Abstract
SCANPS performs iterative profile searching similar to PSI-BLAST but with full dynamic programing on each cycle and on-the-fly estimation of significance. This combination gives good sensitivity and selectivity that outperforms PSI-BLAST in domain-searching benchmarks. Although computationally expensive, SCANPS exploits onchip parallelism (MMX and SSE2 instructions on Intel chips) as well as MPI parallelism to give acceptable turnround times even for large databases. A web server developed to run SCANPS searches is now available at http://www.compbio.dundee.ac.uk/www-scanps. The server interface allows a range of different protein sequence databases to be searched including the SCOP database of protein domains. The server provides the user with regularly updated versions of the main protein sequence databases and is backed up by significant computing resources which ensure that searches are performed rapidly. For SCOP searches, the results may be viewed in a new tree-based representation that reflects the structure of the SCOP hierarchy; this aids the user in placing each hit in the context of its SCOP classification and understanding its relationship to other domains in SCOP.
INTRODUCTION
SCANPS is a program for comparing a protein sequence to a sequence database. It performs iterative profile searching similar to PSI-BLAST (1), but with full dynamic programing on each cycle and on-the-fly estimation of significance. The SCANPS web server has been developed to simplify the running and analysis of SCANPS searches. An innovative aspect of the server is its novel tree-based presentation of results for searches against the SCOP domain database (2). A comparison of a protein domain to the domains in SCOP can be of considerable value in elucidating its structure and function. Facilities for comparing a query sequence to SCOP sequences are also provided by FPS (http://fps.sdsc.edu/), GTOP (http://spock.genes.nig.ac.jp/~genome/grpsblt.html) and CascadeBlast (http://crick.mbu.iisc.ernet.in/~CASCADE/CascadeBlast.html), all of which use PSI-BLAST as the search algorithm. However, interpreting the results of SCOP searches in the context of the classification is hampered by the fact that search methods typically produce a linear table of hits; understanding the relationships between hits to the SCOP database usually requires a manual mapping of the results table onto the SCOP hierarchy. This procedure is tedious and error prone and therefore a browser interface has been developed that maps hits onto the SCOP hierarchy for viewing using a tree-based framework. The new interface allows the user to perform a search of the complete non-redundant SCOP sequence database and view results in a form that allows the information inherent in the SCOP classification to be exploited in interpreting those results.
MATERIALS AND METHODS
Overview of SCANPS
Although SCANPS has been available for over 15 years and has been accessible as a service at the European Bioinformatics Institute (EBI) for 10 years, it has not previously been described in the literature. Accordingly, as background to the new web server, a brief overview of the motivation, novel features and performance of the program is given here.
The basic function of SCANPS is to perform a full Smith–Waterman algorithm (3) comparison of a protein sequence to a protein sequence database with either length dependent or affine gap penalties (4). The program is written in C and effort was put into coding for performance on conventional workstation hardware. This enabled the program to be used routinely for searching large databases on modest computer hardware in contrast to the belief in the early 1990s that Smith–Waterman was too CPU intensive for this task (5). Parallel processing by dynamically splitting the database across multiple processors was demonstrated on a network of five loosely coupled workstations (6), then refined to exploit Symmetric Multi Processing (SMP) hardware via OpenMP (7). The SMP implementation gave near linear speedup on a 24 processor Silicon Graphics Challenge. With the move to commodity PC hardware in the late 1990s, fourway onchip parallelism was implemented on Intel and AMD chips with a speedup over linear code of at least 3× depending on the query and database size. In addition to the onchip parallelism, multiprocessor parallelism was implemented through MPI (8,9). The MPI implementation gave parallel efficiency of over 90% on 16 processors on an Intel PIII cluster connected by 100 MB network.
The speed obtained by parallel processing, coupled with the on-the-fly statistics described below, permitted the implementation of iterative searching. In this mode, a multiple sequence alignment is constructed for sequences that score above a preset significance threshold in the initial search. The multiple alignment is built up by aligning to the query sequence as a template, but using a Position Specific Scoring Matrix (PSSM) as appropriate at each iteration. Alignment columns which contain gaps in the query sequence are deleted; as a result, the alignment is always the same length as the query sequence. A PSSM (10,11) is derived from this alignment and used to re-search the database. Since sequences that are very similar to the query sequence contribute little information to the PSSM, a percentage identity threshold (PIDT) is employed which excludes all sequences from the alignment whose similarity to the query sequence exceeds the threshold. The contributions of each sequence to the PSSM are weighted according to the method of Henikoff and Henikoff (12). The scoring matrix is then constructed at each alignment position similarly to standard log odds matrices (12,13). The process of constructing a PSSM and searching the database is repeated until convergence, or until a preset number of iterations has been completed.
In each database search, the statistical significance of a score between the query and any sequence in the database is assessed by on-the-fly modeling the distribution of query database sequence pair scores. Scores are binned according to the log of the product of the query and database sequence lengths (LPL). Within each LPL bin, an extreme value distribution is fitted to the scores. The extreme-value location and scale parameters are then fitted to exponential and linear equations respectively with respect to the LPL. The resulting extreme-value equations are applied back to all query sequence pairs and the resulting probabilities converted to E-values for ranking and display. This method of estimating significance is similar to those implemented in FASTA/SSEARCH (14); for a full discussion of the similarities and differences between the different fitting schemes and the effect on performance in benchmarks see (15).
Benchmarking of SCANPS
The search performance of SCANPS was compared with that of PSI-BLAST using a benchmark based on SCOP (2). The benchmark dataset comprises 1113 sequences taken from the PDB40D-B dataset constructed by Brenner et al. (16). Single-segment domains whose structure had been determined by X-ray crystallography were selected, representing 479 SCOP superfamilies across 343 SCOP folds. True positives were defined as pairs of sequences belonging to the same SCOP superfamily; true negatives were defined as those pairs in which the sequences belong to different SCOP folds. The resulting set of sequence pairs contain 2528 true positives and 616 923 true negatives, resulting in a total of 618 821 pairs.
Benchmarking was performed by searching the benchmark set with each of the benchmark sequences in turn. Since SCANPS and PSI-BLAST use sequence profiles to enhance search sensitivity, it is necessary to embed the benchmark set in a larger sequence database in order to ensure that there is an adequate set of related sequences to construct the search profile for each benchmark sequence. Accordingly, the benchmark set was embedded in SWALL (17) to create the search database.
Both SCANPS version 2.3.9 and PSI-BLAST version 2.2-17 were run for a maximum of 10 iterations. Search parameters were chosen to reflect the typical use case where a low rate of false positives is acceptable in return for a high rate of true positives found. SCANPS was run with a profile E-value inclusion value of 0.015, a PIDT of 97% and using the BLOSUM50 scoring matrix. Gap penalties for opening and extending gaps were set to 12 and 2, respectively. These parameters have previously been established to provide the optimal combination of good sequence coverage and low error rate (15). To allow direct comparison with PSI-BLAST defaults, a further run of SCANPS was performed with BLOSUM62. PSI-BLAST scans were performed with the default BLOSUM62 scoring matrix and profile E-value cutoff of 0.002. The gap opening and extension penalties were set to 11 and 1, respectively. All other parameters were set to their default values. The E-values for the pairs in the benchmark were collected and ranked in order from lowest to highest and used to calculate coverage versus error-per-query plots.
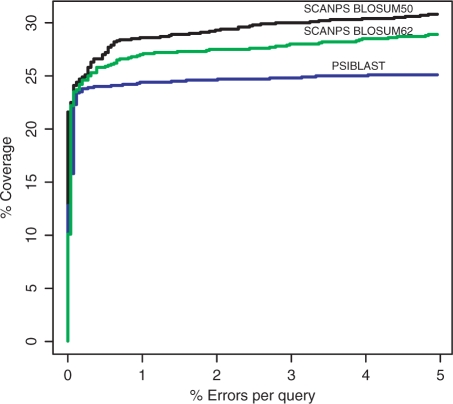
Figure 1 shows plots of percentage true positive pairs versus percentage errors-per-query (EPQ) for SCANPS and PSI-BLAST. SCANPS offers significantly increased coverage versus PSI-BLAST for a given rate of EPQ. For example, at 1% EPQ, SCANPS using the BLOSUM 62 scoring matrix finds 27% of true positives, while PSI-BLAST finds 24.4%. SCANPS when run with the optimal BLOSUM 50 scoring matrix finds 28.6% of true pairs at 1% EPQ.
Figure 1.
Coverage versus error plots for benchmarking of SCANPS and PSI-BLAST. The vertical axis (coverage) represents the number of true positives found divided by the total number of true positives in the benchmark, expressed as a percentage. True positives are those pairs in which the domains belong to the same SCOP superfamily. The horizontal axis (EPQ) represents the number of false positives found divided by the total number of true positives, expressed as a percentage. False positives are those pairs in which the domains belong to different SCOP folds. The black line show results for SCANPS run with the BLOSUM50 scoring matrix; profile inclusion E-value = 0.015, gap opening penalty = 12, gap extension penalty = 2 and profile identity threshold = 97%. The green line show results for SCANPS run with the BLOSUM62 scoring matrix; profile inclusion E-value = 0.015, gap opening penalty = 12, gap extension penalty = 2 and profile identity threshold = 97%. The blue line show results for PSI-BLAST run with the BLOSUM62 matrix; profile inclusion value = 0.002, gap opening penalty = 11 and gap extension penalty = 1. All other parameters for both methods were set to their default values. All runs were for a maximum of 10 iterations and pairs were collected from the final iteration of each search.
The SCANPS web interface
The output from SCANPS consists of tables of hits for each iteration, together with associated pairwise alignments and multiple alignments for all of the hits in a given iteration. This output is usually voluminous, so a major advantage of transforming the raw output into a browsable format is that it becomes much easier to navigate through the results. The web interface also permits crossreferencing to sequence databases and integration of the Jalview viewer (18) for viewing alignments and further analysis. The web interface allows the possibility of more sophisticated representations for SCOP search results as described below.
The server input page (Figure 2) allows the user either to upload a sequence file or paste a sequence directly into a text box. The search database is selected from a pulldown menu, with the current options of UniRef100, UniRef90, UniRef50 (19), PDB (20) and SCOP (2). The UniRef and PDB databases are updated automatically on biweekly and weekly schedules, respectively. The SCOP database is updated manually when a new version of SCOP is released. The SCOP sequences used are generated by ASTRAL (http://astral.berkeley.edu) (21) from the SEQRES records of the corresponding PDB entries and are non-redundant at the level of 100% sequence identity. The user can set all of the search parameters or use the defaults, which, with the exception of iteration number, are those found to be optimum in benchmarking on SCOP superfamily data. Each parameter is documented by a link to the corresponding section of the help documentation. The search is run non-interactively and an Email message provides the user with the URL where the results can be viewed. The search time required depends on the inputs, parameters and the load on the server. Searches of SCOP and PDB are typically returned in within 5 min; UniRef searches may require several hours. The server does not at present use the MPI implementation of SCANPS but it is planned to do so to take full advantage of the large computing cluster that is available to the server.
Figure 2.
The SCANPS server input page. Search sequences can be pasted into the text box or uploaded from a file. Clicking on the name of each parameter displays the appropriate section of the documentation. The parameters displayed are the defaults used for searching.
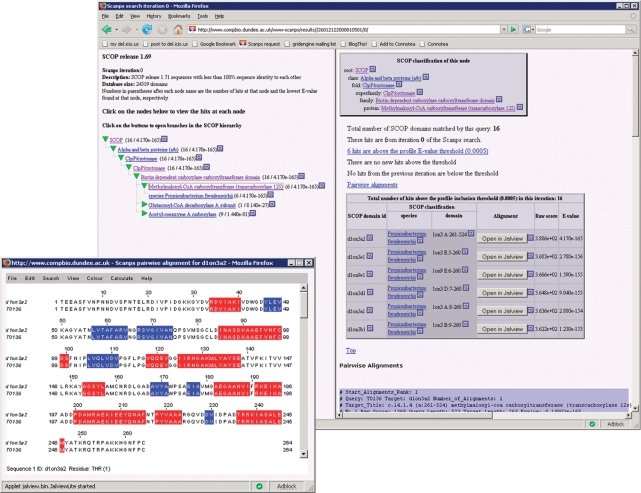
The default output format is a linear presentation of the hits and corresponding pairwise and multiple alignments for each iteration. Hits for each iteration are listed in a table which displays the hit rank, the identifier assigned to the hit in the source database, a descriptive string, the E-value, the raw score and a button to display the pairwise alignment of the query and the hit in Jalview (18). Sequence identifiers are hyperlinked to the corresponding sequence entries on the database website. This is followed by a multiple alignment of the query with all of the hits found in the iteration and then the pairwise alignments. For each pairwise and multiple alignment, a button is provided to view the alignment in the Jalview alignment tool. Jalview provides access to a range of functions for editing and further analysis. When searching against the PDB and SCOP, Jalview uses secondary structure assignments from DSSP (22) to display the secondary structure elements in the database sequences. If a SCOP search has been performed, the results can also be viewed in a hierarchical viewer that maps the results onto the SCOP hierarchy (Figure 3). A separate mapping is generated for each search iteration and can be displayed by clicking a button in the linear interface. The ‘tree frame’ displays a tree-like representation of the SCOP hierarchy, the branches of which may be expanded or collapsed to display particular branches at more fine-grained levels of classification. The nodes in the tree frame are annotated with the number of SCANPS hits at that node and the lowest E-value found for a hit at that node, allowing the viewer to quickly identify those parts of the SCOP hierarchy in which hits are clustered. Clicking on a SCOP node in the tree frame displays the corresponding node page in the node frame. The ‘node frame’ displays, for a given SCOP node, a page (the ‘node page’), which contains a table of the domains in the node and summary information about the node. For each domain, the table lists the domain's SCOP classification and the data returned by the search method.
Figure 3.
Result of a SCANPS search of the SCOP database displayed in the tree-based SCOP results interface, with a pairwise alignment displayed in Jalview in the foreground. The tree frame on the left displays the SCOP hierarchy; the node frame on the right on displays the page for a particular node. The node's SCOP classification is listed at the top of the page. This is followed by summary data from SCANPS and then the domain table. For each domain, the table lists its SCOP classification below the current node, followed by the data for the domain returned by SCANPS. Pairwise alignments for each hit are listed below the domain table; these can also be displayed in Jalview by clicking the appropriate button. The alignment in Jalview is shaded to indicate the positions of strands (blue) and helices (red) in the database structure.
IMPLEMENTATION
The server is implemented as a set of Perl CGI scripts but most of the functionality is contained in a set of common Perl modules used by all the scripts. The SCOP interface is built using an object-oriented Perl library that is designed to facilitate building interfaces for any program that searches SCOP.
CONCLUSION
The SCANPS web server (http://www.compbio.dundee.ac.uk/www-scanps) provides access to SCANPS in the form of a user-friendly web interface with regularly updated databases and postprocessing of results to present them in a form that facilitates analysis and interpretation. The server provides the facility to search the complete SCOP database with a query sequence and display the results in a tree view. This maps hits directly onto the SCOP hierarchy with links to the SCOP database website. It also integrates the Jalview alignment editor for viewing alignments between the hits and the query sequence. Planned enhancements to the server include deploying the MPI implementation of SCANPS to reduce search times further, as well as allowing search of SCOP embedded in UniRef to improve sensitivity for iterative searches.
ACKNOWLEDGEMENTS
Since 1989, SCANPS development has been funded from a variety of sources including the Royal Society (G.J.B.), European Molecular Biology Laboratory (C.W., S.S. and G.J.B.) and the Scottish Funding Council (Scottish Bioinformatics Research Network grant) (T.P.W. and G.J.B.). We thank all our colleagues and SCANPS users for their help and support throughout the development of SCANPS and the new web server interface described here. Funding to pay the Open Access publication charges for this article was provided by the Scottish Funding Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 3.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 4.Barton GJ. Protein sequence alignment techniques. Acta Crystallogr. D. 1998;54:1139–1146. doi: 10.1107/s0907444998008324. [DOI] [PubMed] [Google Scholar]
- 5.Barton GJ. Computer speed and sequence comparison. Science. 1992;257:1609. doi: 10.1126/science.1482492. [DOI] [PubMed] [Google Scholar]
- 6.Barton GJ. Scanning protein sequence databanks using a distributed processing workstation network. Comput. Appl. Biosci. 1991;7:85–88. doi: 10.1093/bioinformatics/7.1.85. [DOI] [PubMed] [Google Scholar]
- 7.Dagum L, Menon R. OpenMP: an industry-standard API for shared memory programming. IEEE Comput. Sci. Eng. 1998;5:46–55. [Google Scholar]
- 8.MPI Forum MPI: A message-passing interface standard. Int. J. Supercomput. Appl. 1994;8:165–416. [Google Scholar]
- 9.Barton GJ, Webber C, Searle SMJ. New developments to SCANPS: high performance parallel iterated protein sequence searching with full dynamic programming and on-the-fly statistics. Presented at the 8th International Conference on Intelligent Systems for Molecular Biology. 2000:19–23. August 2000, La Jolla, California. Available online at: http://www.iscb.org/ismb2000/9_8_pdfs/TuesSA/Barton.pdf. [Google Scholar]
- 10.Barton GJ, Sternberg MJE. Flexible protein sequence patterns, a sensitive method to detect weak structural similarities. J. Mol. Biol. 1990;212:389–402. doi: 10.1016/0022-2836(90)90133-7. [DOI] [PubMed] [Google Scholar]
- 11.Gribskov M, McLachlan AD, Eisenberg D. Profile scanning for three-dimensional structural patterns in protein sequences. Proc. Natl Acad. Sci. USA. 1987;84:4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dayhoff MO, Schwartz RM, Orcutt BC. A model of evolutionary change in proteins. In: Dayhoff MO, editor. Atlas of Protein Sequence and Structure. Washington, DC: National Biomedical Research Foundation; 1978. pp. 345–352. [Google Scholar]
- 14.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webber C. Ph.D. Thesis. Cambridge, UK: EMBL-European Bioinformatics Institute, University of Cambridge; 2003. Protein sequence database searching. [Google Scholar]
- 16.Brenner S, Chothia C, Hubbard T. Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc. Natl Acad. Sci. USA. 1998;95:6073–7068. doi: 10.1073/pnas.95.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 19.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt Reference Clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 20.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner SE, Koehl P, Levitt M. The ASTRAL compendium for sequence and structure analysis. Nucleic Acids Res. 2000;28:254–256. doi: 10.1093/nar/28.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]