Abstract
BIOSMILE web search (BWS), a web-based NCBI-PubMed search application, which can analyze articles for selected biomedical verbs and give users relational information, such as subject, object, location, manner, time, etc. After receiving keyword query input, BWS retrieves matching PubMed abstracts and lists them along with snippets by order of relevancy to protein–protein interaction. Users can then select articles for further analysis, and BWS will find and mark up biomedical relations in the text. The analysis results can be viewed in the abstract text or in table form. To date, BWS has been field tested by over 30 biologists and questionnaires have shown that subjects are highly satisfied with its capabilities and usability. BWS is accessible free of charge at http://bioservices.cse.yzu.edu.tw/BWS.
INTRODUCTION
Biomedical papers provide a wealth of information on genes and proteins and their interactions under different experimental conditions. Today's biologists are able to search through a massive volume of online articles in their research. For example, using NCBI PubMed search, a user can retrieve articles from a database of over 4600 biomedical journals from 1966 to the present, updated daily. However, users of basic search engines, such as PubMed search, may need to further scan or read retrieved articles in more detail to pick out specific information of interest. Needless to say, search services that can identify and mark up key relations, biomedical verbs, entities and terms can save biologists much time. Several advanced search services for biomedical journal articles have already been developed. iHOP (http://www.ihop-net.org) (1), for example, retrieves sentences containing specified genes and labels biomedical entities in them, graphing cooccurrence among all entities. MEDIE (http://www-tsujii.is.s.u-tokyo.ac.jp/medie/) is another advanced search tool that can identify subject–verb–object (syntactic) relations and biomedical entities in sentences.
Our proposed system, BIOSMILE web search (BWS), has similar features to the above systems. It can label biomedical entities in sentences and summarize recognized relations. Before analyzing relations in an article, our system firstly identifies named entities (NE), e.g. DNA, RNA, cell, protein and disease names. However, identifying NEs in natural language is a challenging task, especially with biomedical articles due to the absence of a standard nomenclature and ever evolving range of biomedical terms. To tackle this problem, BWS integrates our previous named entity recognition (NER) system, NERBio (2,3), which was developed for the BioCreAtIvE II Gene Mention (GM) tagging task (4) and BioNLP/NLPBA Bio-Entity Recognition shared task (5). Furthermore, for researchers interested in protein–protein interaction (PPI), BWS classifies articles as PPI-relevant or -irrelevant using a system (3,6) we developed for the BioCreAtIvE II Protein Interaction Article Sub-task (IAS) (7).
In addition to identifying NEs, BWS provides semantic analysis of relations, which is somewhat broader than the relation analysis performed by iHOP and MEDIE. BWS can identify a range of semantic relations between biomedical verbs and sentence components, including agent [deliberately performs the action (e.g. Bill drank his soup quietly)], patient [experiences the action (e.g. The falling rocks crushed the car)], manner, timing, condition, location and extent. These relations can be important for precise definition and clarification of complex biomedical relations. For example, the sentence ‘KaiC enhanced KaiA–KaiB interaction in vitro and in yeast cells’, describes an enhancement relation. BWS can identify the elements in this relation, such as the action ‘enhanced’, the enhancer ‘KaiC’, the enhanced ‘KaiA–KaiB interaction’, and the location ‘in vitro and in yeast cells’. A state-of-art semantic relation analysis technique, semantic role labeling (SRL) (8) [also called shallow semantic parsing (9)], is applied to extract these complex relations. BWS integrates our biomedical semantic role labeler, BIOSMILE (10), to annotate SRL.
To integrate the above features into a sophisticated yet easy-to-use interface, we have turned to the Rich Internet Application (RIA) (11) model for web development. RIA combines the interface functionality of desktop software with the broad reach and low-cost deployment of web applications. The BWS interface is programmed in one popular RIA framework, Flex and the application is run on Microsoft IIS servers.
USAGE
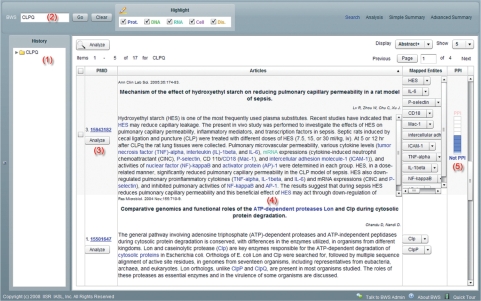
When a user navigates to the BWS site, he or she is presented with a simple search input field at the top of the page. BWS accepts either PubMed identifier (PMID) or keyword input (Figure 1, No. 2), so BWS search queries are compatible with PubMed search. Upon entering a query, users will receive output in the ‘Search’ pane sorted by PMID, including the title, authors and abstract. By clicking on the display pull-down menu at the top of the frame, users can choose to display entire abstract, a brief truncated version or title and author only. The number of displayed results can also be selected in like manner. Recognized NEs, including DNA, RNA, cell, protein and disease, appear in different colored text in the search results (Figure 1, No. 4) and hyperlink to Entrez Gene pages containing more detailed information on them. Entities are mapped to the EntrezGene database. Successfully mapped entities are listed in the ‘Mapped Entities’ column. After clicking on the right down-arrow of each entity, a popup button labeled with ‘EntrezGene’ and its species appears. Clicking on this button now takes users to the entity's corresponding EntrezGene page. A graduated bar meter on the right-hand side of the abstract (Figure 1, No. 5) in the ‘Protein-Protein Interaction’ column indicates PPI relevance. The displayed abstracts can be sorted by PPI relevance by clicking on the column header.
Figure 1.
BWS search interface. The left frame (No. 1) shows the search history. In the search box (No. 2), a user can enter either a PMID or keywords. The rest of the work space shows a table that retrieved abstracts. Each abstract is hyperlinked to the original PubMed abstract (No. 3). For each snippet, BWS annotates gene or protein names in light blue color (No. 4). A graduated bar meter is also presented (No. 5) in the “Protein-Protein Interaction” column to indicate the abstract's relevance to PPI. (BWS search results).
Once search results appear in the table, users can perform relation analysis for a single abstract by clicking the ‘Analyze’ button (Figure 1, No. 3), which appears below the abstract's PubMed ID in the PMID column. For multiple abstracts, they can check off abstracts of interest and then click the ‘Analyze’ button at the top of the search results pane.
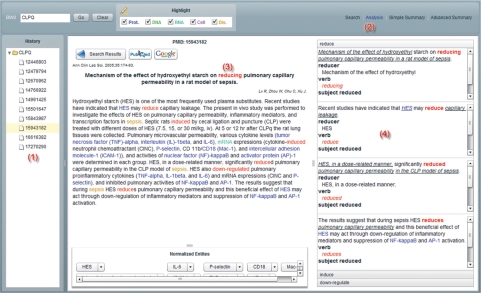
As shown in Figure 2, the results of relation analysis are displayed in the ‘Analysis’ tab pane. Action verbs representing biomedical relations are marked red (Figure 2, No. 3). Clicking on the one of the verbs in the right-hand pane will open a list all the elements of the relation, including agent, patient, location, manner, time, etc. (Figure 2, No. 4). To move between abstracts, users can click on the PMID numbers that appear in the left-hand pane under ‘History’ (Figure 2, No. 1)
Figure 2.
Relation Analysis. The analysis history is displayed in the left pane (No. 1). Analysis results are shown in the Analysis tab pane (No. 2), with biomedical verbs (No. 3) marked in red. The semantic roles related to a verb are listed on the right-hand side in its corresponding accordion pane (No. 4). When users click on verb (No. 3), its pane will automatically expand (No. 4).
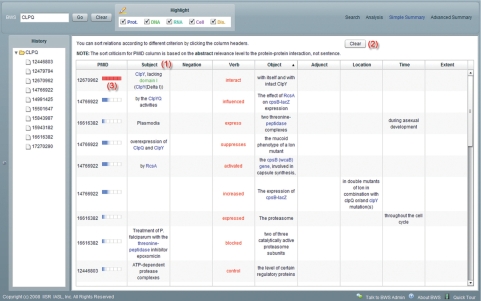
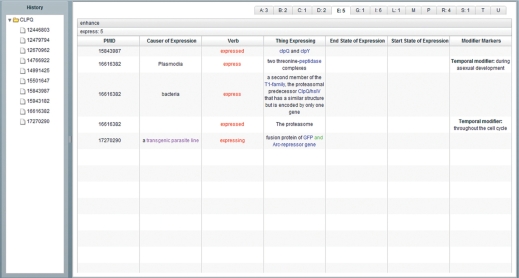
In addition to displaying relations article by article, we provide an analysis summary table that contains all relations in abstracts that appear in the search history. Figure 3 shows the simple version which lists eight major elements in a relation, including subject, negation, verb, adjunct, object, location, time and extent. Here, we use subjects and objects instead of agents and patients. This is because in most cases, agents are subjects and patients are objects of sentences; most users are more familiar with the former two (for users interested in the complete definitions of all predicates, we have provided all SRL framesets in the Supplementary Material). This table provides users a brief summary of relations. Users can sort relations according to different criterion by clicking the column headers (Figure 3, No 1). They can clear all the records by pressing the ‘Clear’ button (Figure 3, No 2). For each extracted relation, the PubMed ID corresponding to the source abstract of this relation is shown in the PMID column. In addition, a graduated bar meter that indicates PPI relevance appears behind the PubMed ID (Figure 3, No 3). The summary table also provides an advanced display (Figure 4), which lists all elements in a relation. The description of each element corresponding to the verb is also displayed. Relations are classified by their main verbs, making it easy to browse through all the relations in an article verb by verb.
Figure 3.
Analysis summary table (simple).
Figure 4.
Analysis summary table (advanced).
METHODS
In the following section, we describe the RIA model, which we used to develop BWS's interface. RIA provides an interactive and user friendly environment while seamlessly incorporating all of BWS's functions. We also introduce the Adobe Flex technology, which is the RIA solution we adopted to implement BWS. Then, we describe BWS's two main modules in details.
BWS as a RIA
RIA is a new model for web development that transfers the processing necessary for the user interface to the web client but keeps the bulk of the data back on the application server. RIAs provide many benefits over traditional web applications: (i) RIAs offer a richer interface that provides a more engaging user experience without the need for page reloads; (ii) RIAs offer real-time feedback and validation to the user, triggered by user events; (iii) the look and feel of a traditional desktop application can be replicated in an RIA; (iv) RIAs can also provide a full multimedia experience, including audio and video and (v) RIAs have capabilities such as real-time chat and collaboration that are either very difficult or simply impossible with traditional web applications.
However, creating a successful RIA based web application is difficult, as many different technologies must interact to make it work, and browser compatibility requires a lot of effort. In order to make the process easier, several open source frameworks have been developed, as well as commercial ones. We have adopted the Adobe Flex framework (http://www.adobe.com/products/flex/) to streamline development. Adobe Flex is an open source framework released by Adobe Systems for building expressive web applications that could deploy consistently on all major browsers. It provides a fast client runtime based on the ubiquitous Adobe Flash Player software.
Module 1: searching and tagging
As shown in Figure 5, this module is depicted in the top block. It comprises three sub-modules, including a search engine, a biomedical NE recognizer and a biomedical PPI abstract classifier.
Figure 5.
The architecture of BIOSMILE Web Search.
The search engine accepts users’ queries and retrieves matching PubMed abstracts. Each query is wrapped as a remote web service call and sent to the NCBI Entrez Utilities Web Service (12). The input of this sub-module can be a PMID or a list of keywords and is compatible with original PubMed Search.
The biomedical NE recognizer, which is based on NERBio (2,3), is employed to label DNA, RNA, cell, protein and disease names in all retrieved abstracts. In NERBio, NER is formulated as a word-by-word sequence labeling task, where the assigned tags delimit the boundaries of any detected NEs. The underlying machine learning model used by NERBio is conditional random fields (13) with a set of features selected by a sequential forward search algorithm.
The PPI abstract classifier assigns each retrieved abstract a score that indicates its relevance to PPI. This score ranges from −1 (least relevant) to +1 (most relevant). This scoring problem is formulated as a document classification task, in which each document is represented by bag-of-words (BoW) features. However, in PPI abstract classification, some words have different levels of information in different contexts. For example, ‘bind’ is more informative when it appears in a sentence that has at least two protein names. Accordingly, we divide the general word bag into several contextual bags. The words in each sentence are bagged according to the number of NEs in the sentence. If there are 0 NEs, the words are put into contextual bag 0; if 1 NE, then bag 1 and if 2 or more NEs, then bag 2. We employ support vector machines (SVM) (14) as the machine learning model to build our PPI abstract classifier (3,6).
Module 2: relation analysis
As shown in Figure 5, this module is depicted in the bottom block. It comprises two sub-modules, including a biomedical semantic relation analyzer and a biomedical full parser (BFP).
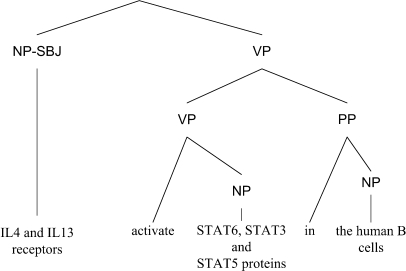
The biomedical semantic relation analyzer extracts relations among phrases and biomedical verbs from sentences. It was developed based on SRL technology. In SRL, sentences are represented by one or more predicate–argument structures (PAS), also known as propositions (15). Each PAS is composed of a predicate (e.g. a verb) and several arguments (e.g. noun phrases and adverbial phrases) that have different semantic roles, including main arguments such as agent or patient, as well as adjunct arguments, such as time, manner or location. Here, the term argument refers to a syntactic constituent of the sentence related to the predicate; and the term semantic role refers to the semantic relationship between a predicate and an argument of a sentence. For example, in Figure 6, the sentence ‘IL4 and IL13 receptors activate STAT6, STAT3, and STAT5 proteins in the human B cells’ describes a molecular activation process. It can be represented by a PAS in which ‘activate’ is the predicate, ‘IL4 and IL13 receptors’ the agent, ‘STAT6, STAT3, and STAT5 proteins’ the patient, and ‘in the human B cells’ the location. Thus, the agent, patient and location are the arguments of the predicate.
Figure 6.
A parsing tree.
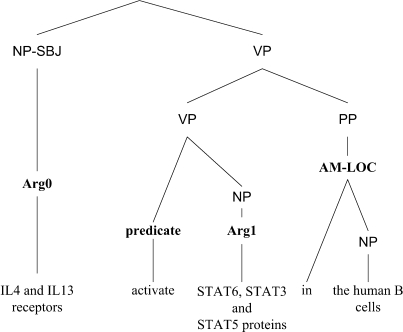
A collection of PASs forms a proposition bank, which is essential in building a machine learning based SRL system. The construction of a proposition bank requires sentences with full parses, and fortunately, the GENIA corpus (16) provides 500 fully parsed biomedical abstracts (Figure 6). In 2006, we constructed the first ever biomedical proposition bank, BioProp (17), by annotating semantic role information on GENIA's full parse trees, as shown in Figure 7. At present, BioProp includes the following 40 predicates: abolish, accompany, activate, alter, associate, bind, block, catalyze, control, decrease, depend, derive, downregulate, enhance, express, generate, increase, induce, influence, inhibit, interact, lead, link, mediate, modify, modulate, phosphorylate, produce, promote, recognize, reduce, regulate, repress, signal, stimulate, suppress, target, trigger, truncate and upregulate. Using BioProp as the training corpus, we then constructed a biomedical SRL system, BIOSMILE (10), which uses the maximum entropy model (18) as its underlying machine learning model. For optimal performance, SRL systems like BIOSMILE require fully parsed sentences. Therefore, to parse PUBMED abstracts retrieved online by BWS, we constructed a BFP, which is based on the Charniak parser (19) with the GENIA corpus (16) as its training data. Its performance is reported in the Results section.
Figure 7.
A parsing tree annotated with semantic roles.
RESULTS
Prediction performance
In this section, we report the prediction performance for our NE recognizer, PPI abstract classifier, BFP and biomedical SRL system. The performance is evaluated in terms of three metrics: precision (P), recall (R) and F-measure (F), which are defined as follows:
 |
We used the datasets provided by the BioCreAtIvE II GM tagging task (4) and IAS task (7) to evaluate our NER system (NERBio) and PPI article classifier, respectively. Details on these two independent training and test datasets can be found at http://biocreative.sourceforge.net. The precision, recall and F-measure of NERBio are 82.59, 89.12 and 85.76%, respectively. Our PPI abstract classifier achieved an F-measure of 80.85% (with a precision of 91.2% and a recall of 78.4%).
To evaluate our BFP, we used the GENIA Treebank (GTB) Beta version for the experimental datasets. The 300 abstracts released on 11 July 2005 are used as the training set, while the 200 abstracts released on 22 September 2004 are used as the test set. Table 1 shows the details of these two datasets. The precision, recall and F-measure of BFP are 85.00, 86.93 and 83.16%, respectively.
Table 1.
Training/Test set of the GENIA Treebank
| Source | Set | Sentences |
|---|---|---|
| GTB 300 | Training | 2768 |
| GTB 200 | Test | 1732 |
Our previous work (10) reported that the precision, recall and F-measure of biomedical SRL (the core technology of biomedical semantic relation analysis) are 87.56, 82.15 and 84.76%, respectively under the condition that gold standard parses are given. To evaluate the actual performance of biomedical SRL on online retrieved sentences, our in-lab biologists annotated a gold-standard dataset that is composed of 100 randomly selected PubMed abstracts with 315 PASs. Table 2 shows the evaluation results for different semantic roles.
Table 2.
The performance of our biomedical SRL system in 100 randomly selected articles
| Semantic Role | P (%) | R (%) | F (%) |
|---|---|---|---|
| Arg0 | 91.86 | 82.93 | 87.18 |
| Arg1 | 91.90 | 76.95 | 83.76 |
| Arg2 | 76.00 | 64.04 | 69.51 |
| ArgM-ADV | 76.00 | 57.58 | 69.51 |
| ArgM-DIS | 100.00 | 95.83 | 97.87 |
| ArgM-LOC | 94.29 | 58.93 | 72.53 |
| ArgM-MNR | 95.65 | 75.00 | 84.08 |
| ArgM-MOD | 94.12 | 94.12 | 94.12 |
| ArgM-NEG | 100.00 | 84.62 | 91.67 |
| ArgM-TMP | 42.86 | 21.43 | 28.57 |
| Overall | 90.06 | 74.85 | 81.75 |
As you can see in Table 2, our biomedical SRL system achieved satisfactory F-measures (87.18 and 83.76%) for Arg0 and Arg1 (in most cases, Arg0 is the subject and Arg1 is the object of a sentence). It shows that we can identify subject–verb–object relations with high accuracy. As to the overall performance, our biomedical SRL system achieved an F-measure of 81.75%, with a precision of 90.06% and a recall of 74.85%, which are slightly lower than the performance achieved under the conditions in which gold-standard parses are given.
Comparison with existing tools
To illustrate the unique functions provided by BWS, we compared it with existing web-based biomedical text analysis tools in Table 3.
Table 3.
Comparison of BWS with existing tools
| Software | Summary | Named entities | PPI abstract classification | Analyzed relation types |
|---|---|---|---|---|
| BWS | (1) Provides an online PubMed search and annotates NEs and PPI relevance for retrieved abstracts. (2) Extracts semantic relations between biomedical verbs and surrounding phrases. |
Protein, DNA, RNA, cell, disease | Yes | Multiple semantic relations, including predicate–agent, predicate–patient, predicate–location, predicate–negation, predicate–extent, predicate–manner, predicate–time, etc. |
| iHOPa | (1) Filters and ranks the retrieved sentence that match the given gene or protein names according to significance, impact factor, date of publication and syntax. (2) Graphs cooccurrence among all entities to allows researchers to explore a network of gene and protein interactions. |
Genes, MeSH-terms, chemical compounds | No | Cooccurrence only |
| MEDIEb | (1) Provides a search interface with subject, verb and object input fields and retrieves S–V–O syntactic relations. (2) Summarizes syntactic relations in table form. |
Genes, diseases | No | Syntactic relations (subject–verb–object) |
User-centered evaluation
In order to evaluate the usability of BWS, we used a system success evaluation framework designed by Dr Hsin-Chun Chen (20,21) to examine users’ self-reported assessments of BWS in terms of user satisfaction, intention to use, system usability, perceived usefulness and perceived ease of use, all of which are critical in system evaluations. Each aspect is described as follows:
User satisfaction: user satisfaction is essential to system evaluation (22). In this work, user information satisfaction refers to the degree that a system can satisfy a user's information needs in the research process.
Intention to use: intention to use is a measure of the likelihood a person will employ BWS (23).
System usability: system usability has been shown to affect user adoption, system usage and satisfaction (24). Several usability instruments have been developed and validated (25). We used the user interaction satisfaction (QUIS) scale (25), which is capable of evaluating a system in five fundamental usability dimensions: overall reaction to the system, screen layout and sequence of system, terminology and system information, learning to use the system and system capabilities.
Perceived usefulness: perceived usefulness is the degree to which a person believes that using a particular system would enhance his or her job performance (26). System usefulness is critical to voluntary use of a new system (26,27) and generally refers to the extent to which an individual considers a system useful in his or her work.
Perceived ease of use: perceived ease of use refers to the degree to which a user believes that using a particular system would be free of effort (26,27). Ease of use represents an essential motivation for individuals’ voluntary use of a system (28), and can affect adoption decisions significantly (27).
User-centered evaluation design
We used previously validated question items (24) to measure user information satisfaction and intention to use using a seven-point Likert scale with 1 being ‘strongly disagree’ and 7 being ‘strongly agree’. We evaluated system usability using the QUIS (25) with nine-point Likert scale. The higher scores represent more agreeable usability (such as easy, wonderful and clear) than lower scores (such as difficult, terrible and confusing), with 9 being most favorable and 1 being the most unfavorable. We also included question items from previous research (26) to measure system usefulness and ease of use, using a seven-point Likert scale with 1 indicating ‘strongly disagree’ and 7 indicating ‘strongly agree’.
Results of user-centered evaluation
A total of 30 subjects participated in this study, including researchers from Taiwan, France and the US. Some of the labs involved in testing include CDC Taiwan, NTU hospital, NTU Microbiology, NTU Department of Agronomy, Hôpital Henri Mondor in France and the Computational Systems Biology Lab in the US. There were 18 male subjects and 12 female subjects, all biomedical researchers (master students, graduates, PhD students and PhDs), who use NCBI-PubMed an average of 1.68 h per week and read on average 1.87 papers per week.
The analysis of our subjects’ self-reported assessments suggested high user satisfaction (mean = 5.39 and SD = 1.01) and high intension (mean = 5.67 and SD = 1.02) to use BWS with 7 indicating ‘strongly agree’. In perceived usefulness and ease of use, our subjects’ responses showed high usefulness (mean = 5.57 and SD = 1.01) and most agreed that BWS is easy to use (mean = 5.62 and SD = 0.68) with 7 indicating ‘strongly agree’. Our subjects also considered BWS reasonably usable, as measured by multiple items listed above (average mean = 6.98 and average SD = 1.24), with 9 indicating ‘strongly agree’ for agreeable usability. Table 4 summarizes all evaluation results.
Table 4.
Summary of descriptive statistics and construct reliability analysis
| Construct | Mean | SD |
|---|---|---|
| User information satisfaction (10 items) | 5.39 | 1.01 |
| Intension to use (3 items) | 5.67 | 1.02 |
| Perceived usefulness (6 items) | 5.57 | 1.01 |
| Perceived ease of use (6 items) | 5.62 | 0.68 |
| System usability | 6.98 | 1.24 |
| Overall reaction towards the system (6 items) | 6.97 | 1.17 |
| Screen layout and sequence of system (4 items) | 6.71 | 1.34 |
| Terminology and system information (6 items) | 6.98 | 1.33 |
| Learning to use the system (6 items) | 7.42 | 1.07 |
| Capabilities of the system (5 items) | 6.81 | 1.32 |
In general, our subjects considered BWS reasonably useful and usable; they also revealed high intention to use our system in the future.
Our subjects considered BWS's processing speed not particularly slow (mean = 5.93 and SD = 1.53). However, we believe speed could be increased by building a local PubMed database to decrease remote retrieval time (currently 50% of processing time) and by increasing the number of servers to prevent system overload from many simultaneous users. We report detailed statistics for processing time in Tables 5 and 6.
Table 5.
PubMed access time comparison
| No. of articles | Average query time | |
|---|---|---|
| Before mirror (s) | After mirror (s) | |
| 5 | 5.05 | 3.24 |
| 10 | 5.83 | 3.48 |
| 20 | 6.73 | 4.06 |
Table 6.
The processing time of NER, PPI abstract classifier and SRL (disable cache)
| NER time | PPI abstract classification time | SRL time |
|---|---|---|
| 0.58 s per sentence | 2.47 s per abstract | 1.99 s per sentence |
Specific usage questionnaire
We designed a follow-up questionnaire with more specific questions related to BWS's unique functions sent to the original 30 subjects. These subjects expressed that BWS's ability to label semantic roles and highlight named entities helped them grasp the main concepts of an abstract (mean = 5.45 and SD = 0.82, with 7 indicating ‘strongly agree’). In addition, they indicated that the summary table helped them efficiently browse biomedical relations across different abstracts (mean = 5.36 and SD = 1.02, with 7 indicating ‘strongly agree’). Some researchers also expressed interest in employing BWS's SRL capabilities to create new applications for specific research purposes (mean = 6.27 and SD = 0.90, with 7 indicating ‘strongly agree’). For example, to find genes in only a specific location.
CONCLUSION
In this article, we have described the development of BWS and its unique features, which include (i) semantic relation analysis of abstracts for selected biomedical verbs and extraction of a wide variety of relational information between sentence components such as agent, patient, negation, extent, location, manner and time; and (ii) PPI relevance ranking for abstracts.
BWS has been tested by many biologists in several countries to measure user satisfaction, usefulness, practicability and ease of use, as well as other aspects of system performance. Overall, our subjects were highly satisfied with BWS's capabilities and found it reasonably usable.
In the near future, BWS will allow users to specify the semantic role of each query term (agent, predicate, patient, etc.) to facilitate searching for specific biomedical relations. The system will also retrieve related sentences instead of entire abstracts to improve readability. In addition, the system will allow users to construct biomedical relation networks from single or multiple retrieved abstracts. Such networks will be presented in a navigable interface to allow visual browsing of complex relations such as biomedical pathways. Since BWS has been programmed as a parallel processing service, we hope to soon install more servers to lower response time. Finally, the BWS interface will be made available in multiple Asian languages, and BWS will integrate a translation service able to translate extracted information such as named entities and relations into Chinese, Japanese and Korean.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This research was supported in part by the National Science Council under grant NSC96-2752-E-001-001-PAE and NSC97-2218-E-155-001 as well as the thematic program of Academia Sinica under grant AS95ASIA02. We especially thank Dr Gary Benson and the NAR reviewers for their valuable comments, which helped us improve the quality of the article.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fernández JM, Hoffmann R, Valencia A. iHOP web services. Nucleic Acids Res. 2007;35:W21–W26. doi: 10.1093/nar/gkm298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai RT-H, Sung C-L, Dai H-J, Hung H-C, Sung T-Y, Hsu W-L. NERBio: using selected word conjunctions, term normalization, and global patterns to improve biomedical named entity recognition. BMC Bioinform. 2006;7(Suppl 5):S11. doi: 10.1186/1471-2105-7-S5-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai H-J, Hung H-C, Tsai RT-H, Hsu W-L. BioCreAtIvE II Workshop. 2007. Sep, IASL Systems in the Gene Mention Tagging Task and Protein Interaction Article Sub-task; pp. 69–79. Madrid, 23–25. [Google Scholar]
- 4.Wilbur J, Smith L, Tanabe LK. BioCreAtIvE II Workshop. 2007. Sep, BioCreative 2 Gene Mention Task; pp. 7–16. Madrid, 23–25. [Google Scholar]
- 5.Jin-Dong K, Tomoko O, Tsuruoka Y, Collier N. Introduction to the bio-entity recognition task at JNLPBA. Proceedings of the International Workshop on Natural Language Processing in Biomedicine and its Applications (JNLPBA-04) 2004:70–75. [Google Scholar]
- 6.Tsai RT-H, Hung H-C, Dai H-J, Lin Y-W, Hsu W-L. Exploiting likely-positive and unlabeled data to improve the identification of protein-protein interaction articles. BMC Bioinform. 2008;9:S3. doi: 10.1186/1471-2105-9-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krallinger M, Valencia A. BioCreAtIvE II Workshop, Madrid. 2007. Sep, Evaluating the Detection and Ranking of Protein Interaction Relevant Articles: the BioCreative Challenge Interaction Article Sub-task (IAS) pp. 29–39. 23–25. [Google Scholar]
- 8.Carreras X, Màrques L. CoNLL-2004. May, Introduction to the CoNLL-2004 Shared Task: Semantic Role Labeling; pp. 89–97. Boston, 6–7. [Google Scholar]
- 9.Pradhan S, Ward W, Hacioglu K, Martin JH, Jurafsky D. Boston, MA, USA: North American chapter of the Association for Computational Linguistics; 2004. Shallow semantic parsing using support vector machines. In; pp. 233–240. Proceedings of the Human Language Technology Conference of the North American chapter of the Association for Computational Linguistics annual meeting, Boston, MA, 2004. [Google Scholar]
- 10.Tsai RT-H, Chou W-C, Su Y-S, Lin Y-C, Sung C-L, Dai H-J, Yeh IT, Ku W, Sung T-Y, Hsu W-L. BIOSMILE: a semantic role labeling system for biomedical verbs using a maximum-entropy model with automatically generated template features. BMC Bioinform. 2007;8:325. doi: 10.1186/1471-2105-8-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loosley C. Keynote Systems. 2006. Technical report. [Google Scholar]
- 12.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;35:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lafferty J, McCallum A, Pereira F. In International Conference on Machine Learning, Williamstown, MA, 2001. Williamstown, MA, USA: International Conference on Machine Learning; 2001. Conditional random fields: probabilistic models for segmenting and labeling sequence data; pp. 282–289. [Google Scholar]
- 14.Joachims T. Proceedings of ECML-98, 10th European Conference on Machine Learning. Springer, Heidelberg, DE: 1998. [Google Scholar]
- 15.Hoernig R, Rauh R, Strube G. CHAPTER 6 Events-II: 1 Modelling event recognition. In: Strube G, Wender KF, editors. The Cognitive Psychology of Knowledge. Amsterdam: Elsevier Science Publishers; 1993. pp. 113–138. [Google Scholar]
- 16.Tateisi Y, Ohta T, Tsujii J.-I. Annotation of predicate-argument structure on molecular biology text. Nature. 1997;386:296–299. [Google Scholar]
- 17.Chou W-C, Tsai RT-H, Su Y-S, Ku W, Sung T-Y, Hsu W-L. ACL Workshop on Frontiers in Linguistically Annotated Corpora. Sydney: 2006. A Semi-Automatic Method for Annotating a Biomedical Proposition Bank; pp. 5–12. 22 July 2006. [Google Scholar]
- 18.Berger AL, Della Pietra VJ, Della Pietra SA. A maximum entropy approach to natural language processing. Comput. Linguist. 1996;22:39–71. [Google Scholar]
- 19.Charniak E. In Proceedings of the first conference on North American chapter of the Association for Computational Linguistics, Seattle, WA, 2000. San Francisco, CA, USA: Morgan Kaufmann Publishers Inc; 2000. A maximum-entropy-inspired parser; pp. 132–139. [Google Scholar]
- 20.Hu PJH, Zeng D, Chen H, Larson C, Chang W, Tseng C, Ma J. System for infectious disease information sharing and analysis: design and evaluation. IEEE Trans. Inform. Technol. Biomed. 2007;11:483–492. doi: 10.1109/TITB.2007.893286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu PJ, Zeng D, Chen H, Larson CA, Tseng C. A web-based system for infectious disease data integration and sharing: evaluating outcome, task performance efficiency, user information satisfaction, and usability. Lect. Notes Comput. Sci. 2007;4506:134. [Google Scholar]
- 22.Ives B, Olson MH, Baroudi JJ. The measurement of user information satisfaction. Commun. ACM. 1983;26:785–793. [Google Scholar]
- 23.Fishbein M, Ajzen I. Belief, Attitude, Intention and Behavior: An Introduction to Theory and Research. Reading, MA: Addison-Wesley; 1975. [Google Scholar]
- 24.Petersen MG, Madsen K, Kjar A. The usability of everyday technology—emerging and fading opportunities. ACM Trans. Comput. Human Int. 2002;9:74–105. [Google Scholar]
- 25.Chin JP, Diehl VA, Norman LK. Development of an Instrument Measuring User Satisfaction of the Human-computer Interface. New York, NY: ACM Press; 1988. [Google Scholar]
- 26.Davis FD. Perceived usefulness, perceived ease of use, and user acceptance of information technology. MIS Quarterly. 1989;13:319–340. [Google Scholar]
- 27.Lee Y, Kozar KA, Larsen KRT. The technology acceptance model: past, present, and future. Commun. Assoc. Inform. Syst. (Volume 12, Article 50) 2003;752:780. [Google Scholar]
- 28.Venkatesh V. Determinants of perceived ease of use: integrating control, intrinsic motivation, and emotion into the technology acceptance model. Inform. Syst. Res. 2000;11:342–365. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.