Abstract
Here we investigate the effects of a myopathy-causing mutation in αB-crystallin, Q151X, upon its structure and function. This mutation removes the C-terminal domain of αB-crystallin, which is expected to compromise both its oligomerization and chaperone activity. We compared this to two other αB-crystallin mutants (450delA, 464delCT) and also to a series of C-terminal truncations (E164X, E165X, K174X, and A171X). We find that the effects of the Q151X mutation were not always as predicted. Specifically, we have found that although the Q151X mutation decreased oligomerization of αB-crystallin and even increased some chaperone activities, it also significantly destabilized αB-crystallin causing it to self-aggregate. This conclusion was supported by our analyses of both the other disease-causing mutants and the series of C-terminal truncation constructs of αB-crystallin. The 450delA and 464delCT mutants could only be refolded and assayed as a complex with wild type αB-crystallin, which was not the case for Q151X αB-crystallin. From these studies, we conclude that all three disease-causing mutations (450delA, 464delCT, and Q151X) in the C-terminal extension destabilize αB-crystallin and increase its tendency to self-aggregate. We propose that it is this, rather than a catastrophic loss of chaperone activity, which is a major factor in the development of the reported diseases for the three disease-causing mutations studied here. In support of this hypothesis, we show that Q151X αB-crystallin is found mainly in the insoluble fraction of cell extracts from transient transfected cells, due to the formation of cytoplasmic aggregates.
The crystallins were first identified as the major proteins of the eye lens and their subsequent classification into α-, β-, and γ-crystallins (1) allowed different functional properties to be assigned to the α- and βγ-crystallin groups (2). The α-crystallins are two distinct proteins derived from two separate genes, αA- and αB-crystallin (2). Whereas αA-crystallin is almost entirely lens specific (3), αB-crystallin is expressed widely in other tissues, most notably astrocytes (4) and muscle (5). In the early 1980s, pioneering work from Klemenz and co-workers (6) and Horwitz (7) laboratories established that αB-crystallin was a protein chaperone and a member of the small heat shock protein (sHSP)2 family, that is now known to comprise 10 different human proteins (8).
When the first mutation (R120G) in αB-crystallin was reported, it was found to cause both cataract and desmin-related myopathy in the affected patients (9). Characteristic histopathological aggregates of the muscle intermediate filament protein desmin were observed (9), strongly suggesting that there is an important functional interaction between intermediate filaments and αB-crystallin. Subsequent analyses showed that the R120G αB-crystallin mutation induced increased binding for desmin intermediate filaments (10), providing an explanation for the formation of the desmin aggregates in the muscles of the affected individuals. Coincidentally, mutations in desmin also induced filament aggregation (11, 12) and these aggregates also contained αB-crystallin (12). Collectively, these data suggest that one of the functions of αB-crystallin is to chaperone intermediate filaments and their networks in cells (10, 13, 14).
Subsequently, three other mutations in the C-terminal extension were reported for αB-crystallin, but in contrast to the R120G mutation, none caused both cataract and myopathy. The first of these was the 450delA mutation that is associated only with cataract and resulted in a 184-residue product where the coding sequence of the C-terminal extension was altered from residue 150 onwards (15). This region is distal to the β-strand 9 and proximal to the conserved hydrophobic IX(I/V) motif (16) that forms part of β-strand 10 in the crystal structure of the archaeal HSP16.5 (17). Two other mutations were then reported (Q151X and 464delCT) that cause myopathy and not cataract (18) that either truncated (Q151X) or substituted the C-terminal extension from residue 155 (464delCT). The fact that all three mutations delete a known substrate interaction site (157–164) (19) and a region important in the oligomerization of αB-crystallin itself (155–166) (20) support the hypothesis that this region in αB-crystallin is important to both the structure and function of the protein. Very recently, a C-terminal point mutation (R157H) (21) was reported, lending further support to this conclusion. Thus far, the effect of such extensive deletions (Q151X) combined with altered C-terminal sequences (450delA and 464CT) upon αB-crystallin structure and function have not been investigated.
In other sHSPs, the C-terminal domain has been shown to be very important to both oligomerization and chaperone function and therefore the expectation is that the C-terminal extension mutations would similarly affect αB-crystallin. Removal of the 17 most C-terminal residues of αA-crystallin reduced its chaperone activity and induced the formation of larger oligomers (22). In two crystal structures of sHSPs (HSP16.5 (17) and HSP16.9 (23)), the C-terminal extension makes contact with a hydrophobic patch on neighboring protein subunits, strengthening the dimer-dimer interaction in the oligomer (23). The sequence between β-strands 9 and 10 of the C-terminal extension contains a hinge that likely contributes to the size polydispersity of the different sHSPs by allowing packing geometries to vary while maintaining common interaction surfaces (23). Some naturally occurring sHSPs lack a C-terminal extension and in such cases the oligomerization is restricted, as seen for Tsp36 from the tapeworm Taenia saginata, which forms dimers and tetramers, but is functional as a chaperone (24). Likewise, removal of the short C-terminal extension from Methanococcus janaschii HSP16.5 resulted in reduced oligomerization, but the retention of chaperone activity (25). This is quite contrary to the closely related lens chaperone αA-crystallin that naturally co-oligomerizes with αB-crystallin in the lens. Stepwise removal of C-terminal sequences from αA-crystallin, including the REEK motif, not only reduced oligomerization, but also resulted in the sequential loss of chaperone activity (26), despite the fact that the IX(I/V) motif was retained in the mutants investigated. Mutations of lysine and glutamic acid residues in the C-terminal extension can improve chaperone activity against amyloid fibril formation providing a potential application of αB-crystallin in the treatment of amyloid-related diseases (27). In αB-crystallin, there has not been any systematic analysis of the role of the C-terminal extension and therefore no potential explanation of whether the three mutations (450delA, Q151X, and 464delCT) cause the various diseases via the loss of chaperone function or perhaps by a different mechanism.
In this study, we have analyzed the effects of the Q151X, 464delCT, and 450delA upon the oligomerization and chaperone activity of αB-crystallin. For comparison, we have also assessed the activity of a series of αB-crystallin constructs truncated at residues Lys174, Ala171, Glu165, and Glu164 to sequentially remove important regions of the C-terminal extension and therefore help us understand the effect of the Q151X mutation. With respect to the role of the C-terminal extension in oligomerization, our data show similar results to those obtained for αA-crystallin (28). Interestingly, the chaperone function of Q151X is actually increased in some in vitro chaperone assays. The ability of Q151X αB-crystallin to resist self-aggregation is, however, very greatly diminished, a trend that is further enhanced by the 464delCT and 450delA mutations, which are only stable in the presence of wild type αB-crystallin. These data show that like other sHSPs, the C-terminal extension is important for oligomerization. Its removal increases the tendency of αB-crystallin to self-aggregate.
EXPERIMENTAL PROCEDURES
Construction of αB-Crystallin Mutants
Primers (Table 1; Sigma Genosys, UK) were designed based on the mRNA sequence for wild type human αB-crystallin (DDBJ/EMBL/GenBank entry NM_001885) and were used to generate the mutants listed, using wild type αB-crystallin as a template for PCR mutagenesis. With the exception of the Q151X and E165X mutations, the amplified products were cloned into the vector pGEM®-T Easy (Promega). After verification of the sequences, the mutant αB-crystallin constructs were subcloned into the NcoI and EcoRI sites of the expression vector pET23d (Invitrogen). The Q151X and E165X mutations were introduced into pET23d by site-directed mutagenesis (Stratagene) using complementary primers (see Table 1). Constructs were sequenced and compared for fidelity to the GenBank™ data base (accession number NM001885).
TABLE 1.
Oligonucleotide primer sequence details used for introduction of the αB-crystallin mutations
The unique restriction enzyme sites incorporated into the primer design are highlighted in bold text. Q151X and E165X were introduced using site-directed mutagenesis (SDM) requiring complementary forward (5′-3′) and reverse (3′-5) primers. The 464delCT, K174X, A171X, and E164X mutant mRNA were amplified using a forward primer overlapping the pET23d insertion site and the 5′ end of the template wild type αB-crystallin. Reverse primers were complementary to the 3′ end of the template wild type αB-crystallin and incorporated the mutation and unique restriction sites to assist screening, using the available software (DNA_Analysis_Frame). A suitable restriction enzyme site was not available for the 464delCT mutation.
| Mutant αB-crystallin | Restriction enzyme site | Primer sequence (5′-3′) |
|---|---|---|
| Q151X | StuI | Forward primer: |
| gtgaatggaccaaggaaataggcctctggccctgagcgcacc | ||
| Reverse primer: | ||
| ggtgcgctcagggccagaggcctatttccttggtccattcac | ||
| E165X | HindIII | Forward primer: |
| attcccatcacccgtgaatagaagcttgctgtcaccgcagccccc | ||
| Reverse primer: | ||
| gggggctgcggtgacagcaagcttctattcacgggtgatgggaat | ||
| 464delCT | NAa | Forward primer: |
| taccatggacatcgccatccaccacc | ||
| Reverse primer: | ||
| tcacgggtgatgggaatggtgcgctcggccagagacctgtttcc | ||
| K174X | SacII | Forward primer: |
| taccatggacatcgccatccaccacc | ||
| Reverse primer: | ||
| ctagggggccgcggtgacagcaggcttc | ||
| A171X | SacI | Forward primer: |
| taccatggacatcgccatccaccacc | ||
| Reverse primer: | ||
| gggcatctatttcttgggagctcaggtgacagcaggcttctc | ||
| E164X | PstI | Forward primer: |
| taccatggacatcgccatccaccacc | ||
| Reverse primer: | ||
| cttgggagctgcggtgactgcaggcttctcttaacgggtgatgggaatggt |
NA, not applicable.
Protein Expression and Purification of αB-Crystallin Mutants
Proteins were recombinantly expressed in Escherichia coli strain BL21 (pLysS). The wild type αB-crystallin, K174X and A171X proteins were prepared essentially as described (29), but adding polyethyleneimine (final concentration 0.1% (w/v)) to remove contaminating DNA (10, 30) before purification by ion exchange chromatography on a Fractogel TMAE column. Peak fractions were pooled and then dialyzed into 20 mm Tris-HCl, 100 mm NaCl, pH 7.4, in preparation for size exclusion chromatography on a Fractogel EMD BioSEC Superformance column (60 × 1.6 cm; VWR, UK).
All other mutant αB-crystallins formed inclusion bodies, and were purified essentially as described (31). These were dissolved in 10 mm Tris-HCl, 6 m urea, 1 mm EDTA, pH 8.0, 0.2 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol and then purified by ion exchange chromatography (29) on a Fractogel TMAE column.
Fractions containing αB-crystallin were pooled and sometimes concentrated using an Amicon Ultra Centrifugal filter device 10K MWCO (Millipore, Bedford, MA). Concentrations of the purified proteins were determined using BCA Protein Assay reagent kit (Pierce) unless otherwise stated.
Refolding of αB-Crystallin Mutants following Inclusion Body Purification
In the Presence of Wild Type αB-Crystallin
The 450delA, 464delCT, and Q151X were refolded in the presence of wild type αB-crystallin. Wild type αB-crystallin was dialyzed for 16 h at 4 °C into 10 mm Tris-HCl, 6 m urea, 1 mm EDTA, pH 8.0, 1 mm dithiothreitol. Protein concentration of the wild type and mutant αB-crystallin were determined by OD280 (extinction coefficients were determined using the Emboss Pepstats program) and then mixed in a 1:1 ratio.
Diluted Refold Protocol (450delA/Wild Type, Q151X/Wild Type, E164X, and Q151X)
The protein mixture was diluted to 0.25 mg/ml in 10 mm Tris-HCl, 6 m urea, 1 mm EDTA, pH 8.0, 1 mm dithiothreitol followed by an additional 6-fold dilution to 1 m urea in refold buffer (10 mm Tris-HCl, 5 mm EDTA, 1 mm EGTA, 150 mm NaCl, pH 7.5) prior to dialysis against the same buffer for 4 h at 16 °C. The dialysate was partially concentrated using Amicon Ultra Centrifugal filter device 10K MWCO (Millipore) and centrifuged for 2 h at 4°C at 250,000 × g in the Beckman Optima MAX Ultracentrifuge MLA-80 rotor (Beckman Instruments Inc., Fullerton, CA). The supernatant was further concentrated to a suitable volume.
Undiluted Refolding Protocol (464delCT/Wild Type, E165X)
The protein mixture was dialyzed into refold buffer for 4 h at 16 °C. The dialysate was centrifuged for 2 h at 4 °C at 250,000 × g in the Beckman Optima MAX Ultracentrifuge MLA-80 rotor (Beckman Instruments Inc.) and the supernatant concentrated to give the desired protein concentration.
Characterization of αB-Crystallin Mutants
Analytical Size Exclusion Chromatography (SEC)
Purified αB-crystallin proteins were analyzed using a Superose 6 column (290 × 10 mm) at a flow rate of 0.2 ml/min at room temperature using a Merck-Hitachi Biochromatography system. The data were analyzed using Chromeleon 6.30 software (Sunnyvale, CA).
Mass Spectrometry for Molecular Weight Determination
Purified proteins were loaded onto a MassPREP desalting cartridge (Waters, Elstree, Herts, UK) to remove buffer contaminants. The protein was eluted with 70% (v/v) acetonitrile, 0.5% (v/v) formic acid at 0.2 ml/min and introduced into a Waters Q-TOF Premier mass spectrometer operating in positive electrospray mode, with a cone voltage of 40 V. Spectra were externally calibrated using sodium formate, then corrected for drift using a leucine enkephalin lockspray. Multiply charged protein ions were deconvoluted using the manufacturer-supplied MaxEnt1 software, to determine protein mass.
CD Spectroscopy
Far UV CD spectra of wild type and mutant αB-crystallin were recorded in a cuvette with 0.1-cm path length at room temperature using a Jasco J-810 spectropolarimeter. Samples were at a concentration of 0.2–0.3 mg/ml, with the exception of Q151X, assayed at 0.1 mg/ml due to the low yield of soluble protein. Samples were assayed in 10 mm Tris, 150 mm NaCl, 5 mm EDTA, 1 mm EGTA, pH 7.5. Four data sets were collected for each sample and averaged, by continuous scanning at 100 nm/min, between wavelengths 190 and 250 nm. The UV CD spectra were baseline corrected using spectra of the buffer. The CD spectra were normalized and expressed as molar ellipticity.
Thermal Stability Assay
Absorbance at 360 nm over a temperature gradient of 25–86 °C at 1 °C/min ramp rate was continuously monitored using the Beckman DU640 spectrophotometer. Samples were assayed at 0.1 mg/ml.
In Vitro Protein Chaperone Assays
Citrate Synthase—A 200-μl volume of citrate synthase from porcine heart (Sigma) was dialyzed for 16 h at 4 °C against 50 mm Tris-HCl, 2 mm EDTA, pH 8.0, and diluted to 0.49 mg/ml. The assay mixture contained citrate synthase and αB-crystallin at a 4:1 ratio of substrate:chaperone in 220 μl. Time-dependant light scattering was measured at 360 nm every 15 s over 30 min at 42 °C using a Beckman DU640 spectrophotometer.
Insulin Assay—A 20-mg aliquot of insulin from bovine pancreas (Sigma) was reconstituted in 100 mm NaSO4, 20 mm NaPO4, pH 6.9 (Insulin buffer), and solubilized by the addition of 30% (v/v) acetic acid and gentle mixing for 15 min. The insulin solution was dialyzed for 16 h at 4 °C against the insulin buffer and the dialysate was centrifuged for 1 min at 13,000 × g in an Eppendorf bench top centrifuge (5417R; Eppendorf, Hamburg, Germany). The insulin concentration was determined from the measured OD280 using its extinction coefficient ((A280/5840) × 5600 = mg/ml). Insulin was mixed with αB-crystallin at a 4:1 ratio and the change in light scattering was measured at 360 nm every 15 s over a period of 15 min at 37 °C using a Beckman DU640 spectrophotometer.
Bis-ANS Fluorescence Measurements
αB-Crystallin variant proteins were incubated in 10 mm Tris, 150 mm NaCl, 5 mm EDTA, 1 mm EGTA, pH 7.5, at a protein concentration of 1 μm and in the presence of a 10-fold molar excess of bis-ANS (4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid, purchased from Invitrogen). Samples were allowed to equilibrate for 1 h at room temperature prior to the acquisition of fluorescence spectra on a Varian Cary Eclipse fluorescence spectrophotometer. The excitation wavelength was 410 nm and emission was collected between 420 and 700 nm with excitation and emission slit widths of 5 nm and a scan speed of 600 nm/min. All spectra are the average of 10 individual scans.
In Vitro Desmin Filament Assembly and Cosedimentation Assay
In vitro assembly and cosedimentation assays were carried out essentially as described (10). Recombinant human desmin was a kind gift from Dr. Ming Der Perng (Durham, UK). Wild type or mutant αB-crystallin was mixed with desmin in low ionic strength buffer at a 1:1 mass ratio at 0.1 mg/ml. After filament assembly, samples were separated into pellet and supernatant fractions by high speed centrifugation (80,000 × g for 30 min at 20 °C in a Beckman TLS-55 rotor (Beckman Instruments Inc., Fullerton, CA)) as described (10) to pellet assembled desmin filament and the associated αB-crystallin. To investigate the ability of the three αB-crystallin mutants and the other C-terminal truncation constructs to prevent filament-filament interactions in vitro, an additional low-speed centrifugation assay was used (2500 × g for 15 min in a bench top centrifuge) as described previously (10). The pellet and supernatant fractions were analyzed by 12% (w/v) SDS-PAGE (32) and protein bands visualized by Coomassie Blue staining. The amount of protein in the supernatant and pellet fractions were analyzed by a luminescent image analyzer (LAS-1000plus; Fuji Film, Tokyo, Japan) and quantified using the Image Gauge software (version 4.0; Fuji Film).
Cell Cultures and Transient Transfection Assays
MCF7 cells were maintained as described previously (10). Wild type and mutant αB-crystallin were subcloned into the mammalian expression vector pcDNA3.1 (Invitrogen) from the bacterial expression vector pET23d, using XbaI and HindIII. Transient transfection of these cells was achieved using Gene-Juice transfection reagent (Novagen) according to manufacturer's protocol. Cells were allowed to recover for 24 h before processing for immunofluorescence microscopy, cell fractionation, and immunoblotting.
Cell Fractionation, SDS-PAGE, and Immunoblotting
Cells grown on 10-cm dishes were washed twice with phosphate-buffered saline before being lysed with 1 ml of extraction buffer (phosphate-buffered saline, 0.5% (v/v) Triton X-100) by repeatedly passing through a 25-gauge needle. Following incubation on ice for 15 min, the lysates were clarified by centrifugation 18,000 × g for 10 min at 4 °C. The resulting supernatant and pellet fractions were solubilized in equal volumes of Laemmli sample buffer before SDS-PAGE and immunoblotting analysis as described (10). Membranes were probed with a panel of purified mouse monoclonal anti-αB-crystallin 2D2B6 and monoclonal anti-HSP27 antibody, ER-D5, as described (10). Antibody labeling was detected by enhanced chemiluminescence using a luminescent image analyzer (LAS-1000plus; Fuji Photo Film (UK), London, United Kingdom).
Immunofluorescence Microscopy
Cells were processed as described previously (10). The primary antibodies used in this study were rabbit polyclonal anti-αB-crystallin, a synthetic peptide corresponding to human αB-crystallin residues 1–10 (MDIAIHHPWI) conjugated to hemocyanin (1:200; Chemicon Ab1546) and monoclonal mouse anti-keratin LE41 (1:1; CRUK). Samples were analyzed using an Axioplan fluorescence microscope (Carl Zeiss, Jena, Germany). Images were obtained using a confocal Axiovert 200M microscope and LSM 510 META software (both Zeiss, Germany). Optical sections were set to ∼1.0 μm. Images were processed and prepared for figures using Adobe Photoshop 8.0 (Adobe Systems). Quantitation of the αB-crystallin phenotypes was by visual assessment of the cells.
RESULTS
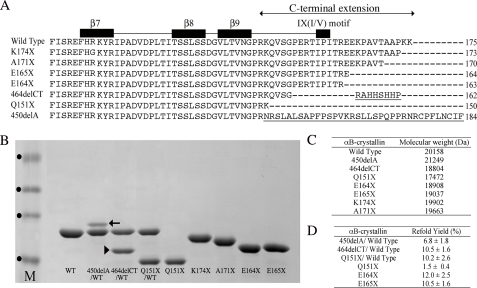
Expression and Purification of the C-terminal Extension αB-Crystallin Mutants—An overview of the C-terminal extension mutants in comparison to wild type αB-crystallin is presented (Fig. 1A). It shows the relationship of the various mutations and truncations to the structural features of the C-terminal extension and also summarizes the consequences of the two frameshift mutations, 450delA and 464delCT, that cause inherited cataract (15) and myofibrillar myopathy (18), respectively. In addition to the three disease-causing mutations (Q151X, 450delA, and 464delCT), a series of C-terminal extension truncation constructs were also expressed and purified (Fig. 1B). All the mutants except A171X and K174X were purified from inclusion bodies. The predicted mass of each of the expressed proteins was confirmed by mass spectrometry measurements (Fig. 1C). Whereas Q151X, E164X, and E165X could be refolded successfully without the addition of wild type αB-crystallin, both 450delA and 464delCT could only be refolded in its presence. When refolded with an equimolar ratio of αB-crystallin, ∼7% of 450delA and 10% of 464delCT remained soluble (Fig. 1D). Although Q151X αB-crystallin could be refolded in the absence of wild type αB-crystallin, the yield could be increased some 7-fold when the wild type protein was included. These data show the disease-causing mutations seriously affect the solubility of αB-crystallin.
FIGURE 1.
Details of the αB-crystallin mutants and their characterization by SDS-PAGE. Structural alignment of the C-terminal extensions of αB-crystallin mutants (A) includes the C-terminal extension from residue 149 (24), and β-sheets and the IX(I/V) motif are indicated as black boxes. The β7 sequence spans residues 118–123 (FHRKYR), β8 sequence spans residues 134–138 (TSSLS), and β9 sequence spans residues 142–148 (VLTVNGP). The highly conserved IX(I/V) motif includes residues 159–161 (IPI). The C-terminal extension consists of residues 149–175 (RKQVSGPERTIPITREEKPAVTAAPKK). B, characterization of the bacterially expressed and purified mutants used in this study by SDS-PAGE. The 450delA (arrow) and 464delCT (arrowhead) mutants could not be produced as individual soluble proteins and therefore were refolded in the presence of wild type αB-crystallin. Molecular weight markers (track M) are indicated (•) in order of increasing relative mobility (40, 33, 24, and 17 kDa). The relative molecular weights of the wild type and mutant αB-crystallins (C) were confirmed by mass spectrometry and compared with values calculated from the amino acid sequences using the Emboss Pepstats program. D, refolding yields for αB-crystallin truncation constructs and mutants purified from inclusion bodies.
The K174X construct removes the two most C-terminal lysine residues and would be expected to retain the secondary structural characteristics and oligomerization characteristics of αB-crystallin (33). The electrophoretic mobility of this K174X mutant was increased relative to the wild type αB-crystallin (Fig. 1B) and is in agreement with previous observations for C-terminal-deleted forms of αB-crystallin (34–37). Removal of the 5 most C-terminal residues generated the A171X mutant and is equivalent to the truncated form of αB-crystallin found in normal rat lenses (38), bovine lenses (39), and in rat models of diabetic cataracts (40). Another C-terminal-truncated form of αB-crystallin (E164X) has 12 residues removed from the C terminus and this has been detected in normal rat lenses (41). This truncation deletes the pair of glutamic acid residues at positions 164 and 165 that are part of the REEK motif in αA-crystallin that have been shown to be important in oligomerization (26). It is also equivalent to the 10-residue C-terminal-truncated form of αA-crystallin (40).
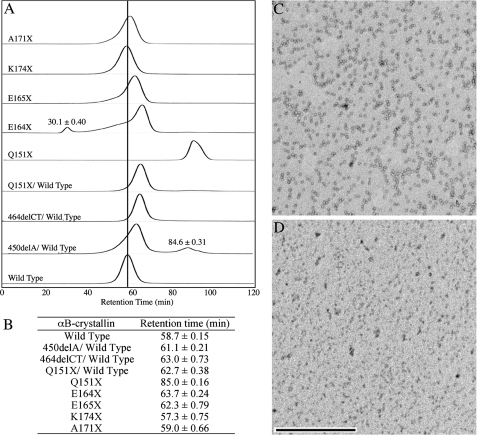
Truncation of the C-terminal Extension Reduces αB-Crystallin Oligomerization—Wild type αB-crystallin formed oligomers that were equivalent to 564 kDa, as calculated from the size exclusion chromatography data, agreeing with previously published data (42). The oligomer population was consistent between different preparations as shown by dynamic light scattering (supplementary data Fig. S1). The sequential truncation of the C-terminal extension of αB-crystallin produces a steady decrease in oligomer size (Fig. 2A). The shortest construct Q151X αB-crystallin produces a peak with very significantly increased elution volume (Fig. 2B) and represents the most dramatic reduction in oligomerization observed for all the constructs studied. This effect on Q151X oligomerization is obvious when the negative stained electron micrographs of wild type and Q151X αB-crystallin are compared (Fig. 2, C and D, respectively). Notice the absence of particles in the Q151X sample (Fig. 2D) relative to the wild type (Fig. 2C).
FIGURE 2.
Effect of the Q151X mutation upon the oligomerization of αB-crystallin. The elution profiles of the various αB-crystallins were determined on a 290 × 10-mm Superose 6 column (A). The elution characteristics of the various mutants are plotted relative to those obtained for wild type αB-crystallin (wild type), as highlighted by the vertical line indicating the retention time of the oligomeric complex, which is the major and only peak resolved in the wild type αB-crystallin sample. Additional minor species seen in E164X and 450delA/wild type are highlighted. A summary of the retention times (B) is shown for the major peak resolved during SEC of the wild typeαB-crystallin and other constructs. The retention times of minor peaks are included in the figure as appropriate. The retention times (±S.D.) were calculated from the average of three cycles of SEC. Negative stained images of wild type (C) and Q151XαB-crystallin (D) were obtained by transmission electron microscopy and illustrate the absence of characteristic particles in the Q151X sample, in agreement with the SEC data. Bar, 500 nm.
Deletion of the two C-terminal lysines (K174X) did not alter significantly the oligomerization of αB-crystallin, as previously reported (33). Removal of 5 C-terminal residues, however, reduced the apparent size of the main peak, as did the removal of both 11 (E165X) and 12 (E164X) residues. In the case of the latter two truncation mutants, there is evidence of increased polydispersity as indicated by the very noticeable shoulder on the main peak for the E165X construct and the presence of an additional peak at the void volume of the column for the E164X construct. The elution volume of the main peak is increased, suggestive of reduced mean size for αB-crystallin oligomers. These data suggest that residues Glu164-Glu165 also contribute to αB-crystallin oligomerization.
The two frameshift mutants 450delA and 464delCT remained insoluble unless they were refolded in the presence of wild type αB-crystallin. As seen from the elution profiles in Fig. 2A, the presence of these mutant proteins decreased the oligomer size of the wild type αB-crystallin. SDS-PAGE analysis (Fig. 1B) revealed that the wild type αB-crystallin is present in excess compared with 450delA αB-crystallin, yet this excess was not sufficient to retain the elution characteristics of the wild type protein. In fact, this proved to be the maximum ratio compatible with protein solubility for the 450delA-wild type αB-crystallin complex and is in stark contrast to 464delCT αB-crystallin, where a 1:1 mixture with wild type αB-crystallin was entirely soluble. The presence of the 450delA αB-crystallin also increased the polydispersity of the main peak, as seen by the shoulder and the additional small peak with a predicted molecular weight equivalent to αB-crystallin dimers.
The 450delA mutation introduces a unique 33-residue polypeptide completely unrelated to the C-terminal extension of wild type αB-crystallin (Fig. 1A) and quite clearly this frame-shift sequence has a significant effect upon the oligomerization of αB-crystallin. It is, however, the Q151X mutation that has the most dramatic effect, with the complete removal of the single peak equivalent to the 564-kDa oligomer and the appearance of a peak with elution characteristics predicted to be equivalent to a monomer. Non-denaturing gel electrophoresis also confirmed that the Q151X mutation dramatically altered its electrophoretic mobility under non-denaturing conditions in line with significantly altered oligomerization (supplementary data Fig. 2).
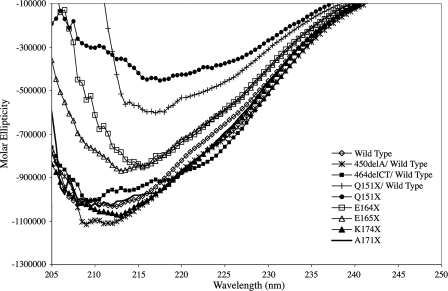
Effect of the C-terminal Mutations upon the Secondary Structure and Oligomerization of αB-Crystallin—CD spectroscopy of wild type αB-crystallin showed a single, broad minima, between 208 and 217 nm, indicative of a protein rich in β sheet (Fig. 3), which is consistent with previously reported data (42–46). The Q151X mutation had the most significant effect upon the far UV CD characteristics of αB-crystallin (Fig. 3), due to a very significant red shift of the minima to 217 nm and a ∼50% reduction in magnitude, suggestive of a significant loss of secondary structure. Similar shift patterns have been seen for C-terminal truncation mutants of αA-crystallin (26). A 1:1 molar mixture of Q151X with αB-crystallin wild type produced a spectrum that was clearly biased toward the Q151X, indicative of the dominant nature of this mutation.
FIGURE 3.
Circular dichroism of wild type αB-crystallin and the C-terminal extension mutants. Far UV CD spectra (205–250 nm) for each protein were the average of four spectra. The far UV CD signal was converted to molar ellipticity expressed as degree/cm2/dmol–1 to normalize for slight differences in molecular mass between the wild type and the other αB-crystallin constructs. The far UV CD spectra of wild type, A171X, and K174X are similar in shape and magnitude and contained broad minima between 208 and 217 nm. The 450delA/wild type and 464delCT/wild type mixtures show increased negative ellipticity and a shift in minima to 208 nm. Truncation of the C-terminal extension results in the loss of secondary structure reflected by a decrease in negative ellipticity and a shift in wavelength minima (E164X, E165X, Q151X, and Q151X/wild type), with the most dramatic effects seen for Q151X.
Of the other mutants studied, the E164X and E165X mutants were intermediate in their effects upon negative ellipticity as compared with the Q151X mutant. This intermediate effect is likely associated with the extent of the C-terminal truncations of E164X, E165X relative to Q151X, and suggests a less significant loss of secondary structure. Interestingly, the E164X mutant showed a red shift in the minimum indicating that the deletion of the second of the two glutamic acid residues in the REEK motif does introduce a detectable change in secondary structure. A recent study showed an R163X mutant with significantly altered secondary structure compared with the wild type (27). The frameshift mutation, 464delCT, introduced a small blue shift of the minimum to 208 nm and the appearance of a shoulder at ∼223 nm, both of which are indicative of increased helicity. This increase may be the result of helix formation by the novel C-terminal sequence. Both the K174X and A171X truncation mutants and the 450delA frameshift mutant have far UV CD spectra very similar to the wild type αB-crystallin.
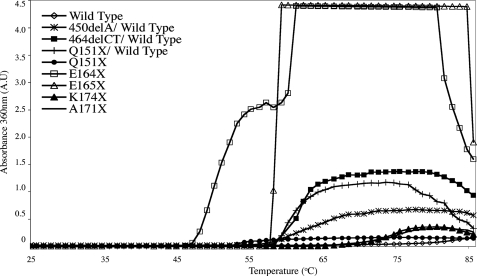
Loss in Heat Stability of αB-Crystallin Correlates with Changes in Secondary Structure—To probe the consequences of the altered secondary sequence characteristics of the αB-crystallin C-terminal extension mutants, their heat stability was determined (Fig. 4). These data show that the two most unstable mutants are E164X and E165X because both the onset and optical signal was very significantly increased compared with wild type αB-crystallin for these two mutants. The E164X mutant began to aggregate at 46 °C, some 30 °C lower than the wild type protein. The E165X mutant was notable for the rapidity of aggregation when initiated at 57 °C, whereas the Q151X mutant was notable in its failure to develop a strong OD360 signal, despite the very significant decrease in stability indicated by the increased turbidity at 53 °C. This signal reached a maximum at 55 °C and remained constant until the conclusion of the assay. The two shorter C-terminal truncations (K174X and A171X), showed decreased stability, but were the most stable of the constructs studied.
FIGURE 4.
Thermal stability of wild type and C-terminal extension mutants of αB-crystallin. The effect of the C-terminal extension mutations on the heat-induced aggregation of αB-crystallin was measured at 360 nm over the temperature range 25–86 °C. The temperature was increased at a 1 °C/min over 1 h. Wild type and mutant αB-crystallins were assayed at 0.1 mg/ml. Notice the significantly reduced stability of the E164X, Q151X, and E165X αB-crystallin mutants.
These data suggest a correlation between altered secondary structure and decreased heat stability. This trend is also seen for the two frameshift mutants (450delA/wild type and 464delCT/wild type) that were more stable than the three shortest constructs, but less stable than either the A171X or K174X truncation constructs. These data correlate with more conservative changes in secondary structure as detected by far UV CD spectroscopy for the two frameshift mutants (Fig. 3) and the largely unchanged spectra for the A171X and K174X mutants.
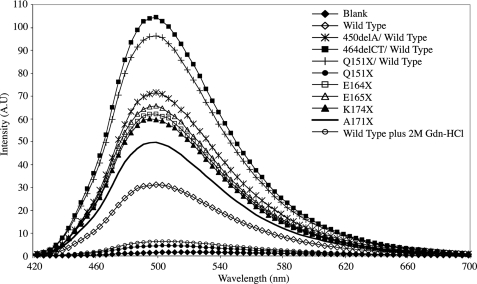
Effect of C-terminal Extension Mutants upon Exposed Hydrophobicity—The probe, bis-ANS, has a low fluorescence quantum yield in aqueous solution, however, this increases dramatically upon binding to surface-exposed hydrophobic regions. As can be seen in Fig. 5, all the αB-crystallin mutants bind more bis-ANS relative to the wild type αB-crystallin, except for the Q151X mutant, which binds substantially less. As all the mutants except Q151X αB-crystallin form oligomers (Fig. 2), we infer from these data that the increase in exposed hydrophobic surfaces arise from changes within their respective oligomeric structures. The reason why Q151X αB-crystallin binds so little bis-ANS is probably due the combined effects of decreased oligomerization and loss of secondary structure. We tested this by examining the bis-ANS binding properties of the wild type protein under mildly denaturing conditions (2 m guanidinium hydrochloride), which have been shown previously to eject αB-crystallin from native α-crystallin oligomers and to cause a significant loss of secondary structure (47). Indeed the presence of the guanidinium hydrochloride greatly reduced the bis-ANS binding to wild type αB-crystallin, with the fluorescence intensity dropping almost as low as that obtained for Q151X αB-crystallin alone.
FIGURE 5.
Bis-ANS fluorescence binding properties ofαB-crystallin mutants. The effect of the C-terminal mutations upon the relative exposed hydrophobicity was performed using bis-ANS to bind to surface-exposed hydrophobic regions on the mutant and wild type αB-crystallin. Each protein was analyzed at 1 μm in the presence of a 10-fold molar excess of bis-ANS. The excitation wavelength was 410 nm and emission spectra were collected between 420 and 700 nm. All spectra are the average of 10 individual scans. The resulting exposed hydrophobicity is increased for all mutants relative to the wild type αB-crystallin, with the exception of the Q151X mutant.
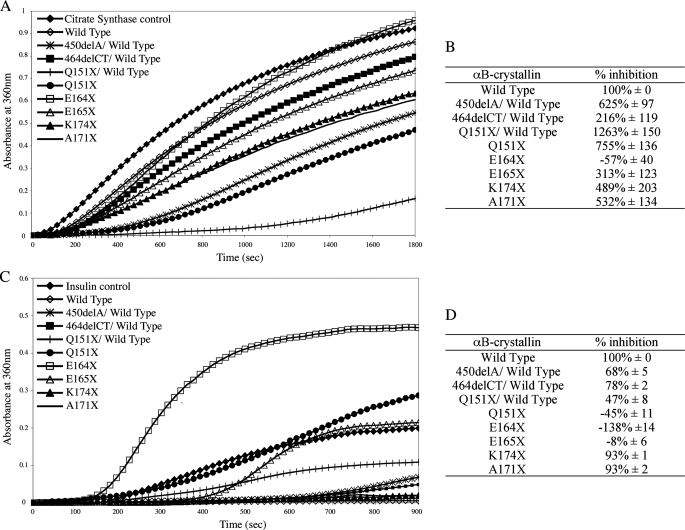
Effect of C-terminal Extension Mutations upon αB-Crystallin Chaperone Activity—Citrate synthase (Fig. 6A) and insulin (Fig. 6C) chaperone assays were used to determine the effects of the αB-crystallin C-terminal extension mutations. Our expectation was that the observed chaperone activity would correlate with the effects of the mutations on protein secondary structure and heat stability. We predicted that E164X, E165X, and Q151X would exhibit the least activity in this assay. The chaperone activity achieved by these mutants in a citrate synthase aggregation assay (Fig. 6B) showed that whereas E164X was indeed the worst of all the αB-crystallin constructs (–57% inhibition) none of the other C-terminal extension constructs performed worse that the wild type protein (100% inhibition). Indeed, the E165X (313%) was slightly better than the wild type protein and Q151X (755%) was one of the best. In fact, the combination of Q151X and wild type αB-crystallin was the best chaperone in the citrate synthase assay (1263% inhibition).
FIGURE 6.
Aggregation of citrate synthase and insulin in the presence of wild type and mutant αB-crystallins. The effect of wild type and mutant αB-crystallin on the thermal aggregation of citrate synthase (A and B), over a 30-min period at 42 °C, and reduction-induced aggregation of insulin (C and D), over a 15-min period at 37 °C was measured using OD360 (OD360). A 4:1 molar ratio of substrate:chaperone was used. The relative inhibition of citrate synthase (B) and insulin (D) aggregation by the C-terminal extension mutants was calculated as a percentage of the level of inhibition achieved by wild type αB-crystallin alone (100% inhibition). These data represented the average (±S.D.) of three independent experiments (see supplementary data Figs. S3 and S4 for the complete data sets).
Another surprise from the citrate synthase chaperone assay was the relative performance of the two frameshift mutants, 450delA and 464delCT, both of which performed better in combination with wild type αB-crystallin than the wild type αB-crystallin alone (625 and 216% inhibition, respectively). These data show that the disease-causing mutations in αB-crystallin (Q151X, 450delA, and 464delCT) all retain significant chaperone activity, surpassing the potential of the wild type protein alone for this client protein in this particular chaperone assay.
In contrast to the results with citrate synthase, all the C-terminal extension mutants performed worse than the wild type αB-crystallin in the insulin-based chaperone assay (Fig. 6C). In this assay, the three most heat-sensitive αB-crystallin mutants, E164X, E165X, and Q151X, increased protein aggregation compared with wild type αB-crystallin (–138, –8, and –45% inhibition, respectively (Fig. 6D)). An equimolar mixture of Q151X and wild type αB-crystallin (47% inhibition) retained significant chaperone activity of Q151X, but it was still one of the poorer chaperones in this assay. The K174X (93%) and A171X (93%) mutants had chaperone activities that most closely resembled the wild type αB-crystallin (100%) and even the two frameshift mutants, 450delA and 464delCT, in their respective combinations with wild type αB-crystallin, still possessed credible activities (68 and 78% inhibition, respectively).
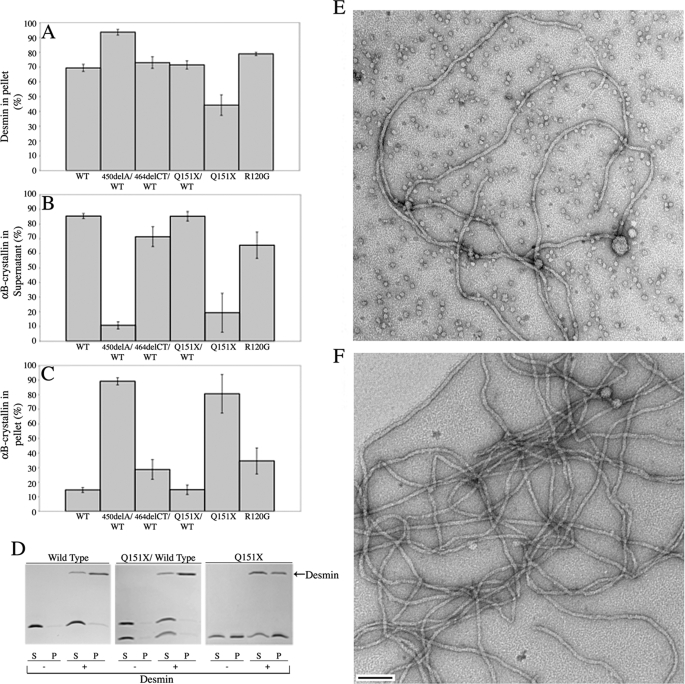
Ability of the Disease-causing αB-Crystallin Mutants to Inhibit Desmin Filament Aggregation—One of the characteristic histopathological features of the human myopathies caused by some of the mutations in αB-crystallin under investigation in this study is the presence of aggregates of desmin filaments (9, 12). It was therefore decided to analyze the ability of the αB-crystallin mutants in this study to prevent desmin filament-filament associations (Fig. 7), using an assay developed to investigate the cell biological effects of the R120G mutation in αB-crystallin (10).
FIGURE 7.
Sedimentation assay to investigate the prevention of desmin filament aggregation in the presence of various αB-crystallins at 37 °C. A–C shows the % desmin pelleted (A), the % αB-crystallin wild type and mutants remaining soluble in the presence of desmin (B), and the % of the total αB-crystallin pelleted in the presence of desmin (C), following low-speed centrifugation (2500 × g). These data were calculated from SDS-PAGE data of the desmin binding assays. D, an example of the data used to calculate these values for wild type, Q151X/wild type, and Q151X αB-crystallin. These data illustrate the ability of the Q151X αB-crystallin to prevent desmin filament aggregation. Images obtained by electron microscopy of negatively stained samples of the assembled filaments for desmin assembled in the presence of wild type (E) and Q151X (F)αB-crystallin are shown to show that the addition of both chaperones has not dramatically altered filament morphology or length. Notice the lack of crystallin particles in the Q151X sample. Bar, 100 nm. S, supernatant; P, pellet.
The assay takes advantage of the fact that intermediate filaments interact with each other in solution (10, 48) and this can be measured by sedimentation assay. The presence of a mutant protein that interacts with desmin filaments, for example, R120G αB-crystallin, activity encourages more filament-filament associations and therefore increases the proportion of pelletable desmin. Both Q151X and 464delCT αB-crystallin when mixed with wild type displayed desmin filament associations similar to wild type alone, whereas the 450delA/wild type protein mixture was significantly more effective than R120G αB-crystallin in inducing this filament association, highlighting the dominant effect of the 450delA mutation over the wild type αB-crystallin (Fig. 7).
In stark contrast to these results, the Q151X αB-crystallin was the most effective chaperone of all the disease-causing mutants and was also better than wild type αB-crystallin in reducing the proportion of pelletable desmin (Fig. 7A). Most of the Q151X mutant co-sedimented with desmin in the pellet fraction (cf. Fig. 7, B and C). Mixing Q151X with wild type αB-crystallin in an equimolar ratio reduced this binding to desmin filaments (cf. Fig. 7, B and C) and restored the levels of pelletable desmin to those obtained in the presence of wild type αB-crystallin (Fig. 7A). In contrast, the 450delA mutant was clearly dominant over the wild type αB-crystallin as there was not only increased binding to the filaments (cf. Fig. 7, B and C), but also a significant increase in the proportion of pelletable desmin (Fig. 7A). We interpret these data to indicate that the increased binding of 450delA αB-crystallin induces more filament-filament interactions resulting in more desmin in the pellet. Interestingly all the mutant αB-crystallins exhibited elevated binding to desmin filaments (Fig. 7C), but under these assay conditions (0.1 mg/ml desmin at 37 °C), the Q151X and 450delA αB-crystallin had opposite effects upon the promotion of filament-filament interactions.
To show that the effects of Q151X αB-crystallin were not due to an inhibition of desmin filament assembly per se, negatively stained samples were analyzed by electron microscopy (Fig. 7E) and the presence of abundant 10-nm filaments confirmed. When compared with desmin assembled in the presence of wild type αB-crystallin (Fig. 7D), the absence of αB-crystallin particles in the Q151X containing sample was also immediately apparent, confirming independently the data in Fig. 2, A and D.
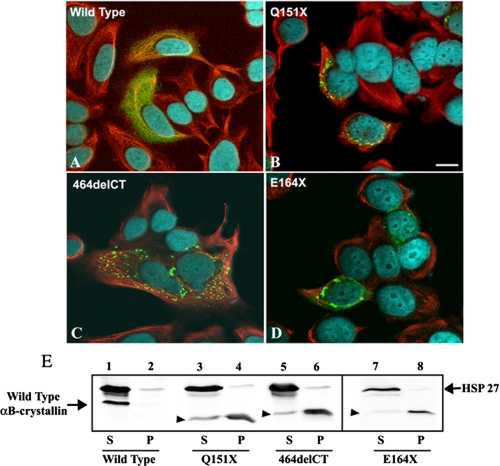
Cytoplasmic Aggregate Formation of Selected αB-Crystallin Mutants in MCF7 Cells—The in vitro assays show that the Q151X mutant increased some chaperone activities, but also significantly destabilized the protein causing it to aggregate. We then decided to investigate the behavior of Q151X αB-crystallin when transiently transfected into tissue culture cells (Fig. 8), using MCF7 cells as established in previous studies investigating desmin (49) and αB-crystallin (10) mutations. MCF7 cells have high endogenous levels of HSP27, a small heat shock protein chaperone that is known to form heterogeneous oligomers with αB-crystallin (50). The cell line also has little or no endogenous αB-crystallin (10, 14), thus avoiding the need to tag the protein for subsequent detection. Cells expressing wild type αB-crystallin (Fig. 8A, green channel), showed a homogeneous cytoplasmic distribution. In contrast, cells expressing the Q151X mutant αB-crystallin were obvious by the presence of cytoplasmic aggregates that were immunopositive for αB-crystallin (Fig. 8B). A similar result was also obtained for two other αB-crystallin constructs, the mutant 464delCT and the C-terminal-truncated αB-crystallin E164X (Fig. 8, C and D). Our previous data (Figs. 2 and 4) suggest that like Q151X, both were destabilized and prone to self-aggregation and the fact that both also form cytoplasmic aggregates supports this conclusion. Our data also complement a paper published as our manuscript was being reviewed, which found that both Q151X and 464delCT αB-crystallin formed aggregates in transfected cells (51). To provide an assessment of the solubility of the different αB-crystallin constructs in populations of transiently transfected MCF7 cells, cell extracts were produced and fractionated into supernatant and pellet fractions by centrifugation prior to analysis by SDS-PAGE and immunoblotting with antibodies to HSP27 and αB-crystallin (Fig. 8E). Transfected wild type αB-crystallin was found almost entirely in the soluble fraction (Fig. 8E, lane 1). Transfected Q151X, 464delCT, and E164X, however, were detected almost entirely in the pellet fractions (Fig. 8E, lanes 4, 6, and 8, respectively) with only a small proportion left in the supernatant fractions (Fig. 8E, lanes 3, 5, and 7, respectively). The endogenous HSP27 was unable to prevent the formation of aggregates by the mutant αB-crystallins and the presence of these aggregates did not cause a significant shift in the solubility of the endogenous HSP27 to the pellet fraction. These data support our hypothesis that the loss of the C-terminal domain, as occurs in the Q151X mutation, dramatically increases the tendency of αB-crystallin to self-aggregate, even in a cellular environment.
FIGURE 8.
Decreased solubility and increased aggregation potential of Q151X αB-crystallin is revealed by transient transfection into MCF7 cells. A–D, MCF7 cells transiently transfected with wild type (A), Q151X (B), 464delCT (C), or E164X (D) αB-crystallin were fixed at 24 h post-transfection and processed for immunofluorescence microscopy. The subcellular distribution of αB-crystallin and keratin were visualized by double labeling with monoclonal anti-keratin (red channel) and polyclonal anti-αB-crystallin antibodies (green channel). A–D are merged images, showing the superimposition of the green, red, and blue (4′,6-diamidino-2-phenylindole staining) channels. Images are individual optical sections acquired using a confocal laser scanning microscope (Zeiss 510). Cells expressing wild type αB-crystallin (A) showed the expected cytoplasmic distribution of αB-crystallin. In contrast, cells expressing mutant αB-crystallin resulted in the formation of cytoplasmic aggregates of αB-crystallin (B–D). Notice that 464delCT aggregates are scattered throughout the cytoplasm, whereas the Q151X and E164X aggregates locate to the perinuclear region of the cell. Bars, 10 μm. Immunoblotting analysis (E) of αB-crystallin transiently transfected in MCF7 cells shows a shift in solubility for the mutant αB-crystallins. At 24 h post-transfection, cells were extracted with detergent buffer followed by centrifugation at 18,000 × g for 10 min at 4 °C. The resulting supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE followed by immunoblotting analysis using anti-HSP 27 (loading control) and αB-crystallin antibodies. The blot was developed using enhanced chemiluminescence. Whereas wild type αB-crystallin was almost entirely soluble (E, lane 1, labeled S), Q151X (lane 4), 464delCT (lane 6), and E164X (lane 8) were found mostly in the pellet fractions (labeled P) of cell extracts. The relative electrophoretic mobility of mutant αB-crystallins is denoted by the arrowhead (▸). Like wild type αB-crystallin, HSP27 was also largely present in the supernatant fractions (E, lanes 2, 4, and 6, labeled P). These data are representative of three experiments.
DISCUSSION
Removal of the C-terminal Extension Reduces αB-Crystallin Oligomerization, but Not Chaperone Activity—The consensus view that has emerged from recent studies on mammalian sHSPs is that the C-terminal extension is important for oligomerization and chaperone function (19, 22, 26, 28). The study presented here is the first to investigate the effects of sequentially truncating this domain (Lys174, Ala171, Glu165, and Glu164) upon both aspects for αB-crystallin. It shows that deletion of the last 5 residues can improve chaperone activity in the citrate synthase assay without dramatically changing the secondary structure or oligomerization of αB-crystallin. Truncations (E164X, E165X) in the conserved REEK motif (26) decreased chaperone activity and oligomerization, although this coincided with more polydispersity for the oligomer as judged by size exclusion chromatography (Fig. 2). The removal of the C-terminal extension by the Q151X mutation produced a construct with the best chaperone activity profile of all the individual protein clients investigated (Figs. 6 and 7) despite the most significant changes in secondary structure (Fig. 3), bis-ANS binding (Fig. 5), and altered oligomerization (Fig. 2).
Whereas these results are in broad agreement with the consensus for other mammalian sHSPs, they demonstrate that the removal of the C-terminal extension from residue 151 prevents oligomerization (at concentrations less than 0.1 mg/ml), but not at the expense of chaperone function for αB-crystallin. Indeed the citroconylation of full-length αB-crystallin achieves similar results and together with these data strongly support the conclusion that oligomerization is not required for αB-crystallin chaperone activity (42).
Deletion of the C-terminal Extension by the Q151X Mutation Increases Chaperone Activity for Some Client Proteins—The somewhat surprising observation is that the Q151X mutation actually improved chaperone activity for some client proteins (desmin and citrate synthase), in stark contrast to the effect of removing a similar region from αA-crystallin where chaperone activity was lost (22, 28).
The C-terminal extension deleted by the Q151X mutation includes the IX(I/V) motif (16), which is part of β-strand 10 (17, 23), that has been shown to be both part of a client protein binding site (19) and an oligomerization sequence (20) in αB-crystallin. This is removed by all three disease-causing mutations, including Q151X, but retained in the other truncation constructs (Fig. 1). Glycine substitutions in the IX(I/V) motif not only increased oligomerization and polydispersity of αB-crystallin, but also significantly increased chaperone activity (52). The data we have presented for Q151X αB-crystallin and the other truncation constructs suggest that the client protein binding site that embraces the IX(I/V) motif (19, 53) is not only dispensable for activity, but that it could actually inhibit chaperone function as its removal actually increased activity (citrate synthase, ∼7.5-fold; desmin, ∼2-fold).
A three-dimensional model of αB-crystallin has suggested that the C-terminal extension potentially acts as a “cap” on an important oligomerization and client protein binding site, namely the β3-β8-β9 interface (46). This region also interacts with the β4 strand, which is important in sHSP oligomerization as seen from the crystal structure of HSP16.9 (23), but it is also critical for chaperone activity (46). In fact reducing αB-crystallin to just its α-crystallin domain spanning residues 57–157, which still includes the β3-β8-β9 surface, retained chaperone activity, but this construct only formed dimers and could not oligomerize (54). It is easy to explain these observations and also our data on the Q151X αB-crystallin mutant, in terms of how the removal of this C-terminal extension cap as proposed in the model (46) could allow substrate to bind more readily and thus enhance chaperone function while also decreasing oligomer size. The data we have presented here support this model and also add an interesting dimension. Our data also suggest that the C-terminal extension actually limits the full potential of αB-crystallin chaperone activity for some client proteins.
Aggregation of αB-Crystallin Encouraged by the Removal of the C-terminal Extension—The strong tendency of Q151X to aggregate at protein concentrations greater than 0.1 mg/ml and its very low refolding yield (Fig. 1D), suggests another potential role played by the C-terminal extension in preventing the uncontrolled self-aggregation of αB-crystallin (55). From the available sHSP crystal structures (17, 23), the C-terminal extension is solvent exposed and contributes to the higher order assembly of the oligomer (23, 56–58). In HSP16.9, the IX(I/V) motif from one subunit lays in a hydrophobic groove between β4 and β8 strands of the other interacting monomer. Although αB-crystallin has similar structural features, including the conserved IX(I/V) motif in its C-terminal extension (Fig. 1A), the oligomers it forms are more polydisperse (55, 59, 60) compared with either HSP16.5 or HSP16.9, suggesting that the precise detail for the interactions of the C-terminal extension in αB-crystallin will vary, not least due to the different hinge sequences between β-strands 9 and 10 (23). The strong tendency of Q151X αB-crystallin to aggregate suggests that the C-terminal extension actually prevents such potential errant associations to favor instead oligomerization.
Reduced Protein Stability and Uncontrolled Self-aggregation Underlies the Molecular Basis of the Q151X, 450delA, and 464delCT Mutations—The loss of the C-terminal extension due to the Q151X mutation in αB-crystallin dramatically altered the secondary (Fig. 3) and tertiary structure of the protein (Fig. 2). This mutant was significantly destabilized as seen from the heat stability assay (Fig. 4) and showed a dramatic increase in the tendency to self-aggregate in vitro (Fig. 1) and in vivo (Fig. 8). Only a small proportion of Q151X αB-crystallin (∼1%, Fig. 1D) remained soluble after refolding. Even in the presence of equimolar wild type αB-crystallin, the mixture had significantly altered secondary structure (Fig. 2), which affected αB-crystallin oligomerization (Fig. 3) and reduced refold yields (Fig. 1D). The Q151X mutation was dominant over the wild type protein in its effects upon heat stability, solubility, and secondary structure. These data suggest that the strong tendency to self-aggregate as a result of reduced stability is potentially an important factor in disease development.
Our data for both 450delA and 464delCT also suggest that it is a similar loss in protein stability that causes cataract and myofibrillar myopathy, respectively. Both mutants can only be refolded in the presence of wild type αB-crystallin and even then the temperature stabilities of the mixtures are reduced by some 20 °C compared with the wild type protein alone. Both the 450delA and 464delCT mutations introduced novel C-terminal peptides that have no homology to the existing C-terminal extension of αB-crystallin (Fig. 1A). Both mutations changed the oligomerization of wild type αB-crystallin and neither mutant/wild type mixture was an effective chaperone in either the desmin (Fig. 7) or insulin (Fig. 6, C and D) assays. Indeed, the 450delA mutation actively promoted filament-filament interactions for desmin, as reported previously for R120G αB-crystallin (10) and, under the assay conditions here, 450delA was more efficient than R120G in this activity. Therefore the loss of chaperone activity coupled with sequestration by client proteins could both contribute to the development of disease in the case of these two mutants. We suggest, however, that the aggregation of αB-crystallin is a key feature that offers potential for gain of function via its amyloidogenic potential (61).
The most difficult observation to explain regarding the Q151X mutation is its muscle-specific effect, which restricts the pathology to myofibillar myopathy (18). In fact all three disease-causing mutations in the C-terminal extension of αB-crystallin (Q151X, 450delA, and 464delCT) are restricted in their pathology to either the lens (450delA (15)) or muscle (Q151X, 464delCT; (18)) and yet αB-crystallin is very highly expressed in both tissues (5). This is a familiar trend for αB-crystallin mutations and even the most recently published mutation, D140N, causes only lens cataract (62). Thus far, R120G αB-crystallin is the only mutation that has produced both lens and muscle pathologies (9), but this is not to say that subclinical pathology can be totally excluded for the other mutations, including Q151X, as tissue biopsies from apparently unaffected tissues were not analyzed. Aggregation is a key histopathological feature of the Q151X mutation (18) concurring with our transient transfection and cell fractionation studies (Fig. 8) and so, this provides a plausible mechanism to support a potential dominant negative effect of the mutation by inhibiting the chaperone function of wild type αB-crystallin via coaggregation with mutant Q151X as implicated from our in vitro studies (Fig. 1).
Supplementary Material
Acknowledgments
We thank Terry Gibbons for technical support and Ming Der Perng for helpful discussions. We thank Cait MacPhee (Dept. of Physics, University of Edinburgh) for discussions and David Dixon for mass spectrometry analyses of recombinant proteins.
This work was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) and ImmunoDiagnostics Systems BBSRC CASE award (to V. H. H.) and National Institutes of Health NINDS Grant P01NS42803 (to R. A. Q.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: sHSP, small heat shock protein; bis-ANS, 4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid; MWCO, molecular weight cut off; SEC, size exclusion chromatography.
References
- 1.Morner, C. (1894) Hoppe-Seyler's Z. Physiol. Chem. 18 61–106 [Google Scholar]
- 2.Bloemendal, H., and de Jong, W. W. (1991) Prog. Nucleic Acids Res. Mol. Biol. 41 259–281 [DOI] [PubMed] [Google Scholar]
- 3.Kato, K., Shinohara, H., Kurobe, N., Goto, S., Inaguma, Y., and Ohshima, K. (1991) Biochim. Biophys. Acta 1080 173–180 [DOI] [PubMed] [Google Scholar]
- 4.Iwaki, T., Kume-Iwaki, A., Liem, R. K., and Goldman, J. E. (1991) Kidney Int. 40 52–56 [DOI] [PubMed] [Google Scholar]
- 5.Kato, K., Shinohara, H., Kurobe, N., Inaguma, Y., Shimizu, K., and Ohshima, K. (1991) Biochim. Biophys. Acta 1074 201–208 [DOI] [PubMed] [Google Scholar]
- 6.Klemenz, R., Frohli, E., Steiger, R. H., Schafer, R., and Aoyama, A. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 3652–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horwitz, J. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 10449–10453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kappe, G., Verschuure, P., Philipsen, R. L., Staalduinen, A. A., Van de Boogaart, P., Boelens, W. C., and De Jong, W. W. (2001) Biochim. Biophys. Acta 1520 1–6 [DOI] [PubMed] [Google Scholar]
- 9.Vicart, P., Caron, A., Guicheney, P., Li, Z., Prevost, M. C., Faure, A., Chateau, D., Chapon, F., Tome, F., Dupret, J. M., Paulin, D., and Fardeau, M. (1998) Nat. Genet. 20 92–95 [DOI] [PubMed] [Google Scholar]
- 10.Perng, M. D., Wen, S. F., van den IJssel, P., Prescott, A. R., and Quinlan, R. A. (2004) Mol. Biol. Cell 15 2335–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldfarb, L. G., Park, K. Y., Cervenakova, L., Gorokhova, S., Lee, H. S., Vasconcelos, O., Nagle, J. W., Semino-Mora, C., Sivakumar, K., and Dalakas, M. C. (1998) Nat. Genet. 19 402–403 [DOI] [PubMed] [Google Scholar]
- 12.Schroder, R., Vrabie, A., and Goebel, H. H. (2007) J. Cell. Mol. Med. 11 416–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholl, I. D., and Quinlan, R. A. (1994) EMBO J. 13 945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perng, M. D., Cairns, L., van den IJssel, P., Prescott, A., Hutcheson, A. M., and Quinlan, R. A. (1999) J. Cell Sci. 112, 2099–2112 [DOI] [PubMed] [Google Scholar]
- 15.Berry, V., Francis, P., Reddy, M. A., Collyer, D., Vithana, E., MacKay, I., Dawson, G., Carey, A. H., Moore, A., Bhattacharya, S. S., and Quinlan, R. A. (2001) Am. J. Hum. Genet. 69 1141–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Jong, W. W., Caspers, G. J., and Leunissen, J. A. (1998) Int. J. Biol. Macromol. 22 151–162 [DOI] [PubMed] [Google Scholar]
- 17.Kim, K. K., Kim, R., and Kim, S. H. (1998) Nature 394 595–599 [DOI] [PubMed] [Google Scholar]
- 18.Selcen, D., and Engel, A. G. (2003) Ann. Neurol. 54 804–810 [DOI] [PubMed] [Google Scholar]
- 19.Ghosh, J. G., Estrada, M. R., and Clark, J. I. (2005) Biochemistry 44 14854–14869 [DOI] [PubMed] [Google Scholar]
- 20.Ghosh, J. G., and Clark, J. I. (2005) Protein Sci. 14 684–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inagaki, N., Hayashi, T., Arimura, T., Koga, Y., Takahashi, M., Shibata, H., Teraoka, K., Chikamori, T., Yamashina, A., and Kimura, A. (2006) Biochem. Biophys. Res. Commun. 342 379–386 [DOI] [PubMed] [Google Scholar]
- 22.Andley, U. P., Mathur, S., Griest, T. A., and Petrash, J. M. (1996) J. Biol. Chem. 271 31973–31980 [DOI] [PubMed] [Google Scholar]
- 23.van Montfort, R. L., Basha, E., Friedrich, K. L., Slingsby, C., and Vierling, E. (2001) Nat. Struct. Biol. 8 1025–1030 [DOI] [PubMed] [Google Scholar]
- 24.Stamler, R., Kappe, G., Boelens, W., and Slingsby, C. (2005) J. Mol. Biol. 353 68–79 [DOI] [PubMed] [Google Scholar]
- 25.Kim, R., Lai, L., Lee, H. H., Cheong, G. W., Kim, K. K., Wu, Z., Yokota, H., Marqusee, S., and Kim, S. H. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 8151–8155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajan, S., Chandrashekar, R., Aziz, A., and Abraham, E. C. (2006) Biochemistry 45 15684–15691 [DOI] [PubMed] [Google Scholar]
- 27.Treweek, T. M., Ecroyd, H., Williams, D. M., Meehan, S., Carver, J. A., and Walker, M. J. (2007) PLoS ONE 2 e1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thampi, P., and Abraham, E. C. (2003) Biochemistry 42 11857–11863 [DOI] [PubMed] [Google Scholar]
- 29.Perng, M. D., Muchowski, P. J., van den IJssel, P., Wu, G. J. S., Clark, J. I., and Quinlan, R. A. (1999) J. Biol. Chem. 274 33235–33243 [DOI] [PubMed] [Google Scholar]
- 30.Horwitz, J., Huang, Q. L., Ding, L., and Bova, M. P. (1998) Methods Enzymol. 290 365–383 [DOI] [PubMed] [Google Scholar]
- 31.Quinlan, R. A., Moir, R. D., and Stewart, M. (1989) J. Cell Sci. 93 71–83 [DOI] [PubMed] [Google Scholar]
- 32.Laemmli, U. (1970) Nature 277 680–685 [DOI] [PubMed] [Google Scholar]
- 33.Plater, M. L., Goode, D., and Crabbe, M. J. (1996) J. Biol. Chem. 271 28558–28566 [DOI] [PubMed] [Google Scholar]
- 34.He, S., Pan, S., Wu, K., Amster, I. J., and Orlando, R. (1995) J. Mass Spectrom. 30 424–431 [Google Scholar]
- 35.Smith, J. B., Shun-Shin, G. A., Sun, Y., Miesbauer, L. R., Yang, Z., Yang, Z., Zhou, X., Schwedler, J., and Smith, D. L. (1995) J. Protein Chem. 14 179–188 [DOI] [PubMed] [Google Scholar]
- 36.Jimenez-Asensio, J., Colvis, C. M., Kowalak, J. A., Duglas-Tabor, Y., Datiles, M. B., Moroni, M., Mura, U., Rao, C. M., Balasubramanian, D., Janjani, A., and Garland, D. (1999) J. Biol. Chem. 274 32287–32294 [DOI] [PubMed] [Google Scholar]
- 37.Colvis, C. M., Duglas-Tabor, Y., Werth, K. B., Vieira, N. E., Kowalak, J. A., Janjani, A., Yergey, A. L., and Garland, D. L. (2000) Electrophoresis 21 2219–2227 [DOI] [PubMed] [Google Scholar]
- 38.Ueda, Y., Duncan, M. K., and David, L. L. (2002) Investig. Ophthalmol. Vis. Sci. 43 205–215 [PubMed] [Google Scholar]
- 39.Van Kleef, S. M., Willems-Thijssen, W., and Hoenders, H. J. (1976) Eur. J. Biochem. 66 477–483 [DOI] [PubMed] [Google Scholar]
- 40.Thampi, P., Hassan, A., Smith, J. B., and Abraham, E. C. (2002) Investig. Ophthalmol. Vis. Sci. 43 3265–3272 [PubMed] [Google Scholar]
- 41.Ueda, Y., Fukiage, C., Shih, M., Shearer, T. R., and David, L. L. (2002) Mol. Cell. Proteomics 1 357–365 [DOI] [PubMed] [Google Scholar]
- 42.Horwitz, J., Huang, Q., and Ding, L. (2004) Exp. Eye Res. 79 817–821 [DOI] [PubMed] [Google Scholar]
- 43.Bova, M. P., Yaron, O., Huang, Q., Ding, L., Haley, D. A., Stewart, P. L., and Horwitz, J. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 6137–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sreelakshmi, Y., and Sharma, K. K. (2006) Mol. Vis. 12 581–587 [PubMed] [Google Scholar]
- 45.Pasta, S. Y., Raman, B., Ramakrishna, T., and Rao, Ch. M. (2003) J. Biol. Chem. 278 51159–51166 [DOI] [PubMed] [Google Scholar]
- 46.Ghosh, J. G., Estrada, M. R., and Clark, J. I. (2006) Biochemistry 45 9878–9886 [DOI] [PubMed] [Google Scholar]
- 47.Doss-Pepe, E. W., Carew, E. L., and Koretz, J. F. (1998) Exp. Eye Res. 67 657–679 [DOI] [PubMed] [Google Scholar]
- 48.Bousquet, O., Ma, L., Yamada, S., Gu, C., Idei, T., Takahashi, K., Wirtz, D., and Coulombe, P. A. (2001) J. Cell Biol. 155 747–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munoz-Marmol, A. M., Strasser, G., Isamat, M., Coulombe, P. A., Yang, Y., Roca, X., Vela, E., Mate, J. L., Coll, J., Fernandez-Figueras, M. T., Navas-Palacios, J. J., Ariza, A., and Fuchs, E. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 11312–11317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zantema, A., Verlaan-De Vries, M., Maasdam, D., Bol, S., and van der Eb, A. (1992) J. Biol. Chem. 267 12936–12941 [PubMed] [Google Scholar]
- 51.Simon, S., Fontaine, J. M., Martin, J. L., Sun, X., Hoppe, A. D., Welsh, M. J., Benndorf, R., and Vicart, P. (2007) J. Biol. Chem. 282 34276–34287 [DOI] [PubMed] [Google Scholar]
- 52.Pasta, S. Y., Raman, B., Ramakrishna, T., and Rao, Ch. M. (2004) Mol. Vis. 10 655–662 [PubMed] [Google Scholar]
- 53.Ghosh, J. G., Houck, S. A., and Clark, J. I. (2008) Int. J. Biochem. Cell Biol. 40 954–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feil, I. K., Malfois, M., Hendle, J., van Der Zandt, H., and Svergun, D. I. (2001) J. Biol. Chem. 276 12024–12029 [DOI] [PubMed] [Google Scholar]
- 55.Aquilina, J. A., Benesch, J. L., Ding, L. L., Yaron, O., Horwitz, J., and Robinson, C. V. (2004) J. Biol. Chem. 279 28675–28680 [DOI] [PubMed] [Google Scholar]
- 56.White, H. E., Orlova, E. V., Chen, S., Wang, L., Ignatiou, A., Gowen, B., Stromer, T., Franzmann, T. M., Haslbeck, M., Buchner, J., and Saibil, H. R. (2006) Structure 14 1197–1204 [DOI] [PubMed] [Google Scholar]
- 57.Kennaway, C. K., Benesch, J. L., Gohlke, U., Wang, L., Robinson, C. V., Orlova, E. V., Saibil, H. R., and Keep, N. H. (2005) J. Biol. Chem. 280 33419–33425 [DOI] [PubMed] [Google Scholar]
- 58.Kim, R., Kim, K. K., Yokota, H., and Kim, S. H. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 9129–9133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haley, D. A., Bova, M. P., Huang, Q. L., McHaourab, H. S., and Stewart, P. L. (2000) J. Mol. Biol. 298 261–272 [DOI] [PubMed] [Google Scholar]
- 60.Haley, D. A., Horwitz, J., and Stewart, P. L. (1998) J. Mol. Biol. 277 27–35 [DOI] [PubMed] [Google Scholar]
- 61.Meehan, S., Knowles, T. P., Baldwin, A. J., Smith, J. F., Squires, A. M., Clements, P., Treweek, T. M., Ecroyd, H., Tartaglia, G. G., Vendruscolo, M., Macphee, C. E., Dobson, C. M., and Carver, J. A. (2007) J. Mol. Biol. 372 470–484 [DOI] [PubMed] [Google Scholar]
- 62.Liu, Y., Zhang, X., Luo, L., Wu, M., Zeng, R., Cheng, G., Hu, B., Liu, B., Liang, J. J., and Shang, F. (2006) Investig. Ophthalmol. Vis. Sci. 47 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.