Abstract
IMGT/V-QUEST is the highly customized and integrated system for the standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) rearranged nucleotide sequences. IMGT/V-QUEST identifies the variable (V), diversity (D) and joining (J) genes and alleles by alignment with the germline IG and TR gene and allele sequences of the IMGT reference directory. New functionalities were added through a complete rewrite in Java. IMGT/V-QUEST analyses batches of sequences (up to 50) in a single run. IMGT/V-QUEST describes the V-REGION mutations and identifies the hot spot positions in the closest germline V gene. IMGT/V-QUEST can detect insertions and deletions in the submitted sequences by reference to the IMGT unique numbering. IMGT/V-QUEST integrates IMGT/JunctionAnalysis for a detailed analysis of the V-J and V-D-J junctions, and IMGT/Automat for a full V-J- and V-D-J-REGION annotation. IMGT/V-QUEST displays, in ‘Detailed view’, the results and alignments for each submitted sequence individually and, in ‘Synthesis view’, the alignments of the sequences that, in a given run, express the same V gene and allele. The ‘Advanced parameters’ allow to modify default parameters used by IMGT/V-QUEST and IMGT/JunctionAnalysis according to the users’ interest. IMGT/V-QUEST is freely available for academic research at http://imgt.cines.fr
INTRODUCTION
IMGT®, the international ImMunoGeneTics information system® (http://imgt.cines.fr) (1), is the international reference in immunogenetics and immunoinformatics. Created in 1989 at the Laboratoire d’ImmunoGénétique Moléculaire (LIGM) (Université Montpellier 2 and CNRS), IMGT® provides a high quality integrated knowledge resource, specialized in the immunoglobulins (IG) or antibodies, T cell receptors (TR), major histocompatibility complex (MHC) of human and other vertebrates and related proteins of the immune system (RPI), which belong to the immunoglobulin superfamily (IgSF) and to the MHC superfamily (MhcSF). IMGT® includes databases, web resources and interactive tools. The accuracy and the consistency of the IMGT® data are based on IMGT-ONTOLOGY, the first ontology for immunogenetics and immunoinformatics (2,3). IMGT/V-QUEST, for ‘V-QUEry and STandardization’, is the first integrated IMGT® tool which has been online since 1997 (4). IMGT/V-QUEST analyses the IG and TR rearranged nucleotide sequences that result from the very complex mechanisms at the origin of antigen receptor diversity (1012 antibodies and 1012 TR per individual) and which include the rearrangements of the variable (V), diversity (D) and joining (J) genes, the N-diversity mechanism and, for IG, the somatic mutations [for review see (5,6)]. IMGT/V-QUEST identifies the V, D and J genes and alleles in rearranged V-J and V-D-J sequences by alignment with the germline IG and TR gene and allele sequences of the IMGT reference directory. It delimits the framework regions (FR-IMGT) and complementarity determining regions (CDR-IMGT) and provides a detailed and accurate characterization of the submitted sequences according to the IMGT Scientific chart rules, based on the IMGT-ONTOLOGY axioms and concepts of description, classification and numerotation (2,3). New functionalities were added to IMGT/V-QUEST through a complete rewrite in Java. Thus, IMGT/V-QUEST analyses batches of sequences (up to 50) in a single run. The analysis has been upgraded with the description of V-REGION mutations, with the identification of the hot spots positions in the closest germline V gene, and with the detection and accurate description of insertions and deletions in the submitted sequences by reference to the IMGT unique numbering (7). IMGT/V-QUEST integrates IMGT/JunctionAnalysis (8) for a detailed analysis of the V-J and V-D-J junctions, and IMGT/Automat (9) for a full annotation of the V-J- and V-D-J-REGION. The interface has been customized to fit the users’ needs: in addition to the standard ‘Detailed view’ which displays the results and alignments for each submitted sequence individually, a new results display ‘Synthesis view’ has been implemented to provide, for a given run, the alignments of the sequences that express the same V gene and allele and, per locus, the results of IMGT/JunctionAnalysis. The ‘Advanced parameters’ allow to modify default parameters used by IMGT/V-QUEST and IMGT/JunctionAnalysis algorithms, according to the users’ interest. IMGT/V-QUEST is currently available for human and mouse rearranged sequences, and partly for 31 other species (nonhuman primates, rat, sheep, teleostei and chondrichthyes). IMGT/V-QUEST is freely available for academic research from the IMGT® Home page (http://imgt.cines.fr).
ALGORITHM AND IMPLEMENTATION
IMGT/V-QUEST was totally rewritten in Java language in order to fully unify the implementation of the different components. The identification of the closest V, D and J genes and alleles of a given receptor type (IG or TR) and of a given species is based on the same principles as previously described (4). IMGT/V-QUEST algorithm was developed using pairwise alignment and sequence comparison of experimental data, expertly annotated and standardized by IMGT® (10). Briefly, the identification of the closest V, D and J genes and alleles is based on global pairwise alignment (two for the V), without insertions nor deletions, of the user sequence with different subsets of the IMGT reference directory, followed by a similarity evaluation. This last step is preceded, for the V region, by the insertion of gaps according to the IMGT unique numbering (7).
The complete rewrite in Java has optimized the processing time and allowed the development and integration of new functionalities which improve the analysis accuracy with score, evaluation, warnings, etc. IMGT/V-QUEST can analyze batches of sequences (up to 50) in a single run. A new option allows to search for potential insertions and deletions in the user sequence. This search follows the identification of the closest V gene and allele. The program proceeds in two alignment steps [Smith and Waterman algorithm (11)] between the user sequence and the closest V genes and alleles, first looking for insertions, then for deletions. If insertions are detected, they are excluded from the user sequence as they disrupt the IMGT numbering but their positions are memorized so they can be displayed in the results page. If deletions are detected, gaps are entered in the user sequence to restore the IMGT numbering. After insertion and/or deletion detection, the steps of V gene and allele identification are performed again. IMGT/V-QUEST provides an accurate localization and description of the mutations in the user sequence by comparison with the closest germline V gene and allele. Nucleotide (nt) mutations are characterized as silent versus nonsilent, and transition versus transversion. Amino acid (AA) changes are qualified according to the IMGT AA classes (12) based on hydropathy, volume and physicochemical characteristics. The tool identifies the mutation hot spots patterns and positions in the closest germline V-REGION. IMGT/V-QUEST integrates IMGT/JunctionAnalysis (8) for the detailed analysis of the junction (performed even if cystein 104 and/or phenylalanine/tryptophan 118 are mutated) (7) and for the optional display of the eligible D genes and alleles, and IMGT/Automat (9) for the full annotation of the V-J-REGION or V-D-J-REGION in an IMGT/LIGM-DB-like format (13).
Regarding the input sequences, IMGT/V-QUEST accepts nucleotide sequences in the International Union of Biochemistry (IUB) code (14) and is able to deal with complementary reverse sequences. The tool trims the ‘n’ nucleotides at the 5′ and 3′ extremities of user sequences since they influence the alignments and the gene and allele identification.
IMGT/V-QUEST SEARCH
An IMGT/V-QUEST search consists of two easy steps: (i) the user selects the antigen receptor (IG or TR) and the species on the IMGT/V-QUEST Home page and (ii) on the next page, the user submits up to 50 nt sequences in FASTA format. By clicking on ‘Start’, the analysis is done automatically with the default parameters.
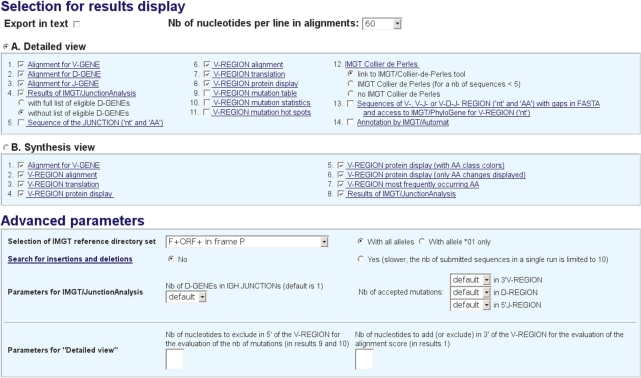
Prior to launching the search, users may customize the results display options in ‘Selection for results display’ (Figure 1). They can export the results in text and choose the ‘Nb of nucleotides per line in alignments’. They can select the option ‘A. Detailed view’ for the display of the results of each analyzed sequence individually (with a choice of 14 different results displays), or the option ‘B. Synthesis view’ for the display of the alignments of sequences that express the same V gene and allele (with a choice of eight different results displays).
Figure 1.
The customizable parameters for an IMGT/V-QUEST search. ‘Selection for results display’ allows the users to select the types of results to be displayed (14 for ‘Detailed view’, and 8 for ‘Synthesis view’). The ‘Advanced parameters’ allow the users to modify the default values.
For sophisticated queries or for unusual sequences, the users can modify the default values in ‘Advanced parameters’. The customizable values are:
‘Selection of IMGT reference directory set’ used for the V, D and J genes and alleles identification and alignments (‘F + ORF’, ‘F + ORF + in-frame P’, ‘F + ORF including orphons’ and ‘F + ORF + in-frame P including orphons’, where F is functional, ORF is open reading frame and P is pseudogene). This allows the user to work with only relevant gene sequences (e.g. orphon sequences are relevant for genomic but not for expressed repertoire studies). The selected set can also be chosen either ‘With all alleles’ or ‘With allele *01 only’.
‘Search for insertions and deletions’. In that case, the number of submitted sequences in a single run is limited to 10.
‘Parameters for IMGT/JunctionAnalysis’: Nb of D-GENEs allowed in the IGH, TRB and TRD junctions and Nb of accepted mutations in 3′V-REGION, D-REGION and 5′J-REGION (default values are indicated per locus in the IMGT/V-QUEST Documentation).
‘Parameters for Detailed View’: ‘Nb of nucleotides to exclude in 5′ of the V-REGION for the evaluation of the nb of mutations’ (to avoid, for example, to count primer specific nucleotides) and/or ‘Nb of nucleotides to add (or exclude) in 3′ of the V-REGION for the evaluation of the alignment score’ (e.g. in case of low or high exonuclease activity).
IMGT/V-QUEST OUTPUT
Detailed view
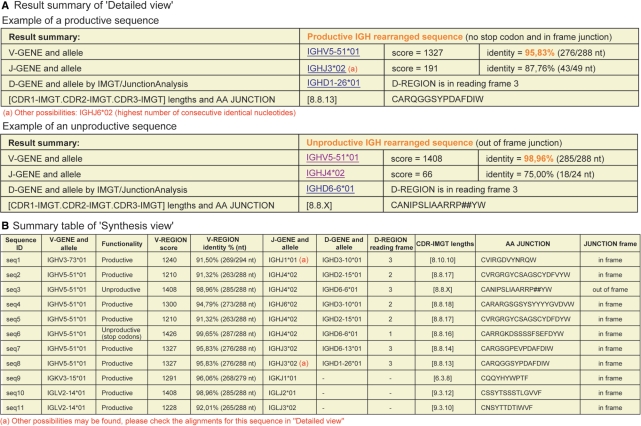
The top of the ‘Detailed view’ results page indicates the number of analyzed sequences with links to individual results. Each individual result comprises the user sequence displayed in FASTA format (a sequence submitted in antisense orientation is shown as complementary reverse sequence that is in V gene sense orientation), a ‘Result summary’ table (Figure 2A) followed, if all parameters were selected, by the 14 different results displays. The ‘Result summary’ provides a crucial feature that is the evaluation of the user sequence functionality performed by IMGT/V-QUEST: productive (if no stop codon and in-frame junction) or unproductive (if stop codons and/or out-of-frame junction). It also summarizes the main characteristics of the analyzed sequence which include the names of the closest ‘V-GENE and allele’ and ‘J-GENE and allele’ with the alignment score and the percentage of identity, the name of the closest ‘D-GENE and allele’ with the D-REGION reading frame, the three CDR-IMGT lengths (shown within brackets, e.g. [8.8.13]) which characterize a V domain and the AA JUNCTION sequence. IMGT/V-QUEST provides warnings that appear, as notes in red to alert the users, if potential insertions or deletions are suspected in the V (sequences with <85% of identity and/or with different CDR1-IMGT and/or CDR2-IMGT lengths compared with the closest germline V-REGION), or if other possibilities for the J gene and allele are identified. If the option ‘Search for insertions and deletions’ was selected, the detection and detailed description of insertions and/or deletions are shown in the ‘Result summary’ first row to capture the user attention. Moreover, insertions appear as capital letters in the FASTA sequence.
Figure 2.
IMGT/V-QUEST output. (A) ‘Result summary’ of ‘Detailed view’. The note (a) represents a warning indicating that there is another possibility for the choice of the J-GENE, the criterion for that choice being between parentheses. In the case of an unproductive sequence due to an out-of-frame junction, the CDR3-IMGT length cannot be determined and is indicated by an ‘X’. The symbol ‘#’ indicates a frameshift. (B) ‘Summary table’ of ‘Synthesis view’. The note (a) represents a warning indicating that there are other possibilities for the results.
Below the ‘Result summary’, the alignments for the V-, D- and J-GENE (4,15,16) comprise the alignment score and the identity percentage with the five closest genes and alleles and, for the V, the length of the V-REGION taken into account for the score evaluation. The ‘Results of IMGT/JunctionAnalysis’ (8,15) include, if selected, the list of eligible D genes and alleles which match >4 nt with the junction, allowing users to visualize the result among other close solutions. ‘Sequence of the JUNCTION (‘nt’ and ‘AA’)’ is given by IMGT/V-QUEST if no results are obtained with IMGT/JunctionAnalysis. Different displays of the V region (‘V-REGION alignment’, ‘V-REGION translation’ and ‘V-REGION protein display’) and of the mutations affecting the V region are all based on the IMGT unique numbering (7) and on the FR-IMGT and CDR-IMGT delimitation. The extensive and standardized characterization of the mutations performed by IMGT/V-QUEST answers the requirements of IG sequence studies, evolution and function analysis, clinical interpretations and antibody engineering. The ‘V-REGION mutation table’ lists the mutations (nt and AA) of the analyzed sequence compared to the closest V-REGION allele. They are described for the V-REGION and for each FR-IMGT and CDR-IMGT, with their positions, and for the AA changes according to the IMGT AA classes (12). For example, c16 > g, Q6 > E (++−) means that the nt mutation (c > g) leads to an AA change at codon 6 with the same hydropathy (+) and volume (+) but with different physicochemical properties (−) classes (12). It is the first time that such qualification of amino acid replacement is provided. The ‘V-REGION mutation statistics’ evaluates the number of silent and nonsilent mutations and the number of transitions and transversions of the analyzed nucleotide sequence, and the number of AA changes of its translated sequence. The ‘V-REGION mutation hot spots’ show the patterns and localization of hot spots in the closest germline V-REGION. The identified hot spots patterns are (a/t)a and (a/g)g(c/t)(a/t) and the complementary reverse motifs are t(a/t) and (a/t)(a/g)c(c/t) (see Lefranc,M-P. and Lefranc,G. Somatic hypermutations, in IMGT Education, http://imgt.cines.fr).
‘IMGT Collier de Perles’ provides a link to the IMGT/Collier-de-Perles tool (17) or directly includes this standardized graphical representation of the user IG or TR rearranged sequence. The ‘Sequences of V-, V-J- or V-D-J-REGION (‘nt’ and ‘AA’) with gaps in FASTA and access to IMGT/PhyloGene for V-REGION (‘nt’)’ provides the analyzed sequence with IMGT gaps, in FASTA format and on one line, and a link to IMGT/PhyloGene (18). The ‘Annotation by IMGT/Automat’ (9) uses the results of the analysis to provide a full automatic annotation of the user sequences for the V-J-REGION or V-D-J-REGION.
Synthesis view
The aim of ‘Synthesis view’, a novel IMGT/V-QUEST result, is to facilitate the comparison of sequences that express the same V gene and allele: it allows to compare the localization of the mutations and the composition of their junctions. The ‘Synthesis view’ comprises a ‘Summary table’ (Figure 2B) and eight different displays (if all were selected). The ‘Summary table’ shows, for each sequence, the name of the closest V gene and allele, the evaluation of the sequence functionality, the V score and percentage of identity, the name of the closest J and D genes and alleles, the D-REGION reading frame, the three CDR-IMGT lengths, the AA JUNCTION and the JUNCTION frame. Warnings appear to alert users on potential insertions or deletions in the V or on other possibilities for the J gene and allele. In such cases, it is strongly recommended to check the individual results of these sequences in ‘Detailed view’.
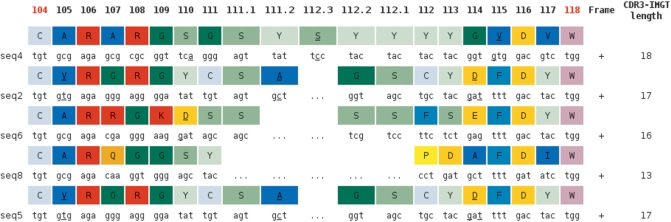
The originality of ‘Synthesis view’ is also to provide alignments of sequences which, in a given run, are assigned to the same V gene and allele. The ‘Alignment for V-GENE’, ‘V-REGION alignment’ and ‘V-REGION translation’ are based on the same characteristics as those of ‘Detailed view’. In addition, the hot spots positions are underlined in the germline V-REGION (for an easy comparison with the mutation localizations) and the name of the closest J gene allele is indicated at the 3′ end of each sequence. The ‘V-REGION protein display’ shows amino acid sequences aligned with the closest V-REGION allele. This protein display is also provided with AA colors according to the IMGT AA classes (12) or with only the AA changes displayed. The ‘V-REGION most frequently occurring AA per position and per FR-IMGT and CDR-IMGT’ table is given for each alignment to highlight the position of conserved AA in sequence batches. The ‘Results of IMGT/JunctionAnalysis’ are displayed per locus (IGH, IGK and IGL for the IG heavy, kappa and lambda sequences, TRA, TRB, TRG and TRD for the TR alpha, beta, gamma and delta sequences) (Figure 3).
Figure 3.
IMGT/JunctionAnalysis translation of ‘Synthesis view’. Results are given per locus, here five IGH sequences are shown. Amino acids of the JUNCTIONs are colored according to the 11 IMGT AA physicochemical classes (12). Underlined nt and AA are mutated compared with the closest germline 3′V-REGION, D-REGION and 5′J-REGION of each JUNCTION.
CONCLUSION
IMGT/V-QUEST is a highly customized and integrated system for the IG and TR standardized V-J and V-D-J sequence analysis. The addition of new features (AA physicochemical characteristics, mutations and insertions/deletions description, ‘Synthesis view’) and ‘Advanced parameters’ provides a wide spectrum of new types of analysis across species. The integration of tools has allowed to answer specific needs: IMGT/JunctionAnalysis for the accurate and extensive description of the V-J and V-D-J junction, IMGT/Automat for the full and detailed user sequence annotation and IMGT/Collier-de-Perles for bridging the gap between sequences and 3D structures. By its wide range of novel customizable and integrated functionalities and by its high level of standardization, IMGT/V-QUEST is unique among the other software programs (19–22). The information provided by IMGT/V-QUEST is of much value for the comparative analysis of the IG and TR sequences (including those of the newly sequenced vertebrate genomes), for statistical analysis of the junctions (23) and of the repertoires, for antibody engineering and therapeutical antibodies (24) (single chain Fragment variable (scFv), phage displays, combinatorial libraries, chimeric, humanized and human antibodies). IMGT/V-QUEST, with an average of 30 000 requests per month, is widely used by the clinicians to analyze the expressed repertoire in normal and pathological immune responses (autoimmune diseases, leukemias, lymphomas, myelomas, infectious diseases, etc.), in diagnostics of clonalities, detection and follow-up of minimal residual diseases.
Given the necessity to have an international and publicly available benchmarch of high quality for clinical data comparison, IMGT® standards based on IMGT-ONTOLOGY have recently been approved by the World Health Organization–International Union of Immunological Societies (WHO–IUIS) subcommittee for IG and TR (25,26). Standardized criteria for the analysis of IG and TR rearranged V-J and V-D-J sequences and 3D structures include IMGT reference directory (available free for academics on the Web site), IMGT gene nomenclature (5,6,10) approved by the HUGO Nomenclature Committee, IMGT label description, IMGT unique numbering for V-DOMAIN (7) and CDR-IMGT delimitations. IMGT/V-QUEST has been recommended by the European Research Initiative on chronic lymphocytic leukemia CLL (ERIC) for the analysis of the IGHV gene mutational status in CLL (27). IMGT/V-QUEST output results evaluation by biologists and feedback from researchers and clinicians are part of the new developments to answer biological and medical needs in an international collaboration (28–30).
AVAILABILITY AND CITATION
Authors who use IMGT/V-QUEST are encouraged to cite this article and to quote the IMGT® Home page (http://imgt.cines.fr). IMGT/V-QUEST is freely available for academic research.
ACKNOWLEDGEMENTS
We are grateful to Vincent Nègre and to the IMGT team for its expertise and constant motivation. IMGT® received funding from Centre National de la Recherche Scientifique CNRS, Ministère de l’Enseignement Supérieur et de la Recherche MESR (University Montpellier 2), Région Languedoc-Roussillon, Agence Nationale de la Recherche ANR (ANR-06-BYOS-0005-01) and European Community (ImmunoGrid, FP6-2004-IST-4). Funding to pay the Open Access publication charges for this article was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lefranc M-P, Giudicelli V, Kaas Q, Duprat E, Jabado-Michaloud J, Scaviner D, Ginestoux C, Clément O, Chaume D, Lefranc G. IMGT, the international ImMunoGeneTics information system®. Nucleic Acids Res. 2005;33:D593–D597. doi: 10.1093/nar/gki065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giudicelli V, Lefranc M-P. Ontology for immunogenetics: IMGT-ONTOLOGY. Bioinformatics. 1999;15:1047–1054. doi: 10.1093/bioinformatics/15.12.1047. [DOI] [PubMed] [Google Scholar]
- 3.Duroux P, Kaas Q, Brochet X, Lane J, Ginestoux C, Lefranc M-P, Giudicelli V. IMGT-Kaleidoscope, the formal IMGT-ONTOLOGY paradigm. Biochimie. 2008;90:570–583. doi: 10.1016/j.biochi.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Giudicelli V, Chaume D, Lefranc M-P. IMGT/V-QUEST, an integrated software program for immunoglobulin and T cell receptor V–J and V–D–J rearrangement analysis. Nucleic Acids Res. 2004;32:W435–W440. doi: 10.1093/nar/gkh412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lefranc M-P, Lefranc G. The Immunoglobulin FactsBook, London: Academic Press; 2001. pp. 1–458. [Google Scholar]
- 6.Lefranc M-P, Lefranc G. The T Cell Receptor FactsBook, London: Academic Press; 2001. pp. 1–398. [Google Scholar]
- 7.Lefranc M-P, Pommié C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 8.Yousfi Monod M, Giudicelli V, Chaume D, Lefranc M-P. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics. 2004;20:i379–i385. doi: 10.1093/bioinformatics/bth945. [DOI] [PubMed] [Google Scholar]
- 9.Giudicelli V, Protat C, Lefranc M-P. The IMGT strategy for the automatic annotation of IG and TR cDNA sequences: IMGT/Automat. In: Christophe C, Lenhof H-P, Sagot M-F, editors. Proceedings of the European Conference on Computational Biology (ECCB 2003) Paris, DKB-31: INRIA (DISC/Spid); 2003. pp. 103–104. [Google Scholar]
- 10.Giudicelli V, Chaume D, Lefranc M-P. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33:D256–D261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 12.Pommié C, Levadoux S, Sabatier R, Lefranc M-P. IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J. Mol. Recognit. 2004;17:17–32. doi: 10.1002/jmr.647. [DOI] [PubMed] [Google Scholar]
- 13.Giudicelli V, Duroux P, Ginestoux C, Folch G, Jabado-Michaloud J, Chaume D, Lefranc M-P. IMGT/ LIGM-DB, the IMGT comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res. 2006;34:D781–D784. doi: 10.1093/nar/gkj088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences; recommendations 1984. Nucleic Acids Res. 1985;10:3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefranc M-P. IMGT, the international ImMunoGenetics information system® http://imgt.cines.fr. In: Lo BKC, editor. Antibody engineering methods and protocols. Methods in Molecular Biology. 2nd edn. Vol. 248. Totowa, NJ, USA: Humana Press; 2004. pp. 27–49. [Google Scholar]
- 16.Giudicelli V, Lefranc M-P. Interactive IMGT on-line tools for the analysis of immunoglobulin and T cell receptor repertoires. In: Veskler BA, editor. New Research on Immunology. New York: Nova Science; 2005. pp. 77–105. [Google Scholar]
- 17.Kaas Q, Lefranc M-P. IMGT Colliers de Perles: standardized sequence-structure representations of the IgSF and MhcSF superfamily domains. Curr. Bioinformatics. 2007;2:21–30. [Google Scholar]
- 18.Elemento O, Lefranc M-P. IMGT/PhyloGene: an on-line tool for comparative analysis of immunoglobulin and T cell receptor genes. Dev. Comp. Immunol. 2003;27:763–779. doi: 10.1016/s0145-305x(03)00078-8. [DOI] [PubMed] [Google Scholar]
- 19.Souto-Carneiro MM, Longo NS, Russ DE, Sun HW, Lipsky PE. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J. Immunol. 2004;172:6790–6802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- 20.Ohm-Laursen L, Nielsen M, Larsen SR, Barington T. No evidence for the use of DIR, D-D fusions, chromosome 15 open reading frames or VH replacement in the peripheral repertoire was found on application of an improved algorithm, JointML, to 6329 human immunoglobulin H rearrangements. Immunology. 2006;119:265–277. doi: 10.1111/j.1365-2567.2006.02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volpe JM, Cowell LG, Kepler TB. SoDA: implementation of a 3D alignment algorithm for inference of antigen receptor recombinations. Bioinformatics. 2006;22:438–444. doi: 10.1093/bioinformatics/btk004. [DOI] [PubMed] [Google Scholar]
- 22.Gaëta BA, Malming HR, Jackson KJ, Bain ME, Wilson P, Collins AM. iHMMune-align: hidden Markov model-based alignment and identification of germline genes in rearranged immunoglobulin gene sequences. Bioinformatics. 2007;23:1580–1587. doi: 10.1093/bioinformatics/btm147. [DOI] [PubMed] [Google Scholar]
- 23.Bleakley K, Giudicelli V, Wu Y, Lefranc M-P, Biau G. IMGT standardization for statistical analyses of T cell receptor junctions: the TRAV-TRAJ example. In Silico Biol. 2006;6:573–588. [PubMed] [Google Scholar]
- 24.Magdelaine-Beuzelin C, Kaas Q, Wehbi V, Ohresser M, Jefferis R, Lefranc M-P, Watier H. Structure–function relationships of the variable domains of monoclonal antibodies approved for cancer treatment. Crit. Rev. Oncol. Hematol. 2007;64:210–225. doi: 10.1016/j.critrevonc.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Lefranc M-P. WHO-IUIS Nomenclature Subcommittee for Immunoglobulins and T cell receptors report. Immunogenetics. 2007;59:899–902. doi: 10.1007/s00251-007-0260-4. [DOI] [PubMed] [Google Scholar]
- 26.Lefranc M-P. WHO-IUIS Nomenclature Subcommittee for Immunoglobulins and T cell receptors report August 2007, 13th International Congress of Immunology, Rio de Janeiro, Brazil. Dev. Comp. Immunol. 2008;32:461–463. doi: 10.1016/j.dci.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Ghia P, Stamatopoulos K, Belessi C, Moreno C, Stilgenbauer S, Stevenson F, David F, Rosenquist R. ERIC recommendations on IGHV gene mutational status analysis in chronic lymphocytic leukemia. Leukemia. 2007;21:1–3. doi: 10.1038/sj.leu.2404457. [DOI] [PubMed] [Google Scholar]
- 28.Belessi CJ, Davi FB, Stamatopoulos KE, Degano M, Andreou TM, Moreno C, Merle-Béral H, Crespo M, Laoutaris NP, Montserrat E, et al. IGHV gene insertions and deletions in chronic lymphocytic leukemia: “CLL-biased” deletions in a subset of cases with stereotyped receptors. Eur. J. Immunol. 2006;36:1963–1974. doi: 10.1002/eji.200535751. [DOI] [PubMed] [Google Scholar]
- 29.Murray F, Darzentas N, Hadzidimitriou A, Tobin G, Boudjogra M, Scielzo C, Laoutaris N, Karlsson K, Baran-Marzsak F, Tsaftaris A, et al. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood. 2008;111:1524–1533. doi: 10.1182/blood-2007-07-099564. [DOI] [PubMed] [Google Scholar]
- 30.Davi F, Rosenquist R, Ghia P, Belessi C, Stamatopoulos K. Determination of IGHV gene mutational status in chronic lymphocytic leukemia: bioinformatics advances meet clinical needs. Leukemia. 2008;22:212–214. doi: 10.1038/sj.leu.2404969. [DOI] [PubMed] [Google Scholar]