells and were subsequently incubated with vehicle or 5 μm PPPA at 37 °C for 30 min. Microsomes were then reisolated, and ACAT activity assay was performed again (Fig. 5B). There was almost no inhibition remaining for the activity of the PPPA-treated samples compared with the vehicle-treated microsomes. We concluded that PPPA interaction with the protein is likely non-covalent. Of note, the specific activity ACAT2 was lower in reisolated microsomes (Fig. 4B compared with Fig. 4A). We suspect that during reisolation of the microsomes by ultracentrifugation, we lost some proteins; thus, there was a reduction in ACAT activity.
FIGURE 5.
Nature of interaction of PPPA with ACAT2 and its effects on the oligomeric state of the protein. A, microsomes were prepared from ACAT2 stable cells incubated with either Me2SO (DMSO) or 5 μm PPPA for 30 min at 37 °C. 50 μg of Me2SO-treated microsomes and 25 μg of PPPA-treated microsomes were used to do a microsomal ACAT assay (described in detail under “Experimental Procedures”). Specific activity was calculated by normalizing activity as dpm of cholesteryl esters formed with the protein mass and the assay run time. Data represent the mean ± S.E. for n = 2. This experiment was repeated twice with similar results. B, 50 μg of ACAT2 stable cell microsomes were incubated with either Me2SO or 5 μm PPPA at 37 °C. This was followed by reisolation of the microsomes by centrifuging at 100 K for 15 min. Microsomal ACAT assay was then performed with the reisolated microsomes. Specific activity was calculated as above. Data represent the mean ± S.E. for n = 2. This experiment was repeated three times with similar result. C, 10 μg of protein in ACAT2 stable cell microsomes and 20 μg of protein in microsomes obtained from AC29 cells transiently transfected with A2Q492L mutant were treated with either Me2SO or 5 μm PPPA at room temperature for 30 min. Samples were then treated with vehicle or disuccinimidyl glutarate (DSG) for another 30 min at room temperature. The cross-linking reaction was quenched by adding Western loading buffer containing 1 m Tris, pH 7.5, to the reaction, and all samples were subjected to Western blotting as described under “Experimental Procedures.” As a control ACAT2 and A2Q492L was run on the gel without any treatment (1st and 4th lane on the gel). Apparent molecular weight of the bands is shown on the right side of the gel.
It has been proposed that ACAT1 can form oligomers in vitro and in intact cells (18). It is possible that this oligomeric state is required for the activity of the enzyme. Because both the ACAT enzymes share a high sequence similarity and catalyze the same chemical reaction, it is possible that ACAT2 also forms a higher order oligomeric state during catalysis. We hypothesized that PPPA interaction may disrupt the oligomeric state of the protein, thereby inhibiting the activity of the enzyme. To test this hypothesis, we performed a cross-linking experiment using disuccinimidyl glutarate, a homobifunctional non-cleavable 7.7-Å arm-length cross-linker. Microsomes prepared from the WT or A2Q492L mutant were incubated with either Me2SO or PPPA followed by another round of incubation with disuccinimidyl glutarate. The oligomeric state of the proteins was visualized by Western blotting (Fig. 5C). This result suggests that PPPA did not alter the overall oligomeric structure of the protein as both the WT and the mutant protein forms similar cross-linked oligomeric forms regardless of vehicle or inhibitor treatment.
DISCUSSION
PPPA is a selective inhibitor of ACAT2 showing 2000-fold higher specificity for inhibition of ACAT2 than ACAT1. In this report we sought to determine the PPPA interaction site within the ACAT2 enzyme protein. We employed a chimeric approach with the hypothesis that if the PPPA-sensitive site of ACAT2 is replaced by the analogous site of ACAT1, the chimeric enzyme will not be inhibited by PPPA. With this approach we identified an ACAT2 sequence, amino acids 480–504, the fifth transmembrane domain of the enzyme, which contains the amino acid sequence responsible for PPPA selectivity of ACAT2. Based on sequence alignment within the fifth transmembrane domain of the two ACAT enzymes, we performed single amino acid mutations where each of the 10 substituted amino acids in ACAT2 was individually changed to its ACAT1 counterpart. Substitution of glutamine 492 with leucine was the specific substitution that eliminated PPPA specificity in ACAT2 inhibition (Fig. 4B). Some loss of selectivity was also seen when the adjacent valine and serine were substituted with leucine and cysteine, respectively. We assume the latter substitutions may be related to steric hindrance surrounding the glutamine residue. In any case the data are clear that PPPA selectivity for ACAT2 inhibition requires Gln-492, which lies within the fifth putative transmembrane domain of ACAT2.
One major problem working with chimeric enzymes is that there might be an altered structure of the protein generated while making the chimera because of the significant changes in its amino acid sequences. Although we cannot completely rule out this possibility for all the chimeras we made, even if there were changes in the structure, it did not affect the level of expression nor the activity or apparent Km for most of the chimeras. Thus, most of our chimeras were comparable in catalytic activity so that we were able to test their activity in the presence of PPPA. When we made A2:1–227 and A2:1–249, there was likely a significant alteration in the three-dimensional structure of the protein possibly as a result of alteration in transmembrane domain utilization. As a result, when attempts to express these two chimeras in AC-29 cells were made, no detectable expression resulted perhaps because misfolded proteins resulted that were rapidly degraded within the cells. In the absence of crystal structure, the chimeric approach to study some of the structural features of ACAT isozymes has proven helpful. We believe this method could also be useful for other proteins where there is a need to document unique isozyme-specific characteristics.
To design the chimeras, we used the ACAT topology model generated by our laboratory (19). To assess membrane topology, a glycosylation reporter gene and FLAG epitope tag sequence was appended to a series of ACAT1 and ACAT2 cDNAs that had been truncated after each of the computer-predicted transmembrane domains. An in vitro translation system was used to express the truncated mutants. Based on glycosylation site utilization and accessibility to exogenous protease, we concluded that both the enzymes span the membrane bi-layer five times with the N-terminal end on the cytoplasmic side and the C-terminal end on the luminal side of the ER membrane. The strength of this approach was that both enzymes were analyzed in concert, so that differences could be detected. Using a different approach, T. Y. Chang and co-workers (20, 21) have predicted ACAT1 has seven transmembrane domains, whereas ACAT2 has only two. Using AC-29 cells they expressed modified human ACAT enzymes in which an epitope tag was inserted after each of the computer-predicted transmembrane domains of the enzyme. Recently, Guo et al. (22) have refined their previous model using a cysteine-scanning mutagenesis approach and reported that human ACAT1 spans the membrane bi-layer nine times. It is important to mention that all methods for membrane topology assessment rely upon modifications of the protein which can change utilization of particular transmembrane domains, depending upon charge density of the amino acid sequences immediately after the transmembrane domain. Thus, it is difficult to ascertain the correct model with certainty. However, when two enzymes are as similar in their amino acid sequence as ACAT1 and ACAT2 and both catalyze the same reaction, it seems unlikely that membrane topology will be so different as having nine transmembrane domains in ACAT1 and only two in ACAT2. The fact that the chimeric enzyme approach used here worked so well also supports the notion that a greater degree of similarity exists between the two enzymes than seven or nine versus two transmembrane domains.
Our evidence shows that Gln-492 is not an active site residue nor is it located within the substrate binding pocket of ACAT2 because when we mutate this residue to leucine, the enzyme is still active, indicating that this residue is not required for activity. This raises the important question, If Gln-492 is not essential for activity of the enzyme, why then does its presence together with the inhibitor affect the activity of the enzyme? According to our topology model, Gln-492 is located within the fifth transmembrane domain of the enzyme. Interestingly, Guo et al. (22) also show that the localization of Leu-514, the analogous residue of Gln-492 in human ACAT1, is localized within a transmembrane domain. Assuming that localization of the Gln-492 and its surrounding residues is indeed within a transmembrane domain of the enzyme, we can speculate about a probable molecular mechanism of inhibition of ACAT2 in the presence of PPPA. Analogous to the proposal for ACAT1, it is possible that ACAT2 also forms a higher order oligomeric structure in vivo that is associated with activity of the enzyme. In fact our cross-linking experiments provide evidence that a higher order oligomeric structure of the WT ACAT2 may occur (Fig. 5C). Transmembrane domains of membrane-bound polytopic proteins often take part in the self-association of the proteins. We speculate that more than one transmembrane domain including the fifth transmembrane domain of ACAT2 is required for forming an oligomeric structure of the protein and that such oligomerization is required for the activity of the enzyme. When PPPA interacts with the enzyme at the Gln-492 residue, the oligomeric structure of the protein may be disrupted to the extent that some of the associations among the transmembrane domain regions of the enzyme are not efficient. This disruption could have the effect of altering the cytoplasmic side conformation near the active site (located just upstream of the fifth transmembrane domain containing Gln-492 in the three-dimensional structure of the protein), resulting in enzyme inhibition. If PPPA interacts only with the fifth transmembrane domain of the enzyme, the oligomerization among the other transmembrane domains might well remain unaffected so that, in our cross-linking experiment, we still see a similarity in apparent higher order oligomeric structure of the WT ACAT2 protein in the presence of PPPA. The hydrophobic nature of the PPPA molecule seems consistent with the possibility that it fits into the membrane where it exerts its effect. All of these speculations are currently under investigation in our laboratory.
In conclusion, in this report we have identified the putative fifth transmembrane domain as the PPPA-sensitive site of ACAT2. Furthermore, we have shown Gln-492 within this transmembrane domain is the primary residue responsible for PPPA interaction within the ACAT2 enzyme protein that results in inactivation of the enzyme. We have repeatedly shown through our animal model studies that liver-specific inhibition of ACAT2 may be beneficial for the treatment of atherosclerosis. Although the molecular mechanism of PPPA-mediated ACAT2 inhibition is not yet clearly understood, our present biochemical study definitely shows that by targeting the fifth transmembrane domain of ACAT2, especially in the region surrounding Gln-492, it is feasible to selectively inhibit ACAT2. These data suggest that specific drug molecules with similarities to PPPA might accomplish a similar selectivity, providing encouragement that selective ACAT2 inhibition is feasible as well as desirable (23).
Supplementary Material
This work was supported by National Institutes of Health Grant NIH-P01-HL49373 (to L. L. R.) and Kakenhi Grant 16073215 from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to H. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. A–C.
Footnotes
The abbreviations used are: ACAT, acyl-CoA:cholesterol acyltransferase (accepted name, sterol o-acyltransferase); AGM, African green monkey; CE, cholesteryl ester; ER, endoplasmic reticulum; PPPA, pyripyropene A; WT, wild type; PNS, post-nuclear supernatant; Bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
S. Sturley, personal communication.
References
- 1.Buhman, K. F., Accad, M., and Farese, R. V. (2000) Biochim. Biophys. Acta 1529 142–154 [DOI] [PubMed] [Google Scholar]
- 2.Rudel, L. L., Lee, R. G., and Cockman, T. L. (2001) Curr. Opin. Lipidol. 12 121–127 [DOI] [PubMed] [Google Scholar]
- 3.Lee, R. G., Willingham, M. C., Davis, M. A., Skinner, K. A., and Rudel, L. L. (2000) J. Lipid Res. 41 1991–2001 [PubMed] [Google Scholar]
- 4.Parini, P., Davis, M., Lada, A. T., Erickson, S. K., Wright, T. L., Gustafsson, U., Sahlin, S., Einarsson, C., Eriksson, M., Angelin, B., Tomoda, H., Omura, S., Willingham, M. C., and Rudel, L. L. (2004) Circulation 110 2017–2023 [DOI] [PubMed] [Google Scholar]
- 5.Rudel, L. L., Parks, J. S., Hedrick, C. C., Thomas, M., and Williford, K. (1998) Prog. Lipid Res. 37 353–370 [DOI] [PubMed] [Google Scholar]
- 6.Carr, T. P., Parks, J. S., and Rudel, L. L. (1992) Arterioscler. Thromb. 12 1274–1283 [DOI] [PubMed] [Google Scholar]
- 7.Rudel, L. L., Davis, M., Sawyer, J., Shah, R., and Wallace, J. (2002) J. Biol. Chem. 277 31401–31406 [DOI] [PubMed] [Google Scholar]
- 8.Meiner, V. L., Cases, S., Myers, H. M., Sande, E. R., Bellosta, S., Schambelan, M., Pitas, R. E., McGuire, J., Herz, J., and Farese, R. V., Jr. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 14041–14046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yagyu, H., Kitamine, T., Osuga, J., Tozawa, R., Chen, Z., Kaji, Y., Oka, T., Perrey, S., Tamura, Y., Ohashi, K., Okazaki, H., Yahagi, N., Shionoiri, F., Iizuka, Y., Harada, K., Shimano, H., Yamashita, H., Gotoda, T., Yamada, N., and Ishibashi, S. (2000) J. Biol. Chem. 275 21324–21330 [DOI] [PubMed] [Google Scholar]
- 10.Buhman, K. K., Accad, M., Novak, S., Choi, R. S., Wong, J. S., Hamilton, R. L., Turley, S., and Farese, R. V., Jr. (2000) Nat. Med 6 1341–1347 [DOI] [PubMed] [Google Scholar]
- 11.Willner, E. L., Tow, B., Buhman, K. K., Wilson, M., Sanan, D. A., Rudel, L. L., and Farese, R. V., Jr. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1262–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, R. G., Kelley, K. L., Sawyer, J. K., Farese, R. V., Jr., Parks, J. S., and Rudel, L. L. (2004) Circ. Res. 95 998–1004 [DOI] [PubMed] [Google Scholar]
- 13.Fazio, S., Major, A. S., Swift, L. L., Gleaves, L. A., Accad, M., Linton, M. F., and Farese, R. V., Jr. (2001) J. Clin. Investig. 107 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell, T. A., III, Brown, J. M., Graham, M. J., Lemonidis, K. M., Crooke, R. M., and Rudel, L. L. (2006) Arterioscler. Thromb. Vasc. Biol. 26 1814–1820 [DOI] [PubMed] [Google Scholar]
- 15.Sliskovic, D. R., Picard, J. A., and Krause, B. R. (2002) Prog. Med. Chem. 39 121–171 [DOI] [PubMed] [Google Scholar]
- 16.Lada, A. T., Davis, M., Kent, C., Chapman, J., Tomoda, H., Omura, S., and Rudel, L. L. (2004) J. Lipid Res. 45 378–386 [DOI] [PubMed] [Google Scholar]
- 17.Duggleby, R. G. (1981) Anal. Biochem. 110 9–18 [DOI] [PubMed] [Google Scholar]
- 18.Yu, C., Chen, J., Lin, S., Liu, J., Chang, C. C. Y., and Chang, T. Y. (1999) J. Biol. Chem. 274 36139–36145 [DOI] [PubMed] [Google Scholar]
- 19.Joyce, C. W., Shelness, G. S., Davis, M. A., Lee, R. G., Skinner, K., Anderson, R. A., and Rudel, L. L. (2000) Mol. Biol. Cell 11 3675–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, S., Lu, X., Chang, C. C. Y., and Chang, T. Y. (2003) Mol. Biol. Cell 14 2447–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, S., Cheng, D., Liu, M. S., Chen, J., and Chang, T. Y. (1999) J. Biol. Chem. 274 23276–23285 [DOI] [PubMed] [Google Scholar]
- 22.Guo, Z. Y., Lin, S., Heinen, J. A., Chang, C. C., and Chang, T. Y. (2005) J. Biol. Chem. 280 37814–37826 [DOI] [PubMed] [Google Scholar]
- 23.Rudel, L. L., Lee, R. G., and Parini, P. (2005) Arterioscler. Thromb. Vasc. Biol. 25 1112–1118 [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson, G. N. (1961) Biochem. J. 80 324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.