Abstract
Medline is a rich information source, from which links between genes and keywords describing biological processes, pathways, drugs, pathologies and diseases can be extracted. We developed a publicly available tool called CoPub that uses the information in the Medline database for the biological interpretation of microarray data. CoPub allows batch input of multiple human, mouse or rat genes and produces lists of keywords from several biomedical thesauri that are significantly correlated with the set of input genes. These lists link to Medline abstracts in which the co-occurring input genes and correlated keywords are highlighted. Furthermore, CoPub can graphically visualize differentially expressed genes and over-represented keywords in a network, providing detailed insight in the relationships between genes and keywords, and revealing the most influential genes as highly connected hubs. CoPub is freely accessible at http://services.nbic.nl/cgi-bin/copub/CoPub.pl.
INTRODUCTION
Analysis and interpretation of microarray data is not a trivial task. Many public and commercial bioinformatics tools have been developed to help scientists interpret the lists of differentially expressed genes that are the result of microarray experiments. For example, Gene Ontology and Pathway Mapping tools (1–4) allow batch input of genes and produce lists of GO terms or pathways that are significantly correlated with the input set of genes (5–13).
The outcome of these tools is based on well-established relationships between the genes and biological processes in which they participate. However, the primary literature contains much more information about the functions of genes than is captured in structured vocabularies or canonical pathways. To extract this additional information on gene function from literature, we used thesaurus-based keyword matching in Medline abstracts to link human, mouse and rat genes to biomedical concepts describing liver pathologies, pathways, GO terms, diseases, drugs and tissues (Table 1). This approach builds on the assumption that co-occurrence of a gene and a biomedical concept in the same abstract is an indication of a functional link between the gene and the concept.
Table 1.
Overview of the 11 thesauri that were generated to search Medline
| Thesaurus category | Number of keywords | Source | URL |
|---|---|---|---|
| Genesa | |||
| Human | 122 425 (24 876 genes) | NCBI's Entrez Gene Database | http://www.ncbi.nlm.nih.gov/sites/entrez |
| Mouse | 130 759 (22 593 genes) | ” | ” |
| Rat | 73 572 (12 296 genes) | ” | ” |
| Gene Ontology (GO) | |||
| Biological processes | 3621 | Gene Ontology Database | http://www.geneontology.org |
| Molecular functions | 961 | ” | ” |
| Cellular components | 216 | ” | ” |
| Liver pathologies | 489 | Textbooks | – |
| Pathways | 817 | KEGG, Reactome, Encyclopedia of Human Genes and Metabolism DB | http://www.genome.jp/kegg/, http://www.genomeknowledge.org, http://humancyc.org |
| Diseases | 4164 | Karolinska Institutet | http://www.mic.ki.se/Diseases/Alphalist.html |
| Drugs | 5796 | RxList database | http://www.rxlist.com/top200.htm |
| Tissues | 1112 | ExPASy Proteomics server | http://www.expasy.org/cgi-bin/lists?tisslist.txt |
aFull gene names, gene symbols, alternative gene names/symbols.
In this article, we describe a tool named CoPub that calculates keyword over-representation for a set of regulated genes in a similar fashion to general GO term over-representation tools, but where the over-represented keywords for the gene set are retrieved directly from Medline by text mining. Several text mining methods for the analysis of microarray data have been published that annotate clustered sets of regulated genes based on their literature profile (14–17), or on their expression profile, often based on subsets of the total Medline repository (18–21). CoPub uses the entire Medline library to calculate robust statistics for gene-keyword co-occurrence, and is not dependent on pre-clustered gene sets to calculate significance for keyword over-representation. In addition to calculating over-represented keywords, CoPub also shows the results graphically in an interactive network, providing an additional level of insight into the biological mechanisms related to a set of regulated genes.
CoPub has two other features: the Gene search and the BioConcept search. The Gene search and the BioConcept search options identify genes and keywords that share occurrences in Medline abstracts with a gene or keyword of interest, which provides a kind of annotation for the gene or keyword of interest.
In an earlier study (22), we successfully applied CoPub for compound toxicity evaluation of a variety of compounds, which shows that CoPub is a useful additional bioinformatics tool for microarray data analysis. CoPub is freely accessible at http://services.nbic.nl/cgi-bin/copub/CoPub.pl.
METHODS
Text mining Medline abstracts
Eleven thesauri were generated to search Medline (Table 1). These thesauri describe genes (human, mouse and rat), Gene Ontology terms, diseases, pathways, drugs, tissues and liver pathologies. The keyword thesauri are based on biological items, which represent an instance of a biological concept (e.g. a gene, a pathway), and may contain one or more keywords (e.g. a gene is assigned a full gene name as well as a gene symbol and gene aliases).
The full Medline baseline XML files (1966 to February 2008) were obtained from the NCBI website (http://www.nlm.nih.gov/bsd/licensee/2008_stats/baseline_doc.html) and extracted to small text files containing title, abstract and substances.
Regular expressions were used to search the compiled Medline text files for the presence of all keywords (∼250 000) from the biological concept thesauri, as described by Alako et al. (23). Keywords that generated a hit in a Medline abstract were stored, together with the PubMed identifiers (IDs) of the Medline records in which the hit occurred. For every biological item, the hits were made non-redundant (note: multiple keywords of a biological item can occur in the same Medline abstract), resulting in a PubMed ID-biological item list. Gene symbols were curated for ambiguity and gene hits of orthologous genes were combined to make the keyword search more comprehensive.
Co-publication of biological items (e.g. a gene with a pathology term) was retrieved from the database by matching common Medline abstract occurrences. For every biological item pair, an R-scaled score, which describes the strength of a co-citation between two keywords given their individual frequencies of occurrence (23), and the literature count, which is the number of co-publications between every biological item pair, was calculated. Both measures were used to describe the strength of the relationship between two keywords.
To link gene expression data to literature data, mappings of Affymetrix probe set identifiers to Entrez Gene identifiers and orthology information were retrieved from Affymetrix human, mouse, rat GeneChip Genome Array annotation files (http://www.affymetrix.com). Mappings of Ensembl identifiers to Entrez Gene identifiers were retrieved from BioMart (http://www.biomart.org).
Keyword enrichment calculation
Keyword enrichment calculation is performed using the Fisher exact test, in which the association of a given keyword with a set of regulated genes (i.e. co-publications in Medline abstracts) is statistically tested against a background set; the set of unchanged genes on the microarray in case Affymetrix probe set identifiers are uploaded, or all other genes in the genome when Entrez Gene identifiers or Ensembl identifiers are uploaded. The calculated P-values are corrected using the Benjamini–Hochberg multiple testing correction method. All statistical tests are done using the R Statistics package (http://www.r-project.org).
To generate literature networks CoPub uses GraphViz (http://www.graphviz.org) to calculate the graph layout (neato), and for exporting the literature network in a scalable vector graphic (SVG) format. Interactivity in generated SVG networks is implemented using Perl and JavaScript.
CoPub WEB SERVER
The CoPub user-interface offers three analysis methods: the Microarray data analysis, the Gene search and the BioConcept search.
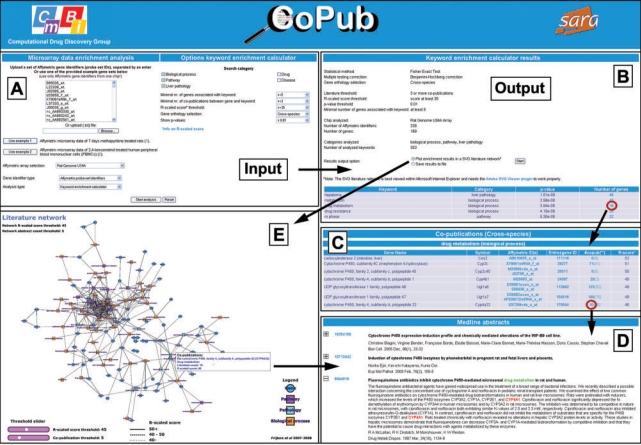
The Microarray data analysis option calculates keyword over-representation for a set of differentially expressed genes, and offers graphical visualization of the analysis results in a literature-based network. Screenshots and workflow of the Microarray data analysis are shown in Figure 1 and described subsequently in more detail.
Figure 1.
Screenshots and workflow of the Microarray data analysis. (A) Input screen for uploading gene identifiers (Affymetrix probe set identifiers, Entrez Gene identifiers or Ensembl identifiers), selection of keyword categories and to specify thresholds (e.g. P-value significance level), with which the keyword over-representation analysis will be performed (sensible defaults are provided). (B) Output screen which reports on significantly linked keywords to the set of submitted genes, ranked on P-values after multiple testing correction. The number of genes that are significantly associated with the analyzed keyword, links to an overview of uploaded genes that share co-publications with the analyzed keyword (C), which provides access to highlighted Medline abstracts in which they co-occur (D). (E) Visualization of the keyword over-representation results in an interactive literature network (as SVG), in which nodes represent genes and keywords, and edges represent links in Medline abstracts. Clicking on an edge retrieves highlighted Medline abstracts in which genes and keywords co-occur (D).
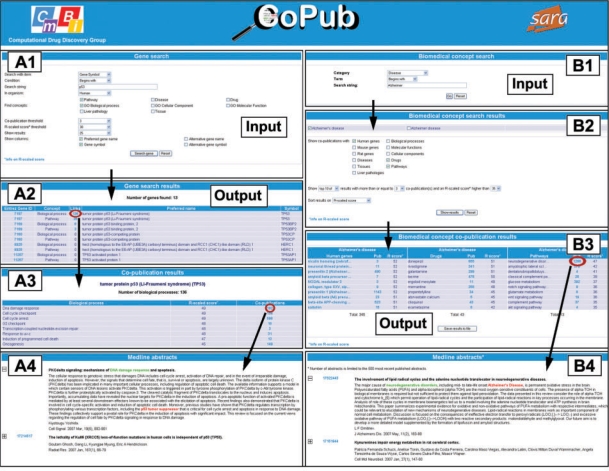
The Gene search identifies genes and keywords that share co-occurrences in Medline abstracts with a gene of interest. It provides answers on a question like; ‘Which diseases and drugs are strongly connected to the gene p53’? In a similar manner, the BioConcept search identifies genes and keywords that share co-occurrences in Medline abstracts with a keyword of interest and provides answers on a question like, ‘Which pathways are associated with Alzheimer's disease?’ Screenshots and the workflows of the Gene search and the BioConcept search are shown in Figure 2 and described below in more detail.
Figure 2.
Screenshots and workflows of the Gene search (A) and the BioConcept search (B). (A1) Input screen of the Gene search, which requires a single gene name as input. Furthermore, the categories of keywords need to be specified for which co-occurrences in literature with the gene of interest will be matched and retrieved. (A2) Output screen of the Gene search, which reports on the number of keywords that co-occur with the gene of interest, and links to an overview of the keywords (A3) and to Medline abstracts in which they co-occur (A4). (B1) Input screen of the BioConcept search, which requires a single keyword as input. (B2) Page to specify the categories of genes and keywords for which co-occurrences in literature with the keyword of interest will be matched and retrieved. (B3) Output screen of the BioConcept search, which reports on genes and keywords that co-occur with the keyword of interest in Medline abstracts, and with links to these abstracts (B4).
Microarray data analysis
Input
The user can select one of two analysis modes for microarray data analysis: the keyword enrichment calculator or the matrix generator. For each of the two analysis modes, the user needs to upload a list of gene identifiers (Affymetrix probe set identifiers, Entrez Gene identifiers or Ensembl identifiers), either by copy–paste or as a text file. Following this, the user must specify the correct Affymetrix microarray chip or species and the categories of keywords used for analysis (Figure 1A). Example gene sets are provided.
For the keyword enrichment calculator, thresholds need to be specified for the P-value significance level, the minimal number of co-publications, the minimal R-scaled score and the minimal number of submitted genes that have linkage with the analyzed keyword in literature (i.e. share abstract occurrences). For the threshold settings, sensible defaults are provided.
Both analysis modes can either be performed with ‘species-specific’ or ‘cross-species’ gene information. In the ‘cross-species’ mode, full gene names as well as gene symbols of human, mouse and rat orthologous genes are combined. This is in contrast to the ‘species-specific’ mode, in which species-specific gene names and symbols are used for analysis.
Output
The matrix generator produces a tab-delimited file in which all co-publication information between the uploaded set of genes and the selected keywords are presented in a matrix format. The values in the matrix can either be the absolute number of co-publications or the R-scaled score between a gene and a keyword. This matrix is provided as a flat-format text file, which can be used for any kind of follow-up analysis, such as clustering the genes on basis of their keyword profile.
The keyword enrichment calculator produces a list of keywords, ranked on P-values (Figure 1B). This list already provides a first impression of the biological processes related to the gene set. The user can drilldown into these results by clicking on the hyperlinked number of genes that are significantly associated with the analyzed keyword (Figure 1B). It links to an overview of uploaded genes that share co-publications with the analyzed keyword (Figure 1C), and provides access to highlighted Medline abstracts in which they co-occur (Figure 1D).
CoPub can also visualize the keyword enrichment results in a SVG format (Figure 1E). In this interactive network, nodes represent over-represented keywords and differentially expressed genes and edges represent literature links. The nodes and edges link to the relevant Medline abstracts in which co-occurring genes and keywords are highlighted. This allows for quick retrieval of relevant literature and interpretation of the data. Threshold sliders for the R-scaled score and the literature count can be used to reduce the size of the network interactively. Alternatively, the literature-network can be re-calculated with new threshold values.
Gene search and BioConcept search
Input
The Gene search option requires a single gene name or symbol, or for the BioConcept search, a single keyword as input (Figure 2). Furthermore, for both the Gene search and the BioConcept search, the categories of keywords need to be specified for which co-occurrences in literature with the gene or keyword of interest will be matched and retrieved.
In addition, thresholds for the minimal number of co-publications and the minimal R-scaled score between keywords/genes can be specified, for which sensible defaults are provided.
Output
Both the Gene search and the BioConcept search return lists of genes and keywords that are strongly linked with the input gene or keyword of interest.
The results on these pages are all hyperlinked. This enables the user to navigate through various pages that report on how many times genes and keywords co-occurred in literature, and provide access to the relevant Medline abstracts (Figure 2).
DISCUSSION AND CONCLUSION
We have developed CoPub, a web-based tool for calculating keyword enrichment in sets of regulated genes. It detects keywords that are significantly linked to a set of genes, using robust co-occurrence statistics of genes and keywords in Medline abstracts. In a study in which gene expression profiles induced by 11 distinct hepatotoxicants were analyzed with CoPub, we were able to accurately describe histopathological findings and the mode of toxicity of these compounds (22). This shows that CoPub is a useful additional tool in the toolbox for the analysis of microarray experiments.
We intend to further develop CoPub by updating its Medline content on a regular basis (once every 2 months), and by adding new and improved keyword thesauri. Furthermore, we will broaden the scope of the keyword over-representation analysis by allowing identifiers from other microarray platforms as input for the keyword over-representation analysis. On the output side, options for multiple graphical output formats, such as png and jpg as well as a connection to Cytoscape, an open source network analysis tool, will be offered, allowing for improved downstream analysis of the results generated by CoPub.
ACKNOWLEDGEMENTS
The authors thank SARA Computing and Networking Services (Amsterdam, The Netherlands) for updating and maintaining our CoPub database and web server, which was made possible by grants received from the Netherlands Bioinformatics Centre (NBIC) under the BioAssist program and from Organon, part of Schering-Plough corporation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cheng J, Sun S, Tracy A, Hubbell E, Morris J, Valmeekam V, Kimbrough A, Cline MS, Liu G, Shigeta R, et al. NetAffx gene ontology mining tool: a visual approach for microarray data analysis. Bioinformatics. 2004;20:1462–1463. doi: 10.1093/bioinformatics/bth087. [DOI] [PubMed] [Google Scholar]
- 2.Nakao M, Bono H, Kawashima S, Kamiya T, Sato K, Goto S, Kanehisa M. Genome-scale gene expression analysis and pathway reconstruction in KEGG. Genome Inform. Ser. Workshop Genome Inform. 1999;10:94–103. [PubMed] [Google Scholar]
- 3.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 4.Mlecnik B, Scheideler M, Hackl H, Hartler J, Sanchez-Cabo F, Trajanoski Z. PathwayExplorer: web service for visualizing high-throughput expression data on biological pathways. Nucleic Acids Res. 2005;33:W633–W637. doi: 10.1093/nar/gki391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 6.Chung HJ, Park CH, Han MR, Lee S, Ohn JH, Kim J, Kim JH. ArrayXPath II: mapping and visualizing microarray gene-expression data with biomedical ontologies and integrated biological pathway resources using scalable vector graphics. Nucleic Acids Res. 2005;33:W621–W626. doi: 10.1093/nar/gki450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey R, Guru RK, Mount DW. Pathway miner: extracting gene association networks from molecular pathways for predicting the biological significance of gene expression microarray data. Bioinformatics. 2004;20:2156–2158. doi: 10.1093/bioinformatics/bth215. [DOI] [PubMed] [Google Scholar]
- 8.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goffard N, Weiller G. PathExpress: a web-based tool to identify relevant pathways in gene expression data. Nucleic Acids Res. 2007;35:W176–W181. doi: 10.1093/nar/gkm261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34:W720–W724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, Muller R, Meese E, Lenhof HP. GeneTrail—advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 13.Huang da W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jelier R, Jenster G, Dorssers LC, Wouters BJ, Hendriksen PJ, Mons B, Delwel R, Kors JA. Text-derived concept profiles support assessment of DNA microarray data for acute myeloid leukemia and for androgen receptor stimulation. BMC Bioinform. 2007;8:14. doi: 10.1186/1471-2105-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaussabel D, Sher A. Mining microarray expression data by literature profiling. Genome Biol. 2002;3:RESEARCH0055. doi: 10.1186/gb-2002-3-10-research0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chagoyen M, Carmona-Saez P, Shatkay H, Carazo JM, Pascual-Montano A. Discovering semantic features in the literature: a foundation for building functional associations. BMC Bioinform. 2006;7:41. doi: 10.1186/1471-2105-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuffner R, Fundel K, Zimmer R. Expert knowledge without the expert: integrated analysis of gene expression and literature to derive active functional contexts. Bioinformatics. 2005;21(Suppl 2):ii259–ii267. doi: 10.1093/bioinformatics/bti1143. [DOI] [PubMed] [Google Scholar]
- 18.Jenssen TK, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat. Genet. 2001;28:21–28. doi: 10.1038/ng0501-21. [DOI] [PubMed] [Google Scholar]
- 19.Rubinstein R, Simon I. MILANO—custom annotation of microarray results using automatic literature searches. BMC Bioinform. 2005;6:12. doi: 10.1186/1471-2105-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blaschke C, Oliveros JC, Valencia A. Mining functional information associated with expression arrays. Funct. Integr. Genomics. 2001;1:256–268. doi: 10.1007/s101420000036. [DOI] [PubMed] [Google Scholar]
- 21.Burkart MF, Wren JD, Herschkowitz JI, Perou CM, Garner HR. Clustering microarray-derived gene lists through implicit literature relationships. Bioinformatics. 2007;23:1995–2003. doi: 10.1093/bioinformatics/btm261. [DOI] [PubMed] [Google Scholar]
- 22.Frijters R, Verhoeven S, Alkema W, van Schaik R, Polman J. Literature-based compound profiling: application to toxicogenomics. Pharmacogenomics. 2007;8:1521–1534. doi: 10.2217/14622416.8.11.1521. [DOI] [PubMed] [Google Scholar]
- 23.Alako BT, Veldhoven A, van Baal S, Jelier R, Verhoeven S, Rullmann T, Polman J, Jenster G. CoPub Mapper: mining MEDLINE based on search term co-publication. BMC Bioinformatics. 2005;6:51. doi: 10.1186/1471-2105-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]