Abstract
The Gene Context Tool (GeConT) allows users to visualize the genomic context of a gene or a group of genes and their orthologous relationships within fully sequenced bacterial genomes. The new version of the server incorporates information from the COG, Pfam and KEGG databases, allowing users to have an integrated graphical representation of the function of genes at multiple levels, their phylogenetic distribution and their genomic context. The sequence of any of the genes can be easily retrieved, as well as the 5′ or 3′ regulatory regions, greatly facilitating further types of analysis. GeConT 2 is available at: http://bioinfo.ibt.unam.mx/gecont.
INTRODUCTION
With more than 660 prokaryotic genomes in the current RefSeq database (1), the need for tools that allow biologists to visualize the genomic context of genes of their interest becomes crucial. Despite the tendency of prokaryotes towards very little overall conservation of gene order (2,3), groups of genes participating in particular functions tend to remain close together across different lineages, either as part of operons (4,5) or as functional neighbourhoods comprising several transcription units (6,7). Genomic context is not limited to genes close to each other. Overall, inference of functional associations from genomic context can be derived from the following kinds of evidence: (i) Gene fusions (8,9), whereby separated genes would be assumed to work together if they are found as a single, fused, gene in another organism; (ii) Conservation of gene order (3,10), where conservation across evolutionarily distant organisms is taken as evidence of functional association and (iii) similarity of phylogenetic profiles (9,11,12), whereby two genes are assumed to work together if their orthologs tend to co-occur, appear and disappear in concert, across different organisms, with the idea that genes working together would both either be present or absent because the presence of a single one of them would be useless without the other. A fourth evidence of functional interactions would be provided by the study of the rearrangement of operons across lineages (13–15). The idea here is that the rearrangements or reorganization of transcription units across genomes might be conservative in the sense that newly formed operons will put genes with related functions together, thus revealing a functional association that would not be apparent in a single organism.
Biologists interested in particular groups of genes or functional modules would be able to find other features by visually inspecting the genomic context or neighbourhood of the genes of their interest. Such experts might be able to interpret these neighbourhoods and find examples of non-orthologous gene displacement (2), or horizontal gene transfers, that might have an effect on the functional module in particular organisms. Further tests of the validity of their findings would be greatly facilitated if the tool used to visualize the gene modules across several genomes would also allow for downloading of meaningful sequences, such as the protein sequences, or the DNA coding for the gene, or the DNA sequences occurring downstream or upstream the gene. There are excellent tools that allow for the retrieval of functional predictions based on genomic context, such as STRING (16) and PROLINKS (17), but such tools restrict the visualization of genomic context to the particular predictions associated to a gene or genes of interest, rather than to any physical neighbours. Also, while protein sequences of predicted interactors can be retrieved from these servers, nucleotide sequences of genes or intergenic regions are not available. In other instances, such as GECO (18), the navigation interface for retrieving this type of information is not simple and the orthology definition is different from any of the most commonly accepted standards. The SEED (19) is a fully automated web resource that analyses the genome context of bacterial and archaeal organisms, however its main purpose is oriented to genome annotation rather than the genome context exploration by a particular user who wants to examine the neighbourhoods of his/her genes of interest. Another useful web server is the comprehensive microbial resource (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi). Although this web server offers a wide variety of tools and resources to highlight differences and similarities between prokaryotic genomes, its comprehensiveness hinders straightforward navigation, further justifying the development of more simplified gene context analysis tools. Here we present Gene Context Tool (GeConT) 2, the second version of GeConT (20), a web-based tool that allows users to visualize their genes of interest, and their genomic context, across all available fully sequenced bacterial genomes. Orthologous domains are highlighted using shared colours. This makes it easy to navigate across the functional neighbourhood of any particular gene and its orthologs.
IMPROVEMENTS
GeConT 2 extends over the previous version in many ways. We have increased the query options to allow one or more of the following: (i) gene ids which can be given as common names, GI numbers as defined in GenBank (21) or SwissProt identifiers (22); (ii) orthologous groups as defined in the COG database (23); (iii) metabolic pathways as described in the KEGG database (24); (iv) protein domains taken from the Pfam database (25); (v) a protein or DNA sequence, from which similarities will be identified using the integrated BLAST (26) search; (vi) complex phrases, using Boolean operators, to allow flexible searches against all the descriptions of the included databases. Since many queries will likely result in hundreds of matches (many of which are likely to be redundant), we implemented a filter that can reduce the display to a user-specified number of non-redundant genomes. This option uses distances calculated from 16S rRNA alignments to select a set of representative genomes specific for each query. Additionally, the user can also restrict the search to particular phylogenetic groups of interest. The genome context can be displayed considering a user-defined number of flanking genes or in accordance to their predicted operon structure (27–29).
In agreement with the increased input flexibility, the output allows visualizing the genes colour-coded according to their COG, Pfam or KEGG assignations. Also new to this version, multiple domains can be visualized as distinct coloured regions within a gene. Domains, genes and intergenic regions are drawn to scale; overlapping genes and non-coding RNA genes are now included. The user can click on any cistron to display relevant information, including descriptions from COG, KEGG and Pfam, as well as the amino acid and nucleotide sequence, and upstream and downstream intergenic flanking regions. GeConT 2 also allows the user to retrieve all the sequence data of the set of genes that have been matched by the input query, facilitating further analysis.
WEB SERVER DESCRIPTION
All the code for GeConT 2 is written in Perl, generating HTML and JavaScript code on the fly, using the GD library for the dynamic creation of images. The server uses fully sequenced genomes downloaded from GenBank. All gene coordinates, DNA strand, names and descriptions are taken from these files. Pfam and COG annotations were computed for all coding sequences using the HMMER package (30). Pfam-A models (25) were directly obtained from ftp://ftp.sanger.ac.uk/pub/databases/Pfam/. COG models were generated by aligning the sequences from every COG with MUSCLE (31) and building the models with hmmerbuild from the HMMER package. KEGG pathway annotations for all genomes were downloaded from ftp://ftp.genome.jp/pub/kegg/pathway/. The resulting annotations for each gene are saved as indexed files that are tied to hashed arrays for faster access. When queried for a particular gene, the server calculates the sizes, distances and neighbours based on the stored information. Once the list of genes to be displayed has been calculated, the server assigns colours starting with the COG, Pfam or KEGG most represented among these genes. In this way, the user can gain a visual insight of the most abundant annotations among the displayed genes. Additionally, the information about any gene can be quickly inspected by placing the mouse over it.
EXAMPLES AND DISCUSSION
With GeConT 2 users will be able to perform fast, integrated and intuitive analyses in fully sequenced genomes. In this section we discuss several examples that help illustrate the functionality of the webserver.
Identifying conserved elements involved in regulating a given pathway
An important feature of GeConT 2 is its potential to do comparative genome analysis of related genes to look for potential conserved regulatory motifs. The gene relationship can be established based on their orthology or biochemical pathway associations as defined in the COG, KEGG and Pfam databases. Since regulatory elements are commonly more conserved in closely related organisms, users can restrict their searches to a particular phylogenetic group. For example, in order to identify likely regulatory elements in methionine metabolism represented by the KEEG pathway 00271 in Firmicutes and Proteobacteria, the user can perform the corresponding searches in these groups by using the ‘Specific taxonomy’ option. The output of two representative organisms of these groups is shown in Figure 1 of the Supplementary Material. Using the operon clustering option, and colouring by COG attributes, there are 17 different operons with enzymes related to methionine metabolism in the Firmicute Bacillus halodurans, while there are 11 operons in the Proteobacteria Caulobacter crescentus. The user can take advantage of the sequence retrieval options in GeConT 2 and get all the 5′ upstream regions for these operons. Using these sequences as the input of motif discovery programs such as MEME (32), the user can verify that the operons involved in this pathway are regulated by the SAM-I and S(MK) riboswitches in Firmicutes and by SAM-II in alpha-Proteobacteria (33–35). It is important to note that redundant information coming from different strains of the same organism might generate over-representation of particular sequences in the data set. To overcome this problem, the user can reduce the number of organisms returned by the ‘maximum genomes to display’ option. Previously we have shown the power of this kind of approach for identifying riboswitches starting from the regulatory regions of genes belonging to a same COG (36). It is now possible, using only web-based tools such as GeConT 2, to perform similar searches using any group of genes or pathways that a user might be interested in.
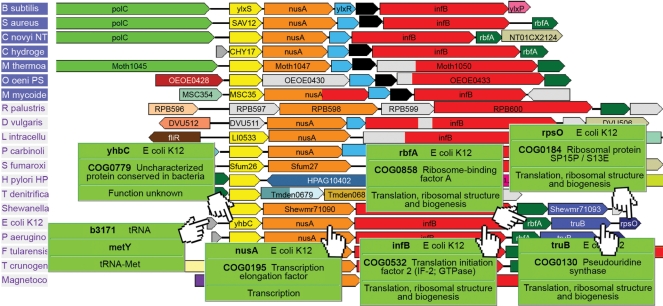
Figure 1.
Genome context of COG0779/Pfam Duf150. Genome context analysis shows that all the conserved neighbouring genes of COG0779 have functions related to translation.
Functional insights for genes of unknown function
Most genes have little or no functional annotation. Even for the most studied bacteria, Escherichia coli, the fraction of genes for which detailed knowledge is available is still low [54% in the latest survey (37)]. Genomic context can give valuable insights into the functional relationships between neighbouring genes, for reasons discussed in the introduction. There are many cases of conserved proteins for which no functional assignation is available in the public databases. Homology searches are of no use, since all the hits also lack function. Context analysis can help solve some of these cases. One such example is annotated in Pfam as Duf150 (Domain of Unknown Function 150) and in the COG database as COG0779 (‘Uncharacterized protein conserved in bacteria’). Figure 1 shows a section of the results when searching for Duf150 in GeConT 2. It is easy to see that the context of this protein is well conserved, and the mouse-over function allows a quick view of the functional assignations of the neighbouring genes, all of which seem to be involved in transcriptional elongation or translation initiation. It is thus quite likely that Duf150/COG0779 members are functionally related to these processes, and this can be considered as a first general function prediction for these previously uncharacterized proteins.
Using context to discover the correct function for paralogs
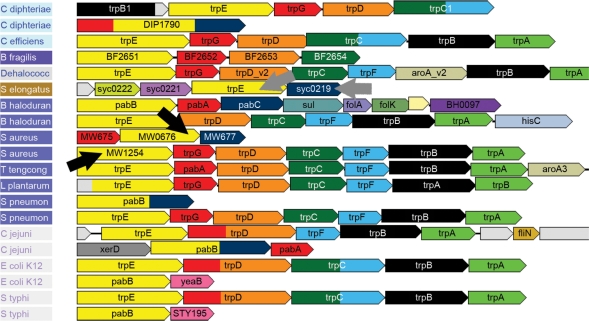
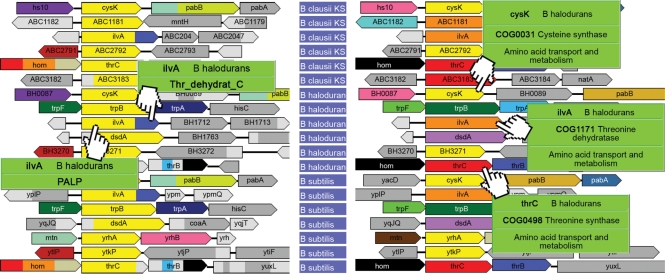
When multiple copies of a gene arise by duplication (paralogs), it can become particularly difficult to assign the correct function, at least by sequence alone. For example, the enzymes TrpE (Anthranilate synthases component I) and PabB (para-aminobenzoate synthases component I) have great sequence similarity and perform very similar reactions. These enzymes use pyruvate as a common substrate, although they participate in different pathways involved in tryptophan and in folate biosynthesis, respectively. Based on genome context, the trpE and pabB genes can easily be distinguished even in un-annotated genomes. With GeConT 2 we can analyse the neighbourhood searching for the other genes of the corresponding metabolic pathways (Figure 2). Another good example of paralogous domains is Palp (Pyridoxal-phosphate-dependant enzyme). Enzymes with this Pfam domain are highly versatile, participating in the biosynthesis of different amino acids such as tryptophan, cysteine, serine and threonine. Again, the context as well as the COG annotations allow us to easily distinguish between the different pathways and correctly identify the specific function of each gene (Figure 3).
Figure 2.
Identification of gene function among paralogous genes, based on the operon structure and COG annotations. The trpE genes can be differentiated from pabB (both in yellow) since the first one is transcribed with other genes of the tryptophan biosynthetic pathway, such as trpD (orange) trpC (dark green), trpF (light blue), while pabB is part of operons carrying genes of the folate biosynthetic pathway, such as pabC (dark blue). In Staphylococcus aureus MW2 trpE and pabB are not annotated (black arrows), yet we can clearly distinguish them from their context. We can also see that in Synechococcus elongatus, trpE is incorrectly annotated since this gene is co-transcribed with pabC (grey arrows).
Figure 3.
Genome context of the Pfam Palp domain in Firmicutes. One of the most salient features of GeConT 2 is the possibility of displaying contexts of multiple instances of a domain within a single genome, besides corresponding contexts in other genomes. On the left, genes are coloured by Pfam domains, with the yellow one corresponding to the Pfam Palp domain. On the right, the same genes are coloured by COG. This analysis shows how this catalytic domain can be used for different purposes by enzymes involved in the biosynthesis of different amino acids.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We wish to thank Shirley Ainsworth for bibliographical assistance and Abel Linares, Arturo Ocadiz, Juan Manuel Hurtado, Alma Martinez and Nancy Mena for computer support. G.M-H. acknowledges computer facilities of the Shared Hierarchical Academic Research Computing Network (SHARCNET). Funding was provided by Natural Sciences and Engineering Research Council of Canada (NSERC) to G.M-H. Sanger Institute Postdoctoral Fellowship to C.A-G. Consejo Nacional de Ciencia y Tecnología (CONACyT) [60127-Q] and PAPIIT IN212708 grants to E.M. Macroproyecto de Tecnologías de la información y la computación-UNAM to E.M. Funding to pay the Open Access publication charges for this article was provided by CONACYT [60127-Q].
Conflict of interest statement. None declared.
REFERENCES
- 1.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koonin EV, Mushegian AR, Bork P. Non-orthologous gene displacement. Trends Genet. 1996;12:334–336. [PubMed] [Google Scholar]
- 3.Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, Yuan Y. Predicting function: from genes to genomes and back. J. Mol. Biol. 1998;283:707–725. doi: 10.1006/jmbi.1998.2144. [DOI] [PubMed] [Google Scholar]
- 4.Ermolaeva MD, White O, Salzberg SL. Prediction of operons in microbial genomes. Nucleic Acids Res. 2001;29:1216–1221. doi: 10.1093/nar/29.5.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreno-Hagelsieb G, Trevino V, Perez-Rueda E, Smith TF, Collado-Vides J. Transcription unit conservation in the three domains of life: a perspective from Escherichia coli. Trends Genet. 2001;17:175–177. doi: 10.1016/s0168-9525(01)02241-7. [DOI] [PubMed] [Google Scholar]
- 6.Tamames J, Casari G, Ouzounis C, Valencia A. Conserved clusters of functionally related genes in two bacterial genomes. J. Mol. Evol. 1997;44:66–73. doi: 10.1007/pl00006122. [DOI] [PubMed] [Google Scholar]
- 7.Galperin MY, Koonin EV. Who's your neighbor? New computational approaches for functional genomics. Nat. Biotechnol. 2000;18:609–613. doi: 10.1038/76443. [DOI] [PubMed] [Google Scholar]
- 8.Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 9.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 10.Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 12.Gaasterland T, Ragan MA. Microbial genescapes: phyletic and functional patterns of ORF distribution among prokaryotes. Microb. Comp. Genomics. 1998;3:199–217. doi: 10.1089/omi.1.1998.3.199. [DOI] [PubMed] [Google Scholar]
- 13.Rogozin IB, Makarova KS, Murvai J, Czabarka E, Wolf YI, Tatusov RL, Szekely LA, Koonin EV. Connected gene neighborhoods in prokaryotic genomes. Nucleic Acids Res. 2002;30:2212–2223. doi: 10.1093/nar/30.10.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snel B, Bork P, Huynen MA. The identification of functional modules from the genomic association of genes. Proc. Natl Acad. Sci. USA. 2002;99:5890–5895. doi: 10.1073/pnas.092632599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janga SC, Collado-Vides J, Moreno-Hagelsieb G. Nebulon: a system for the inference of functional relationships of gene products from the rearrangement of predicted operons. Nucleic Acids Res. 2005;33:2521–2530. doi: 10.1093/nar/gki545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowers PM, Pellegrini M, Thompson MJ, Fierro J, Yeates TO, Eisenberg D. Prolinks: a database of protein functional linkages derived from coevolution. Genome Biol. 2004;5:R35. doi: 10.1186/gb-2004-5-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuenne CT, Ghai R, Chakraborty T, Hain T. GECO—linear visualization for comparative genomics. Bioinformatics. 2007;23:125–126. doi: 10.1093/bioinformatics/btl556. [DOI] [PubMed] [Google Scholar]
- 19.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciria R, Abreu-Goodger C, Morett E, Merino E. GeConT: gene context analysis. Bioinformatics. 2004;20:2307–2308. doi: 10.1093/bioinformatics/bth216. [DOI] [PubMed] [Google Scholar]
- 21.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium TU. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno-Hagelsieb G, Collado-Vides J. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics. 2002;18(Suppl. 1):S329–S336. doi: 10.1093/bioinformatics/18.suppl_1.s329. [DOI] [PubMed] [Google Scholar]
- 28.Janga SC, Moreno-Hagelsieb G. Conservation of adjacency as evidence of paralogous operons. Nucleic Acids Res. 2004;32:5392–5397. doi: 10.1093/nar/gkh882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreno-Hagelsieb G. Operons across prokaryotes: genomic analyses and predictions 300+ genomes later. Curr. Genomics. 2006;7:163–170. [Google Scholar]
- 30.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 31.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuchs RT, Grundy FJ, Henkin TM. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 35.Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 36.Abreu-Goodger C, Ontiveros-Palacios N, Ciria R, Merino E. Conserved regulatory motifs in bacteria: riboswitches and beyond. Trends Genet. 2004;20:475–479. doi: 10.1016/j.tig.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, et al. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.