Abstract
CS23D (chemical shift to 3D structure) is a web server for rapidly generating accurate 3D protein structures using only assigned nuclear magnetic resonance (NMR) chemical shifts and sequence data as input. Unlike conventional NMR methods, CS23D requires no NOE and/or J-coupling data to perform its calculations. CS23D accepts chemical shift files in either SHIFTY or BMRB formats, and produces a set of PDB coordinates for the protein in about 10–15 min. CS23D uses a pipeline of several preexisting programs or servers to calculate the actual protein structure. Depending on the sequence similarity (or lack thereof) CS23D uses either (i) maximal subfragment assembly (a form of homology modeling), (ii) chemical shift threading or (iii) shift-aided de novo structure prediction (via Rosetta) followed by chemical shift refinement to generate and/or refine protein coordinates. Tests conducted on more than 100 proteins from the BioMagResBank indicate that CS23D converges (i.e. finds a solution) for >95% of protein queries. These chemical shift generated structures were found to be within 0.2–2.8 Å RMSD of the NMR structure generated using conventional NOE-base NMR methods or conventional X-ray methods. The performance of CS23D is dependent on the completeness of the chemical shift assignments and the similarity of the query protein to known 3D folds. CS23D is accessible at http://www.cs23d.ca.
INTRODUCTION
Over the past 20 years, nuclear magnetic resonance (NMR) spectroscopy has emerged as one of the most powerful physical methods to study the structure and dynamics of proteins. It combines the strengths of such methods as fluorescence and circular dichroism for characterizing proteins in solution with the power of X-ray crystallography to perform these characterizations atom by atom. Aside from X-ray crystallography, NMR is the only other method available that allows the 3D structure of proteins to be determined to atomic resolution (1). Furthermore, NMR is the only known method that allows protein structures and dynamics to be determined in solution (i.e. near physiological conditions). To date, more than 7000 peptide and protein structures have been determined by NMR and deposited into the PDB (2). Each one of these structures was determined using a three-step process pioneered by Kurt Wuthrich and colleagues (3) in the early 1980s. This process involves: (i) determining the chemical shift assignments of the target protein; (ii) measuring the inter- and intra-residue 1H NOEs (nuclear Overhauser enhancements) to generate short-range distance constraints and (iii) using the NOE-derived constraints to perform molecular dynamics or distance geometry to calculate the 3D structure of the protein. Over the years, slight improvements to this process have occurred with the inclusion of more experimental constraints such as heteronuclear J-couplings (4) and residual dipolar couplings (5) or the introduction of improved conformational sampling protocols such as simulated annealing (6). Nevertheless, the central concept of using NOE-based methods to determine the 3D structure of proteins has remained essentially unchanged for nearly a quarter century.
While NOE-based methods are generally robust and well-proven, they are not without their faults. In particular, the measurement of 1H NOEs is both time-consuming and error-prone. Furthermore, NOEs becomes progressively less useful and more difficult to measure as the size of the proteins increases. This constraint means that the determination of 3D structures by NMR for proteins larger than 200 residues is very difficult and infrequent. With the emergence of structural genomics and structural proteomics, there have been a number of efforts aimed at accelerating the NMR structure determination process or extending the upper size limits for which NMR can be used. Almost all of these have focused on either eliminating the chemical shift assignment step (7,8) or automating the assignment of NOEs (9,10).
Rather than looking at ways of improving NOE measurements, a small minority of NMR researchers have been looking at ways of skipping the NOE step altogether and going directly from chemical shift assignments to 3D structures (11–14). Unlike NOEs, chemical shifts are easy to measure and not particularly sensitive to protein size restraints. Furthermore, chemical shifts provide exquisitely detailed information about the covalent structure of atoms and molecules. Indeed, when properly analyzed, chemical shifts can be used to infer secondary structure, flexibility, dihedral angles, side-chain orientations, hydrogen-bonding interactions and electrostatic or ionic interactions (11). Recently, several papers have appeared which suggest that reasonably accurate protein structures can be determined directly from chemical shifts (11–14). Of particular interest is the recent work by Cavalli et al. (13) and Shen et al. (14), both of which showed that accurate 3D structures for a number of small (<120 residue) proteins could be calculated using a combination of NMR chemical shifts, fragment assembly and heuristic potential functions. However, the computational effort required for this process is quite extreme, with individual structures requiring 1000s of CPU hours of calculation.
Here, we wish to describe a simple web server, called CS23D (chemical shift to 3D structure), that allows the rapid (∼15 min on 1 CPU) and accurate determination of protein structures using only assigned chemical shifts as input. CS23D builds on nearly 15 years of research in our lab related to using chemical shifts to identify secondary structures (15), to predict torsion angles (16), to identify protein folds (11), to predict protein flexibility (17), to refine protein structures (11) and to correct chemical shift referencing (18). In particular, CS23D combines a technique we call maximal subfragment assembly with other techniques such as chemical shift threading, de novo structure generation (via Rosetta), chemical shift-based torsion angle prediction and chemical shift refinement to generate and refine the protein coordinates. Tests conducted on >100 proteins from the BioMagResBank (for which chemical shifts and 3D structures are available) indicate that CS23D converges (i.e. finds a solution) for about 95% of protein queries. The resulting structures have a backbone RMSD <2 Å of the known structure and generally exhibit better geometry and chemical shift agreement than conventionally determined NMR structures. Additional details about CS23D are given subsequently.
PROGRAM DESCRIPTION
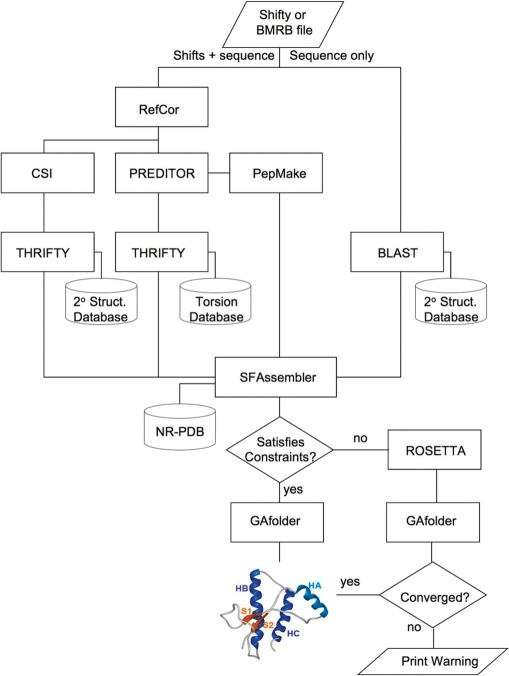
CS23D is composed of two parts, a front-end web interface (written in Perl and HTML) and a back-end consisting of eight different alignments, structure generation and structure optimization programs (written in Java, Perl, Python and C/C++) along with three local databases (Figure 1). The front-end accepts both SHIFTY (18) and BMRB (19) formatted chemical shift files. The shift files may be either pasted or typed into the text box or uploaded through a file browse button. The output for a typical CS23D structure calculation consist of a set of 10 lowest energy PDB coordinates in a simple, downloadable text format. A hyperlink to view the single lowest energy structure through the WebMol viewer (20) is also provided. In addition, details about the overall energy score (prior to and following energy minimization), chemical shift correlations (between the observed and calculated shifts) and torsion angle violations is provided at the top of the output page. If the structure calculation failed to converge to a reasonable value, a warning is printed at the top of the page. Details about the CS23D energy function, reasonable values for chemical shift ‘energies’ and reasonable values for torsion angle violations is provided in the Documentation link on the CS23D home page.
Figure 1.
A flow chart outlining the general structure of the CS23D web server and the programs that it calls to generate protein structures from chemical shift data. The specific function of each of the named programs (THRIFTY, PEPMAKE, ROSETTA, etc.) is explained in the text.
ALGORITHMS, DATABASES AND TESTING
A flow chart describing the processing logic used in CS23D is shown in Figure 1. As seen in this diagram, the input chemical shift file is initially partitioned into two parts, a sequence-only file and a sequence plus chemical shift file. The sequence file is searched against a nonredundant database of PDB sequences and secondary structures from PPT-DB (21) using BLAST (22) with a length-dependent Expect cutoff, ranging from 10−1 (for <11 residues) to 10−5 (for >50 residues). This step is done to identify short sequence fragments of known protein structures that exhibit good (>35% over 20+ residues) sequence identity to the query sequence. This step allows CS23D to find a series of maximum-length sequence fragments that matches the query sequence.
At the same time, the chemical shift file is submitted to three different chemical shift analysis programs: RefCor (17,18), CSI (15) and PREDITOR (16). The RCI program is used to re-reference the input chemical shifts, the CSI program is used to calculated secondary structure locations from the input chemical shifts and PREDITOR is used to calculate backbone and side-chain torsion angles. Checking chemical shift files for proper referencing is critical to obtaining accurate information about protein structures. Furthermore, many operations in CS23D (including CSI and PREDITOR) depend on having correctly referenced chemical shifts.
PREDITOR is a locally developed program that uses chemical shift similarity over short protein fragments and a database of known protein torsion angles to generate backbone and side-chain torsion angles. It has been shown to be very fast (10 sec) and accurate (85% of residues within 15°). The backbone torsion angles derived from PREDITOR are then mapped into nine different regions in Ramachandran space, each of which are assigned specific letters. The details of the torsion-angle-letter mapping scheme are shown in the CS23D Documentation web page.
By converting pairs of torsion angles into letters, it is possible to use a third program called THRIFTY (11) to perform chemical shift threading. THRIFTY uses the ‘sequence’ of shift-derived torsion angles generated by PREDITOR and searches against a database of ∼18 500 nonredundant PDB structures that have had their structures converted to the previously described nine-letter Ramachandran code. Once again, BLAST, with similar scoring cutoffs to that used in the sequence alignment step, is used to identify fragments of varying length in the CS23D torsion-angle database that maximally match the query torsion angles. This chemical threading process can also be used at a global level to identify potential protein folds that match the chemical shift information contained in the query protein assignments. THRIFTY is also used to perform a secondary structure alignment between the secondary structure of the query protein (calculated by CSI) and a large database of known protein secondary structures maintained at the PPT-DB (21). This secondary structure threading is used to evaluate and select subfragments that will be used to assemble the final 3D structure.
Subfragments identified by the sequence alignment and chemical shift threading schemes are then compared, weighted and assembled into an initial 3D structure using a program called SFassembler (subfragment assembler). SFassembler evaluates each of the fragments found by the sequence alignment, secondary structure threading and torsion angle threading steps. Four cases are considered: (i) if the same or similar fragments are found in all three cases (with sequence or secondary structure sharing >50% identity), SFassembler uses the coordinates of the fragment from the sequence-matched PDB file; (ii) if there are significant differences (<50% secondary structure identity) between the secondary structures found for the sequence-derived match and the shift-derived match, SFassembler uses the coordinates of the shift-matched PDB file; (iii) if no significantly matching sequence fragment is found, but a shift-based threading fragment match is found, the coordinates from the shift-matched fragment are used by SFassembler and (iv) if no fragment is found that has either a significant sequence match or a significant chemical shift match, then the torsion angles calculated from PREDITOR are used to generate the coordinates (via PEPMAKE). After these checks have been performed, SFassembler takes all selected PDB fragments or PREDITOR-generated coordinate files and concatenates them together into a single 3D backbone structure.
For instance, if one fragment of the query protein (say residues 1–55) meets criteria #1, another fragment (say residues 56–93) meets criteria #3 and a third fragment (say residues 94–106) meets criteria #4, SFassembler will generate coordinates and concatenate all three segments together. Backbone gaps are filled in using a technique called cyclic coordinate descent (23), while side chains are added using standard homology modeling techniques. Because no length limits are placed on the size of the matching subfragments, SFassembler can sometimes function as a homology modeling program, particularly if a single large and contiguous region of sequence similarity is found. However, unlike conventional homology modeling programs, SFassembler is also capable of generating structures for tandem, multi-domain proteins, for chimeric proteins, for proteins that fall far below sequence thresholds required for standard homology modeling, and most importantly, for generating structural elements for which no structure template exists (see the ‘Gallery’ page on the CS23D home page for examples).
The initial structure generated by SFassembler is evaluated by calculating a weighted correlation coefficient between the observed shifts and the calculated shifts using SHIFTX (24). If this correlation coefficient is too low or if the concatenated structure has <40% of the structure assembled by PREDITOR-generated coordinates (criteria #4), the structure is accepted and refined using a locally developed chemical shift/energy refinement program called GAfolder. After refinement, the PDB coordinates of the 10 lowest energy structures are mailed to the user along with the details of their evaluation and energy minimization. If the resulting structure fails these checks, it is discarded and a new structure is generated using Rosetta (25).
Rosetta is used by CS23D as a fail-safe procedure to generate potential 3D folds when the query protein exhibits absolutely no known sequence or chemical shift similarity over any part to any previously characterized protein. This will occur in about 5% of the cases handled by CS23D. Briefly, Rosetta is a public domain, open source, ab initio protein structure prediction program developed by David Baker's lab over the last decade (25). It uses the concept of fragment-based assembly similar to that employed by SFassembler, but at a much lower level of sequence identity and over much shorter fragment lengths. CS23D's implementation of Rosetta uses a local fragment generation routine constrained by PREDITOR-generated torsion angles. It employs Rosetta's default values for generating and evaluating protein structures, but these candidate structures are further evaluated using a combination of weighted chemical shift correlation coefficients (a chemical shift ‘energy’). The weightings used in the chemical shift evaluation step are provided in the CS23D Documentation pages. To limit the time taken in this step, CS23D limits the number of Rosetta-generated candidate structures to 300. The structure exhibiting the lowest chemical shift energy is identified and refined using GAfolder. If after the refinement step, the structure has a positive energy value, CS23D generates the following warning message ‘Structure Did Not Converge’. However, the coordinates of the highest scoring structure are still presented in the output so that the user may attempt to use these as starting coordinates for a more conventional NOE-based structure determination.
For its energy minimization and chemical shift refinement step, CS23D employs a torsion-angle-based energy minimizer called GAfolder that uses a genetic algorithm to sample conformation space. The method is similar to that employed by GENFOLD (26), although GAfolder uses cyclic coordinate descent (23) to perform more efficient writhe operations and it uses coordinates generated by the PREDITOR step (Figure 2) to perform segment swapping operations. The GAfolder potential energy function is a knowledge-based potential that includes information on predicted/known secondary structure, radius of gyration, hydrogen-bond energies, number of hydrogen bonds, allowed backbone and side-chain torsion angles, atom contact radii (bump checks), disulfide bonding information and a modified threading energy based on the Bryant and Lawrence potential (27). The chemical shift component of the GAfolder potential uses weighted correlation coefficients calculated between the observed and SHIFTX (24) calculated shifts of the structure being refined. The weighting coefficients for all of the parameters are given in the CS23D Documentation web page. Evaluations using nearly 44 000 3D structure predictions from the CASP7 collection showed that the GAfolder potential (without the chemical shift component) was able to identify the native or near-native (<2.5 Å RMSD) structure in 80/90 (89%) of the CASP7 targets.
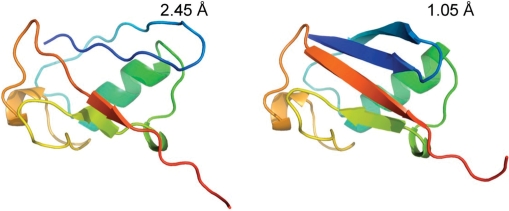
Figure 2.
An illustration of how chemical shift minimization can improve the structure of a poorly modeled initial structure. This shows the improvement seen in a deliberately distorted model of ubiquitin. In this case, the protein regains much of its lost secondary structure and the RMSD converges from 2.5 to 1.0 Å from the native structure.
RESULTS AND EVALUATION
CS23D was evaluated in six different ways: (i) assessing its ability to refine input structures; (ii) assessing the quality of its structures relative to known X-ray and NMR structures; (iii) assessing it capacity or robustness in generating (and refining) randomly selected structures from the BMRB; (iv) assessing its capacity to generate and refine very recently submitted BMRB assignments; (v) assessing its capability to generate and refine de novo structures and (vi) comparing its performance to previously described programs (13,14). In the first assessment, the quality of CS23D's chemical shift/energy refinement routine was investigated by looking at how distorted or nonnative structures could be refined towards near-native or native structures. This was tested on a collection of eight proteins for which complete chemical shifts and high-quality X-ray structures were available. These structures were then distorted or damaged using random torsion angle displacement. After this distortion step the structures were run through GAFolder for 3000 iterations and the resulting structures were assessed in terms of the RMSD between the calculated structure and the native structures as well as the energy difference between the native and the calculated structure. Table 1 in the CS23D Documentation web page shows the results of these calculations. On an average, the RMSD was lowered by 1.0 Å, with the final structure having an average RMSD of 1.5 Å relative to the native X-ray structure. Figure 2 shows an example of how GAFolder was able to correct a distorted (2.5 Å RMSD) structure of ubiquitin and bring it back to within 1.0 Å RMSD of the ‘true’ ubiquitin structure.
Table 2 (Documentation web page) illustrates the results of comparing the X-ray structure, NMR structure and CS23D-derived structure for nine proteins for which both X-ray and NMR structures exist. As seen in this table, the CS23D-derived structures have about one-third of the number of Ramachandran violations and omega angle violations found in their conventionally determined NMR counterparts. Likewise, the proportion of hydrogen bonded residues, hydrogen-bond energies and average chemical shift correlation coefficients are about 5–10% higher for CS23D-dervied structures than conventionally determined NMR structures. In fact, the CS23D structures are generally closer in structure quality characteristics to the corresponding high-resolution X-ray structures.
To assess CS23D's effectiveness at generating high-quality structures from standard BMRB files, 62 BMRB files of monomeric proteins were randomly selected from the RefDB database (18). In order to avoid having the query match itself, the PDB structure of each of these query proteins was removed from CS23D's sequence/structure databases. After this database-editing step, each of the 62 proteins was submitted to the CS23D server using its default settings. The structure of each of the lowest energy CS23D generated structures was evaluated by comparing its RMSD to that of the native structure. CS23D was able to generate a converged structure in 97% (60/62) of the cases. Further, the average Ca RMSD between the known structure and the CS23D generated structures was 1.1 Å RMSD (the range was 0.1–3.1 Å). The average time to generate and refine each structure was 10.2 min. This varied depending on the size, sequence similarity to known structure and structure generation methods (homology modeling/fragment assembly, chemical shift threading, de novo structure generation) used by CS23D. Interestingly, the majority of cases (92%) were solved by CS23D using large fragment (i.e. homology) modeling supplemented with fragment concatenation.
The high level of returned results probably exaggerates the real frequency with which CS23D would generate converged structures. This is because many of the proteins in RefDB (and by default the BMRB) are well-studied proteins for which many homologues have been characterized. To better assess the effectiveness of CS23D among newly deposited BMRB entries (for which novel folds or structural genomics entries may be more common), we downloaded all the 3 April 2008 ‘Recent Releases’ from the BMRB chemical shift repository and processed them through CS23D (total = 48 files). As before, the PDB structure of each query protein was removed from CS23D's sequence/structure database prior to structure generation. Of the 39 files for which structures were available in the PDB (for comparative purposes), CS23D succeeded in generating converged structures for 36 of them (Web Table 4), with approximately the same proportion generated via homology modeling/fragment assembly (82%), chemical shift threading (14%) and de novo structure generation (4%) as seen in Web Table 3. Interestingly, several independent estimates of the reported frequency of completely novel folds being deposited into the PDB or being solved by NMR suggest that only about 5% of all structures have this characteristic (28). Based on these data, we would estimate that CS23D, under routine use by the NMR community, would be able to generate a converged structure at least 95% of the time.
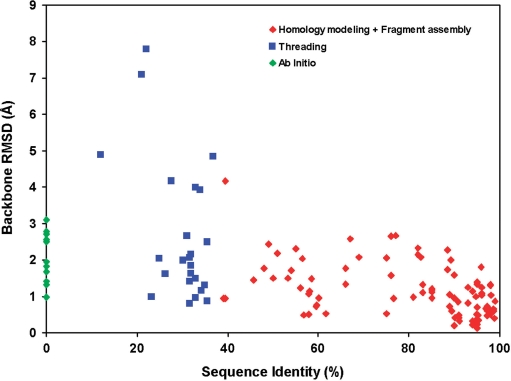
To assess CS23D's capabilities in generating and refining de novo structures, we conducted several tests (Tables 5 and 6 on the CS23D Documentation page). In the first instance, 10 small proteins (<130 residues) were processed through CS23D with the subfragment assembly and chemical shift threading options turned off (Table 5). This forced the program to use only its ab initio folding components (torsion angle constraints + Rosetta). The average backbone RMSD for this set of ab initio CS23D-folded proteins was 2.1 Å. Table 6 illustrates the performance of CS23D relative to the level of sequence identity and subfragment or chemical shift threading matches. In preparing Table 6, we used ubiquitin as the query and progressively removed each of the matching homologues from CS23D's structure databases. In total, ubiquitin had nine proteins that exhibited sequence identity of >40% and BLAST expect scores of <10−10 over some significant portion of the ubiquitin sequence. The backbone RMSD between the CS23D structure and the ‘true’ ubiquitin structure ranged from ∼0.5 Å (at 90–100% identity) to 0.9 Å (at 40% sequence identity). After these were removed, we found an additional 14 proteins in the PDB that exhibited secondary structure or torsion-angle identity of >25% and BLAST scores of <10−10. The RMSD between the CS23D structure and the ‘true’ ubiquitin structure in these cases ranged from 0.8 Å (at 35–55% ‘torsion’ identity) to 4.2 Å (at 25–35% ‘torsion’ identity). After these were removed from the PDB, we found that CS23D was still able to generate moderately good models of ubiquitin (within 2.8 Å RMSD) in 3/10 attempts. Figure 3 summarizes the results from Web Tables 3–6 in terms of the performance of CS23D's three different structure generation schemas (subfragment assembly + homology modeling, chemical shift threading and ab initio structure generation) relative to the level of sequence identity of the matching templates or subfragments.
Figure 3.
A scatter plot showing the performance of CS23D's three approaches to structure generation (subfragment assembly + homology modeling, chemical shift threading and ab initio structure generation) relative to the level of sequence identity of the matching templates or subfragments. Data from Tables 3–6 in the CS23D web Documentation pages were used to assemble this graph.
Finally, to compare CS23D's performance to previously described programs—CHESHIRE (13) and CS-Rosetta (14)—we ran CS23D on a number of the testing/training proteins used in these papers. This included 12 proteins used in their initial training and testing (Table 7a of the Web Documentation page), seven proteins that failed to converge for CS-Rosetta (Table 7b) and six proteins that were part of a blinded test for CS-Rosetta and for which chemical shifts were available (Table 7c). As seen in Table 7a, the average backbone RMSD for CHESHIRE, CS-Rosetta and CS23D predictions is 1.52, 1.48 and 1.64 Å, respectively. In other words, there is little to distinguish between the three methods. As shown in Table 7b, CS23D was able to generate good quality structures for four of the seven structures (57%) that CS-Rosetta could not generate. Interestingly, both programs failed for 1JW3, 1TVG and 2GDT. Table 7c shows that the structures generated by CS-Rosetta for ‘blinded’ structural genomics targets and generally better than those generated by CS23D (1 Å RMSD versus 3 Å RMSD). This highlights the fact that in trying to make CS23D a rapid structure generation tool, we have compromised some of its accuracy and capacity to generate de novo folds. Nevertheless, CS23D appears to perform as well as or better than CS-Rosetta and CHESHIRE in most other structure determination tasks. Furthermore, CS23D is approximately 10 000 times faster.
CONCLUSIONS
CS23D is designed to address a continuum of structure generation queries. At one extreme, if a submission has 99% sequence identity and exhibits >90% secondary structure conservation to a protein in the PDB, CS23D essentially functions as a homology modeling server with a (unique) chemical shift refinement step. In this case, the structures generated by CS23D are somewhat better than those generated by conventional NMR methods (as measured by Ramachandran violations, chemical shift correlations, hydrogen bonding and other structure evaluation tools). At the other extreme, if a submission has <5% sequence identity and exhibits <30% secondary structure conservation to a protein in the PDB, then CS23D essentially functions as an ab initio 3D structure predictor that employs an extra chemical shift refinement step. Between these extremes, CS23D is able to exploit a variety of other techniques to assemble, extend and refine selected protein fragments (ranging from 20 to 200 residues) to routinely (95% of the time) create high-quality 3D structures that are often better than those generated by conventional methods.
Obviously, CS23D is not without some limitations and there are certainly areas where improvements could be made. In particular, smarter use of chemical shift constraints for the Rosetta portion of the program could certainly increase CS23D's frequency of convergence for queries having completely novel folds. Additionally, CS23D could be improved by allowing it to accept NOE, J-coupling or residual dipolar coupling constraints as part of its input data. This could be particularly useful for modeling protein complexes or proteins with ligands—two areas where chemical shift constraints are essentially useless. Despite these limitations, we believe the speed, accuracy, frequency and reliability with which CS23D can generate (and refine) 3D protein structures—using only chemical shifts and sequence data—should make it a very useful addition to the current arsenal of structure generation and refinement tools available to biomolecular NMR spectroscopists.
ACKNOWLEDGEMENTS
Funding for this project was provided by the Alberta Prion Research Institute, PrioNet, NSERC and Genome Alberta. Funding to pay the Open Access publication charges for this article was provided by the Alberta Prion Research Institute (APRI).
Conflict of interest statement. None declared.
REFERENCES
- 1.Wuthrich K. NMR - this other method for protein and nucleic acid structure determination. Acta Crystallogr., D. 1995;51:249–270. doi: 10.1107/S0907444994010188. [DOI] [PubMed] [Google Scholar]
- 2.Kouranov A, Xie L, de la Cruz J, Chen L, Westbrook J, Bourne PE, Berman HM. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34(Database issue):D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wuthrich K. NMR of Proteins and Nucleic Acids. New York, NY: John Wiley & Sons; 1986. [Google Scholar]
- 4.Schmidt JM, Löhr F, Rüterjans H. Heteronuclear relayed E.COSY applied to the determination of accurate 3J(HN,C') and 3J(H beta,C') coupling constants in Desulfovibrio vulgaris flavodoxin. J. Biomol. NMR. 1996;7:142–152. doi: 10.1007/BF00203824. [DOI] [PubMed] [Google Scholar]
- 5.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 6.Holak TA, Nilges M, Oschkinat H. Improved strategies for the determination of protein structures from NMR data: the solution structure of acyl carrier protein. FEBS Lett. 1989;242:218–224. doi: 10.1016/0014-5793(89)80473-9. [DOI] [PubMed] [Google Scholar]
- 7.Atkinson RA, Saudek V. The direct determination of protein structure by NMR without assignment. FEBS Lett. 2002;510:1–4. doi: 10.1016/s0014-5793(01)03208-2. [DOI] [PubMed] [Google Scholar]
- 8.Grishaev A, Llinas M. CLOUDS, a protocol for deriving a molecular proton density via NMR. Proc. Natl Acad. Sci. USA. 2002;99:6707–6712. doi: 10.1073/pnas.082114199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linge JP, O’Donoghue SI, Nilges M. Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol. 2001;339:71–108. doi: 10.1016/s0076-6879(01)39310-2. [DOI] [PubMed] [Google Scholar]
- 10.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 11.Wishart DS, Case DA. Use of chemical shifts in macromolecular structure determination. Methods Enzymol. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
- 12.Gong H, Shen Y, Rose GD. Building native protein conformation from NMR backbone chemical shifts using Monte Carlo fragment assembly. Protein Sci. 2007;16:1515–1521. doi: 10.1110/ps.072988407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Protein structure determination from NMR chemical shifts. Proc. Natl Acad. Sci. USA. 2007;104:9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl Acad. Sci. USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 16.Berjanskii MV, Neal S, Wishart DS. PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 2006;34(Web Server issue):W63–W69. doi: 10.1093/nar/gkl341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berjanskii M, Wishart DS. NMR: prediction of protein flexibility. Nat. Protocols. 2006;1:683–688. doi: 10.1038/nprot.2006.108. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Neal S, Wishart DS. RefDB: a database of uniformly referenced protein chemical shifts. J. Biomol. NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 19.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, et al. BioMagResBank. Nucleic Acids Res. 2008;36(Database issue):D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walther D. WebMol—a Java-based PDB viewer. Trends Biochem. Sci. 1997;22:274–275. doi: 10.1016/s0968-0004(97)89047-0. [DOI] [PubMed] [Google Scholar]
- 21.Wishart DS, Arndt D, Berjanskii M, Guo AC, Shi Y, Shrivastava S, Zhou J, Zhou Y, Lin G. PPT-DB: the protein property prediction and testing database. Nucleic Acids Res. 2008;36(Database issue):D222–D229. doi: 10.1093/nar/gkm800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canutescu AA, Dunbrack R.L., Jr. Cyclic coordinate descent: a robotics algorithm for protein loop closure. Protein Sci. 2003;12:963–972. doi: 10.1110/ps.0242703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neal S, Zhang H, Nip AM, Wishart DS. Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J. Biomol. NMR. 2003;26:215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- 25.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 26.Bayley MJ, Jones G, Willett P, Williamson MP. GENFOLD: a genetic algorithm for folding protein structures using NMR restraints. Protein Sci. 1998;7:491–499. doi: 10.1002/pro.5560070230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bryant SH, Lawrence CE. An empirical energy function for threading protein sequence through the folding motif. Proteins. 1993;16:92–112. doi: 10.1002/prot.340160110. [DOI] [PubMed] [Google Scholar]
- 28.Levitt M. Growth of novel protein structural data. Proc. Natl Acad. Sci. USA. 2007;104:3183–3188. doi: 10.1073/pnas.0611678104. [DOI] [PMC free article] [PubMed] [Google Scholar]