Abstract
DNA and RNA oligomers are used in a myriad of diverse biological and biochemical experiments. These oligonucleotides are designed to have unique biophysical, chemical and hybridization properties. We have created an integrated set of bioinformatics tools that predict the properties of native and chemically modified nucleic acids and assist in their design. Researchers can select PCR primers, probes and antisense oligonucleotides, find the most suitable sequences for RNA interference, calculate stable secondary structures, and evaluate the potential for two sequences to interact. The latest, most accurate thermodynamic algorithms and models are implemented. This free software is available at http://www.idtdna.com/SciTools/SciTools.aspx.
INTRODUCTION
Synthetic oligonucleotides are widely employed in various molecular biology applications, e.g. polymerase chain reaction (PCR), molecular beacons, microarrays, mutagenesis, RNAi, antisense and de novo gene construction (1–7). Published bioinformatics algorithms can predict biophysical properties of oligonucleotides from their sequence and estimate performance of oligonucleotides in specific assays both singly and together with other sequences (8,9). Here, we describe an online suite of computational software tools that enable molecular biologists to design, evaluate and make informed decisions about the properties of nucleic acid sequences. The IDT SciTools receives over 7000 unique visitors and 1.5 million hits every month. The web servers consist of several independent applications summarized in Table 1. Instructions and help to each software tool can be found at the top of web input forms. The code is regularly updated when more accurate models and algorithms are published. New applications will be added in the future.
Table 1.
Summary of IDT SciTools
| Software tool | Features |
|---|---|
| OligoAnalyzer | Comprehensive oligonucleotide analysis (molecular weight, extinction coefficient, melting temperature, folding and hybridization of strands, effects of mismatches). |
| PrimerQuest | Select optimal probes and primers for PCR assays. |
| LNA design | Tool to design LNA modified probes and primers having specific duplex stability. |
| ddRNAi design | Design sequences for DNA-directed RNA interference. |
| RNAi design | Tool to design siRNA duplex oligomers. |
| TriFECTa RNAi Kits | Predesigned dicer-substrate RNAi duplexes, gene knockdown kits. |
| Antisense design | Antisense oligonucleotide selection tool. |
| mFold | Prediction of oligonucleotide secondary structure. |
| DilutionCalc | Calculate volumes to dilute oligonucleotide to the desired concentration. |
| ResuspensionCalc | Calculate volumes to dissolve a dry lyophilized nucleic acid to the desired concentration. |
ONLINE TOOLS
OligoAnalyzer 3.1
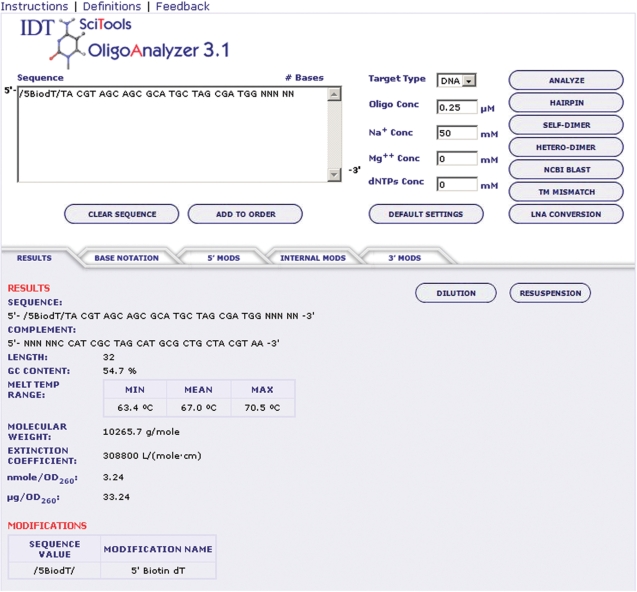
The OligoAnalyzer is the central calculator where various kinds of information about an oligonucleotide sequence can be predicted. The interface is presented on Figure 1. A user can input a nucleotide sequence and conditions, i.e. the concentrations of DNA, Na+, K+, Mg2+ and deoxynucleoside triphosphates. Melting temperature is predicted under these conditions for the duplex where the oligonucleotide hybridizes to the complementary sequence. This complementary strand can be either RNA or DNA; this is selected using the Target Type option. The oligonucleotide sequence can be modified with over 150 different labels and chemical groups (e.g. biotin, phosphorothioate, fluorescent dyes) using symbols listed in the tabbed sections below the sequence box. Seven different analyses can be performed when the specific button is selected on the right side of the interface. Selection of the ANALYZE button results in the physical properties of the oligonucleotide, such as a complementary sequence, oligonucleotide length, content of G and C bases, melting temperature, extinction coefficient at 260 nm and molecular weight (Figure 1). Published nearest-neighbor parameters are employed to calculate the extinction coefficient (10–12). Using values obtained from the published literature or coefficients estimated at Integrated DNA Technologies, the effects of modifications are included in the oligonucleotide extinction coefficient.
Figure 1.
Interface of OligoAnalyzer 3.1. Instructions and help files are at the top of the calculator. The sequence and conditions of experiments are entered in the upper region. Many chemical modifications can be added to a sequence using symbols from tabbed sections in the middle segment. The results are displayed in the lower left segment.
The oligonucleotide molecular weight also includes the weights of any chemical modifications. These weights have been experimentally validated (± 2 g/mol) for thousands of synthesized sequences by electrospray-ionization liquid chromatography mass spectrometry (13).
Melting temperatures are calculated from the nearest-neighbor model (14–16) and the duplex is assumed to melt in two-state fashion,
| 1 |
Oligonucleotide concentration, [S1], is assumed to be significantly larger (at least 6 ×) than the concentration of the complementary target, [S2], as this is seen in many molecular biology assays. In that case, Coligo is equal to [S1] and the concentration of the target can be neglected (17). If [S1] is not significantly larger than [S2], but [S1] ≥ [S2], the following concentration should be entered into the calculator,
| 2 |
If [S2] < [S1], Equation (2) is valid when the designation of strands is switched. Transition enthalpy, ΔH°, and entropy, ΔS°, are calculated from the latest nearest-neighbor parameters for DNAs (15,16) and RNAs (18,19). The effects of counterions are modeled using the improved corrections for monovalent ions (20) and magnesium ions (53),
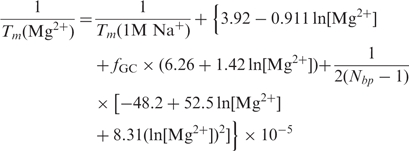 |
3 |
This unique biophysical model employed for various counterions is not implemented elsewhere (21–23). Thermodynamic parameters are not available for many modifications that were demonstrated to change duplex stability (e.g. 2′-O-methyl RNA, 2-aminopurine, Cy3 dye) (13), and their effects on melting temperature are therefore neglected in the current version. When these parameters are published, they will be implemented in the predictive algorithm. If a sequence contains degenerate bases, the minimum and the maximum melting temperatures for the mixture of sequences are also estimated (Figure 1). The thermodynamic algorithm was validated using an independent set of over 100 different sequences ranging in length from 8 to 60 base pairs that were not used to derive the algorithm (20).
Selection of the HAIRPIN button will present the user with the input form for predicting oligonucleotide secondary structures. This tool uses the mFold algorithm (9,24–26) that is described later. The SELF-DIMER and HETERO-DIMER buttons allow the user to examine possible duplexes when oligonucleotide anneals to itself or another target sequence. Predicted structures from most stable to least stable are shown. These cross-hybridization analyses are important for PCR assays where primer–primer interactions can decrease the efficiency of the reaction and cause secondary by-products. Selection of the NCBI BLAST button sends the sequence to the NCBI website for searching various databases using the short nearly exact matches method (27). This analysis can provide predicted annealing sites of the oligonucleotide within a genome or other group of candidate sequences.
Selection of the TM MISMATCH button will allow the user to examine the effects of single base mismatches on duplex stability and oligonucleotide hybridization. Several published sets of nearest-neighbor parameters from SantaLucia's lab are employed to make these predictions (16,28–32). Dangling unpaired bases usually stabilize the duplex, so the predictive algorithm also takes these effects into account (33). If a red target base is clicked, a dropdown box will appear and allow the user to select the desired base mismatch. The target concentration can be set to zero when the target concentration is negligible in comparison with the oligonucleotide concentration. Results will show melting temperatures of perfectly matched and mismatched duplexes as well as the fractions of oligonucleotide bound to the targets.
The LNA CONVERSION button will be described later. The tool allows the user to position LNA modifications within a sequence, so that the desired melting temperature of the duplex sequence is achieved.
PrimerQuestSM
Primer and probe selection for the PCR-based assays are important activities in molecular biology. Several software packages were therefore designed for this procedure (34–39). PrimerQuestSM is based on the Primer3 code (37). However, the selection method was improved and a graphical user interface was created. The algorithm finds sequences having desired oligonucleotide length, GC content, melting temperature, content of consecutive GC base pairs and sequence stability at the 3′ end. The intramolecular secondary structures, long repeats of the same bases and cross-hybridization between primers and the probes are minimized in the primer selection model. Furthermore, the oligonucleotide melting temperature is calculated using the same thermodynamic model employed in the OligoAnalyzer. Once the nucleotide sequence is entered in the sequence box, the name and design criteria for the sequence of interest can be set using the appropriate fields under the basic, standard and advanced tabs. The basic interface exposes the minimal information that needs to be entered and hides detailed criteria. These basic settings are suitable for typical PCR experiments. The standard and advanced tabs show increasing amounts of settings that an advanced user can configure to customize their predictive model. The CALCULATE button submits the data for the prediction of primers with the desired properties.
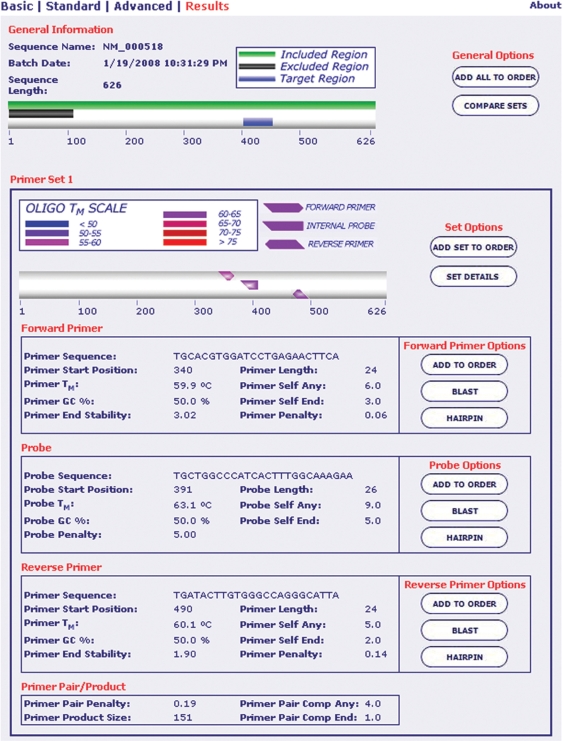
Results show several sets of probes and primers that were found to be optimal (Figure 2). The predicted sets are ranked from best to next best. A graphical representation of the sequence is displayed with color bars for included, excluded and targeted regions. The biophysical properties of the primers and probes are also reported.
Figure 2.
Example of PrimerQuestSM results. Primers and a probe were selected for beta hemoglobin mRNA. Included and excluded regions are displayed as horizontal colored graphic bars along the sequence. The target region is included in the amplicon. The primers and the probe are shown as horizontal arrows where the location and direction of the arrow represent hybridization area and orientation. Properties of the primers and the probe are reported below graphical illustrations.
LNA design
Chimeric probes containing locked nucleic acid residues were demonstrated to increase duplex stability, specificity and mismatch discrimination (40,41). These properties improve genotyping and microarray assays. The LNA design tool suggests positions within a specific sequence, where LNA modifications can be introduced to produce the desired biophysical properties. A user enters desired number of LNA residues and the melting temperature of LNA-modified duplex. The software will attempt to decrease the length of the sequence and introduce LNA modifications, so that the desired melting temperature is achieved. The LNA residues are indicated with ‘+’ symbol in front of the base. Melting temperatures are predicted using the nearest-neighbor two-state model (Equation 1), the latest thermodynamic parameters (16,42), and improved salt corrections for the effects of monovalent and magnesium ions (20,53). The algorithm was tested with a published set of melting data for LNA modified oligomers (40,42).
Antisense design
Expression of specific genes can be suppressed with antisense oligonucleotides (43). Software can be used to select the most effective antisense oligonucleotides based on a model that discriminates between effective and ineffective antisense sequences. The nucleotide sequence of a gene or other target candidate for antisense-based knockdown can be retrieved from NCBI databases using GenBank ID or RefSeq ID. Antisense DNA oligomers are typically from 19 to 26 bases long and modified with phosphorothioates for nuclease resistance. Optionally, 2′-O-methyl RNA residues can be introduced to increase duplex stability and resistance against nucleases. The general algorithm for predicting active sites includes or excludes 3 and 4 base long motifs that are correlated with antisense activity responses (44,45). Sequences with the best score for the number of positive motifs (CCAC, TCCC, ACTC, GCCA, CTCT) are most likely to show antisense activity. A user can modify search criteria and include or exclude various motifs.
RNAi design, ddRNAi design and TriFECTa RNAi kits
In addition to antisense-based gene knockdown, the use of short interfering RNAs to induce RNA interference is a powerful strategy to suppress gene expression in vivo (5,6). These three tools assist in the design of effective siRNAs, as there are several properties that discriminate between effective and ineffective siRNA duplexes (46,47). The ddRNAi tool helps to design siRNAs, which are expressed directly from DNA transfected into cells to make the siRNA (48–50).
The RNAi design software tool allows users to predict effective short synthetic 27-mer siRNA duplexes that are delivered to target cells (6). A user can specify criteria for the siRNA duplex and overhangs, e.g. desired duplex length, strand content of G and C bases and various sequence motifs at specific positions. Mixed bases can be introduced and different weights can be assigned to each motif. The algorithm searches a target gene sequence, calculates scores for sequence candidates and ranks optimal siRNA duplexes. The duplexes can have symmetrical overhangs up to 3 bases long. Results show properties of selected siRNA duplexes and their location within the target sequence.
SiRNA–TriFECTa sequences are a collection of predesigned dicer-substrate siRNA sequences that have been found to be optimal using dicer-substrate siRNA design criteria. Besides incorporating siRNA activity criteria into the design algorithm, additional analyses are performed, so that chosen siRNA sequences do not target alternatively spliced exons and do not include known polymorphic sites. Gene sequences from the NCBI reference sequence set within eight organisms can be selected and displayed.
mFold
This software tool predicts the most stable secondary structure of an oligonucleotide by minimizing folding free energy (51). Suboptimal energetic secondary structures having free energies close to minimal ΔG° can be predicted as well. The mFold software was developed by and implemented in collaboration with Prof. Michael Zuker. The algorithm has been well tested and described in published sources (9,24–26). A user can input a nucleotide sequence and the conditions, including temperature and ionic concentrations. The folded structures are predicted at the specified temperature. The results show both a dot-plot diagram of possible base pairings and predicted secondary structures. These structures are ranked from the highest to lowest probability using transition free energies. Melting temperature is estimated using a two-state model. The connectivity table for each base and details of the energetics (each loop and stack ΔG° contributions) can also be obtained.
Dilution and resuspension calculations
Oligonucleotide dilutions can be calculated using the dilution calculator. A user inputs initial concentration, volume and desired final concentration. The calculator returns volumes used to mix solutions. Various concentration formats and units are accepted. Similarly, the resuspension calculator determines the volume of solution needed to achieve a specific concentration when known moles or mass of dry oligonucleotide are dissolved. Both calculators can be brought directly from the OligoAnalyzer results. In that case, oligonucleotide properties predicted by OligoAnalyzer are transferred automatically to these calculators.
Biophysics.idtdna.com
Subdomain http://biophysics.idtdna.com contains advanced, unique calculators that are being developed. Software is stable and tested, but it has yet to be included into IDT SciTools. In the current version, the extinction coefficients and UV spectrum from 215 to 310 nm can be predicted for both single-stranded and double-stranded DNA oligomers. The models, parameters and their accuracy tests have been recently published (12). A user can also choose to apply Cavaluzzi–Borer correction for extinction coefficients of DNA bases at 260 nm (52). The predicted UV spectrum is plotted and extinction coefficients at each wavelength are tabulated.
Webservices
Additional tools are also freely available, but the software requires a free login username and password to manage the user's information between sessions due to the computational time requirements. For example, oligonucleotides suitable for creating a gene expression microarray can be designed with the Workbench application. However, the design and storage of possibly thousands of target sequences is not amenable to be done during a single web browser session. The use of a free user login allows this information to be associated with a single user. Finally, for the programmatic incorporation of some of these software engines, IDT also provides a set of web service based interfaces to allow external software developers access to these functions (http://www.idtdna.com/AnalyzerService/AnalyzerService.asmx).
CONCLUSIONS
IDT SciTools web server provides useful predictions of oligonucleotide properties under various experimental conditions. The software tools help to select oligonucleotides that are most likely to exhibit the best performance in biological applications.
ACKNOWLEDGEMENTS
Funding to pay the Open Access publication charges for this article was provided by Integrated DNA Technologies, Inc.
Conflict of interest statement. The authors are or have been employed by Integrated DNA Technologies, Inc., (IDT). IDT is not a publicly traded company and has filed, or may have filed, at least one patent application on the materials or methods described in this article. IDT also offers oligonucleotides for sale similar to the oligonucleotides described in this article. The authors do not own any shares or equity in IDT.
REFERENCES
- 1.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet G, Tyagi S, Libchaber A, Kramer FR. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat. Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 4.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 6.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 7.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 8.Horne MT, Fish DJ, Benight AS. Statistical thermodynamics and kinetics of DNA multiplex hybridization reactions. Biophys. J. 2006;91:4133–4153. doi: 10.1529/biophysj.106.090662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuker M. Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol. 2000;10:303–310. doi: 10.1016/s0959-440x(00)00088-9. [DOI] [PubMed] [Google Scholar]
- 10.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 11.Fasman G. Handbook of Biochemistry and Molecular Biology. Cleveland, OH: CRC Press; 1975. [Google Scholar]
- 12.Tataurov AV, You Y, Owczarzy R. Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids. Biophys. Chem. 2008;133:66–70. doi: 10.1016/j.bpc.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Moreira BG, You Y, Behlke MA, Owczarzy R. Effects of fluorescent dyes, quenchers, and dangling ends on DNA duplex stability. Biochem. Biophys. Res. Commun. 2005;327:473–484. doi: 10.1016/j.bbrc.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 14.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 15.Owczarzy R, Vallone PM, Gallo FJ, Paner TM, Lane MJ, Benight AS. Predicting sequence-dependent melting stability of short duplex DNA oligomers. Biopolymers. 1997;44:217–239. doi: 10.1002/(SICI)1097-0282(1997)44:3<217::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 16.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owczarzy R. Melting temperatures of nucleic acids: Discrepancies in analysis. Biophys. Chem. 2005;117:207–215. doi: 10.1016/j.bpc.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Xia T, SantaLucia J., Jr., Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 19.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 20.Owczarzy R, You Y, Moreira BG, Manthey JA, Huang L, Behlke MA, Walder JA. Effects of sodium ions on DNA duplex oligomers: improved predictions of melting temperatures. Biochemistry. 2004;43:3537–3554. doi: 10.1021/bi034621r. [DOI] [PubMed] [Google Scholar]
- 21.Herold KE, Rasooly A. Oligo Design: a computer program for development of probes for oligonucleotide microarrays. Biotechniques. 2003;35:1216–1221. doi: 10.2144/03356bc02. [DOI] [PubMed] [Google Scholar]
- 22.Kibbe WA. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35:W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panjkovich A, Norambuena T, Melo F. dnaMATE: a consensus melting temperature prediction server for short DNA sequences. Nucleic Acids Res. 2005;33:W570–W572. doi: 10.1093/nar/gki379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaeger JA, Turner DH, Zuker M. Improved predictions of secondary structures for RNA. Proc. Natl Acad. Sci. USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker M. Prediction of RNA secondary structure by energy minimization. Methods Mol. Biol. 1994;25:267–294. doi: 10.1385/0-89603-276-0:267. [DOI] [PubMed] [Google Scholar]
- 26.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Allawi HT, SantaLucia J., Jr. Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry. 1997;36:10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 29.Allawi HT, SantaLucia J., Jr. Nearest-neighbor thermodynamics of internal A.C mismatches in DNA: sequence dependence and pH effects. Biochemistry. 1998;37:9435–9444. doi: 10.1021/bi9803729. [DOI] [PubMed] [Google Scholar]
- 30.Allawi HT, SantaLucia J., Jr. Thermodynamics of internal C.T mismatches in DNA. Nucleic Acids Res. 1998;26:2694–2701. doi: 10.1093/nar/26.11.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allawi HT, SantaLucia J., Jr. Nearest neighbor thermodynamic parameters for internal G.A mismatches in DNA. Biochemistry. 1998;37:2170–2179. doi: 10.1021/bi9724873. [DOI] [PubMed] [Google Scholar]
- 32.Peyret N, Seneviratne PA, Allawi HT, SantaLucia J., Jr. Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 33.Bommarito S, Peyret N, SantaLucia J., Jr. Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res. 2000;28:1929–1934. doi: 10.1093/nar/28.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haas S, Vingron M, Poustka A, Wiemann S. Primer design for large scale sequencing. Nucleic Acids Res. 1998;26:3006–3012. doi: 10.1093/nar/26.12.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olson SA. EMBOSS opens up sequence analysis. European Molecular Biology Open Software Suite. Brief Bioinform. 2002;3:87–91. doi: 10.1093/bib/3.1.87. [DOI] [PubMed] [Google Scholar]
- 36.Podowski RM, Sonnhammer EL. MEDUSA: large scale automatic selection and visual assessment of PCR primer pairs. Bioinformatics. 2001;17:656–657. doi: 10.1093/bioinformatics/17.7.656. [DOI] [PubMed] [Google Scholar]
- 37.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 38.Womble DD. GCG: The Wisconsin Package of sequence analysis programs. Methods Mol. Biol. 2000;132:3–22. doi: 10.1385/1-59259-192-2:3. [DOI] [PubMed] [Google Scholar]
- 39.Xu D, Li G, Wu L, Zhou J, Xu Y. PRIMEGENS: robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics. 2002;18:1432–1437. doi: 10.1093/bioinformatics/18.11.1432. [DOI] [PubMed] [Google Scholar]
- 40.You Y, Moreira BG, Behlke MA, Owczarzy R. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006;34:e60. doi: 10.1093/nar/gkl175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (locked nucleic acids): synthesis of adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerization, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 42.McTigue PM, Peterson RJ, Kahn JD. Sequence-dependent thermodynamic parameters for locked nucleic acid (LNA)-DNA duplex formation. Biochemistry. 2004;43:5388–5405. doi: 10.1021/bi035976d. [DOI] [PubMed] [Google Scholar]
- 43.Izant JG, Weintraub H. Constitutive and conditional suppression of exogenous and endogenous genes by anti-sense RNA. Science. 1985;229:345–352. doi: 10.1126/science.2990048. [DOI] [PubMed] [Google Scholar]
- 44.Matveeva OV, Tsodikov AD, Giddings M, Freier SM, Wyatt JR, Spiridonov AN, Shabalina SA, Gesteland RF, Atkins JF. Identification of sequence motifs in oligonucleotides whose presence is correlated with antisense activity. Nucleic Acids Res. 2000;28:2862–2865. doi: 10.1093/nar/28.15.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McQuisten KA, Peek AS. Identification of sequence motifs significantly associated with antisense activity. BMC Bioinform. 2007;8:184. doi: 10.1186/1471-2105-8-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peek AS. Improving model predictions for RNA interference activities that use support vector machine regression by combining and filtering features. BMC Bioinform. 2007;8:182. doi: 10.1186/1471-2105-8-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peek AS, Behlke MA. Design of active small interfering RNAs. Curr. Opin. Mol. Ther. 2007;9:110–118. [PubMed] [Google Scholar]
- 48.Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, Salvaterra P, Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 49.Scherr M, Morgan MA, Eder M. Gene silencing mediated by small interfering RNAs in mammalian cells. Curr. Med. Chem. 2003;10:245–256. doi: 10.2174/0929867033368493. [DOI] [PubMed] [Google Scholar]
- 50.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 51.Nussinov R, Jacobson AB. Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc. Natl Acad. Sci. USA. 1980;77:6309–6313. doi: 10.1073/pnas.77.11.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Owczarzy R, Moreira BG, You Y, Behlke MA, Walder JA. Biochemistry. 2008. Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. [Epub ahead of print; doi:10.1021/bi702363u] [DOI] [PubMed] [Google Scholar]