Abstract
The generation of expressed sequence tag (EST) libraries offers an affordable approach to investigate organisms, if no genome sequence is available. OREST (http://mips.gsf.de/genre/proj/orest/index.html) is a server-based EST analysis pipeline, which allows the rapid analysis of large amounts of ESTs or cDNAs from mammalia and fungi. In order to assign the ESTs to genes or proteins OREST maps DNA sequences to reference datasets of gene products and in a second step to complete genome sequences. Mapping against genome sequences recovers additional 13% of EST data, which otherwise would escape further analysis. To enable functional analysis of the datasets, ESTs are functionally annotated using the hierarchical FunCat annotation scheme as well as GO annotation terms. OREST also allows to predict the association of gene products and diseases by Morbid Map (OMIM) classification. A statistical analysis of the results of the dataset is possible with the included PROMPT software, which provides information about enrichment and depletion of functional and disease annotation terms. OREST was successfully applied for the identification and functional characterization of more than 3000 EST sequences of the common marmoset monkey (Callithrix jacchus) as part of an international collaboration.
INTRODUCTION
In spite of considerable genome sequencing efforts during the last years there is still a large number of poorly characterized organisms, which serve as models for the investigation of different phenotypes and diseases. Sequencing of cDNA libraries offers a low-cost approach to obtain information about the protein-coding genes of these organisms. End-sequencing of cDNA clones with standard primers results in expressed sequence tag (EST) libraries with up to thousands of DNA fragments. The dbEST at the NCBI (1), one of the largest resources, contains more than 48 000 000 EST sequences from 1500 organisms. In addition, EST data are used to validate gene models on genome sequences, to analyse results from proteome experiments and to construct organism-specific microarrays (2).
High-throughput in silico analysis of thousands of EST sequences for the identification of corresponding gene products and associated information requires an automated EST analysis tool which should fulfil several requirements: (i) it should be designed in a way that it can be operated without in-depth bioinformatics skills; (ii) it should allow the analysis of ESTs from organisms with different phylogenetic background; (iii) the tool has to identify gene products that correspond to ESTs with high accuracy; (iv) in order to provide the user with a primary characterisation about the dataset there is a demand of a systematic functional annotation of the dataset and (v) statistics about functional characteristics that are significantly over- or under-represented within the dataset. There are a number of EST processing systems existing like ESTAnnotator (3), ESTAP (4), ESTExplorer (5), PartiGene (6) or EST2uni (7). However, many of the existing tools require local installation and maintenance of the latest versions of the tools and databases. This hampers an immediate analysis for occasional users and requires investments for disc requirements, database installation and administration.
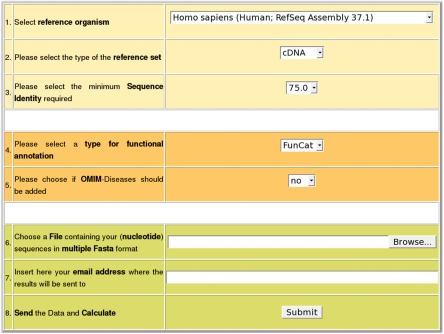
Here, we present OREST (Figure 1), a web-based EST analysis pipeline for gene assignment and systematic functional annotation of large amounts of DNA sequences. OREST allows mapping of user data to the fungal model organism Saccharomyces cerevisiae as well as to several mammalian datasets. Automated functional assignment of the gene products can be performed via FunCat or GO annotation schemes. Mapping to the human dataset predicts also the association of the ESTs with diseases. Over- und under-represented features from functional annotation and disease relevance are obtained through a statistical analysis. Advantage and usability of the OREST EST analysis pipeline has been shown in a successful analysis of more than 3000 ESTs of the common marmoset monkey (Callithrix jacchus) within an international scientific consortium (2).
Figure 1.
Screenshot of the OREST index page.
OVERVIEW OF OREST
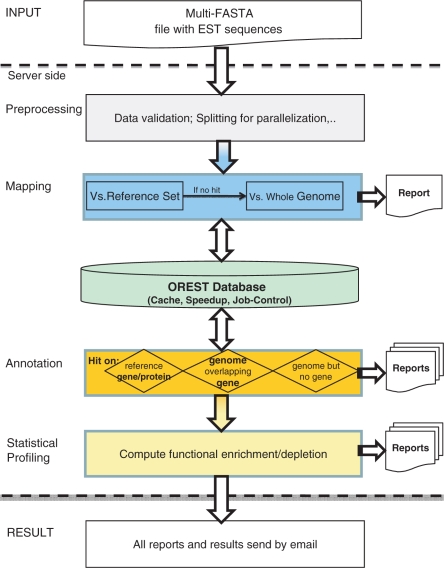
The main function of the OREST server (http://mips.gsf.de/genre/proj/orest/index.html) is the EST annotation pipeline, whose processes can be separated into four consecutive steps: selection of parameters for analysis, data pre-processing and EST mapping, functional annotation and statistical analysis. The workflow of the pipeline is depicted in Figure 2.
Figure 2.
OREST workflow. OREST accepts multiple FASTA files as input. Results are calculated in four steps: pre-processing, mapping, annotation and statistical profiling.
Selection of parameters
The first step of EST analysis in OREST is the upload and validation of the DNA sequences as Fasta file and selection of appropriate parameters. Uploaded files can contain up to 50 000 sequences for cDNA/genome mapping and 30 000 for protein mapping. As reference dataset for EST analysis OREST offers a selection of different mammalian model organisms (human, mouse, rat and five other mammals) as well as bakery's yeast S. cerevisiae for the analysis of fungal EST libraries. Depending on the phylogenetic relationship between sample and organism of the reference dataset the user can select a suitable minimum sequence similarity.
The type of reference set determines whether the data are mapped against a genomic/transcript dataset or, after six-frame translation, against a protein reference dataset. Organism specific datasets for a protein sequence based analysis are obtained from UniProt (8), and for DNA-based analysis OREST uses weekly downloadable entries from RefSeq (1). Predicted gene models from RefSeq are omitted.
For functional annotation, the user can select between FunCat annotation (9) and GO annotation (10). Analysis of fungal ESTs is only possible with FunCat. If human is selected as reference organism the annotation can be supplemented with disease association of gene products via Morbid Map (OMIM) (1).
EST pre-processing and mapping
If the software for trimming of EST sequences and removal of vector sequences is not provided by the vendor of the DNA-sequencer, respective web-based tools like WebTraceMiner (11) can be used. EST sequences shorter than 100 bases are discarded by OREST. For mapping against a proteomic reference set, the input sequences are translated into protein sequences in all six reading frames. The analysis is not performed with the complete translated sequence but only with the putative N-terminus or C-terminus of the protein sequence given that the partial orf has a length of at least 20 amino acids.
For the sequence similarity comparison the Blat software (12) is used. Blat is specifically designed to perform EST and mRNA alignments with genomic DNA. Compared to other existing tools Blat was shown to be 500 times faster (12). Blat is also able to detect splice sites in vertebrate DNA sequences, which enable the extraction of the protein coding part of partially spliced ESTs that is not possible with a BlastX search.
In contrast to other EST annotation tools, the mapping is performed by two consecutive steps. The first step consists of the mapping of the pre-processed input sequences against the chosen reference set (genetic or proteomic) and the identification of insignificant hits. If no result was observed for mapping on cDNAs or protein sequences, the analysis is performed with the reverse complementary sequence. In this first step, there are two frequent reasons for insignificant results: low sequence quality or incompleteness of available reference datasets. ESTs, which would be discarded because of the latter reason can be saved by the second step. Here, EST data are mapped against the whole genome. This step is done by calculating overlaps of the mapped ESTs and RefSeq (valid coordinates)/CYGD entries on the genome. If the exonic area of a gene is covered, the EST is preserved, otherwise the sample is assigned with the label ‘no hit’. Including the genome information, in a collaborative project (2), additional 13% of the ESTs could be annotated. Calculations with gene product reference sets as first step is required, in order to minimize the computational expensive mapping against the whole genome. As mentioned above, a cut-off value for the significance of hits can be selected with a minimum similarity between 50% and 90% for cDNA and genome reference sets. For mapping against protein sequences a value of 80% is fixed in order to avoid spurious hits.
Functional annotation
Exploitation of the large amount of data generated by EST experiments requires systematic annotation of the individual sequences. For functional annotation of eukaryotic gene products, two heavily used annotation schemes exist, the GO (10) and the FunCat (9). For mammalian EST datasets, the user can select between GO and FunCat, for yeast OREST only supports FunCat annotation.
The GO annotation is obtained from the corresponding RefSeq/SwissProt entries. If the user prefers a hierarchical annotation scheme for downstream analysis the GO information is mapped to the FunCat (9), which is used many times for analyses of high-throughput data sets. If yeast is the model organism, the FunCat annotation is directly retrieved from CYGD (13).
For functional annotation in OREST, ESTs are separated in two result sets: those having a hit against a reference transcript and those mapped to the genome. ESTs with a hit against a reference gene or protein are annotated with GO identifiers and GO descriptions of the corresponding RefSeq or SwissProt/UniProt entries. GO annotation with the label IEA (inferred by electronic annotation) are not included since it tends to be error-prone. Only the best three hits per EST are provided as result. For EST sequences without corresponding gene the coordinates on the genome will be provided.
Using the human reference set for analysis also allows to annotate or to predict the association of genes with diseases. For this additional annotation, SwissProt and RefSeq entries are mapped to corresponding Morbid Map (OMIM) (1) disease identifiers. The mapping is performed using the annotated gene names and synonyms. To prevent mapping of ambiguous gene names, a minimum length of three letters is required. Such instances occur for identical synonyms that exist in different genes like ‘TF’ for transferrin (NM_001063) and coagulation factor III (thromboplastin, tissue factor, NM_001993).
Statistics
Do cancer-associated ESTs show non-random enrichment of certain functional families? How are the ESTs of interest related to molecular function? Can we detect EST markers pointing to diseases? All these questions can be addressed by OREST based on a rigour statistical evaluation and solid data basis. The latter is achieved via utilizing the latest Gene-Ontology, FunCat and OMIM disease-information of the genes of which the ESTs originate. Statistical enrichment and depletion of annotation terms is performed using the PROMPT (14) framework. Briefly, for all available annotation terms found to be attributed to the user supplied ESTs, the over- or under-representation is computed in comparison to the whole genome of the respective organism. Statistical significance is assessed by an e-score representing the likelihood that the difference would be found by random. The e-score is calculated as described in Castilo-Davis and Hartl (15) using a hypergeometric distribution with conservative Bonferroni correction. As a result, OREST provides a convenient integrated profiling of disease and functional annotations along with statistical significance determination for all processed ESTs.
Output
The output of the complete analysis of OREST will be sent via Email and consists of two different kinds of reports—the annotation results and statistical reports based on the annotation. Annotation results are differentiated according to their mapping quality. Statistical results are provided for the annotated genes, respectively. If OMIM-diseases where selected another two reports are included. For file formats see Supplementary Data S1.
Use case
The OREST pipeline has still been used in an international project with several cooperation partners for the analysis of cDNAs from marmoset monkey (2). The project started about one and a half years ago and encompassed more than 3000 EST sequences. Thus, OREST has proved its performance and value not only with test data but in a real collaborative project.
IMPLEMENTATION/SOFTWARE DESIGN-ENVIRONMENT
The OREST website runs in a J2EE environment using a Sun Java System Application server version 9.1 and is seamlessly integrated within the MIPS Genome Research Environment (GenRE) (16). The server consist of two independent pipelines for the different reference set types (genomic/transcript, protein), whereas all computation tasks are distributed on 40 CPUs [13 Linux machines (AMD64 Processor, 2-4 CPUs, 4 GB RAM per CPU)] using the Sun Grid Engine (www.sun.com) and thus ensures scalability and fast processing. Caching of processed data and results, as well as job control is aided by a MySQL database backend. The OREST pipeline is completely implemented in Java version 1.5. Mappings are performed using BLAT version 34 (64bit version).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
Funding to pay the Open Access publication charges for this article was provided by Helmholtz Zentrum München.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Datson NA, Morsink MC, Atanasova S, Armstrong VW, Zischler H, Schlumbohm C, Dutilh BE, Huynen MA, Waegele B, Ruepp A, et al. Development of the first marmoset-specific DNA microarray (EUMAMA): a new genetic tool for large-scale expression profiling in a non-human primate. BMC Genomics. 2007;8:190. doi: 10.1186/1471-2164-8-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hotz-Wagenblatt A, Hankeln T, Ernst P, Glatting KH, Schmidt ER, Suhai S. ESTAnnotator: a tool for high throughput EST annotation. Nucleic Acids Res. 2003;31:3716–3719. doi: 10.1093/nar/gkg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao C, Cushman JC, May GD, Weller JW. ESTAP—an automated system for the analysis of EST data. Bioinformatics. 2003;19:1720–1722. doi: 10.1093/bioinformatics/btg205. [DOI] [PubMed] [Google Scholar]
- 5.Nagaraj SH, Deshpande N, Gasser RB, Ranganathan S. ESTExplorer: an expressed sequence tag (EST) assembly and annotation platform. Nucleic Acids Res. 2007;35:W143–W147. doi: 10.1093/nar/gkm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parkinson J, Anthony A, Wasmuth J, Schmid R, Hedley A, Blaxter M. PartiGene—constructing partial genomes. Bioinformatics. 2004;20:1398–1404. doi: 10.1093/bioinformatics/bth101. [DOI] [PubMed] [Google Scholar]
- 7.Forment J, Gilabert F, Robles A, Conejero V, Nuez F, Blanca JM. EST2uni: an open, parallel tool for automated EST analysis and database creation, with a data mining web interface and microarray expression data integration. BMC Bioinform. 2008;9:5. doi: 10.1186/1471-2105-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang C, Wang G, Liu L, Ji G, Liu Y, Chen J, Webb JS, Reese G, Dean JF. WebTraceMiner: a web service for processing and mining EST sequence trace files. Nucleic Acids Res. 2007;35:W137–W142. doi: 10.1093/nar/gkm299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guldener U, Munsterkotter M, Kastenmuller G, Strack N, Van Helden J, Lemer C, Richelles J, Wodak SJ, Garcia-Martinez J, Perez-Ortin JE, et al. CYGD: the comprehensive yeast genome database. Nucleic Acids Res. 2005;33:D364–D368. doi: 10.1093/nar/gki053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt T, Frishman D. PROMPT: a protein mapping and comparison tool. BMC Bioinform. 2006;7:331. doi: 10.1186/1471-2105-7-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castillo-Davis CI, Hartl DL. GeneMerge—post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- 16.Mewes HW, Frishman D, Mayer KF, Munsterkotter M, Noubibou O, Pagel P, Rattei T, Oesterheld M, Ruepp A, Stumpflen V. MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res. 2006;34:D169–D172. doi: 10.1093/nar/gkj148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.