Abstract
Analysis of protein–ligand complexes and recognition of spatially conserved physico-chemical properties is important for the prediction of binding and function. Here, we present two webservers for multiple alignment and recognition of binding patterns shared by a set of protein structures. The first webserver, MultiBind (http://bioinfo3d.cs.tau.ac.il/MultiBind), performs multiple alignment of protein binding sites. It recognizes the common spatial chemical binding patterns even in the absence of similarity of the sequences or the folds of the compared proteins. The input to the MultiBind server is a set of protein-binding sites defined by interactions with small molecules. The output is a detailed list of the shared physico-chemical binding site properties. The second webserver, MAPPIS (http://bioinfo3d.cs.tau.ac.il/MAPPIS), aims to analyze protein–protein interactions. It performs multiple alignment of protein–protein interfaces (PPIs), which are regions of interaction between two protein molecules. MAPPIS recognizes the spatially conserved physico-chemical interactions, which often involve energetically important hot-spot residues that are crucial for protein–protein associations. The input to the MAPPIS server is a set of protein-protein complexes. The output is a detailed list of the shared interaction properties of the interfaces.
INTRODUCTION
Proteins, which are essential to all biological systems, function by interacting with other molecules. Consequently, multiple alignment of the protein binding regions can help in defining the properties that are essential for the interaction with certain binding partners and in inferring the function. Here, we consider two related problems of multiple alignments: that of protein-binding sites and of protein–protein interfaces, PPIs, which are formed between pairs of interacting binding sites.
Multiple sequence and structural alignment have become a common practice (1). Yet, a dissimilarity in the global properties does not necessarily imply different functions; indeed, it has been shown that convergent evolution of binding sites is not a rare phenomenon (2). Several methods have been developed to identify specific 3D patterns of protein catalytic residues (3–7). However, many binding sites of small molecules such as ATP and estradiol do not share common patterns of amino acids (8–10); rather, they present a set of surface regions with similar physico-chemical properties and shapes. While several approaches have been proposed for recognition and pairwise alignment of such functional sites (10–13), no multiple alignment methods are available. Since pairwise alignments may contain a large number of features that are not essential for the binding, multiple alignment methods are required to determine the smallest set of features, a consensus, that is necessary to achieve a desired biological consequence.
Consideration of pairs of interacting binding sites, which form PPIs, provide additional valuable information of the actual interactions formed between the molecules. Analysis of a set of protein–protein complexes helped in gaining important insights toward deciphering the principles of protein–protein interactions (14–20) and their modular architecture (21). Previous PPI alignment methods, which considered the backbone Cα atoms (22) or the physico-chemical binding patterns (23,24) aligned only pairs of PPIs. However, multiple alignment methods are required for recognition of conservation of the spatial interaction patterns formed between the molecules.
In this article, we present two webservers for multiple spatial alignment of protein-binding sites and PPIs. The difference between the two methods is in the input representation. While the first method, MultiBind (25), looks at the binding site surface of one molecule, the second method, MAPPIS (26), constructs interaction edges between two interacting proteins. MultiBind aligns a set of binding sites and recognizes the common spatial arrangements of their physicochemical properties. On the other hand, MAPPIS performs multiple alignments of PPIs and recognizes the spatially conserved interaction patterns. Both methods consider the physico-chemical properties formed by groups of atoms and are independent of the overall similarity in the protein sequences or folds.
MULTIBIND: MULTIPLE ALIGNMENT OF PROTEIN-BINDING SITES
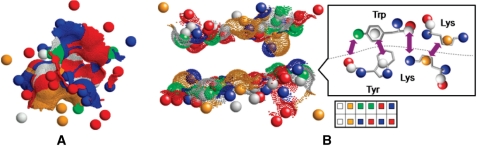
Given a set of binding sites that bind the same small molecule, our goal is to reveal the common physico-chemical pattern that may be responsible for the binding. Figure 1A illustrates the binding site representation, which is crucial for the description of the chemistry of the recognized patterns. Each binding site is determined by the solvent accessible surface points (27) that are located < 4Å from the surface of the binding partner. Following the definition of Schmitt et al. (28), each amino acid in a binding site is represented by points in 3D space termed pseudocenters. Each pseudocenter represents one of the following properties important for protein–ligand interactions: hydrogen-bond donor (DON), hydrogen-bond acceptor (ACC), mixed donor/acceptor (DAC), hydrophobic aliphatic (ALI) and aromatic contacts (PI). We considered all the pseudocenters with at least one surface exposed atom. The pseudocenters and the surfaces are assigned such attributes as charge, normal vectors of the surface direction, ring plane orientation as well as surface patch size and curvature (25,29).
Figure 1.
Physico-chemical representation of binding sites and PPIs. (A) Representation of a binding site by its pseudocenters (balls). Hydrogen bond donors are blue, acceptors are red, donors/acceptors are green, hydrophobic aliphatic are orange and aromatic are white/gray. The surface is represented as dots and are colored according to the property of the corresponding, surface exposed, pseudocenters. (B) An interface as a pair of interacting binding sites. The surfaces and the pseudocenters are colored as in (A). The rightmost figure illustrates the definition of pseudocenters and the bar at the bottom illustrates the complementarity of the pseudocenter properties.
MultiBind (25) is an efficient method, which achieves this goal by local multiple alignment of protein binding sites, which are not assumed to share any sequence or fold similarity. MultiBind utilizes a time efficient Geometric Hashing method (30), which allows recognition of the candidate 3D transformations that align pairs of structures. Then, by applying a branch-and-bound procedure, MultiBind recognizes a combination of multiple 3D transformations that give the highest scoring common 3D pattern. The score of a pattern is the sum of similarity scores of the matched pseudocenters. These are measured by a scoring function that compares properties like spatial proximity (after the superimposition), charge, surface curvature as well as aromatic ring plane orientation.
The input to the MultiBind webserver consists of set of protein—small molecule complexes (defined by the PDB codes or uploaded files). The binding sites of each complex are automatically extracted according to the ligands bound to the input structure. The output of MultiBind is a set of physico-chemical properties shared by all the input binding sites. We provide the details of the properties and the amino acids that contribute to these as well as the 3D transformation that superimposes the binding sites in 3D space. We provide a PDB file with the spatial superimposition of the input complexes, which can be viewed online with a Jmol script that visualizes the shared patterns (Figure 2).
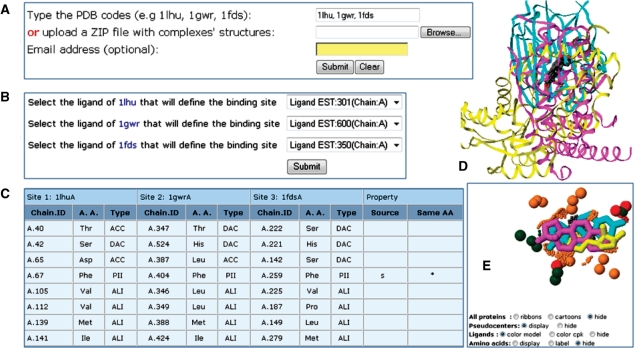
Figure 2.
Web interface and output of MultiBind. (A) Entrance webserver page. (B) Selection of the binding sites of interest by the description of the bound ligands. Only ligands listed as HETATM records with more than seven non-hydrogen atoms are considered. (C) Example of an output page, which details the matched pseudocenters of the common pattern. Each three columns present the details of a specific pseudocenter: (i) chain identifier and residue number; (ii) residue type; (iii) pseudocenter type. Although the pseudocenters are not required to have the same amino acid identity or origin (backbone or side chain), we indicate the conservation of these (b/s or *, respectively). (D) A default Jmol visualization of the superimposed complexes. (E) A Jmol visualization of the common pattern. The shared pseudocenters are represented as balls, colored as in Figure 1. The ligands, represented as sticks, are not considered by MultiBind, but their spatial alignment supports the correctness of the solution. The buttons at the bottom detail the web page options that should be selected to obtain this visualization automatically.
MAPPIS: MULTIPLE ALIGNMENT OF PPIs
Given a set of protein–protein complexes, our goal is to align them in 3D space and recognize the shared spatially conserved interaction patterns. Similarly to multiple sequence and structure alignment, the main motivation is the assumption that an interaction common to a number of interfaces is functionally more significant than a similar interaction found in a single or a pair of PPIs. The uniqueness of MAPPIS lies in its ability to detect spatially conserved patterns of interactions even when there is no sequence or fold similarity between the corresponding proteins. Recently, we have applied MAPPIS to different families of PPIs and observed that most of the conserved physico-chemical interactions are contributed by the hot spot residues, and consequently, MAPPIS predicts hot spots with a high success rate (29).
Figure 1B, illustrates the PPI representation by its physico-chemical properties and interactions. Specifically, an interaction across PPIs is defined by a pair of close enough pseudocenters, one from each side of the interface, possessing complementary physico-chemical properties (hydrogen bond donors are complementary to acceptors, while hydrophobic aliphatic and aromatic centers can interact with similar ones). MAPPIS calculates a set of transformations, which superimpose the PPIs according to their similar interactions that can be of the following three types: hydrogen bonds, hydrophobic aliphatic and aromatic (π) contacts. Two interactions are considered similar if they are created by similar pseudocenters that are superimposed to nearby spatial locations (e.g. within 3Å). The similarity of interactions from two different PPIs is scored according to the similarity of the corresponding pseudocenters and the complementarity of their properties. Specifically, we measured the complementarity in terms of the pseudocenter proximity, charge complementarity, surface fit as well as aromatic ring orientations (favoring perpendicular and parallel π stacking). MAPPIS finds a set of transformations that superimpose the input PPIs in 3D space in a way that maximizes the spatial and chemical similarity of their interactions and pseudocenters.
The input to MAPPIS is a set of protein–protein complexes with at least one pair of interacting protein chains. The interacting chains, which define the PPI, can be either specified by the user or selected from a list of automatically recognized interactions. The chain definition is followed by the automatic construction of the PPIs and their multiple alignment with MAPPIS. The output of MAPPIS is a set of the physico-chemical interactions shared by all the PPIs. We provide a PDB file with the superimposed complexes, which can be viewed online with a Jmol script that visualizes the shared properties and interactions (Figure 3).
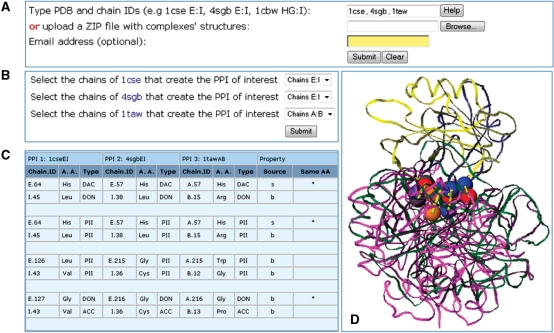
Figure 3.
Web interface and output of MAPPIS. (A) Entrance webserver page. (B) An option for the automatic selection of the interacting protein chains (if not specified manually). Two protein chains are considered as interacting if there are at least five atoms of one chain that are within the distance of 6.0 Å from the other. (C) Example of an output page, which details the common pattern of matched interactions. Each pair of rows details a shared interaction, which is defined by the interacting pseudocenters from two different chains of a PPI. As in MultiBind each three columns present the details of a specific pseudocenter of a PPI. (D) A default Jmol visualization of the aligned complexes. The shared physico-chemical properties are represented as in Figure 1. The corresponding common interactions are represented as yellow sticks.
PERFORMANCE AND AVAILABILITY
The webservers of MultiBind and MAPPIS are available from http://bioinfo3d.cs.tau.ac.il. Although the running times of each algorithm are several minutes, the server overload may lead to longer running times. Consequently, the user has an option to supply an email address to which the link to the output page will be sent upon the completion. Users who are interested in performing large scale database analysis and classification are advised to download the freely available software packages. The packages contain the Linux executable programs as well as user manuals and a set of scripts for the extraction of binding sites and PPIs.
ACKNOWLEDGEMENTS
The research of A.S.P. was supported by the Clore PhD Fellowship. The research of H.J.W. has been supported in part by the Israel Science Foundation (grant no. 281/05), by the NIAID, NIH (grant No. 1UC1AI067231), by the Binational US-Israel Science Foundation (BSF) and by the Hermann Minkowski-Minerva Center for Geometry at TAU. This publication has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract NO1-CO-12400. This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. Funding to pay the Open Access publication charges for this article was provided by SAIC-Frederick, Inc.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wolfson HJ, Shatsky M, Schneidman-Duhovny D, Dror O, Shulman-Peleg A, Ma B, Nussinov R. From structure to function: methods and applications. Curr. Prot. Pep. Sci. 2005;6:171–183. doi: 10.2174/1389203053545435. [DOI] [PubMed] [Google Scholar]
- 2.Bock ME, Garutti C, Guerra C. Convergent evolution of enzyme active sites is not a rare phenomenon. J. Mol. Biol. 2007;372:817–845. doi: 10.1016/j.jmb.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 3.Artymiuk PJ, Poirrette AR, Grindley HM, Rice DW, Willett P. A graph-theoretic approach to the identification of three-dimensional patterns of amino acid side-chains in protein structures. J. Mol. Biol. 1994;243:327–344. doi: 10.1006/jmbi.1994.1657. [DOI] [PubMed] [Google Scholar]
- 4.Wallace AC, Laskowski RA, Thornton JM. Derivation of 3D coordinate templates for searching structural databases: application to Ser-His-Asp catalytic triads in the serine proteinases and lipases. Protein Sci. 1996;5:1001–1013. doi: 10.1002/pro.5560050603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell R. Detection of protein three-dimensional side-chain patterns: new examples of convergent evolution. J. Mol. Biol. 1998;279:1211–1227. doi: 10.1006/jmbi.1998.1844. [DOI] [PubMed] [Google Scholar]
- 6.Spriggs RV, Artymiuk PJ, Willett P. Searching for patterns of amino acids in 3d protein structures. J. Chem. Inf. Comput. Sci. 2003;43:412–421. doi: 10.1021/ci0255984. [DOI] [PubMed] [Google Scholar]
- 7.Binkowski T, Adamian L, Liang J. Inferring functional relationship of proteins from local sequence and spatial surface patterns. J. Mol. Biol. 2003;232:505–526. doi: 10.1016/s0022-2836(03)00882-9. [DOI] [PubMed] [Google Scholar]
- 8.Moodie SL, Mitchell JBO, Thornton JM. Protein recognition of adenylate: an example of a fuzzy recognition template. J. Mol. Biol. 1996;263:486–500. doi: 10.1006/jmbi.1996.0591. [DOI] [PubMed] [Google Scholar]
- 9.Denessiouk KA, Rantanen V, Johnson M. Adenine recognition: a motif present in ATP-, CoA-, NAD-, NADP-, and FAD-dependent proteins. Proteins. 2001;44:282–291. doi: 10.1002/prot.1093. [DOI] [PubMed] [Google Scholar]
- 10.Shulman-Peleg A, Nussinov R, Wolfson HJ. Recognition of functional sites in protein structures. J. Mol. Biol. 2004;339:607–633. doi: 10.1016/j.jmb.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jambon M, Imberty A, Deleage G, Geourjon C. A new bioinformatic approach to detect common 3d sites in protein structures. Proteins. 2003;52:137–145. doi: 10.1002/prot.10339. [DOI] [PubMed] [Google Scholar]
- 12.Brakoulias A, Jackson RM. Towards a structural classification of phosphate binding sites in protein-nucleotide complexes: an automated all-against-all structural comparison using geometric matching. Proteins. 2004;56:250–260. doi: 10.1002/prot.20123. [DOI] [PubMed] [Google Scholar]
- 13.Bock ME, Garutti C, Guerra C. Discovery of similar regions on protein surfaces. J. Comput. Biol. 2007;14:285–299. doi: 10.1089/cmb.2006.0145. [DOI] [PubMed] [Google Scholar]
- 14.Jones S, Thornton JM. Principles of protein-protein interactions. Proc. Natl Acad. Sci. USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 16.Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 17.Bahadur RP, Chakrabarti P, Rodier F, Janin J. A dissection of specific and non-specific protein-protein interfaces. J. Mol. Biol. 2004;336:943–955. doi: 10.1016/j.jmb.2003.12.073. [DOI] [PubMed] [Google Scholar]
- 18.Valdar WS, Thornton JM. Protein-protein interfaces: analysis of amino acid conservation in homodimers. Proteins. 2001;42:108–124. [PubMed] [Google Scholar]
- 19.Sheinerman FB, Norel R, Honig B. Electrostatic aspects of protein-protein interactions. Curr. Opin. Struct. Biol. 2000;10:153–156. doi: 10.1016/s0959-440x(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 20.Nooren IMA, Thornton JM. Diversity of protein-protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reichmann D, Rahat O, Albeck S, Meged R, Dym O, Schreiber G. The modular architecture of protein-protein binding interfaces. Proc. Natl Acad. Sci. USA. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keskin A, Tsai CH, Wolfson HJ, Nussinov R. A new, structurally non-reduntant, diverse dataset of protein-protein interfaces and its implications. Protien Sci. 2004;13:1043–1055. doi: 10.1110/ps.03484604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shulman-Peleg A, Mintz S, Nussinov R, Wolfson H. Protein-protein interfaces: recognition of similar spatial and chemical organizations. In: Jonassen I, Kim J, editors. Workshop on Algorithms in Bioinformatics, Springer, Lec. Notes in Comp. Sci. Vol. 3240. Heidelberg, Germany: 2004. pp. 194–205. [Google Scholar]
- 24.Mintz S, Shulman-Peleg A, Wolfson HJ, Nussinov R. Generation and analysis of a protein-protein interface dataset with similar chemical and spatial patterns of interactions. Proteins. 2005;61:6–20. doi: 10.1002/prot.20580. [DOI] [PubMed] [Google Scholar]
- 25.Shatsky M, Shulman-Peleg A, Nussinov R, Wolfson H. The multiple common point set problem and its application to molecule binding pattern detection. J. Comput. Biol. 2006;13:407–442. doi: 10.1089/cmb.2006.13.407. [DOI] [PubMed] [Google Scholar]
- 26.Shulman-Peleg A, Shatsky M, Nussinov R, Wolfson H. Berthold M, editor. MAPPIS: multiple 3D alignment of protein-protein interfaces. Complife, Konstanz, Germany, Springer Lec. Notes in Comp. Sci. 2005;Vol. 3695:91–103. [Google Scholar]
- 27.Connolly M. Analytical molecular surface calculation. J. Appl. Cryst. 1983;16:548–558. [Google Scholar]
- 28.Schmitt S, Kuhn D, Klebe G. A new method to detect related function among proteins independent of sequence or fold homology. J. Mol. Biol. 2002;323:387–406. doi: 10.1016/s0022-2836(02)00811-2. [DOI] [PubMed] [Google Scholar]
- 29.Shulman-Peleg A, Shatsky M, Nussinov R, Wolfson H. Spatial chemical conservation of hot spot interactions in protein-protein complexes. BMC Biol. 2007;5:43. doi: 10.1186/1741-7007-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfson HJ. Heidelberg, Germany: Springer LNCS; 1990. In Proceedings of the 1st European Conference on Computer Vision (ECCV) pp. 526–536. [Google Scholar]