Abstract
Basic Local Alignment Search Tool (BLAST) is a sequence similarity search program. The public interface of BLAST, http://www.ncbi.nlm.nih.gov/blast, at the NCBI website has recently been reengineered to improve usability and performance. Key new features include simplified search forms, improved navigation, a list of recent BLAST results, saved search strategies and a documentation directory. Here, we describe the BLAST web application's new features, explain design decisions and outline plans for future improvement.
INTRODUCTION
Basic Local Alignment Search Tool (BLAST) is a sequence similarity search program that can be used via a web interface or as a stand-alone tool to compare a user's query to a database of sequences (1,2). Several variants of BLAST compare all combinations of nucleotide or protein queries with nucleotide or protein databases. BLAST is a heuristic that finds short matches between two sequences and attempts to start alignments from these ‘hot spots’. In addition to performing alignments, BLAST provides statistical information about an alignment; this is the ‘expect’ value or false-positive rate.
BLAST is one of the most widely used bioinformatics research tools, yet until recently, its web interface had numerous usability problems. The first phase of the redesign, on which we report here, defines consistent navigation between pages, offers new features such as the ability to save search parameter sets, and provides easy access to formatting controls, recent results and documentation.
BLAST INTERFACE USABILITY PROBLEMS
The legacy BLAST web interface grew incrementally as a group of web forms acting as a front-end to a growing collection of BLAST algorithms and programs. The resulting gradual accretion of features caused a host of usability problems. Form design was inconsistent and page navigation was difficult. There was no way to get a list of recent searches, so when a browser window was closed, the search results were effectively lost. The number of parameters made repeating searches error-prone. BLAST forms displayed a complex set of arcane input parameters that did not always correspond to the chosen program. Search strategies (i.e. sets of form parameters) could only be saved as browser bookmarks, tying them to a specific browser and machine. Users had to remember the meaning of program names (e.g. tblastx), and documentation was scattered and often out-of-date. The legacy forms also did not take advantage of recent improvements in web technologies and improved browser support for web standards.
The redesign replaces the old forms and navigation pages with an integrated web application that addresses all of these usability concerns.
OVERVIEW
The key BLAST pages now have a consistent design and structure. Each page has a header that contains links to the NCBI home page and a sign-in box for NCBI's login and customization interface, My NCBI. Just below the header is a list of links (called ‘breadcrumbs’) that shows the current page's location and provides navigation to related pages, Figure 1. Also, in the header are tabs that provide access to the main application pages, as follows:
Home: navigation to BLAST forms, organism-specific databases, specialized tools, tips and news.
Saved Strategies: filled-in BLAST forms that have been saved to My NCBI.
Recent Results: links to unexpired BLAST results.
Help: a documentation directory.
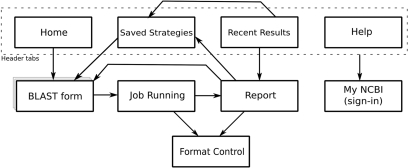
When the user initiates a new job from a BLAST form, BLAST immediately presents the Job Running page, which reports the status of a running job and an estimate of how long it will take to complete. The formatting parameters for a BLAST job may be changed on the Format Control page as the job runs, since formatting only occurs after search and alignment. When the job completes, BLAST presents the BLAST Report. From the Report, the user may now re-format the current job, run another BLAST job using the same parameters as a starting point, or navigate to one of the other application pages. The Recent Results page shows the status and some of the parameters of the user's unexpired BLAST jobs, and links directly to the BLAST Report for each job. A page flow map of these steps is presented in Figure 2. Each box in the figure represents a page in the BLAST web application.
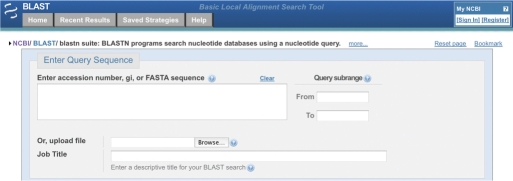
Figure 1.
Query sequence section of nucleotide blast form. The blue header provides links to the NCBI home page (left-most double helix) as well as tabs that can take a user to the BLAST home page, recent search results for a user, strategies saved via My NCBI and a help directory. On the far right is the My NCBI sign in box. Immediately below the header on the left side are bread-crumbs for navigation. The top part of the form is common to the major BLAST pages. As shown, this is followed by a form allowing the user to enter his/her query sequence and associated data. See text for details.
Figure 2.
BLAST screen flow map. Each box represents a different page in the BLAST web application. A user will normally enter through the ‘Home’ page and from there select a ‘BLAST form’ to submit a search. After the search is submitted the ‘Job running’ page is shown until the search is done, after which the ‘Report’ page is shown. From the ‘Report’ page the user may reformat, modify the current search and resubmit, or save the search strategy in My NCBI.
The following sections describe these new features in detail.
APPLICATION PAGES
Home page
The BLAST home page is always available from each page header's Home tab. Along the right side of the page are tips and news about BLAST. The top section of the page links to several organism-specific BLAST pages (which have not yet been incorporated into the redesign), in order of how often they are used as species limits in BLAST searches. Other species-specific BLAST pages are available from the ‘list all genomic databases’ link, which temporarily leads to the MapViewer home page. The MapViewer features a taxonomic directory that includes links to species- and group-specific BLAST pages, where they exist. Users have found this link to the MapViewer home page confusing, so a more usable solution is under development.
The middle section of the home page links to and describes the five general BLAST form types: Nucleotide BLAST, Protein BLAST, blastx, tblastn and tblastx. Nucleotide BLAST subsumes standard blastn, megablast and discontiguous megablast, presenting these three options as alternative algorithms for searching nucleotide databases with a nucleotide query. Similarly, Protein BLAST subsumes blastp, PSI-BLAST and PHI-BLAST.
The bottom section of the home page lists specialized BLAST types, such as searches for SNPs or gene expression profiles, and tools that use BLAST as an enabling technology, such as bl2seq (‘BLAST two sequences’), which uses BLAST for alignment but not for search.
BLAST form
All of the generic BLAST forms linked from the home page now share a common design. Only the options corresponding to the selected program type and algorithm appear on each form.
The Enter Query Sequence section at the top of the form (Figure 1) provides a place to enter one or more query sequences, either by accession or gi number, or as IUPAC sequence in FASTA format. Supported IUPAC characters are documented in BLAST help at http://www.ncbi.nlm.nih.gov/BLAST/blastcgihelp.shtml. The optional Query Subrange boxes limit the search to a subrange of the query sequence. As an alternative to cut/pasting sequence into a text box, you may also upload the query sequence(s) from a local disk file.
The new Job Title is the job name that appears in Saved Strategies and Recent Results, as well as at the top of every BLAST report. The title also appears in the title bar of the browser window or tab for the report, and as the default title of any bookmark to the report. The default title for a job is the query sequence definition line (in FASTA, the line beginning with ‘>’), but you may type over the default title to label the job in any way you like. When the input sequence is an accession or gi number, the BLAST web interface automatically looks up the definition line in GenBank without reloading the page. If multiple sequences are present, an appropriate descriptive title is generated (e.g. ‘5 nucleotide sequences’).
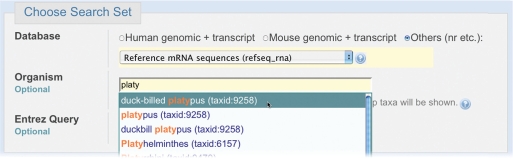
The Choose Search Set section of the BLAST form selects the BLAST database to be searched and applies limiting criteria, such as organism or Entrez query. Searches may be limited to a specific organism (species or taxonomic group) by typing the scientific name, common name or taxid (the integer id for the taxon in the NCBI Taxonomy database). As the user types the organism name, the Organism entry box prompts the user with a drop-down list of potential completions (Figure 3.) At any time, the user may hit the down-arrow key to scroll through the list of choices, and/or hit the Return key to choose the selected taxon. The list is limited to 20 items, and is sorted in reverse order of how often each taxon appears in GenBank, placing more commonly studied organisms at the top of the list. This ‘autocomplete’ feature both helps users know what organism names are available, and prevents spelling and typing errors.
Figure 3.
Potential completions for organism names are suggested as the user types. The first 20 matches to the user's query are presented, with matches anywhere in the matching organism allowed (e.g. plat finds ‘duck billed platypus’ even though ‘plat’ is not at the beginning of the target text). See text for details.
The limits and other values specified on each BLAST form remain in effect for the duration of the browser session, or until they are reset by the user. If the user signs in to My NCBI, they remain in effect across browser sessions.
The nucleotide BLAST form has additional search set options. The nucleotide Database section provides three common choices: Human genomic + transcript, Mouse genomic + transcript and Other. The genomic + transcript databases contain only NCBI reference sequences. They contain both genomic sequences and mRNAs for the organism, so both sequence types appear on the resulting report. Other contains the previously-available databases in a drop-down list. If the user selects a database from that list, Other is chosen automatically.
The genomic + transcript databases make it easier to search human and mouse sequences, and they automatically show transcript alignments to the genome. The human and mouse data sets use a new fast indexed search algorithm that decreases time-to-completion of a typical search by a factor of four (Morgulis,A. et al., manuscript in preparation). Searches for organisms other than human or mouse require simply selecting an alternate database, and an optional Organism limit. Within a browser session, each BLAST form automatically selects the database the user last chose, so an alternate database must be chosen only once.
The Program Selection section of the BLAST form selects the algorithm used for search and alignment. For nucleotide searches, the choices are megablast (default), discontiguous megablast and blastn. For protein searches, the options are blastp (default), PSI-BLAST and PHI-BLAST. The help link for this section leads to the BLAST program selection guide, which describes the algorithms and the criteria for choosing among them.
At this point in the form, most users will simply press the BLAST button to initiate a new search. BLAST previously opened results in a new window by default, which many users found annoying and disorienting. The new default behavior is for results to appear in the same window as the form (thereby replacing the form). The user may request results in a new window by checking the checkbox next to the BLAST button.
Detailed parameters for tuning the chosen program remain on the form, but they are now collapsed under a link entitled Algorithm Parameters, since only a tiny fraction of users ever use them. Clicking the link reveals the parameter controls. Of course, once the link is clicked, the parameters remain visible for the rest of the browser session. These parameters change depending upon the algorithm selected.
On the nucleotide form the available algorithms are megablast, discontiguous megablast and blastn. Choosing megablast selects a large word size (currently 28) and optimizes reward and penalty (1 and −2) for alignments of about 95% identity (3). Discontiguous megablast and blastn have parameters more suitable for inter-species comparisons, with a smaller word size (11) and reward and penalty (2, −3) that optimize for alignments of about 85% identity (3).
On the protein form the available choices are blastp, PSI-BLAST and PHI-BLAST. Choosing PSI-BLAST instead of blastp displays more target sequences, and allows the user to select sequences to build the PSSM for the next PSI-BLAST iteration. Both of these cases use ‘conditional compositional score matrix adjustments’ (4). PHI-BLAST does not support compositional adjustments, so the option disappears if PHI-BLAST is selected.
One new advanced feature has been added: BLAST now detects short input sequences for the nucleotide and protein search forms, and adjusts parameters to improve the chance of finding relevant matches. For short sequences (up to 30 residues for proteins, 50 bases for nucleotides), BLAST now automatically decreases word size (to seven for nucleotides, two for proteins), increases expect value (to 1000), and turns off low-complexity filtering. In addition, proteins use the PAM30 scoring matrix for short sequences as suggested by Altschul (5). This feature can be turned off in the Algorithm Parameters section of the form.
Job running
The user submits a new BLAST job by pressing the BLAST form button. BLAST immediately presents the Job Running page, which reports some statistics about the job, and provides an estimate of completion time. The Job Running view periodically refreshes itself, effectively polling the server while the job runs. BLAST automatically displays the BLAST report when the job completes. A link to the Format Control page (described below) can be used to set formatting parameters as the job runs.
Format control
The Format Control page specifies formatting parameters for a specific BLAST job. It provides a few simplifications of and additions to the previous design. Alignments formatted as XML or ASN.1, and Bioseqs (ASN.1 only) now produce a file download, instead of encoded text displayed in the browser. Limit controls (i.e. the Descriptions, Graphical Overview and Alignments counts; the Organism and Entrez limits; and the expect value range) limit the items shown on the report for a completed job, rather than limiting the search set, as they do on the BLAST form. The Format Control form has a text input for the Request ID (RID), allowing the user to format the current job, or any other known RID. Clicking the View Report button displays the requested job's Report page or, for incomplete jobs, the Job Running page.
Report page
The current BLAST report pages are basically the same as the previous design, with a reformatted header and some new features. To the right of the breadcrumbs are three links:
Reformat these results leads to the Format Control page,
Edit and Resubmit leads to the original BLAST form, with the current parameters selected and
Save Search Strategy saves the search parameters for the job so the user can run the same job again later with identical parameters. This option is available only if the user is signed in to My NCBI, since saved strategies are user-specific.
The Report Page [see Chapter 6 of (6) or http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=handbook.chapter.ch16 for details) is divided into four sections:
The Summary section provides links to alternate report formats: the taxonomy report (hits clustered taxonomically), the link to the MapViewer's ‘Genome View’ (hits shown on a genomic sequence map), and a new tree view (hits clustered by similarity).
The Graphical Overview section presents a graphic of the regions of the result set that aligned to the query (called ‘high-scoring pairs’, or HSPs), plotted against the query sequence. The graphic is unchanged from the previous design.
The Descriptions section is a table of the sequences that matched the query, sorted by increasing expect value. When the ‘Advanced view’ box is checked on the Format Control form, the Descriptions table can be resorted by clicking the header columns and more of each result sequence definition line is visible.
The Alignments section presents the alignments of the HSPs, either as a series of pairwise alignments (default), or as a single block of all HSPs anchored to the query. These formats are described in previous web server issues (7,8). Web log analysis has shown that the links from subject sequences to other databases, particularly to Gene, are underutilized, so now each alignment contains an informative link to Gene, where such a link exists.
Recent Results
The Recent Results page displays a list of links to unexpired BLAST jobs for the current browser session. Each item in the list provides a link (via the RID) to the Format Control page for the corresponding job. Also displayed are the time and date the job was submitted and will expire, the job status (Running, Done or Error), the BLAST program name, the job title, the query sequence length, the BLAST database used and links to save the search strategy for the job (if signed in) or to remove the item from the list. Removing the item from the list does not remove the results from the server; the results can still be retrieved by RID. Currently, results are removed from the server only by expiration.
The Recent Results list is available even if the user is not signed in to My NCBI, but then the list is available only on one machine, and restarting the browser or clearing the browser cache clears the list. If the user signs in to My NCBI, the list becomes available on other machines and in other browsers, and will survive reboots, browser restarts and cache clears.
Recent Results also provides a text box that looks up any BLAST job by RID. BLAST RIDs are case-insensitive, alphanumeric strings that avoid certain letters that could be confused with digits. They have been shortened to 11 characters (previously 37) making them easier to type, format, print, jot down on paper or send in an email. BLAST RIDs contain a randomly generated part, making valid RIDs very difficult to guess.
Saved Strategies
Users who sign in to My NCBI can save the search strategy of a BLAST job for later use. Search strategies may be saved by clicking the “save” link on a Recent Results item, or by clicking the ‘Save Search Strategy’ link on a BLAST report. A saved search strategy comprises a title (by default, the title of the original job), the program name, and all program parameters used to run the job. The query sequence is also saved if either the query was entered as an accession or gi number, or if the total sequence length is <10 kb. Saved BLAST search strategies do not expire.
FUTURE DIRECTIONS
The present redesign mostly addressed usability problems with input forms and results navigation. Future work will focus on better integration of more BLAST databases, more reporting options, support for batch and interactive operations, better formatting control and improved interpretation discovery.
For historical reasons, many of the BLAST databases available on the NCBI site are not consistently organized. Additional database types, including organism- or taxonomic group-specific databases, environmental samples, WGS records, traces and HTGS databases, will soon be reorganized to improve user experience.
The existing standard BLAST report will be supported and gradually improved. The Taxonomy, Genome View and Tree View reports will be better integrated into the new design. Additional report types, not yet designed, will become available that will take advantage of the more interactive features available in today's web technology. For example, a BLAST report type that initially shows only hit descriptions, and displays alignments only on demand, could provide quicker performance and easier navigation than the current, often multi-megabyte, all-in-one page download. Batch operations on groups of selected sequences, including printing, sequence downloading, batch linking, multiple alignment and PSI-BLAST iteration, are currently awkward or require ad hoc cut-and-paste operations in other programs. Upcoming features will focus on operations on selected subsets of results.
The Format Control form will be more easily accessible from BLAST reports, and the form will be further extended and refined. Sets of formatting parameters will be savable as named stylesheets, and usable directly from the reports. Dynamic HTML techniques will simplify measuring and recording intervals within and between sequence coordinate systems.
Future work will also improve and extend BLAST-related programs such as bl2seq (‘BLAST 2 sequences’), which use BLAST as an enabling technology.
Finally, BLAST will further catalyze discovery by displaying more about sequences and their relationships to other data. While BLAST mostly works in sequence space, the real value of BLAST lies in interpretation of the alignments. In the future, BLAST will increasingly offer, when appropriate, additional information about the matched sequences and sequence ranges themselves (such as sequence composition, motifs and other annotated features), as well as links to publication, gene, expression and other related data available at NCBI.
The BLAST team is very interested in how users apply BLAST to their daily work, and input is solicited. Please send suggestions to blast-help@ncbi.nlm.nih.gov.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Richa Agarwala, Stephen Altschul, Kevin Bealer, Christiam Camacho, Peter Cooper, George Coulouris, Susan Dombrowski, Mike Gertz, David Lipman, Wayne Matten, Alexander Morgulis, Jim Ostell, Jason Papadopoulos, Eric Sayers, Alejandro Schaffer, Tao Tao, David Wheeler, Vahram Avagyan, Melissa Landrum, Greg Schuler, Kim Pruitt, Yuri Wolf and Kira Makarova for helpful conversations and supporting work that made this website possible. The authors would like to thank Sergey Kurdin for assistance in producing the figures. This research was supported by the Intramural Research Program of the NIH, National Library of Medicine. Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.States DJ, Gish W, Altschul SF. Improved sensitivity of nucleic acid database searches using application-specific scoring matrices. METHODS. 1991;3:66–70. [Google Scholar]
- 4.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu YK. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul SF. Amino acid substitution matrices from an information theoretic perspective. J. Mol. Biol. 1991;219:555–565. doi: 10.1016/0022-2836(91)90193-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korf I, Yandell M, Bedell J. BLAST. Sebastopol, CA: O’Reilly and Associates; 2003. [Google Scholar]
- 7.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye J, McGinnis S, Madden TL. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006;34:W6–W9. doi: 10.1093/nar/gkl164. [DOI] [PMC free article] [PubMed] [Google Scholar]