Abstract
KEGG Atlas is a new graphical interface to the KEGG suite of databases, especially to the systems information in the PATHWAY and BRITE databases. It currently consists of a single global map and an associated viewer for metabolism, covering about 120 KEGG metabolic pathway maps and about 10 BRITE hierarchies. The viewer allows the user to navigate and zoom the global map under the Ajax technology. The mapping of high-throughput experimental data onto the global map is the main use of KEGG Atlas. In the global metabolism map, the node (circle) is a chemical compound and the edge (line) is a set of reactions linked to a set of KEGG Orthology (KO) entries for enzyme genes. Once gene identifiers in different organisms are converted to the K number identifiers in the KO system, corresponding line segments can be highlighted in the global map, allowing the user to view genome sequence data as organism-specific pathways, gene expression data as up- or down-regulated pathways, etc. Once chemical compounds are converted to the C number identifiers in KEGG, metabolomics data can also be displayed in the global map. KEGG Atlas is available at http://www.genome.jp/kegg/atlas/.
INTRODUCTION
The continuous development of high-throughput experimental technologies generates large-scale data sets, now including metagenomes and personal genomes, which necessitates renewed efforts to develop computational technologies for better integration and biological interpretation. One key element is a computerized resource for higher-level knowledge on metabolism and other cellular processes as functional aspects of the biological systems. The KEGG resource (http://www.genome.jp/kegg/) (1) has been developed to represent such knowledge in terms of the molecular interaction and reaction networks and has enabled ‘pathway mapping’, which is to map large-scale data sets to the KEGG reference pathways for understanding higher-level functional meanings. In KEGG and other pathway databases (2,3), experimental knowledge is organized and diagramed as smaller networks, and web interfaces and visualization tools have been developed to overview and analyze computationally generated global networks (3–6).
Recently, a manually created global map for metabolism has been released in KEGG (1), combining about 120 existing pathway maps and corresponding to about 2000 genes. The global map is part of KEGG Atlas, a new graphical interface providing a global view and zoom-in capability to reach and integrate existing resources. The KEGG Atlas viewer allows the user to navigate the global map under the Asynchronous JavaScript and XML (Ajax) technology. The global map is created as a Scalable Vector Graphics (SVG) formatted file rather than a raster graphics image, which makes it much easier and faster to perform manipulations on the map, e.g. in terms of coloring and thickening. KEGG Atlas also provides the mapping interface to color objects in the global map with the user's own data. Thus, it enables the user to view and compare the entire metabolism, such as by mapping genomic, transcriptomic, metagenomic and metabolomic data. In addition, the user can reconstruct the global metabolic pathway from genomic or metagenomic data via the KAAS web server (http://www.genome.jp/kegg/kaas/) (7), which provides automatic annotation based on the KEGG Orthology (KO) system.
KEGG ATLAS
Global metabolism map
The global metabolism map is a single connected metabolic pathway, manually created by combining about 120 existing KEGG metabolic pathway maps excluding some secondary metabolism maps. First, a ‘net-element’ is manually defined as a segment lacking branches in the existing pathway map and is given the N number identifier after removing duplicates in multiple maps. Two-thirds of the net-elements are single reactions connecting substrates and products, but the rest corresponds to multiple reaction steps in the original maps and contains ‘hidden’ compounds. Then, the net-element is drawn on the global map as a line (curved or straight) connecting two compounds or a group of partially overlapping lines connecting three or more compounds (multiple substrates and products). The compounds are drawn as circles and identified by the C numbers. Because the global map is a connection diagram of main compounds, the same line may correspond to multiple net-elements, for example, with different cofactors.
The global map is created as an SVG file. The file is structured as a comprehensible format and consists of four layers: net-element, compound, placemark and category layers. Lines and circles in the global map are the objects in the net-element and compound layers, respectively. The placemark layer contains objects for linking to existing KEGG pathway maps and BRITE hierarchies, which appear as text information on the global map. The category layer is used to distinguish the KEGG pathway category as shown by color coding on the global map. The current version of the global map consists of 1534 net-elements and 934 compounds. The global map is continuously updated in accordance with the addition and modification of the existing pathway maps.
Global map viewer
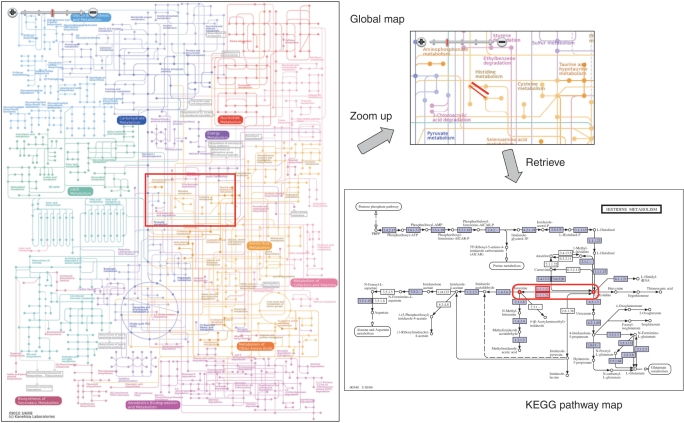
We have developed a global map viewer using the Ajax technology, and made it available at the KEGG Atlas website (http://www.genome.jp/kegg/atlas/). To make the viewer usable with different web browsers in different platforms, the SVG file is converted to PNG image files at five levels of resolution, each containing one to 256 pieces of small tiled images. The user can move the map by dragging the mouse and zoom the map by controlling the slider button (Figure 1). Because Ajax provides an interactive interface without web page reloading, only the necessary images are transferred into the user's web browser each time the user drags the map. When the mouse cursor is put on a line/circle object on the global map, the corresponding object is highlighted. This also indicates that the line/circle object is clickable to retrieve the corresponding N/C number entry. When the line is clicked, the user can follow the link to retrieve the KEGG pathway map highlighting the genes (proteins) and compounds that constitute the net-element (Figure 1). When the circle is clicked, the corresponding KEGG COMPOUND entry is retrieved. The text for placemarks can be clicked to zoom into existing KEGG pathway maps and BRITE hierarchies.
Figure 1.
KEGG Atlas global metabolism map viewer.
MAPPING
Organism-specific global map
There are over 700 organisms with complete genome sequences currently available in KEGG. Genes in each organism are annotated with the KEGG Orthology (KO) identifiers, or the K numbers, representing manually defined ortholog groups that correspond to the nodes in the KEGG pathway map or the bottom leaves in the BRITE hierarchy (1). The net-element in the global map designates a set of reactions, and it also represents a set of KO entries for the enzymes that catalyze the reactions. Thus, as a default the global map viewer displays the reference pathway linked to KO. The genomic content of enzyme genes can then be mapped to this reference global map through the conversion of gene identifiers to K numbers. The global map viewer contains a mechanism to select an organism and manipulate an organism-specific global map in which lines and edges presumed to be absent are dimmed out. The organism-specific global map is generated on the fly from the SVG file, and it is linked to the existing organism-specific pathway maps, which are still precomputed GIF image files. The conversion of existing maps into SVG files is underway.
Mapping the user's data
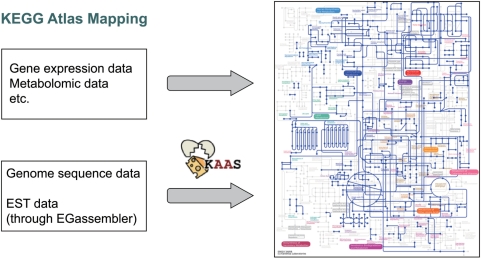
The KEGG Atlas website provides a mapping interface to allow mapping of genes/compounds as colored lines/circles in the global map (Figure 2). Taking advantage of this function, the user can enter the data into the boxed textarea or upload a file containing the data and obtain the colored global map. This is particularly useful for mapping gene expression profile data, where a color chart represents the level of expression. The originally colored global map can be examined using the Ajax-based viewer or downloaded as a high-resolution PNG file. When the reference global map (default) is selected, the KEGG Atlas mapping interface accepts K numbers and C numbers as input data for coloring lines and circles, respectively. When an organism-specific map is selected, gene identifiers can be used in place of K numbers. Each identifier is to be given one per line, optionally followed by color specification. Because line coloring is based on the net-element, which may consist of multiple K numbers or gene identifiers, an option is provided either to allow partial matches or to limit to complete matches requiring a full set of identifiers.
Figure 2.
KEGG Atlas mapping from the web interface or the KAAS annotation server.
Via KAAS automatic annotation server
In the case of an organism stored in KEGG, the user can simply select the organism to view and manipulate the organism-specific global map. For an organism not stored in KEGG, the automatic annotation server KAAS (7) is available for generating an organism-specific global map (Figure 2). Given a set of amino acid sequences for genes in the complete genome, KAAS performs BLAST comparisons against a manually curated set of KEGG GENES. The assignment of KO identifiers (K numbers) to individual genes is based on best-hit relationships as well as sequence-similarity scores. The publicly available version of KAAS is already implemented with the feature to generate an organism-specific global map by mapping assigned KOs to the reference global map.
POTENTIAL APPLICATIONS
The global map enables data viewing and extraction protocols to be developed such as the one reported here or others that are being developed (8). Besides genomic and transcriptomic data, metabolomic data can be mapped to the global metabolism map, perhaps with color coding as a quantitative measure. The global map should also be useful to compare the entire metabolism in different organisms or conditions, such as dynamical changes of gene expressions in a time-series microarray experiment, changes of metabolic capabilities seen in metagenomics data under different environmental conditions, and correlations and complementarities of metabolic capabilities in symbiotic relationships.
AVAILABILITY
KEGG Atlas is made available as a component of the KEGG system and can be accessed at http://www.genome.jp/kegg/atlas/.
ACKNOWLEDGEMENTS
This work was supported by the Institute for Bioinformatics Research and Development of the Japan Science and Technology Agency, the 21st Century COE program ‘Genome Science’, and a grant-in-aid for scientific research on the priority area ‘Comprehensive Genomics’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The computational resource was provided by the Bioinformatics Center, Institute for Chemical Research, Kyoto University. Funding to pay the Open Access publication charges for this article was provided by the grant-in-aid for scientific research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, Paley S, Rhee SY, Shearer AG, Tissier C, et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2008;36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vastrik I, D’Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu Z, Ng DM, Yamada T, Chen C, Kawashima S, Mellor J, Linghu B, Kanehisa M, Stuart JM, DeLisi C. VisANT 3.0: new modules for pathway visualization, editing, prediction and construction. Nucleic Acids Res. 2007;35:W625–W632. doi: 10.1093/nar/gkm295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kono N, Arakawa K, Tomita M. MEGU: pathway mapping web-service based on KEGG and SVG. In Silico Biol. 2006;6:621–625. [PubMed] [Google Scholar]
- 6.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letunic I, Yamada T, Kanehisa M, Bork P. iPath: interactive exploration of biochemical pathways and networks. Trends Biochem. Sci. 2008;33:101–103. doi: 10.1016/j.tibs.2008.01.001. [DOI] [PubMed] [Google Scholar]