Abstract
The CGView Server generates graphical maps of circular genomes that show sequence features, base composition plots, analysis results and sequence similarity plots. Sequences can be supplied in raw, FASTA, GenBank or EMBL format. Additional feature or analysis information can be submitted in the form of GFF (General Feature Format) files. The server uses BLAST to compare the primary sequence to up to three comparison genomes or sequence sets. The BLAST results and feature information are converted to a graphical map showing the entire sequence, or an expanded and more detailed view of a region of interest. Several options are included to control which types of features are displayed and how the features are drawn. The CGView Server can be used to visualize features associated with any bacterial, plasmid, chloroplast or mitochondrial genome, and can aid in the identification of conserved genome segments, instances of horizontal gene transfer, and differences in gene copy number. Because a collection of sequences can be used in place of a comparison genome, maps can also be used to visualize regions of a known genome covered by newly obtained sequence reads. The CGView Server can be accessed at http://stothard.afns.ualberta.ca/cgview_server/
INTRODUCTION
Despite continual advances in sequence analysis and annotation programs, manual visualization of sequence characteristics remains an important part of understanding gene structure, function and evolution (1). For many fully sequenced genomes, web-based genome browsers offer graphical maps that are integrated with underlying databases of sequences, annotations and analyses (2–5). Genome browsers allow the simultaneous display of the genome sequence together with numerous annotation tracks, such as known genes, predicted genes, ESTs, mRNAs and contigs. In addition, genome browsers provide a window into comparative genomics by displaying similarity information, obtained using a variety of searching and alignment approaches. In cases where a particular genome sequence is not yet available online, comparisons can be performed using more specialized tools. For example, PipMaker (6) and ACT (7) can be used to visualize the similarity between user-supplied sequences, and offer more flexibility than genome browsers in terms of how sequences are compared. PipMaker is a web server that generates a percent identity plot (pip), which shows the position and percent identity of gap-free alignment segments. Feature information can be included in the graphical output, by supplying an optional features file. ACT (Artemis Comparison Tool) is a stand-alone Java program that can be used in conjunction with BLAST to compare two DNA sequences. When supplied with a BLAST results file (the user must perform the BLAST comparison separately), ACT connects regions of similarity between the sequences using coloured lines. These lines can reveal which segments of the genomes are conserved, and can highlight differences in genome organization, such as changes in gene order, or gene duplications. If GenBank or EMBL files are used as the input for ACT, the features described in the files are displayed along with the BLAST results.
Although PipMaker and ACT can accept sequences from any source species, neither generates the circular maps that are popular for visualizing bacterial and organellar genomes. Several programs for creating circular maps are available, including CGView (8), GenomePlot (9), GenoMap (10) and the Microbial Genome Viewer (11). Here we describe the CGView Server, which represents our efforts to integrate many of the capabilities of PipMaker, ACT and BLAST with CGView. The CGView Server generates graphical maps that can be used to visualize sequence conservation in the context of sequence features, imported analysis results, open reading frames and base composition plots. Publication-quality customizable maps can be generated, showing the full sequence, or a more detailed view of a region of interest. Sample maps and data sets further illustrating applications of the CGView Server are available at http://stothard.afns.ualberta.ca/cgview_server/
PROGRAM DESCRIPTION
Data is submitted to the CGView Server via a simple web interface. The minimum information required to obtain a map is a DNA sequence and an email address. Four formats for the sequence are accepted: raw, FASTA, GenBank and EMBL. If either of the latter two formats is used, gene annotations in the file will appear on the map. An email address is required, since the map, which may take several minutes to generate, is returned as an email attachment. All fields in the submission form include a context-sensitive help icon, which can be used to access a description of the options available or the information required.
Additional feature information pertaining to the primary DNA sequence can be supplied in the form of a GFF (General Feature Format) file (http://www.sanger.ac.uk/Software/formats/GFF/). GFF is a format for describing genes and other features associated with nucleic acid and protein sequences. This ‘features’ file can be used to supply gene positions for inclusion on the map that are not given in the primary sequence file. If the GFF file contains single-letter COG functional categories in the ‘feature’ column, the CGView Server will colour the features according to COG category (12). Alternatively, the features can be coloured according to gene type (CDS, tRNA, rRNA or other). GFF files are available from several analysis programs, or they can be assembled manually in spreadsheet programs like Excel. Quantitative measurements can be added to the map using a second ‘analysis’ GFF file. This file can be used to visualize scores or measurements arising from analysis programs, or from laboratory experiments.
In addition to the required primary DNA sequence, up to three comparison sequences can be provided. These can be in raw, FASTA or multi-FASTA format. The multi-FASTA format allows a collection of sequences to be used for a single comparison. Potential collections include all the members of a protein family, or the set of proteins encoded by a particular bacterial genome. For each comparison sequence there is a set of options for specifying the search type and search parameters. These allow searches to be conducted at the DNA or protein level, and hits to be filtered based on significance (e-value), alignment length and percent identity.
The final section of the CGView Server interface provides options for controlling the display of features calculated directly from the primary sequence (GC content, GC skew, ORFs, start and stop codons), and for adjusting the organization and appearance of the map. For example, BLAST hits can be arranged according to the reading frame of the query (for tbastx and blastx searches). This capability can be useful for identifying which ORFs in an overlapping group are conserved. BLAST hits can also be drawn with partial opacity such that regions of the primary sequence producing multiple overlapping hits can easily be identified. Other options include the ability to draw a zoomed view of the map, feature labels, a feature legend and a title.
Data submitted to the CGView Server enters an analysis queue. A Perl program checks the queue periodically, and processes jobs sequentially. Processing begins with the formatdb program (included with BLAST), which is used to convert any comparison sequences into BLAST databases. The primary sequence, serving as the query, is first split into smaller sub-sequences of a user-defined size before calling standalone BLAST. The primary sequence file, BLAST results, GFF files and user options are passed to another Perl script, which builds an XML file for the CGView map-drawing program (8). CGView generates a PNG image, and the image and a description of the submitted files and settings are emailed to the user.
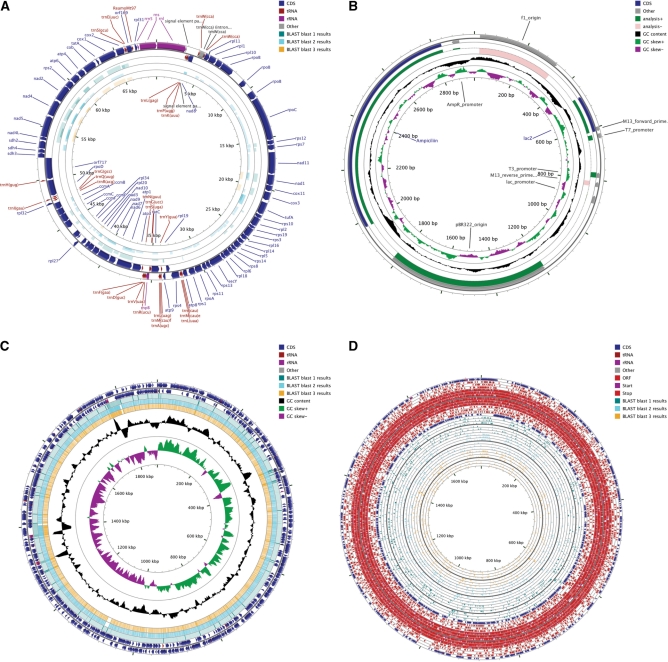
The maps generated by the CGView Server consist of concentric feature rings (Figure 1). Depending on the selected settings, these rings are used to display gene information read from the primary sequence file, features or analysis results from the GFF files, base composition plots, ORFs, start and stop codons, and BLAST results (Figure 2). Features are coloured according to type, and in some cases the height of the feature is adjusted to reflect its properties. BLAST hits, for example, are drawn with a height that is proportional to the percent identity of the hit. Similarly, score values are used to determine the height of features in the analysis GFF file. An optional legend can be used to identify all features based on colour. Labels can be drawn for features read from the primary sequence record or ‘features’ GFF file. A sequence ruler, drawn inside of the innermost feature ring, allows the approximate positions of features to be determined.
Figure 1.
Sample output from the CGView Server. (A) Comparison of a mitochondrial genome with three other genomes using blastx. (B) Visualizing analysis scores for features of a plasmid. (C) Comparison of a bacterial genome with reads from a 454 sequencer using blastn. (D) Visualizing features, ORFs, start and stop codons of a bacterial genome and comparing the sequence with proteins encoded by three other bacteria.
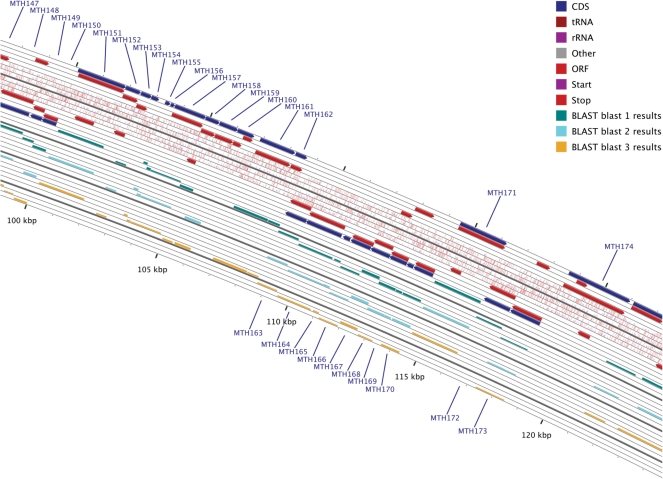
Figure 2.
Example of a zoomed map produced by the CGView Server. A 40× zoomed view of the sequence depicted in Figure 1D, centered on base 110 000. The contents of the feature rings (starting with the outermost ring) are as follows. Ring 1: forward strand features read from the primary sequence GenBank file. Rings 2,3,4: forward strand ORFs in reading frames 3,2,1. Rings 5,6,7: forward strand start and stop codons in reading frames 3,2,1. Rings 8,9,10: reverse strand start and stop codons in reading frames 1,2,3. Rings 11,12,13: reverse strand ORFs in reading frames 1,2,3. Ring 14: reverse strand features read from the primary sequence GenBank file. Rings 15,16,17,18,19,20: BLAST hits obtained from blastx search of bacterial genome 1 proteins, in which the query was translated in reading frames 3,2,1,−1,−2,−3. Rings 21,22,23,24,25,26: BLAST hits obtained from blastx search of bacterial genome 2 proteins, in which the query was translated in reading frames 3,2,1,−1,−2,−3. Rings 27,28,29,30,31,32: BLAST hits obtained from blastx search of bacterial genome 3 proteins, in which the query was translated in reading frames 3,2,1,−1,−2,−3.
CONCLUSION
The CGView Server is a comparative genomics tool for circular genomes (plasmid, bacterial, mitochondrial and chloroplast) that allows sequence feature information to be visualized in the context of sequence analysis results and sequence similarity plots. The server seamlessly integrates several sequence analysis procedures and tools with the CGView genome visualization program. The server accepts a variety of commonly used data formats, and generates high-quality, fully labelled graphical maps.
One drawback of the CGView Server compared to standalone tools like ACT is that the server returns static images. Although these images are suitable for publication, ACT may be more useful for in-depth exploration of sequences and BLAST results. To partially overcome the limitations of providing static images, the CGView Server includes an option for generating zoomed maps. Another limitation for some users may be the inability of the CGView Server to generate more conventional linear maps. The web-based Microbial Genome Viewer can be used to generate circular or linear maps, and may be more appropriate for some users.
Despite these limitations, maps generated by the CGView Server can be used to aid in the identification of conserved or diverged genome segments, instances of horizontal gene transfer, and differences in gene copy number. Because a collection of sequences can be used in place of a comparison genome, maps can be used to identify sequences that are part of a particular family, or to visualize regions of a known genome covered by newly obtained sequence reads. Sample maps and data sets further illustrating applications of the CGView Server are available at http://stothard.afns.ualberta.ca/cgview_server/
ACKNOWLEDGEMENT
Funding to pay the Open Access publication charges for this article was provided by Alberta Livestock Industry Development Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1.Stothard P, Wishart DS. Automated bacterial genome analysis and annotation. Curr. Opin. Microbiol. 2006;9:505–510. doi: 10.1016/j.mib.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spudich G, Fernández-Suárez XM, Birney E. Genome browsing with Ensembl: a practical overview. Brief. Funct. Genom. Proteomics. 2007;6:202–219. doi: 10.1093/bfgp/elm025. [DOI] [PubMed] [Google Scholar]
- 4.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: the Artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 8.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 9.Gibson R, Smith DR. Genome visualization made fast and simple. Bioinformatics. 2003;19:1449–1450. doi: 10.1093/bioinformatics/btg152. [DOI] [PubMed] [Google Scholar]
- 10.Sato N, Ehira S. GenoMap, a circular genome data viewer. Bioinformatics. 2003;19:1583–1584. doi: 10.1093/bioinformatics/btg195. [DOI] [PubMed] [Google Scholar]
- 11.Kerkhoven R, van Enckevort FH, Boekhorst J, Molenaar D, Siezen RJ. Visualization for genomics: the microbial genome viewer. Bioinformatics. 2004;20:1812–1814. doi: 10.1093/bioinformatics/bth159. [DOI] [PubMed] [Google Scholar]
- 12.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]