Abstract
We have developed PVS (Protein Variability Server), a web-based tool that uses several variability metrics to compute the absolute site variability in multiple protein-sequence alignments (MSAs). The variability is then assigned to a user-selected reference sequence consisting of either the first sequence in the alignment or a consensus sequence. Subsequently, PVS performs tasks that are relevant for structure-function studies, such as plotting and visualizing the variability in a relevant 3D-structure. Neatly, PVS also implements some other tasks that are thought to facilitate the design of epitope discovery-driven vaccines against pathogens where sequence variability largely contributes to immune evasion. Thus, PVS can return the conserved fragments in the MSA—as defined by a user-provided variability threshold—and locate them in a relevant 3D-structure. Furthermore, PVS can return a variability-masked sequence, which can be directly submitted to the RANKPEP server for the prediction of conserved T-cell epitopes. PVS is freely available at: http://imed.med.ucm.es/PVS/.
INTRODUCTION
Multiple sequence alignments (MSAs) of homologous proteins encompass unique patterns of conserved and variable residues. The functional relevance of conserved residues is widely acknowledged. Indeed, functionally important residues such as those defining interacting sites, substrate binding sites or simply relevant to protein-structure integrity, display a low rate of substitution. This observation is predicted by the neutral evolution model (1), which also indicates that variable residues are somehow less important. Consequently, many methods have been developed to look for general and subfamily conservation patterns (2–8) as a key to identify functionally important residues. Moreover, some of these approaches are available for public use through the web (9–11). While these methods and related servers are very useful to identify functionally relevant residues, they generally underestimate the variability in the MSAs and certainly dismiss the significance of variable sites.
Variable residues in proteins can however be functionally relevant. Indeed, sequence variability is widely used by biological systems to generate functional heterogeneity. Thus, the hypervariable residues in the T-cell receptors (TCR) and Immunoglobulins match the antigen-binding residues (12). Likewise, the most polymorphic (variable) residues in the human leukocyte antigens (HLAs) are located on their binding groove, explaining the distinct peptide-binding specificities of the HLA allelic variants (13,14). Therefore, having a direct estimate of the sequence variability in an MSA is important to fill gaps in structural knowledge and to offer insight for function-structure studies. Indeed, long before the first antigen-bound immunoglobulin crystal structures were solved (15–17), Kabat (18) was able to anticipate that highly variable segments in immunoglobulin molecules match the antigen contact sites. Importantly, the estimation of sequence variability in rapidly evolving protein antigens from pathogens that use sequence variation for immune evasion (19–21) provides a mean to identify conserved antigenic determinant targets (epitopes), and consequently it is useful for epitope-vaccine design.
For all the above, we have developed PVS, a web server that provides absolute sequence variability estimates ‘per site’ in an MSA as determined by the Shannon Entropy (22), the Simpson Diversity Index (23) and the Wu-Kabat Variability Coefficient (18). The Wu-Kabat's coefficient, perhaps the most popular sequence variability metric, is effective in resolving the highest diversity positions, but as it has been noted, underestimates the diversity in the MSA (24). In comparison, Shannon and Simpson methods are statistically more sound for quantifying a system diversity, and are widely used in ecology and sequence analyses (25). Following the variability computations, PVS can plot the variability in the MSA and display it in a relevant 3D-structure. PVS can also return the selected reference sequence with the variable positions masked, as well as the sequence fragments (minimum length selected by the user) containing only nonvariable residues, as determined by a user-provided variability threshold. Within the PVS output page, the user can also locate the conserved fragments in the provided 3D-structure, and submit the variability-masked sequence to the RANKPEP server (26,27) for the prediction of conserved T-cell epitopes. Here we will show that these features are particularly relevant for epitope discovery-driven design of vaccines against pathogens displaying large sequence variability.
SYSTEMS AND METHODS
Automated generation of MSAs
Automated MSAs are obtained from the protein sequence of a Protein Data Bank (PDB) file following a BLAST (28) search against the SWISSPROT database. The BLAST search is performed using an E value of 1e–20 and a maximum of 250 hits are considered. Subsequently, the relevant sequence hits are aligned using MUSCLE (29).
Computation of sequence variability
The Shannon Diversity Index (Shannon Entropy) (22), the Simpson Diversity Index (23) and the Wu-Kabat Variability Coefficient (30) are used to estimate the sequence variability ‘per site’ (V) in MSAs.
The Shannon Diversity Index (H) is given by
| 1 |
where, pi is the fraction of residues of amino acid type i, and M represents the total number of amino acid types in a given site. H ranges from 0 (only one amino acid type is present at that position) to 4.322 (all 20 amino acids are equally represented in that position). Note, that for a site including gaps the maximum value of H will be 4.39.
We estimate the Simpson Diversity Index (D) using the following equation:
| 2 |
where, ni is the number of residues of type i, N is the total number of residues and S is the number of different symbols ‘per site’. From Equation (2) it follows that 0 ≤ D ≥ 1. Those sites with D values near 1 are highly variable and those with D values near 0 are almost constant.
The Wu-Kabat Variability Coefficient (W) is given by:
| 3 |
Here, N is the number of sequences in the MSA, k is the number of different amino acids at a given position and n is the frequency of the most common amino acid at that position. The minimum value of W is 1. Unlike for H and D, W maximum value increases with the number of sequences in the MSA.
Mapping sequence variability onto a 3D-structure
Given a relevant PDB file with the coordinates of a 3D-structure, the V in an MSA is mapped onto the 3D-structure by simply replacing the B-factor of the relevant residues in the PDB with the computed V values.
Implementation
PVS is implemented on an Apache Web server running under the Mac OSX operating system. The PVS functional core consists of a PERL CGI (Common Gateway Interface) script that handles the input, executes several subroutines implementing the above outlined methods, and then assembles and displays the results. PVS uses GNUPLOT (http://www.gnuplot.info) to plot the variability and the Bioperl Bio::Graphics module (http://www.bioperl.org) to generate sequence graphs with features. For displaying 3D-structures, PVS uses Jmol, an open-source Java molecular viewer for three-dimensional chemical structures (http://www.jmol.net).
DESCRIPTION AND USAGE OF THE SERVER
Web interface
The PVS web interface will dynamically change to present only those fields that apply to the user made selections. This is done using JavaScript. Moreover, the web interface is divided into the INPUT, SEQUENCE VARIABILITY OPTIONS and OUTPUT TASKS sections which overall facilitate an intuitive use of the server. The web interface also provides links to help pages, and specific information regarding the elements featured by the server can be obtained from the question mark icons. A description of the server usage, including the input and output follows here.
Input and variability options
The main input data for PVS can either be (i) an MSA or (ii) a PDB and users have to select one type or another from the INPUT section. Once a selection is made, the PVS web interface will show only the fields relevant to the selected input type. Thus, for the MSA option, the user can either paste or upload the alignment, which can be in CLUSTALW, GCG or FASTA formats. For the PDB input option, the user can either upload a PDB file or supply a PDB code and PVS will retrieve the corresponding PDB file from the Brookhaven database (http://www.rcsb.org/). Next, an MSA will be built from the sequence of the PDB chain—specified by the user—as detailed in ‘Systems and methods’ section. If no chain is provided, the first chain in the PDB file will be taken by default. Currently, PVS will only process MSAs with less than 400 sequences and 250 000 symbols. Also, automated MSAs will only be generated from PDB protein sequences shorter than 400 residues. If such limits are exceeded, the server will return an error.
Subsequently, PVS will subject the MSA to a sequence variability analysis using several methods that can be selected by the user from the ‘Sequence variability options’ section. The default method, ‘Shannon’, uses the Shannon Diversity Index as the variability metric [Systems and methods section, (Equation (1)]. Additionally, users can also select the ‘Wu-Kabat’ Variability Coefficient [Systems and methods, Equation (2)] and the ‘Simpson’ Diversity Index [Systems and methods, Equation (3)].
Output
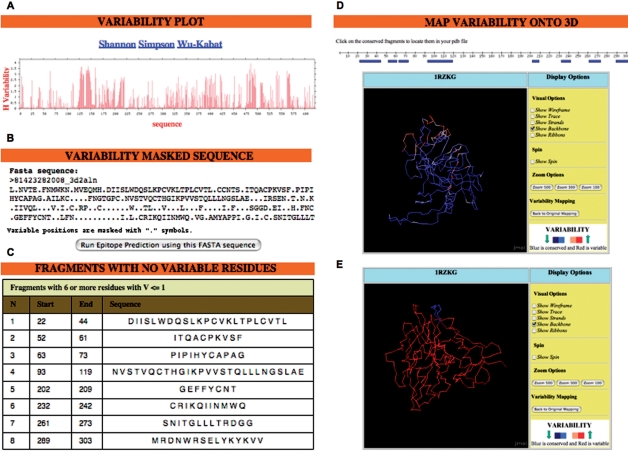
The output for PVS will be determined by the user-selected options in the ‘Output tasks’ section. By default, PVS will ‘plot the variability’ in the MSA—computed for each selected variability method—against a reference sequence selected by the user (Figure 1A). The reference sequence can either be a consensus sequence (default) or the first sequence in the MSA. Additionally, the following tasks can be performed by PVS: (i) ‘Mask sequence variability’; (ii) ‘Return conserved fragments’ and (iii) ‘Map structural variability’. The outputs and restrictions resulting from selecting these tasks are discussed below.
Figure 1.
PVS output. The figure shows a composition with the possible outputs of PVS. Results were obtained using an MSA corresponding to the HIV1 glycoprotein gp120 (residues 31–183 in gp160 from HIV-1 strain H2XB2). The MSA was generated from 359 representative sequences of the HIV-1 clades A (73), B (85), C (85), D (51) and 01_AE (65) using the program MUSCLE (29). The MSA is available at http://imed.med.ucm.es/PVS/supplemental/gp120_aln.html. The sequence variability was computed using the ‘Shannon’, ‘Simpson’ and ‘Wu-Kabat’ methods, and from the ‘sequence variability options’, a reference ‘consensus sequence’ and the default ‘variability threshold of 1.0’ were selected. (A) ‘Variability plot’. Users can change the variability metric (‘Shannon’, ‘Simpson’ and ‘Wu-Kabat’) by clicking on the relevant links. (B) ‘Variability masked sequence’. The sequence is returned in FASTA and T-cell epitope predictions can be obtained by clicking on the ‘Run Epitope Prediction’ bottom. (C) ‘Conserved fragments with no variable residues’. In this example, a ‘minimal fragment length’ of eight was selected. (D) ‘Structural variability mapping’. Sequence variability in the alignment was mapped onto the 3D-coordinates of gp120 (chain G of PDB 1RZK). The output allows the visualization of the variability in several user-selected renderings of the 3D structure. PVS can also display a graph of the protein sequence with the conserved fragments shown in blue. By clicking on a fragment, the user will locate it on the 3D-structure as shown in (E) with fragment 2. The output used to make this figure is available at: http://imed.med.ucm.es/PVS/supplemental/gp120_pvs.html.
Mask sequence variability
This option returns the selected reference sequence so that those residues with V greater or equal than the selected variability threshold are masked using a ‘.’ symbol. The variability-masked sequence is returned in FASTA format (Figure 1B), and it can be submitted to RANKPEP (26,27), the only T-cell epitope prediction tool that can anticipate conserved T-cell epitopes from a variability-masked sequence.
Return conserved fragments
This option identifies those fragments (minimum length selected by user) in the selected reference sequence consisting only of consecutive residues with V below the set variability threshold (Figure 1C). These fragments are returned, sorted in a table by their position in the MSA. For options (i) and (ii), the variability threshold must be between 0 and 4.3 in the case of the Shannon Entropy and between 0 and 1 for the Simpson Diversity Index (See Systems and methods section), otherwise PVS will return an error message. The default ‘variability threshold’ is 1.0 for the ‘Shannon’ Entropy method and 0.46 for the ‘Simpson’ Diversity Index, values which are regarded as indicatives of low variability (24). If the Shannon and Simpson methods were selected, PVS will proceed considering the variability threshold as for Shannon. Note that unlike the Shannon and Simpson Diversity Index, the upper value of the Wu-Kabat Variability Coefficient increases with the number of sequences in the MSA (see Systems and methods section). Therefore, since the ‘variability threshold’ must be entered prior to submitting the job, the options of masking the variability and returning conserved fragments are not available if the Wu-Kabat Variability Coefficient is the only variability metric selected.
Map structural variability
The sequence variability in the MSA is mapped onto a 3D-structure through a B-factor (see Systems and methods section). If an MSA was entered in PVS, the user must upload a relevant PDB to map the sequence variability onto it. Obviously, if the input was a PDB, PVS will map the sequence variability onto that same 3D structure. Note that when the ‘Map structural variability’ option is selected the variability is only computed for the positions in the MSA that map with the PDB. The resulting 3D structure is displayed using an interactive Jmol applet (JavaScript must be enabled in the browser) that allows the user to visualize the variability over several structural renderings, in a color scale that goes from blue for constant residues to red for highly variable residues (Figure 1D). In addition, if the ‘Return Conserved fragments’ task had also been selected, PVS will display a graph of the protein sequence with the conserved fragments shown in blue. By clicking on a fragment, the user will locate it on the 3D structure (Figure 1E).
Limitations
Proper computation of sequence variability from MSAs is contingent on the quality of the alignments. Therefore, we suggest evaluating the reliability of MSAs using the corresponding applications implemented in the TCOFFEE web server (http://www.igs.cnrs-mrs.fr/Tcoffee/) (31). This evaluation is particularly relevant when working with MSAs of distantly related proteins. However, the users should not have problems with the quality of MSAs built from very similar sequences (e.g. allelic and antigen variants). Likewise, we do not anticipate quality problems on the automated MSAs generated by the server because they are built considering only highly similar protein sequences. Finally, while the methods implemented in PVS are for computing sequence variability from MSAs, other methods do exist that can estimate sequence variability without the need of an MSA (32–34).
COMPARISON WITH AVAILABLE SERVERS
Sequence variability or conservation analyses, particularly when combined with mapping the variability onto a relevant 3D-structure, are useful to explore structure–function relationships and to reveal functionally relevant residues. Not surprisingly, some servers are already available (summarized in Table 1) that given an MSA can perform related tasks, such as providing a consensus sequence as ‘Consensus’, or plotting the relative sequence variability as in ‘WebVar’ (35). Other servers such as ‘Conseq’ and ‘TreeDet’ (20) carry out sophisticated conservation analyses to identify functionally relevant residues, and ‘Consurf’ (20), using the same phylogeny-dependent algorithms as ‘Conseq’ (9), maps the conservation scores onto a relevant 3D-structure. The ‘Conservancy’ (36) server is another related tool that from a set of user-provided predefined epitopes, identifies their conservation as determined by a percentage of identity. In comparison, PVS can handle more input types (PDBs or MSAs) and formats (MSAs can be in FASTA, CLUSTAW and GCG) that most of the related servers, and offers the largest set of functional tasks (Table 1). In any case, despite all these servers being related to some extent, they differ with regard to their methods and specific objectives, and indeed PVS is unique for using sequence variability analyses to help with epitope-vaccine design.
Table 1.
Web servers related to PVS
| Web server | Input: formats | Output and tasks | Ref |
|---|---|---|---|
| • PVS http://imed.med.ucm.es/PVS/ | • MSA: CLUSTAL, FASTA, GCG/MSF • PDB: Uploaded or retrieved • MSA and PDB |
1. Compute sequence variability 2. Plot sequence variability 3. Map and display variability in 3D structures 4. Mask sequence variability 5. T-cell epitope prediction 6. Return conserved fragments 7. Locate conserved fragments into 3D structures/B-cell epitope prediction |
|
| • SVS* http://bio.dfci.harvard.edu/Tools/svs.html | • MSA: CLUSTAL | 1. Compute sequence variability as given by Shannon Entropy 2. Plot sequence variability 3. Return conserved fragments |
|
| • SiteVarProt http://159.149.109.16/Tools/SiteVarProt.php | • MSA: FASTA | 1. Compute relative sequence variability 2. Plot sequence variability |
(35) |
| • Consensus http://coot.embl.de/Alignment//consensus.html | • MSA: CLUSTAL and GCG/MSF | 1. Consensus sequence at various thresholds with amino acid groupings | |
| • Conseq http://conseq.bioinfo.tau.ac.il/ | • SEQUENCE: FASTA • MSA: NBRF/PIR, EMB, FASTA, GDE, CLUSTAL, GCG/MSF and RSF |
1. Compute conservation scores 2. Compute solvent accessibility 3. Return color-coded sequence with calculations |
(9) |
| • Consurf http://consurf.tau.ac.il/ | • PDB: Uploaded or retrieved • MSA and PDB |
1. Compute conservation scores 2. Map and display conservation scores in 3D structures |
(11) |
| • TreeDet http://www.pdg.cnb.uam.es/Servers/treedet/ | • MSA: CLUSTAL, FASTA, MSF and PIR | • Predicts and display functionally relevant residues | (10) |
| • Conservancy http://tools.immuneepitope.org/tools/conservancy | • SEQUENCES: FASTA | • Computes per site sequence identity of epitopes in protein sources | (36) |
PVS is an enhanced version of SVS, a server previously developed by Dr Reche. SVS has >85 000 hits since it started running in 2002.
PVS RELEVANCE FOR EPITOPE DISCOVERY: WORKED EXAMPLES
Sequence variability analyses are commonly applied to infer evolutive and functional information in systems where functional diversity is achieved through sequence variation. For example, we previously applied a sequence variability analysis to human class I and class II MHC molecules (13), which, when correlated with the available structural information, clearly showed that the majority of the polymorphisms exhibited by these molecules are related with their differential peptide-binding specificity. In addition, we could also identify some other polymorphisms that could determine the restriction by their cognate T-cell receptors. While these classic structure–function studies can be carried in PVS, we will focus here on illustrating the use of PVS in the context of epitope-vaccine design.
PVS results are in fact tuned to facilitate the design of vaccines driven by epitope discovery against pathogenic organisms such as HIV-1, where sequence variation largely contributes to immune evasion, and sequence variability analyses are needed to identify conserved epitopes (37). The discovery of conserved T-cell epitopes (antigenic peptides recognized by the T cells when bound and displayed by MHC molecules in the cell surface of target cells) is facilitated in PVS by providing variability-masked sequences that can be submitted directly to the RANKPEP web server. Subsequently, RANKPEP will only return predicted conserved T-cell epitopes, thus also reducing the number of T-cell epitopes that have to be considered for experimental epitope confirmation. For example, from the gp120 variability masked sequence shown in Figure 1, RANKPEP will return two conserved T-cell epitopes restricted by the HLA I molecule A*0201 (KLTPLCVTL and PVVSTQLLL) as judged by their above-threshold binding score to A*0201 and by the predicted proteasomal cleavage. These predictions can be obtained from the gp120 PVS result page at: http://imed.med.ucm.es/PVS/supplemental/gp120_pvs.html. In comparison, the corresponding gp120 sequence of HIV-1 H2XB2 strain will yield 10 epitopes, a 5-fold increase in the epitope number (data not shown). Therefore, regardless of the predictive power of RANKPEP, this strategy saves the time, effort and resources one would need to consume confirming nonconserved T-cell epitopes that are not as suitable for vaccine design.
PVS results can also be helpful for the identification of conserved B-cell epitopes, the antigenic determinants of antibodies (Abs). As an example, we were able to detect seven highly conserved fragments of six or more residues (Table 2) from an MSA of the ectodomain of HIV-1 gp41 (details in Table 2 legend), which is the target of various broadly neutralizing Abs (38). Interestingly, fragments 5 and 7 encompass the antigenic determinants (B-cell epitopes) of the monoclonal antibodies CL3 and ZE10, respectively, both broadly neutralizing (38). Abs, however, only recognize solvent-exposed epitopes and most of them are conformational but can also be linear. Consequently, when used as immunogens, the majority of these conserved fragments will fail to yield Abs cross-reacting with the native antigen. However, one can also use PVS to locate the conserved fragments in the 3D-structure (when available), and select those that are surface exposed. Under such scenario, the chance of producing Abs that are cross-reactive with the native antigen and broadly neutralizing will be greatly increased. For example, in Figure 1E we have chosen to display the conserved fragment 2 (ITQACPKVSF) from HIV-1 gp120, which is readily accessible to the solvent. Moreover, from the PVS results obtained from the gp120 MSA (http://imed.med.ucm.es/PVS/supplemental/gp120_pvs.html) one could also see that fragment 3 and significant portions of fragments 1, 4 and 6 are also accessible to the solvent.
Table 2.
Conserved fragments in ectodomain of HIV-1 gp41
| N | Start | End | Sequence |
|---|---|---|---|
| 1 | 1 | 7 | S T M G A A S |
| 2 | 9 | 25 | T L T V Q A R Q L L S G I V Q Q Q |
| 3 | 27 | 55 | N L L R A I E A Q Q H L L Q L T V W G I K Q L Q A R V L A |
| 4 | 62 | 67 | D Q Q L L G |
| 5 | 69 | 74 | W G C S G K |
| 6 | 87 | 92 | S W S N K S |
| 7 | 153 | 158 | W L W Y I K |
Fragments were selected to have six or more consecutive residues with H ≤ 1, and were obtained from an MSA of the HIV-1 gp41 ectodomain (residues 528–674 in gp160 from HIV-1 strain H2XB2). The MSA includes 359 representative sequences of HIV-1 clades A (73), B (85), C (85), D (51) and 01_AE (65) that were aligned using MUSCLE (29). The MSA is available at http://imed.med.ucm.es/PVS/supplemental/gp41_ecto_aln.html
CONCLUSIONS AND FUTURE DIRECTIONS
PVS is a user-friendly and versatile web server where sequence variability computations are exploited to facilitate structure-function studies and, unlike any other related server, de novo epitope discovery. In the future, we plan to include additional variability and conservation scores. Moreover, we will implement solvent accessibility calculations, which should enhance the potential of PVS in structure–function studies and B-cell epitope discovery.
ACKNOWLEDGEMENTS
This work was supported by a Ramón y Cajal Grant (‘convocatoria 2005’) and by grant SAF2006-07879 from the ‘Ministerio de Educación y Ciencia’ (M.E.C) of Spain, both to P.A.R. The authors wish to thank Dr Jose R. Regueiro for corrections and thoughtful comments. Funding to pay the Open Access publication charges for this article was provided by M.E.C of Spain (SAF2006-07879).
Conflict of interest statement. None declared.
REFERENCES
- 1.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, pp. 34–55: Cambridge University Press; 1983. [Google Scholar]
- 2.del Sol Mesa A, Pazos F, Valencia A. Automatic methods for predicting functionally important residues. J. Mol. Biol. 2003;326:1289–1302. doi: 10.1016/s0022-2836(02)01451-1. [DOI] [PubMed] [Google Scholar]
- 3.Hannenhalli SS, Russell RB. Analysis and prediction of functional sub-types from protein sequence alignments. J. Mol. Biol. 2000;303:61–76. doi: 10.1006/jmbi.2000.4036. [DOI] [PubMed] [Google Scholar]
- 4.Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J. Mol. Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 5.Madabushi S, Yao H, Marsh M, Kristensen DM, Philippi A, Sowa ME, Lichtarge O. Structural clusters of evolutionary trace residues are statistically significant and common in proteins. J. Mol. Biol. 2002;316:139–154. doi: 10.1006/jmbi.2001.5327. [DOI] [PubMed] [Google Scholar]
- 6.Mihalek I, Res I, Lichtarge O. A family of evolution-entropy hybrid methods for ranking protein residues by importance. J. Mol. Biol. 2004;336:1265–1282. doi: 10.1016/j.jmb.2003.12.078. [DOI] [PubMed] [Google Scholar]
- 7.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18:S71–S77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 8.Thibert B, Bredesen DE, del Rio G. Improved prediction of critical residues for protein function based on network and phylogenetic analyses. BMC Bioinformatics. 2005;6:213. doi: 10.1186/1471-2105-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–1324. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
- 10.Carro A, Tress M, de Juan D, Pazos F, Lopez-Romero P, del Sol A, Valencia A, Rojas AM. TreeDet: a web server to explore sequence space. Nucleic Acids Res. 2006;34:115. doi: 10.1093/nar/gkl203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paul WE. 5th edn. Philadelphia: Lippincott Williams & Wilkins; 1998. Fundamental Immunology. pp 47–59, pp. 227–259. [Google Scholar]
- 13.Reche PA, Reinherz EL. Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J. Mol. Biol. 2003;331:623–641. doi: 10.1016/s0022-2836(03)00750-2. [DOI] [PubMed] [Google Scholar]
- 14.Stern LJ, Wiley DC. Antigen peptide binding by class I and class II histocompatibility proteins. Structure. 1994;2:245–251. doi: 10.1016/s0969-2126(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 15.Padlan EA, Silverton EW, Sheriff S, Cohen GH, Smith-Gill SJ, Davies D. Structure of an antibody-antigen complex: crystal structure of the HyHEL-10 Fab-lysozyme complex. Proc. Natl Acad. Sci. USA. 1989;86:5938–5942. doi: 10.1073/pnas.86.15.5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rose DR, Strong RK, Margolies MN, Gefter ML, Petsko GA. Crystal structure of the antigen-binding fragment of the murine anti-arsonate monoclonal antibody 36-71 at 2.9-A resolution. Proc. Natl Acad. Sci. USA. 1990;87:338–342. doi: 10.1073/pnas.87.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stanfield RL, Fieser TM, Lerner RA, Wilson IA. Crystal structures of an antibody to a peptide and its complex with peptide antigen at 2.8 A. Science. 1990;248:712–719. doi: 10.1126/science.2333521. [DOI] [PubMed] [Google Scholar]
- 18.Kabat EA. Antigenic determinants and antibody complementarity. Folia Allergol. 1970;17:425. [PubMed] [Google Scholar]
- 19.Mendis KN, David PH, Carter R. Antigenic polymorphism in malaria: is it an important mechanism for immune evasion? Immunol. Today. 1991;12:A34–A37. doi: 10.1016/S0167-5699(05)80010-6. [DOI] [PubMed] [Google Scholar]
- 20.Phillips RE, Rowland-Jones S, Nixon DF, Gotch FM, Edwards JP, Ogunlesi AO, Elvin JG, Rothbard JA, Bangham CR, Rizza CR, et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 21.Weber F, Elliott RM. Antigenic drift, antigenic shift and interferon antagonists: how bunyaviruses counteract the immune system. Virus Res. 2002;88:129–136. doi: 10.1016/s0168-1702(02)00125-9. [DOI] [PubMed] [Google Scholar]
- 22.Shannon CE. The mathematical theory of communication. Bell Sys. Tech. J. 1948;27:379–423. 623–656. [Google Scholar]
- 23.Simpson EH. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- 24.Stewart JJ, Lee CY, Ibrahim S, Watts P, Shlomchik M, Weigert M, Litwin S. A Shannon entropy analysis of immunoglobulin and T cell receptor. Mol. Immunol. 1997;34:1067–1082. doi: 10.1016/s0161-5890(97)00130-2. [DOI] [PubMed] [Google Scholar]
- 25.Baczkowski AJ, Joanes DN, Shamia GM. Range of validity of alpha and beta for a generalized diversity index H (alpha, beta) due to Good. Math. Biosci. 1998;148:115–128. doi: 10.1016/s0025-5564(97)10013-x. [DOI] [PubMed] [Google Scholar]
- 26.Reche PA, Glutting J.-P, Reinherz EL. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics. 2004;56:405–419. doi: 10.1007/s00251-004-0709-7. [DOI] [PubMed] [Google Scholar]
- 27.Reche PA, Glutting JP, Reinherz EL. Prediction of MHC class I binding peptides using profile motifs. Hum. Immunol. 2002;63:701–709. doi: 10.1016/s0198-8859(02)00432-9. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. Print 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu TT, Kabat EA. An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J. Exp. Med. 1970;132:211–250. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirot O, O'Toole E, Notredame C. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calhoun JR, Kono H, Lahr S, Wang W, DeGrado WF, Saven JG. Computational design and characterization of a monomeric helical dinuclear metalloprotein. J. Mol. Biol. 2003;334:1101–1115. doi: 10.1016/j.jmb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Dahiyat BI, Mayo SL. De novo protein design: fully automated sequence selection. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 34.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 35.Mignone F, Horner DS, Pesole G. WebVar: A resource for the rapid estimation of relative site variability from multiple sequence alignments. Bioinformatics. 2004;20:1331–1333. doi: 10.1093/bioinformatics/bth076. [DOI] [PubMed] [Google Scholar]
- 36.Bui HH, Sidney J, Li W, Fusseder N, Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinformatics. 2007;8:361. doi: 10.1186/1471-2105-8-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reche PA, Keskin DB, Hussey RE, Ancuta P, Gabuzda D, Reinherz EL. Elicitation from virus-naive individuals of cytotoxic T lymphocytes directed against conserved HIV-1 epitopes. Med. Immunol. 2006;5:1. doi: 10.1186/1476-9433-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat. Rev. Immunol. 2004;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]