Abstract
Enamelin is critical for proper dental enamel formation, and defects in the human enamelin gene cause autosomal dominant amelogenesis imperfecta. We used gene targeting to generate a knock-in mouse carrying a null allele of enamelin (Enam) that has a lacZ reporter gene replacing the Enam translation initiation site and gene sequences through exon 7. Correct targeting of the transgene was confirmed by Southern blotting and PCR analyses. No enamelin protein could be detected by Western blotting in the Enam-null mice. Histochemical 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) staining demonstrated ameloblast-specific expression of enamelin. The enamel of the Enam+/- mice was nearly normal in the maxillary incisors, but the mandibular incisors were discolored and tended to wear rapidly where they contacted the maxillary incisors. The Enam-/- mice showed no true enamel. Radiography, microcomputed tomography, and light and scanning electron microscopy were used to document changes in the enamel of Enam-/- mice but did not discern any perturbations of bone, dentin, or any other tissue besides the enamel layer. Although a thick layer of enamel proteins covered normal-appearing dentin of unerupted teeth, von Kossa staining revealed almost a complete absence of mineral formation in this protein layer. However, a thin, highly irregular, mineralized crust covered the dentin on erupted teeth, apparently arising from the formation and fusion of small mineralization foci (calcospherites) in the deeper part of the accumulated enamel protein layer. These results demonstrate ameloblast-specific expression of enamelin and reveal that enamelin is essential for proper enamel matrix organization and mineralization.
Enamel crystals have a unique shape and organization. They are many times longer than they are wide and are organized into rods (prisms), each comprising about 10,000 individual crystallites (1, 2). Each rod is the product of a single ameloblast in which crystals are formed under the control of specialized enamel proteins secreted by ameloblasts (3). These proteins assemble immediately subjacent to the secretory pole of the ameloblast cell membrane such that mineralization occurs immediately at this cell-matrix interface (4). Because enamel crystals grow longer at this mineralization front, the ameloblasts are displaced away from the growing tooth as the enamel layer as a whole thickens. Once the enamel crystals are fully elongated and the enamel layer has reached its final thickness, the secretion of enamel proteins is terminated or greatly reduced, accumulated extracellular proteins are degraded and reabsorbed, and the crystals grow in width and thickness until adjacent crystals come into contact. Thus, the secretion of enamel proteins is associated with the early part of crystal formation, when the crystals are growing primarily in length and the growing crystal tips are in close proximity to the secretory surface of the ameloblast.
There are three major secretory stage enamel proteins: amelogenin (5, 6), ameloblastin (7), and enamelin (8, 9). All belong to the secretory calcium-binding phosphoprotein (SCPP) gene family (10), and all are critical for proper dental enamel formation. Amelogenin is the most abundant secreted enamel protein and is expressed from the X or the X and Y chromosomes depending upon the organism (11, 12). AMELX (Xp22.3) mutations cause X-linked amelogenesis imperfecta in humans, an inherited disorder that affects only the enamel layer of the dentition (13, 14). Amelx-null mice also exhibit an amelogenesis imperfecta phenotype (15). Although amelogenin expression has been detected in many tissues, including the brain (16, 17), no defects in non-dental tissues have been noted in families with X-linked amelogenesis imperfecta. No cases of amelogenesis imperfecta caused by human ameloblastin (AMBN, 4q13.3) mutations have been reported, but Ambn-null mice lack enamel (18). Human enamelin (ENAM, 4q13.2) gene mutations cause autosomal dominant amelogenesis imperfecta (19-21). ENAM mutations show a dose effect, so that a single mutant allele causes a mild form of amelogenesis imperfecta, whereas having defects in both alleles eliminates the enamel layer (22, 23). Mutations in the mouse enamelin gene have been induced with the mutagen N-ethyl-N-nitrosourea, and four separate Enam point mutations have been identified: p.S55I, pE57G, the splice donor site in exon 4, and pQ176X (24, 25). The resulting enamel phenotypes include a rough and pitted enamel surface in heterozygous mice and enamel agenesis in the null condition.
In addition to the three secretory calcium-binding phosphoproteins, two proteases are secreted into the developing enamel layer: matrix metalloproteinase 20 (MMP-20)2 and kallikrein 4 (KLK4) (26, 27). MMP-20 is secreted in the early (secretory) stage along with amelogenin, ameloblastin, and enamelin, whereas KLK4 is expressed in the later (maturation) stage (28, 29). Defects in the genes encoding these enzymes cause autosomal recessive amelogenesis imperfecta in humans (30-32). Mmp20-null mice have a thin enamel that is softer than normal and tends to chip off the crown surface (33, 34). MMP-20 expression appears to be restricted to developing teeth (35), whereas KLK4 (19q13.4) is expressed in many tissues, but most notably it is expressed in developing teeth and in prostate gland (36-38). It is clear that amelogenin, ameloblastin, enamelin, MMP-20, and KLK4 are all necessary for proper dental enamel formation, but they are not sufficient. Defects in AMELX, ENAM, MMP20 (11q22.3), and KLK4 account for only about one-fourth of all cases of amelogenesis imperfecta (39). Despite the large number of amelogenesis imperfecta cases of unknown etiology, the proteins resident in the enamel layer of developing teeth have been thoroughly analyzed, and it is likely that most, if not all, of the major extracellular components have been identified (40).
The secretion of proteases along with the enamel proteins means that intact enamelin and ameloblastin cannot be isolated from developing teeth. These enamel proteins have multiple glycosylation sites and other post-translational modifications, even hydroxyproline (41, 42), and efforts to generate recombinant forms with the appropriate modifications have failed to produce them in any significant quantity (43). Therefore, it is currently impossible to reconstitute the secretory stage extracellular matrix and recapitulate the formation of enamel crystals in vitro. To gain insights into the molecular mechanisms regulating dental enamel formation and to decisively characterize the normal temporal and spatial patterns of Enam expression, we have used gene targeting to knock out normal Enam expression, while replacing the 5′ Enam code with that of a β-galactosidase (β-gal) reporter in mice. We have demonstrated here that Enam is expressed solely and specifically by ameloblasts and that Enam-null mice do not make enamel because of a complete failure to mineralize at the appropriate place, resulting in the lack of a mineralization front at the secretory surface of the ameloblast.
EXPERIMENTAL PROCEDURES
Animal Protocol—All procedures involving animals were reviewed and approved by the University Committee on Use and Care of Animals at the University of Michigan.
Knock-in Targeting Construct—Construction of the targeting vector started with pLOz (Ozgene Pty. Ltd., Murdoch, Western Australia), a modified pBlueScript SK vector containing a lacZ (β-galacatosidase) reporter with a mouse nuclear localization signal (NLS) upstream of a phosphoglycerine kinase (PGK) promoter driving a neomycin (Neo) selection marker, all inserted into the unique XbaI site in the multiple cloning region of the vector. The PGK-Neo code was flanked by loxP sites so that it could be deleted later by Cre recombinase (Cre). The Enam 5′-homology arm (5244 bp) was generated by PCR using 129Sv/j genomic DNA as template and ligated into the SalI site of pLOz. The Enam 3′ homology arm (5180 bp) starting in intron 7 and ending in intron 8 was inserted into the BamHI site. The final targeting construct (Fig. 1A) was characterized by DNA sequencing and restriction mapping, linearized by digestion with AclI, and transfected into embryonic stem cells by electroporation.
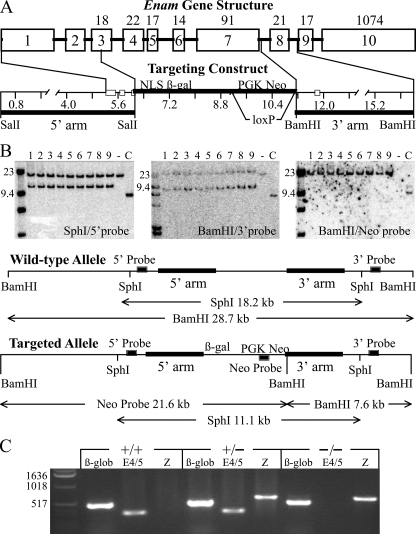
FIGURE 1.
Gene targeting strategy of the Enam mouse model. A, depiction of the mouse Enam gene and the targeting construct. The ten Enam exons are indicated by numbered boxes. Exons 3-10 are coding, with the number of codons shown above each exon. The targeting construct was designed to replace the first five Enam coding exons with NLS β-gal, so that the Enam promoter would drive reporter expression instead of enamelin. The 5′ homology arm ended in exon 3, slightly upstream of the Enam translation initiation codon. The 3′ homology arm started in intron 7 and ended in intron 8. The translation initiation codon of LacZ with a mouse nuclear localization signal (NLS β-gal) in the hybrid exon 3 was positioned where the Enam initiation codon had been. Following the reporter gene were the selection genes (PGK Neo) bracketed by loxP recombination signals so that they could be deleted later by mating with mice expressing Cre recombinase. B, Southern blots demonstrated proper integration of the targeting construct. DNA isolated from tail biopsies of 9 putative Enam+/lacZ mice (from three different founders) were tested (lanes 1-9) alongside wild type (-) and a positive probe control (+). Left, DNA digested with SphI and probed with the 5′-probe shows wild type (18.2 kb) and targeted allele (11.1 kb) bands. Center, DNA digested with BamHI and probed with the 3′-probe shows wild type (28.7 kb) and targeted allele (7.6 kb) bands. Right, the BamHI filter rehyribidized with the Neo probe demonstrating that there was no random integration of the targeting construct, as only the 21.6-kb band predicted for the targeted allele was observed. C, reverse transcription-PCR genotyping to detect β-globin (β-glob; 494 bp), lacZ (Z; 610 bp), and enamelin exons 4 and 5 (E4/5; 324 bp).
Generation of Knock-in Mice—Successful homologous recombination in the embryonic stem cells that survived G418/gancyclovier positive-negative selection was screened for by Southern blot hybridization. Three correctly targeted clones were identified and injected into C57BL/6 blastocysts. The resultant female chimeras were mated to C57BL/6 males, and the offspring were analyzed for germ line transmission by Southern blot hybridization (Fig. 1B) using three probes. The 5′-probe (546 bp) was generated with primers 5′-CTTGCTGGTGACACTGAAGTTCC-3′ and 5′-ATTGGGGGTATGCTGTATGCC-3′ and hybridized against SphI-digested genomic DNA. Both the wild type (18.2 kbp) and the knock-in (11.1 kbp) bands were observed. The 3′-probe (564 bp) was generated with primers 5′-GGAGGAGCCCAGTGAAATAAGC-3′ and 5′CAGAAAGCAGAATGCCTTGGTG-3′ and hybridized against BamHI-digested genomic DNA. Both the wild type (28.7 kbp) and the knock-in (7.6 kbp) bands were observed. A knock-in-specific neomycin (Neo) probe (620 bp) was generated with primers 5′-GACTGGGCACAACAGACAATC-3′ and 5′-CCAAGCTCTTCAGCAATATCAC-3′ and hybridized against BamHI-digested genomic DNA to check for randomly integrated targeting vector. Only the band expected for the correctly targeted recombination (21.6 kbp) was observed. Five heterozygous offspring of the germ line chimera were mated with B6 Cre deleter mice to remove the PGK-Neo code. Three female and two male heterozygous mice were generated that survived. The presence or absence of a Neo cassette or Cre deleter sequence was determined by PCR analyses (Table 1). Wild type (Enam+/+), heterozygous (Enam+/lacZ or Enam+/-), and homozygous (EnamlacZ/lacZ or Enam-/-) were distinguished by PCR genotyping of genomic DNA obtained by tail biopsy (Fig. 1C) using primer pairs for β-globin, Enam exons 4/5, and lacZ. β-Globin was a positive control demonstrating adequate DNA template; a positive Enam band at 324 bp demonstrated the presence of at least one Enam allele. A positive lacZ band at 610 bp demonstrated the presence of at least one knock-in gene. The heterozygous Enam+/lacZ mice were backcrossed onto C57BL/6 for six generations, which established a breeding colony. All mice were maintained on moistened chow.
TABLE 1.
PCR primers for genotyping
| Target | Primer sequence | Size |
|---|---|---|
| bp | ||
| LacZ | TSF2: AAGTTTTGGGATTTGGCTCA | 610 |
| TSR2: GTTGCACCACAGATGAAACG | ||
| Exons 4 and 5 | E4/5F: GCCCAAAGCACAGTCATTTT | 324 |
| E4/5R: TAGGACCTGGCACGTGTCTC | ||
| Exon 6 | E6F: CATATGTGCAGGCAAACACC | 351 |
| E6R: ACAAACACAGCCCAGCTTCT | ||
| Exon 7 | E7F: GGGACGAAACTGGACTTCAA | 610 |
| E7R: ACAGGTGGGGACCTCTCTTT | ||
| Neo | Neo P1: AGGATCTCCTGTCATCTCACCTTGCTCCTG | 492 |
| Neo P2: AAGAACTCGTCAAGAAGGCGATAGAAGGCG | ||
| Neo | Neo P3: AGACAATCGGCTGCTCTGAT | 351 |
| Neo P4: ATACTTTCTCGGCAGGAGCA | ||
| Cre | Cre F1: CGTACTGACGGTGGGAGAAT | 261 |
| Cre R1: TGCATGATCTCCGGTATTGA | ||
| β-Globin | Fwd: CCAATCTGCTCACACAGGATAGAGAGGGCAGG | 494 |
| Rev: CCTTGAGGCTGTCCAAGTGATTCAGGCCATCG |
Physical Assessment and Blood Chemistry—The three mouse enamelin genotypes (+/+, +/-, -/-) were evaluated from birth for appearance, physical activity, size discrepancy, rate of growth, food intake, and reproductive physiology. Peripheral blood samples (n = 3) were collected from 6-week-old mice for each of the three genotypes, and a comprehensive panel of blood chemistry tests was performed at the Animal Diagnostic Laboratory of the University of Michigan.3
β-Galactosidase (lacZ) Expression Assay—Mouse heads were collected from newborn mice up to 21 days of age, fixed in 4% paraformaldehyde overnight, and processed for β-galactosidase staining (44-46). The dissected tissues were fixed in 4% paraformaldehyde for 30 to 90 min according to the age of the mice and size of the tissue block, washed with phosphate-buffered saline (PBS) three times for 15 min each, and then decalcified in 10% EDTA for up to 7 days according to the age of the mice (47). The decalcified tissues were washed three times in PBS (for 15 min) and embedded in Tissue-Tek® O.C.T. compound (ProSciTech, Queensland, Australia). Tissue blocks were sectioned at 16-μm thickness, post-fixed for 5 min in 4% paraformaldehyde, washed three times in PBS (for min), and incubated at 45 °C for 3 h in freshly prepared X-gal staining buffer, pH 8.0, containing 1 mg/ml X-gal, 100 mm Hepes, 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, 1 mm MgCl2, 2% Triton X-100, and 1 mm dithiothreitol (48). Tissue sections were rinsed and stored in PBS for counterstaining with hematoxylin and then observed under a dissection microscope (Nikon SMZ1000) or a light microscope (Nikon Eclipse E600). All images were captured using a digital camera (Nikon DXM1200) and Act1 imaging software (Mager Scientific, Dexter, MI).
Extraction and Analysis of Tooth Proteins—Day 7 mouse pups were decapitated, and the mandibles were removed and placed on ice in a Petri dish. The incisors and first molars were extracted using fine point tissue forceps under a dissecting microscope, and the soft tissues (pulp and enamel organ) were removed and placed in PBS on ice. Incisors and molars for each enamelin genotype were pooled, weighed, and extracted with 0.8 ml of 0.5 m acetic acid containing 0.5 mm benzamidine and 1,10-phenanthrolin while shaking at 4 °C overnight. On days 2 and 3 the teeth were pelleted by centrifugation and again extracted with 0.5 m acetic acid. The supernatants were combined, desalted using a PD-10 column (GE Healthcare), lyophilized, and weighed, and 5 μg of protein was dissolved in 15 μl of SDS sample buffer. Each sample was separated by SDS-PAGE on three different gels. One gel was stained with Coomassie Brilliant Blue; the others were transblotted to membranes and blocked with milk proteins for Western blot analyses. One membrane was immunostained using polyclonal rabbit antibodies (1:2000) against rM179 amelogenin (49); the other membrane was immunostained using affinity-purified rabbit antibodies (1:1000) raised against the enamelin peptide EQDFEKPKEKDPPK and designated Enam221. After washing with PBS, the blots were incubated with anti-rabbit IgG secondary antibody (1:10,000) conjugated to horseradish peroxidase (Bio-Rad). Amelogenin was detected by 3,3′-diaminobenzidine tetrahydrochloride staining, and enamelin was detected by enhanced chemiluminescence (ECL; GE Healthcare Biosciences).
Eruption Rate Determination—The eruption rates of mandibular incisors from each of the three genotypes at ages 5-6 weeks were determined (50, 51). Mice received inhalation anesthesia, isoflurane, at the surgical room at Dental Unit for Laboratory Animal Medicine for the incisor notching procedure. One dose of analgesic, buprenorphine (0.2 mg/25 gm body weight), was given intramuscularly immediately before the procedure for pain control. A dental handpiece with a sterile, round bur size (¼) was used to create a circular lesion 1 mm in diameter in the mandibular buccal plate and penetrate the enamel and dentin of the incisor. Hemostasis was achieved by using a cotton pellet, direct pressure, and buffered aluminum chloride epinephrine-free hemostatic liquid (Hemodent™, Primer USA). The mice were euthanized after 10 days, and the hemi-mandibles were dissected and radiographed at ×5 magnification. The incisors were traced on a view box, and the distance between the notch on the bone and the notch on the incisor was measured with a Vernier Caliper three times per sample. The distance between the stationary mark in the bone and the mark on the erupting incisor was divided by the amount of time that had elapsed since the marks were made, providing the eruption rate. Statistical analysis using one-way analysis of variance was performed.
Radiography—Hemi-mandibles of 2-week-old Enam+/+, Enam+/-, and Enam+/- mice were freed from soft tissues and radiographed using a Faxitron™ x-ray cabinet (model MX-20, Faxitron X-ray Corp., Wheeling, IL) operating at 30 kV. Mandibles from three mice from each of the three genotypes were evaluated and compared using digital micrographs obtained under identical Faxitron settings.
Microcomputed Tomography—Microcomputed tomography was performed on hemi-mandibles at the level of the first molar from three samples of each genotype (model 1072; Skyscan, Kontich, Belgium). The x-ray source was operated at maximum power (80 KeV) and at 100 μA. Images were captured using a 12-bit, cooled, charge-coupled device camera (1024 × 1024 pixels) coupled by a fiber optic taper to the scintillator. Using a rotation step of 0.9°, total scanning time was 35 min for each rotated sample, after which ∼300 sections (slice-to-slice distance of 16.5 μm) were reconstructed using Skyscan tomography software based on triangular surface rendering to give a three-dimensional distribution of the calcified tissue. Appropriate imaging planes selected to show three-dimensionalized longitudinal “sections” (segments) of the first molar and cross-sections of the underlying incisor were selected using a limited number of acquired x-ray slices.
Histologic Evaluation of Mandibular First Molars at 7 Days—Light microscopy of undecalcified mandibles at the level of the unerupted first molar was performed on wild type, heterozygous, and enamelin-null mice at 1 week of age. Mandibles were fixed with 4% paraformaldehyde/0.1% glutaraldehyde, dehydrated in ethanol, and embedded in LR White acrylic resin (London Resin Co., Berkshire, UK). Heat-polymerized blocks were sectioned with a diamond knife on an ultramicrotome (model Reichert Ultracut E, Leica, Wetzlar, Germany) to produce 0.5 μm-thick sections. Von Kossa staining for mineral was performed by applying 3% silver nitrate to the sections and exposing them to bright light for 30 min. Sections were counterstained for tissue and cell morphology using toluidine blue. Light micrographs were obtained using a Sony DXC-950 3-CCD camera (Sony, Tokyo, Japan) mounted on an optical microscope (model Leitz DMRBE, Leica).
Scanning Electron Microscope (SEM) Evaluation of Mandibular First Molars at 7 Days—Mandibles from 1-week-old mice (n = 3 for each of the three genotypes) were fractured with a razor blade at the level of the first molar, critical-point dried, and mounted with conductive carbon cement onto metallic stubs. Samples were sputter-coated with a 20-nm-thick AuPd film to increase conductivity and imaged with a field-emission gun scanning electron microscope (model S-4700, Hitachi High Technologies America, Pleasanton, CA) operating in the secondary electron detection mode at an accelerating voltage of 3-5 kV.
SEM Evaluation of Molars and Incisors at 7 Weeks—Rehydrated freeze-dried mandibular jaws from 7-week-old wild type, heterozygous, and homozygous knock-in mice were washed briefly in diluted sodium hypochlorite solution, rinsed in deionized water, and examined without coating in backscatter mode using a JEOL-JSM6460LV (JEOL Ltd., Japan) SEM operated at 20 kV. Incisor and molar samples were fractured and mounted and coated with AuPd film and evaluated using a JEOL JSM 6300.
Incremental Mineral Contents of Incisor Enamel at Week 7—Male and female mice at 7 weeks of age were randomly selected and euthanized for tissue collection. Hemi-mandibles and hemi-maxillae were removed, quickly cleaned of adhering soft tissues, and flash-frozen in prechilled containers immersed in dry ice. The tissues were then freeze-dried for 48 h at -55 °C. The bone and enamel organs covering the freeze-dried incisors were removed, and the exposed enamel surfaces were wiped gently with dry Kimwipes (Kimberly-Clark Corp., Roswell GA). The enamel layer on each maxillary and mandibular incisor was transected with a scalpel blade into a series of 1-mm-long strips from the apical toward the incisal ends. Each strip was removed, placed in a separate small aluminum pan, and dried overnight at 45 °C. The strips were cooled to room temperature and weighed on a SC2 microbalance (Sartorius AG, Goettingem, Germany) to obtain the “total dry weight” of the enamel strip. Each strip was transferred to a separate crucible and then placed inside an Isotemp muffle furnace (Fisher Scientific) and heated at 575 °C for 18 h. The crucibles were cooled, and each enamel strip was reweighed to obtain the “ashed weight” of the strip. This procedure vaporized organic material and any bound water, leaving only mineral contained in each enamel strip (52). For purposes of this study, the protein content of a strip was defined as the difference between starting dry weight minus the final ashed weight. The percent mineral by weight in each strip was calculated by the equation (ashed weight/total dry weight) × 100. Data from strip dissections were collected from a minimum of 12 maxillary and 12 mandibular incisors per genotype (72 or more incisors overall) and analyzed in terms of three primary stages of enamel development (secretory, early maturation, and nearly mature) using version 7 of Statistica for Windows (Statsoft Inc., Tulsa, OK). Preliminary analyses indicated that neither raw weight data nor transformed values followed normal distributions, so statistical tests of mean differences were done by nonparametric methods.
Enamelin and Crystal Growth in Vitro—To test the effect of enamelin on crystal growth habit, hydroxyapatite (HA) crystals were grown in a steady-state agarose gel system (53) in the presence of 0, 10, 25, or 50 μg/ml purified porcine 32-kDa enamelin. Crystals were harvested from the gels using 5% sodium hypochlorite and subjected to x-ray diffraction line-broadening analysis with a Rigaku x-ray diffractometer (XRD) using Cu Kα radiation and a highly crystalline fluoroapatite standard (54).
RESULTS
Generation of the Enam-null/lacZ Knock-in Mouse—We used gene targeting to generate a mouse strain carrying a null allele of enamelin (Enam) that has a lacZ reporter gene inserted into the 5′-region of the Enam locus (Fig. 1A). Mice hosting the targeting construct in both Enam alleles are alternatively referred to as Enam-/- or as EnamlacZ/lacZ to emphasize either the loss of Enam expression or the introduction of NLS β-gal expression within the context of the enamelin gene. The lacZ coding sequence, which expresses bacterial β-galactosidase, was modified to include a mouse NLS and inserted into the first Enam coding exon (exon 3), replacing the Enam translation initiation site. All of the upstream Enam coding sequence (exons 3 through 7) was deleted, so that the lacZ insert blocked enamelin expression, including the remaining downstream part of Enam (exons 8 through 10). The coding sequences for the selection markers (PGK-Neo) were bracketed with loxP sites and later removed by mating Enam+/lacZ mice with Cre deleter mice. Correct targeting and integration were confirmed by Southern blot analyses (Fig. 1B) and reverse transcription-PCR genotyping using primer pairs specific for β-globin, Enam exons 4 and 5, and the lacZ reporter (Fig. 1C). By replacing the Enam translation initiation site and 5′-code with the NLS β-gal translation initiation site and code, we simultaneously knocked out Enam expression while knocking in NLS β-gal expression in its place. Because the NLS β-gal code is positioned in the exact genomic context as wild type Enam, NLS β-gal expression provides a sensitive reporter for native Enam expression.
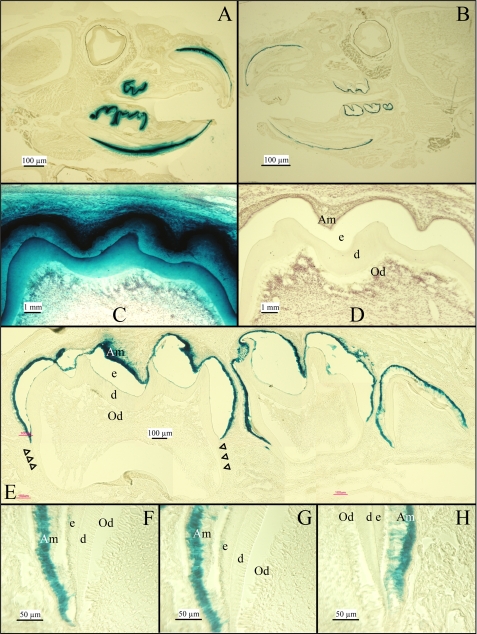
Tissue-specific Expression of NLS β-gal—Replacing the Enam 5′ coding region with the NLS β-gal code established it as a specific reporter for enamelin expression. The NLS β-gal is readily distinguished from native β-gal activity because of its nuclear localization and its high pH optima. Mouse β-galactosidase is a lysosomal enzyme that is only marginally active at pH 7.5, whereas bacterial β-galactosidase can be strongly detected above pH 8 (55). X-gal is a substrate analogue that is cleaved by β-galactosidase to yield 5-bromo-4-chloro-3-hydroxyindole, an insoluble blue product. The strength of the signal on a section can be controlled by varying the length of the X-gal incubation period. The optimal incubation time for the purpose of demonstrating tissue-specific expression leaves a blue stain restricted to the nucleus. Longer incubation times are useful to demonstrate the absence of marginal reporter activity in tissues separated from the main sites of expression, with the only disadvantage being leakage of the blue signal beyond the nucleus and into immediately adjacent structures. We assayed for the expression of NLS β-gal expressed from the enamelin promoter in 2-week-old Enam+/lacZ knock-in mice by histochemistry at pH 8 (Fig. 2). Extended X-gal staining of mouse sections shows intensive signal over ameloblasts, the enamel-forming cells in developing teeth (Fig. 2, A, C, and E). No signal was observed in any other tissue in the head (Fig. 2A) or the rest of the body (data not shown). Reducing the X-gal incubation time limited the signal in developing teeth to ameloblasts (Fig. 2, B and F-H). Odontoblasts were negative. Extended incubation in wild type teeth showed no signal (Fig. 1D). These results correlate well with previous studies of enamelin expression by in situ hybridization (9, 56). The only difference is that maturation stage ameloblasts are positive for X-gal staining but negative for in situ hybridization. This difference is the likely result of the NLS β-gal protein being more stable than Enam mRNA, allowing it to persist in the nucleus (and generate positive X-gal staining) after expression of the protein has terminated. Enam expression was detected only by ameloblasts, and by no other cell types, using the highly sensitive β-gal reporter. The close correlation between the lacZ staining in the Enam+/lacZ mice and normal ameloblast-specific expression enamelin mRNA as determined by in situ hybridization (56) provides additional confirmation that the knock-in construct was targeted correctly.
FIGURE 2.
Expression of the NLS β-gal Enam+/lacZ mice at 2 weeks. A and B, β-galactosidase histostaining of mouse heads sectioned through the dentition show positive signal restricted to the teeth even when overstained. C and D, β-gal overstaining signal was observed in ameloblasts of molar (C), whereas the negative control of wild type mouse shows no histostaining (D). E, modestly overstained developing mandibular molars show signal concentrated over the nuclei of ameloblasts. Staining stops at the cemento-enamel junction; no staining was observed along the developing root (arrowheads). F-H, β-gal histostaining near the cervical loop of developing mandibular molars shows specific staining localized to the nuclei of ameloblasts. Odontoblasts are negative. Am, ameloblasts; e, enamel; d, dentin; Od, odontoblasts.
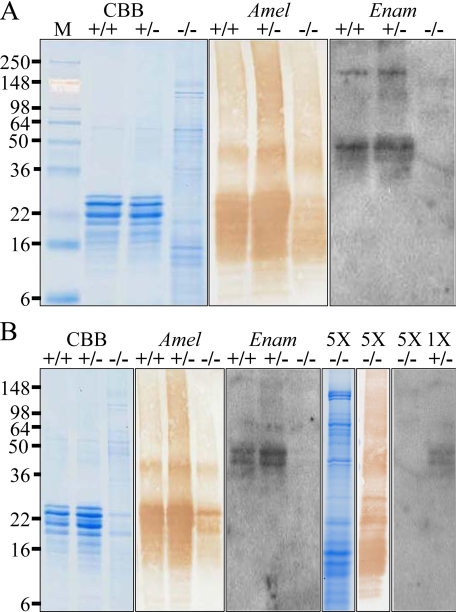
Western Blot Analyses—Additional evidence supporting the conclusion that Enam was successfully disrupted by homologous recombination with the targeting construct is provided by the results of Western blot analyses. Enamel proteins were extracted from day 7 incisors (Fig. 3A) and first molars (Fig. 3B), and equal quantities of proteins from Enam+/+, Enam+/-, and Enam-/- mice were analyzed by SDS-PAGE and Western blotting. Enamelin protein was easily detected in extracts from Enam+/+ and Enam+/- mice but not from the enamelin null mice (Fig. 3). Even when the proteins from the Enam-/- were loaded in five times the amount that could be detected in Enam+/- mice, no enamelin protein could be detected (Fig. 3B).
FIGURE 3.
Western blots of amelogenin and enamelin in day 7 enamel extracts. Shown are the matrix protein profiles of mandibular incisors (A) and molars (B) from wild type (+/+), heterozygous (+/-), and homozygous (-/-) mice observed on Coomassie Brilliant Blue (CCB)-stained SDS-PAGE and Western blots stained with polyclonal antibodies against recombinant mouse amelogenin (Amel) and anti-peptide antibodies raised against enamelin (Enam). One μg of protein from extracted unerupted teeth was applied to each lane. The protein profiles were similar for the wild type and heterozygous mouse samples. No enamelin was detected in the Western blot analyses of the molars or incisors of the Enam-/- mice. In the Enam-null mouse there was significantly less accumulated amelogenin (based upon the Western blot analysis), so the experiments were repeated using 5 μg of null-mouse molar sample (5X). No enamelin was detected on Western blots of null mouse tooth extracts, even when the lane was heavily loaded.
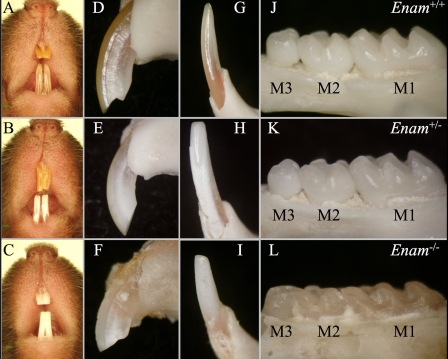
Gross Tooth Morphology and Appearance—Photographic evaluation of the mouse dentition at 7-weeks documents changes in tooth shape and color between the wild type and Enam mice (Fig. 4). All teeth in the Enam-/- mice lack enamel (Fig. 4, C, F, I, and L), whereas the Enam+/- mice exhibit defects that are obvious without magnification. The mandibular incisors of the Enam+/- mice are consistently chalky white in the erupted portion (Fig. 4, B and H), whereas the maxillary incisors are more variable, ranging from near normal (Fig. 4B) to chalky white (Fig. 4E). The mandibular incisor enamel near the functional part of the incisal edge is always missing, apparently being rapidly abraded during function, leaving a prominent wear facet on the labial surface. Enamel abrasion is also evident on the molar cusps of the Enam+/- mice (Fig. 4K). The degree of occlusal wear is most pronounced on the Enam-/- molars (Fig. 4L), which are flattened and rounded at the cusp tips.
FIGURE 4.
Photographic examination of the wild type (Enam+/+; top row), heterozygous (Enam+/-; middle row), and null (Enam-/-; bottom row) mouse dentitions at 7 weeks. The color of the incisor enamel displayed an enamelin dose effect, with an increasingly chalky appearance with less enamelin expression (A-C). The mandibular and maxillary incisors of wild type mice are brownish yellow in color (A). The mandibular incisors in the heterozygous mice are chalky white (B). The mandibular and maxillary incisors in the null mice are both chalky white (C). The surfaces of the maxillary and mandibular incisors of wild type mice (D and G) and the maxillary incisor of heterozygous mice (E) are smooth and unbroken. The surfaces of the maxillary and mandibular incisors of the enamelin-null mice (F and I) and the mandibular incisors of heterozygous mice (H) are rough and broken. The crowns of the first (M1), second (M2), and third (M3) molars were examined (J-L); the crown morphology of heterozygous mouse molars appeared normal, except for a subtle thinning and pitting of the enamel and hypoplasia of the M3 distal cusp (K). This contrasts with null mouse molars (L), which showed altered cusp morphology due to enamel hypoplasia or aplasia and severe coronal wear into the underlying dentin.
The maxillary incisors of Enam+/- mice often appeared normal although some teeth seemed to have less yellow-brown pigment in the enamel compared with others, and the wear patterns on the incisal tips of some teeth were not always uniform or as deeply grooved lingually as seen on the maxillary incisors of wild type animals. The mandibular incisors of Enam+/- mice, in contrast, appeared dramatically different, with loss of translucency and pigmentation and the presence of chalky white enamel covering along the labial sides of the incisors. Occlusal wear patterns on these teeth were also altered, with severely abraded enamel and exposed dentin evident along the central labial side in addition to the usual lingual side on erupted portions of the teeth. This resulted in incisal tips that had a blunted chisel shape rather than the evenly pointed shovel shape typical of wild type incisors.
Abnormalities of enamel were evident on both the maxillary and the mandibular incisors of Enam-/- mice. These teeth had a white opaque appearance and showed severe dentin abrasion along the labial and lingual sides of the teeth. The erupted portions of the incisors were coated with a thin layer of calcified material that felt gritty and sandpaper-like in consistency. The crowns of the molars were coated with a thin layer of similar calcified material and showed overt occlusal wear. Large nodules of calcified material were also seen in clusters along the palatal and lingual sides of some molar crowns. Some animals showed severe periodontal problems around the mandibular molars, with obvious bone loss, widened interdental spaces, and bifurcation openings in relation to the roots of the first and sometime second molars. There seemed to be abundant connective tissue infiltration within in the lamina propria under the gingival epithelium. There was also evidence for connective tissue infiltration in relation to the embedded portions of the incisors. Abundant pigment was seen in the enamel organ cells all across the maturation stage, but none was present in the calcified material covering these teeth. Underneath the cells the enamel surfaces were lumpy, often more so laterally than mesially. This calcified material was frail and easily dislodged from the tooth. Large nodular ridges and valleys of calcified material were seen in repeating waves across the maturation stage area, mostly at the central and mesial sides along the labial surface. There was a small apical area on the incisors where the cells looked normal.
Physical and Chemistry Assessments—The Enam+/- and Enam-/- mice could not be distinguished from their wild type littermates in terms of their physical appearance, level of activity, rate of growth, and overall body size. All animals were maintained on soft chow, and the heterozygous and homozygous Enam-deficient mice did not display any delay in growth or development when compared with their wild type littermates. There was no statistically significant difference among mice of the three genotypes in a panel of blood chemistry tests, including the serum levels of calcium, phosphate, alkaline phosphatase, and glucose, factors known to be involved in critical mineralization and physical activities (Table 2).
TABLE 2.
Blood chemistry tests a 6 weeks
| Test | Genotype | Result | Reference |
|---|---|---|---|
| Calcium | Enam+/+ | 9.45 ± 1.06 | 5.9-9.4 mg/dl |
| Enam+/− | 9.60 ± 0.75 | ||
| Enam−/− | 9.58 ± 0.56 | ||
| Phosphate | Enam+/+ | 15.95 ± 6.72 | 6.1-10.1 mg/dl |
| Enam+/− | 12.08 ± 2.09 | ||
| Enam−/− | 17.30 ± 6.83 | ||
| Alkaline phosphatase | Enam+/+ | 237.0 ± 200.8 | 62-209 units/liter |
| Enam+/− | 237.8 ± 155.6 | ||
| Enam−/− | 279.5 ± 87.07 | ||
| Glucose | Enam+/+ | 126.00 ± 33.94 | 90-192 mg/dl |
| Enam+/− | 119.60 ± 34.24 | ||
| Enam−/− | 157.75 ± 96.08 |
The conclusion that the targeting construct had knocked out Enam while knocking in lacZ was supported by Southern blot analyses, PCR genotyping, histochemistry, and the dental phenotype in the Enam+/- and Enam-/- mice. The blood chemistry results suggest that there was no systemic disturbance in calcium and phosphate homeostasis, whereas the β-gal histochemistry emphasized the specific expression of Enam by ameloblasts. Given the strong evidence that the targeting construct had specifically ablated Enam, we carried out a more rigorous characterization of the dental phenotype.
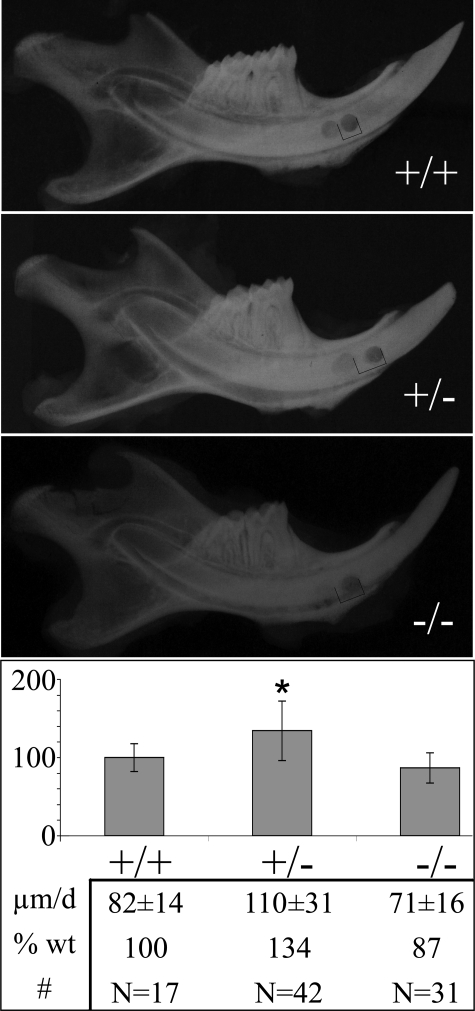
Incisor Eruption Rates—Rodent incisors continuously erupt as they form, and increasing the eruption rate leaves less time for the enamel layer to mature (harden) (57). Removing an incisor from occlusion with the opposing tooth (unimpeded growth) can double its rate of eruption (58). Although enamelin is not believed to play a role in tooth eruption, an increased eruption rate might result from faster occlusal wear or a failure of opposing teeth to contact in occlusion. As changes in eruption rate can affect the degree of mineralization, mandibular incisor eruption rates were measured in 5-6-week-old mice (Fig. 5). Typically, eruption rate studies notch the enamel and adjacent bone and measure the distance between the two notches after a week of eruption; however, it was not possible to notch the enamel in this study because of the fragility of the enamel layer in the Enam mice, so a circularly bored hole was made laterally through the bone and incisor, and the subjects were subsequently maintained on analgesics to minimize potential pain during mastication. Only minor differences in eruptions rates between the wild type and enamelin mice were observed. The average eruption rate for mandibular incisors of wild type mice was 82 μm/day. The rate for the Enam-null mice was 71 μm/day. The eruption rate for the heterozygous (Enam+/-) mice was somewhat higher (110 μm/day). All three rates were lower than those measured previously for mouse mandibular incisors (160 μm/day) (50), perhaps because of the use of soft diets and analgesics.
FIGURE 5.
Eruption rate determination. The eruption rates for the mandibular incisors from wild type (n = 17), heterozygous (n = 42), and homozygous (n = 31) mice at age 5-6 weeks were determined. The eruption rate of individual incisors from the heterozygous and the homozygous knock-in mice were normalized against the mean eruption rate of the wild type incisors. On average, the heterozygous incisors erupt at a rate of 134% of the wild type incisors, whereas the homozygous knock-in incisors erupt at an average of 87% of the wild type rate. The asterisk denotes statistical significance in Enam+/+ versus Enam+/- and Enam+/- versus Enam-/- mice at p < 0.001.
The somewhat accelerated mandibular incisor eruption rate determined for the Enam+/- mice may relate to differences in the way the maxillary and mandibular incisors are affected. Most maxillary incisors of Enam+/- mice appear normal (Fig. 4B). The lower incisors of Enam+/- mice, in contrast, consistently display a loss of translucency and pigmentation, with severely abraded enamel and exposed dentin at the contact point (Fig. 4H). Perhaps the weaker mandibular incisors of the Enam+/- mice functioning against harder maxillary incisors increases the wear rate of the mandibular incisors and this attrition allows for their faster eruption. The modest differences in eruption rates among the Enam+/+, Enam+/-, and Enam-/- mice are not likely to cause significant differences in the degree of mineralization of the various experimental mice under investigation.
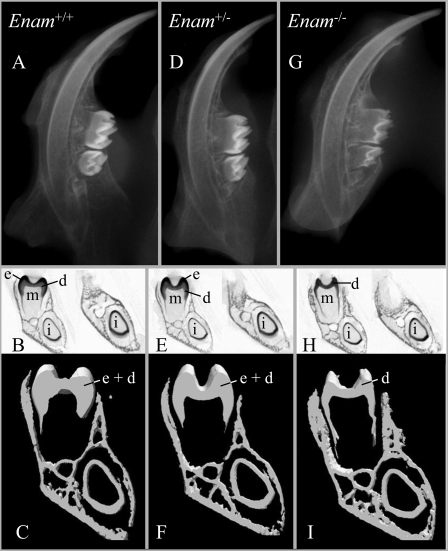
Radiography—Dental radiographs demonstrated that the radio-opacity of the mouse mandibular dental crowns at 2 weeks decreases progressively from the wild type (Fig. 6A) to the Enam+/- (Fig. 6D) to the Enam-/- mice (Fig. 6G), with the reduction in tooth mineralization being most obvious in the molars. The mineralized crowns of the Enam-/- mice were demonstrably thinner and less dense than in the wild type, suggesting that the Enam-null mice might lack enamel altogether. Microcomputed tomography scans (Fig. 6, B, E, and H) at the level of the first mandibular molar and in the diastema region (between the molars and incisors) show that the mineralized enamel layer in the Enam-/- mice is either too thin to be detected or absent (Fig. 6H), whereas enamel is clearly present in the Enam+/+ (Fig. 6B) and Enam+/- (Fig. 6E) mice. Microcomputed tomography (μCT) analyses of mandibular frontal sections through the mesial and buccal cusps of M1 followed by three-dimensionalized reconstructions (Fig. 6C, F, and I) provided images that could be used to assess the mineralized tissue volume and macroscopic tooth morphology of the Enam mice. The μCT reconstructions revealed a thinning of the mineralized tissue (from a lack of mineralized enamel) in the molar crown and incisor crown from Enam-/- mice (Fig. 6I) relative to the wild type (Fig. 6C) and Enam+/- (Fig. 6F) mice, whereas no differences were observed in bone and dentin. The radiography and μCT analyses suggest a total or near total lack of dental enamel in the Enam-/- mice. No differences in volume or tooth morphology could be discerned between the wild type and Enam+/- mice.
FIGURE 6.
Radiographs, μCT x-ray single scans, and three-dimensional reconstructions of mandibles from 2-week-old Enam+/+, Enam+/-, and Enam-/- mice. Radiography of hemi-mandibles (A, D, and G) shows reduced tooth mineralization in the Enam-/- mice, particularly in the molars. Single μCT scans (B, E, and H) at the level of the first molar (left images) and more incisally in the diastema region (right images) show that the mineralized enamel layer is either too thin to be detected or is absent in the Enam-/- mice (H), whereas it is clearly present in the Enam+/+ (B) and Enam+/- (E) mice where the enamel (e) can be readily discriminated from dentin (d). Microcomputed tomography analyses of mandibular frontal sections through the mesial and buccal cusps of M1 followed by three-dimensionalized reconstructions (C, F, and I) show mineralized tissue volume and macroscopic tooth morphology, revealing a thinning of mineralized tissue (from a lack of mineralized enamel) in the molar crown (m) and incisor crown (i) analog from Enam-/- mice (I) relative to Enam+/+ (C) and Enam+/- (F) mice where differences are not distinguishable. No differences were observed in bone.
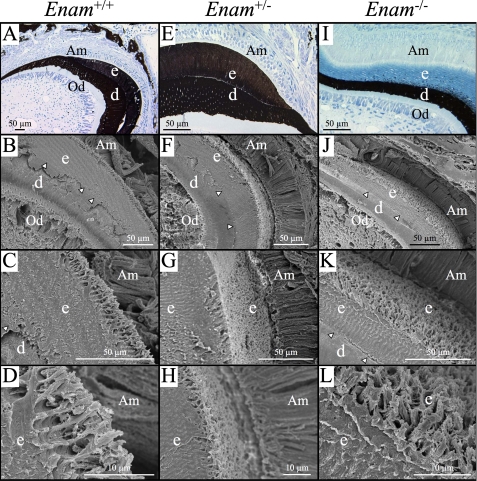
Microscopic Analyses—Developing mandibular molars of the 1-week-old mice were examined histologically after von Kossa staining (Fig. 7, A, E, and I), which stains mineralized tissues brown to black but does not stain osteoid or predentin. A dramatic finding was that the secretory stage enamel layer of the Enam-/- mice was von Kossa-negative, except for small nodules (mineralization foci) that increased in number near the dentino-enamel junction (DEJ) (Fig. 7I). No von Kossa staining was observed at the mineralization front, which is along the secretory aspect of the distal membrane of the ameloblasts (at the surface of the enamel layer). The secretory stage enamel of the Enam+/+ and Enam+/- mice stained throughout, although the enamel of the heterozygous mice did not stain as deeply as that of the wild type mice. This finding strongly suggests that without enamelin, enamel proteins accumulate in the enamel space but cannot catalyze the extension of enamel crystals. Therefore, none of the mineral covering dentin in the Enam-/- mice is true enamel, which normally forms by the elongation of thin crystal ribbons (about 10,000 beneath each cell) at the mineralization front that subsequently grow in width and thickness to harden the enamel (1).
FIGURE 7.
Histology (A, E, and I) and scanning electron micrographs (B-D, F-H, and J-L) of Enam+/+, Enam+/-, and Enam-/- mice at 7 days. A, E, and I, histology sections of unerupted, undecalcified molars after von Kossa staining to show mineralization of alveolar bone, dentin, and enamel in the various genotypes, as indicated. Whereas well developed and mineralized enamel (e) and dentin (d) were present in both wild type (A) and heterozygous (E) mice, only small, punctate foci of mineralization were detected within the enamel layer near the dentino-enamel junction, despite there being a thick accumulation of organic material in the enamel layer, presumably amelogenin and other enamel matrix constituents (I). SEM images of similar regions in each of the genotypes are shown at successively increasing magnification in each column. The forming enamel in the heterozygous mice (F-H) appears generally similar to that in wild type (B-D) mice. The forming enamel in the Enam-/- mouse (J-L), despite being unmineralized, shows remarkable similarity to normal enamel matrix structure. Am, ameloblasts; Od, odontoblasts; arrowheads indicate the dentino-enamel junction.
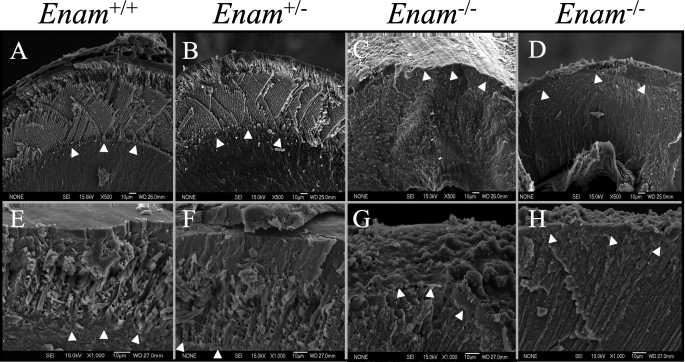
Scanning electron micrographs of 1-week mouse molars and incisors revealed an early enamel layer with the distinctive pattern of inner and outer enamel (59). Unexpectedly, the minimally calcified accumulated enamel matrix of the Enam-null mice also showed patterns suggestive of enamel rods that even changed patterns between the inner and outer enamel (Fig. 7, J-L). An SEM of teeth at 7 weeks (Fig. 8) showed enamel in the maturation stage of development. Organized enamel rods were clearly evident in the Enam+/+ and Enam+/- mice (Fig. 8, A and B), but were completely absent from the Enam-null mice. In the Enam-/- mice the accumulated enamel proteins were absent, and only a very thin (0-20 μm) layer of mineral capped the dentin. The surface of the crown was very rough and looked to comprise isolated or fused calcospherites (Fig. 8G). The molars in Enam+/- mice grossly appeared normal, but SEM imaging indicated that the enamel on some teeth appeared thinner than normal, and there were focal areas of roughening and pitting of the enamel surface that were most often noticeable along the distal lingual side of the second molar and mesial lingual side of the third molar.
FIGURE 8.
SEM analysis of erupted mouse incisors and molars at 7 weeks. SEM was used to examine fractured sections of mouse incisors (A-D) and molars (E-H). The enamel of wild type (A and E) and heterozygous (B and F) both showed a thick enamel layer with well defined rod (prism) structures. In contrast, the enamel of Enam-null mice (C, D, G, and H) was extremely thin and irregular, with a rough surface. In some places (G), the enamel did not even form sufficiently to complete the DEJ. Arrowheads delineate the DEJ.
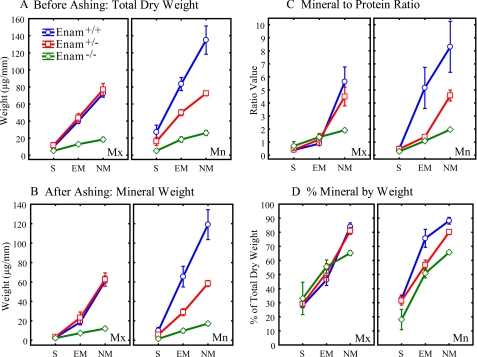
Mineral and Organic Content of Incisor Enamel—Whole pieces of developing enamel of equally sized length were removed sequentially from the apical (early) to the incisal (late) ends of mouse incisors. Each strip was dried and weighed giving its total dry weight (mineral plus protein). Each increment was ashed, which vaporized the organic material, and reweighed, giving the ashed weight (mineral only) of the strip. The protein content is the total dry weight minus the ashed weight. The percent mineral equals the (ashed weight/total dry weight) × 100. The data were averaged by major developmental stages (see supplemental table) and plotted for the Enam+/+, Enam+/-, and Enam-/- mice (Fig. 9). The total dry weights and mineral weights of enamel strips on maxillary incisors from the Enam+/+ and Enam+/- mice were indistinguishable. In contrast, the mandibular incisors of Enam+/- mice had total dry weights (mineral plus protein) and mineral weights that were almost exactly half of those measured for enamel strips at equivalent developmental stages in wild type mice (Fig. 9, A and B). The mineral to protein ratio in enamel on mandibular incisors of the Enam+/- mice (Fig. 9C) was normal during the secretory stage, markedly below normal during early maturation when enamel crystals start growing in volume, and still 50% below normal at a time when the enamel in wild type mice was almost too hard to cut with a scalpel blade (nearly mature). Similar trends were noted for the percent mineral by weight (Fig. 9D). It was possible to remove enamel strips from the erupted portions of these teeth, indicative of continued below normal hardness compared with enamel on mandibular incisors of wild type mice (data not shown).
FIGURE 9.
Mineral and protein content of incisors at 7 weeks. The developing enamel covering continuously growing maxillary and mandibular incisors of wild type (Enam+/+, blue circles), heterozygous (Enam+/-, red squares), and null (Enam-/-, green diamonds) mice was transected into a series of sequential strips from the apical toward the incisal ends. Strips associated with three main stages of enamel development (S, secretory; EM, early maturation; NM, nearly mature enamel) were pooled together from at least a dozen teeth per genotype to compute means ± 95% confidence intervals for the total dry weight before ashing (A), mineral weight after ashing (B), mineral-to-protein ratio (C), and percent mineral by weight (D). No differences in mineral content of developing enamel were observed on maxillary incisors of Enam+/+ and Enam+/- mice in contrast to Enam-/- mice, where enamel mineral content was dramatically less and reached a maximum of only about 66% mineral by weight (D). The enamel on the mandibular incisors of Enam+/- mice was softer compared with Enam+/+ mice, and strips could be cut along the entire length of these teeth. Overall, the mandibular incisors showed the clearest indication of a “dose effect” between wild-type, heterozygous, and Enam-null mice.
We were able to remove strips of calcified material along the entire length of the maxillary and mandibular incisors in Enam-null mice. The maxillary and mandibular incisors both showed dramatically lower values for total dry weight and mineral weight across the lengths of the incisors (Fig. 9, A and B), measuring only about 15% of the values determined for wild type mice. The mineral to protein ratios were only 25% of normal (Fig. 9C), and the percent mineral by weight at an advanced stage of development was 66%. In contrast, the nearly mature enamel in wild type mice was 85-90% mineral by weight (Fig. 9D). The mineral to protein ratio and percent mineral by weight for the Enam-null mice were almost identical to values measured independently for normal compact bone (data not shown).
Enamelin and Crystal Growth—Most of the 186-kDa porcine enamelin protein is degraded and reabsorbed from the enamel matrix, with the exception of a 32-kDa cleavage product that accumulates to about 1% of total enamel protein (60). HA crystals were grown in agarose gels in the presence of 0-50 μg/ml porcine 32-kDa enamelin. X-ray diffraction line-broadening analysis showed that crystal length (d002) was 537 ± 22.2 nm in the absence of 32-kDa enamelin and increased with protein concentration to 668 ± 23.0 nm at 50 μg/ml. HA crystal cross-section (d310) did not change with enamelin concentration (265 ± 8.20 nm and 262 ± 5.19 nm at 0 and 50 μg/ml, respectively). Therefore, 32-kDa enamelin significantly increased the length of crystals grown in agarose gels, whereas not affecting the crystal cross-section. This suggests that enamelin is promoting HA growth along the crystallographic c-axis.
DISCUSSION
Amelogenin protein dominates the enamel matrix of developing teeth. The non-amelogenin proteins (ameloblastin and enamelin) are in low abundance and are much more difficult to isolate and characterize. Because of its large teeth and availability from the food industry, the pig is the main animal model used for studying enamel proteins (61). Pig enamelin is secreted as a 186-kDa (1104 amino acids) glycoprotein that is rapidly processed, probably by MMP-20, in the extracellular space (21). The secreted protein is processed from its C terminus to generate 155-, 145-, and 89-kDa (Met1-Trp627) N-terminal cleavage products, but these cleavage products do not last long and are found only near the enamel surface (41, 62). Secretion, proteolytic processing, and reabsorption of selected cleavage products back into ameloblasts determine the makeup of the organic matrix that controls mineralization. The C-terminal portion of enamelin is particularly short-lived, so that anti-peptide antibodies specific for the enamelin C-terminal region show intense immunostaining only at the mineralization front where they are secreted, and no signal is observed deeper in the enamel (8). In contrast, antibodies raised to other regions of the enamelin protein stain the entire thickness of enamel (60, 64, 65). The specific localization of the intact enamelin protein (by the C-terminal antibody) at the mineralization front has long suggested that enamelin is critical for enamel crystal elongation. Despite the accumulation of a thick layer of enamel proteins (presumably mostly amelogenin) in the extracellular space beneath secretory ameloblasts in the Enam-/- mice, no mineralization occurs at the mineralization front. This strongly suggests that enamelin is a critical component of the mineralization front that promotes or catalyzes the extension of enamel crystals along their c-axes (66). Furthermore, because the initial enamel crystals that appear suddenly at the DEJ are virtually identical to those extending at the mineralization front (4, 67) and because the rapid onset of enamel crystal formation appears to be absent in the Enam-/- mice, it may be that the initiation and elongation of enamel crystallites occurs by the same or a related enamelin-dependent mechanism. Ultrastructural analyses of the DEJ and mineralization fronts of un-demineralized sections of developing teeth would help clarify the validity of these conclusions, a work that is in progress.
The small amount of calcified material that covers dentin in the Enam-/- mice seems to be more bone-like than true enamel, although it is of course not bone and is the product of ameloblasts (not osteoblasts). Both the mineral to protein ratio and percent mineral by weight are identical to values calculated for compact bone. We concluded that there is no true enamel covering the dentin in the Enam-/- mice. The mineral that forms does so by a completely different mechanism than for true enamel. It appears to precipitate deep within the unmineralized enamel protein layer as sporadic calcospherites that expand and eventually fuse, forming a weak crust covering the dentin that easily crumbles during function. These findings emphasize the importance of the mineralization front located at the secretory surface of the ameloblast distal membrane as being central to the formation of true enamel. The ribbon-like shape of the enamel crystals and their organization into rods is established at the mineralization front and is enamelin-dependent.
Besides the secreted, intact glycoprotein, enamelin cleavage products accumulate as the enamel layer expands and may also play important roles in amelogenesis. Most of the diverse, poorly characterized enamelin cleavage products have little affinity for enamel crystals and accumulate in the sheath space between the enamel rods, along with the N-terminal fragments of ameloblastin (62). The most abundant enamelin cleavage product is the 32-kDa enamelin glycoprotein (Leu136-Arg241) that accumulates among the crystallites (in enamel rods) throughout the entire thickness of secretory stage enamel (60, 64). This 106-amino acid domain contains two phosphoserines and three N-linked glycosylations (68, 69). The 32-kDa enamelin does not engage in protein-protein interactions with amelogenin (70) but shows relatively strong affinity for binding to hydroxyapatite (71). This part of enamelin accumulates to relatively high levels because its extensive post-translational modifications make it resistant to further degradation by MMP-20 (72), which is the predominant proteolytic activity in secretory stage enamel (73). In vitro the 32-kDa enamelin increases the length but not the cross-section of HA crystals grown in agarose gels. These results suggest that the 32-kDa enamelin might play a role in shaping enamel crystallites in vivo.
The NLS β-gal reporter expressed in place of enamelin vividly demonstrates the highly restricted pattern of Enam expression. If enamelin is expressed by any cell other than ameloblasts, it must indeed be at very low levels relative to secretory ameloblasts. This highly restricted pattern of expression contrasts with amelogenin, which is strongly expressed by ameloblasts but can be detected in many other tissues. This seems to suggest that marginal enamelin expression in other tissues might be deleterious and selected against, whereas marginal amelogenin expression may be more readily tolerated. Using transgenic mouse models, it was determined that a 4.5-kb Enam promoter segment upstream of the translation initiation site can drive tissue-specific expression of the mouse enamelin gene (although at lower than normal levels); however, a 3.2-kb Enam promoter construct was expressed predominantly by osteoblasts in the periosteum and not by ameloblasts. The promoter sequence between 3.2- and 4.5-kb region might contain DNA sequence motifs for transcription factor binding that direct the tooth-specific gene expression of enamelin (63).
In summary, we have generated and characterized an Enam knock-out/NLS β-gal knock-in mouse. The results show that enamelin is specifically expressed by ameloblasts and that enamelin expression is necessary for proper functioning of the mineralization front associated with the ameloblast secretory surface; without enamelin there is no true enamel. Enamelin defects are dose-dependent in mice, as shown previously for humans. Understanding enamelin function is central to understanding the molecular mechanisms regulating enamel formation during tooth development.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Saunders (University of Michigan), Dr. Ashok Kulkarni (NIDCR/National Institutes of Health), and Lydia Malynowsky and Zoe Venetis (both of McGill University) for their insightful consultations and technical support. X-ray diffraction analysis was performed by Dr. Marc Grynpas at Mount Sinai Hospital, Toronto. Final design and fabrication of the knock-in construct and successful establishment of the mouse founders were accomplished by Ozgene Ltd., Australia.
This study was supported by NIDCR/National Institutes of Health Grant Project DE 11301. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental table.
Footnotes
The abbreviations used are: MMP-20, matrix metalloproteinase 20; β-gal, β-galactosidase; X-gal, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; KLK4, kallikrein 4; Neo, neomycin; NLS, nuclear localization signal; PBS, phosphate-buffered saline; PGK, phosphoglycerine kinase; DEJ, dentino-enamel junction; HA, hydroxyapatite; μCT, microcomputed tomography; SEM, scanning electron microscopy.
For detailed information on these analyses, contact the authors.
References
- 1.Daculsi, G., and Kerebel, B. (1978) J. Ultrastruct. Res. 65 163-172 [DOI] [PubMed] [Google Scholar]
- 2.Nanci, A. (ed) (2003) in Ten Cate's Oral Histology Development, Structure, and Function, 6th Ed., pp. 218-224, Mosby, St. Louis, MO
- 3.Fincham, A. G., Moradian-Oldak, J., and Simmer, J. P. (1999) J. Struct. Biol. 126 270-299 [DOI] [PubMed] [Google Scholar]
- 4.Ronnholm, E. (1962) J. Ultrastruct. Res. 6 249-303 [DOI] [PubMed] [Google Scholar]
- 5.Snead, M. L., Zeichner-David, M., Chandra, T., Robson, K. J., Woo, S. L., and Slavkin, H. C. (1983) Proc. Natl. Acad. Sci. U. S. A. 80 7254-7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snead, M. L., Lau, E. C., Zeichner-David, M., Fincham, A. G., Woo, S. L., and Slavkin, H. C. (1985) Biochem. Biophys. Res. Commun. 129 812-818 [DOI] [PubMed] [Google Scholar]
- 7.Krebsbach, P. H., Lee, S. K., Matsuki, Y., Kozak, C. A., Yamada, K., and Yamada, Y. (1996) J. Biol. Chem. 271 4431-4435 [DOI] [PubMed] [Google Scholar]
- 8.Hu, C.-C., Fukae, M., Uchida, T., Qian, Q., Zhang, C. H., Ryu, O. H., Tanabe, T., Yamakoshi, Y., Murakami, C., Dohi, N., Shimizu, M., and Simmer, J. P. (1997) J. Dent. Res. 76 1720-1729 [DOI] [PubMed] [Google Scholar]
- 9.Hu, C. C., Hart, T. C., Dupont, B. R., Chen, J. J., Sun, X., Qian, Q., Zhang, C. H., Jiang, H., Mattern, V. L., Wright, J. T., and Simmer, J. P. (2000) J. Dent. Res. 79 912-919 [DOI] [PubMed] [Google Scholar]
- 10.Kawasaki, K., and Weiss, K. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4060-4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau, E. C., Mohandas, T. K., Shapiro, L. J., Slavkin, H. C., and Snead, M. L. (1989) Genomics 4 162-168 [DOI] [PubMed] [Google Scholar]
- 12.Salido, E. C., Yen, P. H., Koprivnikar, K., Yu, L. C., and Shapiro, L. J. (1992) Am. J. Hum. Genet. 50 303-316 [PMC free article] [PubMed] [Google Scholar]
- 13.Wright, J. T. (2006) Am. J. Med. Genet. A 140 2547-2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu, J. C., Chun, Y. H., Al Hazzazzi, T., and Simmer, J. P. (2007) Cells Tissues Organs 186 78-85 [DOI] [PubMed] [Google Scholar]
- 15.Gibson, C. W., Yuan, Z. A., Hall, B., Longenecker, G., Chen, E., Thyagarajan, T., Sreenath, T., Wright, J. T., Decker, S., Piddington, R., Harrison, G., and Kulkarni, A. B. (2001) J. Biol. Chem. 276 31871-31875 [DOI] [PubMed] [Google Scholar]
- 16.Li, Y., Yuan, Z. A., Aragon, M. A., Kulkarni, A. B., and Gibson, C. W. (2006) Eur. J. Oral. Sci. 114 Suppl. 1, 190-193, 201-192, 381 [DOI] [PubMed] [Google Scholar]
- 17.Deutsch, D., Haze-Filderman, A., Blumenfeld, A., Dafni, L., Leiser, Y., Shay, B., Gruenbaum-Cohen, Y., Rosenfeld, E., Fermon, E., Zimmermann, B., Haegewald, S., Bernimoulin, J. P., and Taylor, A. L. (2006) Eur. J. Oral. Sci. 114 Suppl. 1, 183-189, 201-182, 381 [DOI] [PubMed] [Google Scholar]
- 18.Fukumoto, S., Kiba, T., Hall, B., Iehara, N., Nakamura, T., Longenecker, G., Krebsbach, P. H., Nanci, A., Kulkarni, A. B., and Yamada, Y. (2004) J. Cell Biol. 167 973-983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajpar, M. H., Harley, K., Laing, C., Davies, R. M., and Dixon, M. J. (2001) Hum. Mol. Genet 10 1673-1677 [DOI] [PubMed] [Google Scholar]
- 20.Kim, J. W., Seymen, F., Lin, B. P., Kiziltan, B., Gencay, K., Simmer, J. P., and Hu, J. C. (2005) J. Dent. Res. 84 278-282 [DOI] [PubMed] [Google Scholar]
- 21.Hu, J. C., and Yamakoshi, Y. (2003) Crit. Rev. Oral Biol. Med. 14 387-398 [DOI] [PubMed] [Google Scholar]
- 22.Hart, T. C., Hart, P. S., Gorry, M. C., Michalec, M. D., Ryu, O. H., Uygur, C., Ozdemir, D., Firatli, S., Aren, G., and Firatli, E. (2003) J. Med. Genet. 40 900-906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozdemir, D., Hart, P. S., Firatli, E., Aren, G., Ryu, O. H., and Hart, T. C. (2005) J. Dent. Res. 84 1036-1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masuya, H., Shimizu, K., Sezutsu, H., Sakuraba, Y., Nagano, J., Shimizu, A., Fujimoto, N., Kawai, A., Miura, I., Kaneda, H., Kobayashi, K., Ishijima, J., Maeda, T., Gondo, Y., Noda, T., Wakana, S., and Shiroishi, T. (2005) Hum. Mol. Genet. 14 575-583 [DOI] [PubMed] [Google Scholar]
- 25.Seedorf, H., Klaften, M., Eke, F., Fuchs, H., Seedorf, U., and Hrabe de Angelis, M. (2007) J. Dent. Res. 86 764-768 [DOI] [PubMed] [Google Scholar]
- 26.Bartlett, J. D., Simmer, J. P., Xue, J., Margolis, H. C., and Moreno, E. C. (1996) Gene 183 123-128 [DOI] [PubMed] [Google Scholar]
- 27.Simmer, J. P., Fukae, M., Tanabe, T., Yamakoshi, Y., Uchida, T., Xue, J., Margolis, H. C., Shimizu, M., DeHart, B. C., Hu, C. C., and Bartlett, J. D. (1998) J. Dent. Res. 77 377-386 [DOI] [PubMed] [Google Scholar]
- 28.Hu, J. C., Sun, X., Zhang, C., Liu, S., Bartlett, J. D., and Simmer, J. P. (2002) Eur. J. Oral Sci. 110 307-315 [DOI] [PubMed] [Google Scholar]
- 29.Simmer, J. P., Sun, X., Yamada, Y., Zhang, C. H., Bartlett, J. D., and Hu, J. C.-C. (2004) in Proceedings of the 8th International Symposium on Biomineralization, Niigata, Japan, September 25-28, 2001 (Kobayashi, I., and Ozawa, H., eds) pp. 348-352, Tokai University Press, Hadano, Japan
- 30.Hart, P. S., Hart, T. C., Michalec, M. D., Ryu, O. H., Simmons, D., Hong, S., and Wright, J. T. (2004) J. Med. Genet. 41 545-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, J. W., Simmer, J. P., Hart, T. C., Hart, P. S., Ramaswami, M. D., Bartlett, J. D., and Hu, J. C. (2005) J. Med. Genet. 42 271-275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozdemir, D., Hart, P. S., Ryu, O. H., Choi, S. J., Ozdemir-Karatas, M., Firatli, E., Piesco, N., and Hart, T. C. (2005) J. Dent. Res. 84 1031-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caterina, J. J., Skobe, Z., Shi, J., Ding, Y., Simmer, J. P., Birkedal-Hansen, H., and Bartlett, J. D. (2002) J. Biol. Chem. 277 49598-49604 [DOI] [PubMed] [Google Scholar]
- 34.Bartlett, J. D., Beniash, E., Lee, D. H., and Smith, C. E. (2004) J. Dent. Res. 83 909-913 [DOI] [PubMed] [Google Scholar]
- 35.Bartlett, J. D., Ryu, O. H., Xue, J., Simmer, J. P., and Margolis, H. C. (1998) Connect. Tissue Res. 39 405-413 [DOI] [PubMed] [Google Scholar]
- 36.Nelson, P. S., Gan, L., Ferguson, C., Moss, P., Gelinas, R., Hood, L., and Wang, K. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 3114-3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu, J. C., Zhang, C., Sun, X., Yang, Y., Cao, X., Ryu, O., and Simmer, J. P. (2000) Gene 251 1-8 [DOI] [PubMed] [Google Scholar]
- 38.Obiezu, C. V., Shan, S. J., Soosaipillai, A., Luo, L. Y., Grass, L., Sotiropoulou, G., Petraki, C. D., Papanastasiou, P. A., Levesque, M. A., and Diamandis, E. P. (2005) Clin. Chem. 51 1432-1442 [DOI] [PubMed] [Google Scholar]
- 39.Kim, J. W., Simmer, J. P., Lin, B. P., Seymen, F., Bartlett, J. D., and Hu, J. C. (2006) Eur. J. Oral. Sci. 114 Suppl. 1, 3-12 [DOI] [PubMed] [Google Scholar]
- 40.Yamakoshi, Y., Hu, J. C.-C., Ryu, O. H., Tanabe, T., Oida, S., Fukae, M., and Simmer, J. P. (2003) in Proceedings of the 8th International Symposium on Biomineralization, Niigata, Japan, September25-28, 2001 (Kobayashi, I., and Ozawa, H., eds) pp. 338-342, Tokai University Press, Hadano, Japan
- 41.Fukae, M., Tanabe, T., Murakami, C., Dohi, N., Uchida, T., and Shimizu, M. (1996) Adv. Dent. Res. 10 111-118 [DOI] [PubMed] [Google Scholar]
- 42.Hu, C.-C., Fukae, M., Uchida, T., Qian, Q., Zhang, C. H., Ryu, O. H., Tanabe, T., Yamakoshi, Y., Murakami, C., Dohi, N., Shimizu, M., and Simmer, J. P. (1997) J. Dent. Res. 76 648-657 [DOI] [PubMed] [Google Scholar]
- 43.Iwata, T., Yamakoshi, Y., Hu, J. C., Ishikawa, I., Bartlett, J. D., Krebsbach, P. H., and Simmer, J. P. (2007) J. Dent. Res. 86 153-157 [DOI] [PubMed] [Google Scholar]
- 44.Lobe, C. G., Koop, K. E., Kreppner, W., Lomeli, H., Gertsenstein, M., and Nagy, A. (1999) Dev. Biol. 208 281-292 [DOI] [PubMed] [Google Scholar]
- 45.Chai, Y., Jiang, X., Ito, Y., Bringas, P., Jr., Han, J., Rowitch, D., Soriano, P., McMahon, A., and Sucov, H. (2000) Development (Camb.) 127 1671-1679 [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Z., Song, Y., Zhao, X., Zhang, X., Fermin, C., and Chen, Y. (2002) Development (Camb.) 129 4135-4146 [DOI] [PubMed] [Google Scholar]
- 47.Rosen, D. (1981) Neuropathol. Appl. Neurobiol. 7 331-340 [DOI] [PubMed] [Google Scholar]
- 48.Young, D. C., Kingsley, S. D., Ryan, K. A., and Dutko, F. J. (1993) Anal. Biochem. 215 24-30 [DOI] [PubMed] [Google Scholar]
- 49.Simmer, J. P., Lau, E. C., Hu, C. C., Aoba, T., Lacey, M., Nelson, D., Zeichner-David, M., Snead, M. L., Slavkin, H. C., and Fincham, A. G. (1994) Calcif. Tissue Int. 54 312-319 [DOI] [PubMed] [Google Scholar]
- 50.Ness, A. R. (1965) Arch. Oral Biol. 10 439-451 [DOI] [PubMed] [Google Scholar]
- 51.Hwang, W. S., and Tonna, E. A. (1965) J. Dent. Res. 44 42-53 [DOI] [PubMed] [Google Scholar]
- 52.Smith, C. E., Chong, D. L., Bartlett, J. D., and Margolis, H. C. (2005) J. Bone Miner. Res. 20 240-249 [DOI] [PubMed] [Google Scholar]
- 53.Pampena, D. A., Robertson, K. A., Litvinova, O., Lajoie, G., Goldberg, H. A., and Hunter, G. K. (2004) Biochem. J. 378 1083-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grynpas, M. D., and Hunter, G. K. (1988) J. Bone Miner. Res. 3 159-164 [DOI] [PubMed] [Google Scholar]
- 55.Weiss, D. J., Liggitt, D., and Clark, J. G. (1999) Histochem. J. 31 231-236 [DOI] [PubMed] [Google Scholar]
- 56.Hu, J. C., Sun, X., Zhang, C., and Simmer, J. P. (2001) Eur. J. Oral Sci. 109 125-132 [DOI] [PubMed] [Google Scholar]
- 57.Robinson, C., Kirkham, J., and Nutman, C. A. (1988) Cell Tissue Res. 254 655-658 [DOI] [PubMed] [Google Scholar]
- 58.Risnes, S., Septier, D., and Goldberg, M. (1995) Connect. Tissue Res. 32 183-189 [DOI] [PubMed] [Google Scholar]
- 59.Moinichen, C. B., Lyngstadaas, S. P., and Risnes, S. (1996) J. Anat. 189 325-333 [PMC free article] [PubMed] [Google Scholar]
- 60.Tanabe, T., Aoba, T., Moreno, E. C., Fukae, M., and Shimizu, M. (1990) Calcif. Tissue Int. 46 205-215 [DOI] [PubMed] [Google Scholar]
- 61.Hu, J. C., Yamakoshi, Y., Yamakoshi, F., Krebsbach, P. H., and Simmer, J. P. (2005) Cells Tissues Organs 181 219-231 [DOI] [PubMed] [Google Scholar]
- 62.Dohi, N., Murakami, C., Tanabe, T., Yamakoshi, Y., Fukae, M., Yamamoto, Y., Wakida, K., Shimizu, M., Simmer, J. P., Kurihara, H., and Uchida, T. (1998) Cell Tissue Res. 293 313-325 [DOI] [PubMed] [Google Scholar]
- 63.Hu, Y., Papagerakis, P., Ye, L., Feng, J. Q., Simmer, J. P., and Hu, J. C.-C. Eur. J. Oral. Sci., in press [DOI] [PMC free article] [PubMed]
- 64.Uchida, T., Tanabe, T., Fukae, M., and Shimizu, M. (1991) Arch. Histol. Cytol. 54 527-538 [DOI] [PubMed] [Google Scholar]
- 65.Uchida, T., Tanabe, T., Fukae, M., Shimizu, M., Yamada, M., Miake, K., and Kobayashi, S. (1991) Histochemistry 96 129-138 [DOI] [PubMed] [Google Scholar]
- 66.Simmer, J. P., and Fincham, A. G. (1995) Crit. Rev. Oral Biol. Med. 6 84-108 [DOI] [PubMed] [Google Scholar]
- 67.Nanci, A. (ed) (2008) in Ten Cate's Oral Histology Development, Structure, and Function, 7th Ed., pp. 141-190, Mosby, St. Louis, MO
- 68.Yamakoshi, Y. (1995) Calcif. Tissue Int. 56 323-330 [DOI] [PubMed] [Google Scholar]
- 69.Yamakoshi, Y., Pinheiro, F. H., Tanabe, T., Fukae, M., and Shimizu, M. (1998) Connect. Tissue Res. 39 39-46, 63-37 [DOI] [PubMed] [Google Scholar]
- 70.Yamakoshi, Y., Hu, J.-C., Fukae, M., Tanabe, T., Oida, S., and Simmer, J. (2004) in Biomineralization (Biom2001): Formation, Diversity, Evolution and Application (Kobayashi, I., and Ozawa, H., eds) pp. 326-332, Tokai University Press, Kanagawa, Japan
- 71.Yamakoshi, Y., Tanabe, T., Oida, S., Hu, C. C., Simmer, J. P., and Fukae, M. (2001) Arch. Oral Biol. 46 1005-1014 [DOI] [PubMed] [Google Scholar]
- 72.Yamakoshi, Y., Hu, J. C.-C., Fukae, M., Iwata, T., and Simmer, J. P. (2006) Eur. J. Oral Sci. 114 Suppl. 1, 266-271 [DOI] [PubMed] [Google Scholar]
- 73.Ryu, O. H., Fincham, A. G., Hu, C. C., Zhang, C., Qian, Q., Bartlett, J. D., and Simmer, J. P. (1999) J. Dent. Res. 78 743-750 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.