Abstract
The Gene Ontology (GO) vocabulary has been extensively explored to analyze the functions of coexpressed genes. However, despite its extended use in Biology and Medical Sciences, there are still high levels of uncertainty about which ontology (i.e. Molecular Process, Cellular Component or Molecular Function) should be used, and at which level of specificity. Moreover, the GO database can contain incomplete information resulting from human annotations, or highly influenced by the available knowledge about a specific branch in an ontology. In spite of these drawbacks, there is a trend to ignore these problems and even use GO terms to conduct searches of gene expression profiles (i.e. expression + GO) instead of more cautious approaches that just consider them as an independent source of validation (i.e. expression versus GO). Consequently, propagating the uncertainty and producing biased analysis of the required gene grouping hypotheses. We proposed a web tool, Onto-CC, as an automatic method specially suited for independent explanation/validation of gene grouping hypotheses (e.g. coexpressed genes) based on GO clusters (i.e. expression versus GO). Onto-CC approach reduces the uncertainty of the queries by identifying optimal conceptual clusters that combine terms from different ontologies simultaneously, as well as terms defined at different levels of specificity in the GO hierarchy. To do so, we implemented the EMO-CC methodology to find clusters in structural databases [GO Directed acyclic Graph (DAG) tree], inspired on Conceptual Clustering algorithms. This approach allows the management of optimal cluster sets as potential parallel hypotheses, guided by multiobjective/multimodal optimization techniques. Therefore, we can generate alternative and, still, optimal explanations of queries that can provide new insights for a given problem. Onto-CC has been successfully used to test different medical and biological hypotheses including the explanation and prediction of gene expression profiles resulting from the host response to injuries in the inflammatory problem. Onto-CC provides two versions: Ready2GO, a precalculated EMO-CC for several genomes and an Advanced Onto-CC for custom annotation files (http://gps-tools2.wustl.edu/onto-cc/index.html).
INTRODUCTION
High-throughput experimental techniques, such as microarrays, produce large amounts of data and knowledge about gene expression levels. Frequently the output of such analysis consists of a list of significant or ranked differentially expressed genes that may lead to clusters of tens to hundreds of them. These data are of little use if it is not possible to interpret the results in a biological context (1). To alleviate this problem, the Gene Ontology Consortium provides consistent descriptions of gene products. This biological knowledge is organized as hierarchical, structured and controlled vocabularies named Gene Ontologies (GOs) (2), which describe gene products in terms of their associated molecular functions (MF), biological processes (BP) and cellular components (CC). Nowadays, the GO Consortium provides GO annotations for many different organisms (2).
Several tools have been developed to identify clusters of GO terms that can explain sets of coexpressed genes from microarray experiments (3). These approaches often search for overrepresented GO terms describing a group of genes using different statistical approaches such as Fisher's exact test [FatiGO (4)], χ2 or binomial distribution [Onto-Express (5)], or calculating z-scores under the hypergeometric distribution [MAPPFinder (6)].
One of the principal problems when identifying biologically meaningful clusters in the GO database is that the quality of the annotations is based on the available knowledge. For example, some biological processes are studied in more detail than others, thus generating long branches with very specific GO terms while other branches remain almost undescribed. To address this uncertainty, most of currently available tools ask the user to select a custom level of specificity (e.g. level 3) for the retrieved terms, often constraining found GO terms (e.g. all biological processes) to the same levels, retrieving not only limited but too general or too specific information. Moreover, most of the available clustering methods search each ontology independently, thus, missing relevant relationships among terms from different ontologies.
The crucial drawback shared by these methods is that their subjacent clustering algorithm is not originally designed to deal with hierarchically organized information (7). This constrains their ability to search through the complex relationships underlying structural data contained in the Directed acyclic Graph (DAG) of the GO database (Appendix A, Figure 1). A structural database can be viewed as a graph containing nodes, which represent objects; and the relationships among these objects can be represented by edges. In this case, a substructure corresponds to a subgraph of the GO DAG (Supplementary Figure 2) (8). Conceptual clustering techniques have been successfully applied to structural databases by searching through a predefined space of potential hypothesis (i.e. substructures) for those that best fits the training examples (8,9). However, searching for conceptual clusters in a graph-based structure such as the GO DAG, would result in the generation of many substructures with small extent, as it is easier to model smaller data subsets than larger representative ones (10).
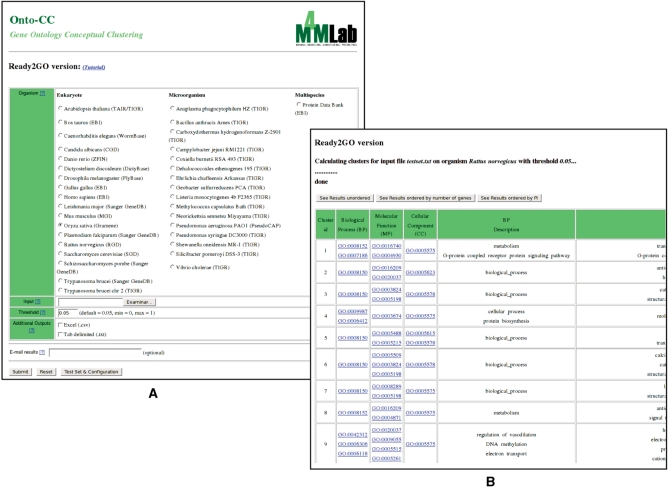
Figure 1.
Ready2GO web interface. (A) Snapshot of the input form. Several genomes are available, along with two multispecies databases. (B) Snapshot of the output results. In addition to the HTML table, two output files are also available to download the obtained results: .csv (comma separated version, suitable for MS Excel) and .txt (tab separated).
The usefulness of existing functional profiling approaches is impacted by the annotation bias present in the GO database, as well as by the constraints imposed by the clustering methods. Therefore, to extract better-defined concepts, Onto-CC uses the CC methodology (11) inspired on conceptual clustering techniques, which obtain sets of optimal clusters based on their specificity, diversity and number of retrieved gene product. These are conflicting criteria that can be approached as an optimization problem. The basic challenge is to avoid the potential bias caused by weighting the objectives (10), which always derives in the convergence to solutions corresponding to single or limited regions of the search space (i.e. GO DAG). This problem is noteworthy because typical data mining approaches, particularly in computational biology, tend to emphasize consensus or most frequent patterns (7) that often conceal rather than reveal novel and useful knowledge about the problem (12,13).
METHODOLOGY
Onto-CC server searches for explanations and functional validation of a group of genes, provided by the user, potentially related (e.g. coexpressed genes). Different subsets from the query are statistically compared with independently precalculated clusters from the GO database of the selected organism. These clusters share common sets of features (i.e. GO terms) hierarchically organized at distinct levels of specificity in a structural database (i.e. GO DAG). Indeed, Onto-CC considers the three different ontologies simultaneously. The groups resulting from the former relationships (i.e. conceptual clusters) should be optimal, avoiding redundancy, but permitting descriptions of the genes from different points of view. In other words, one gene can belong to different conceptual clusters characterized by different sets of features (14). Summarizing, this web tool allows the users to validate their hypothesis about sets of gene products by establishing relationships between them and GO clusters, which were identified by a conceptual clustering inspired algorithm. Onto-CC does not only retrieve clusters of genes, but also performs a differential feature selection for each cluster (15).
The precalculated clusters, termed substructures in a DAG database, are obtained following these steps: (i) Given a GO annotation file from a specific genome, the algorithm randomly create potential substructures harboring distinct features (i.e. GO terms) defined at different specificity levels and ontologies. Onto-CC does not select a priori one specificity level in the GO DAG, like most of the state of the art tools do (e.g. level 3). Yet, it searches through different specificity levels through the composite DAG space for potential substructures using an evolutionary algorithm (EA) (16). (ii) The initial substructures evolve guided by a multiobjective/multimodal optimization approach based on two objectives: the degree of matching between the terms contained in the substructure and the GO terms that characterize a subset of gene products (i.e. specificity) and the number of gene products described by the substructure (i.e. support) (see Methodology Details in Appendix A). These are contradictory objectives, since when the specificity increases, the support usually decreases and vice versa. Particularly, the goal is to select substructures that satisfy a tradeoff between specificity and support. (iii) The final set of clusters is achieved when the maximum number of Genetic Algorithm generations is reached. These results are non-dominated clusters, which are salient groups of genes/GO terms that are not worst than any other final solution in both objectives (see Methodology Details in Appendix A). These groups consist of all possible optimal variations of GO terms defined at different specificity levels, ontologies and gene products.
Onto-CC server provides two services: Ready2GO and Advanced Onto-CC version. The Ready2GO service is a precalculated version of conceptual clusters for over 30 different genomes annotated by the GO Consortium. The Advanced Onto-CC is thought to be for users working with not fully described systems, genome custom annotation or genomes that are still not annotated by the GO Consortium. In this case, the conceptual clusters will be calculated on the fly based on the annotation files provided by the user.
IMPLEMENTATION
The mapping script is written in perl using bioperl modules and accessing several web services (e.g. biomaRt resources, genome home pages and UniProt ID mapping web interface). Onto-CC was developed in Eiffel v6.0 (Eiffel is an ISO standardized, object-oriented programing language based on the design by contract paradigm).
Execution times vary depending on the number of input accession numbers combined with the size of the genome annotation data, for the Ready2GO version. The test file with default values spends ∼1 min on a 64-bit computer with 2 GHz processors. For the advanced version, Step 1 takes several minutes for a standard file. This time consumption does not only depend on the number of input accession numbers and annotations, but also on the size of the EA population and number of evaluations to perform. We recommend saving the Step 1 output results in order to reuse them for Step 2 without having to rebuild the clustering. Test file with default values spends <15 s for Step 1 and <5 s for Step 2 using the previous machine specifications.
WEB INTERFACE
The web server is available being implemented using CGI scripts that communicate with several perl scripts and the EMO-CC unix executable. Each of the Onto-CC versions, Ready2GO and Advanced, has a tutorial available along with example test files. The tutorials explain which parameters can be tuned and between which ranges they can be modified. Default settings should be adopted for beginners. The online tutorials cover the following help topics: organism, annotation, input, threshold value, EMO-CC parameters, additional outputs, Email results and output results. The query starts by clicking the ‘submit’ button. Results are provided as HTML for visual inspection and can also be received by Email. In case of error, a human readable message is displayed.
Databases
Standard protein databases are used to query GO terms from accession number lists provided by the user. The database accession numbers that can be used are: UniProt (accession number or ID) (17), RefSeq (18), Ensembl (19), Vega (20), GI (21), Gene name, Dictybase (22), CGD (16), Flybase (23), GeneDB (24), TIGR (25), MGD (26), RGD (27), SGD (28), PseudoCAP (29), TAIR (30), Wormbase (31), ZFIN (32) and/or PDB (33). For each organism exists a precalculated mapping between all those databases and the GO project. We update the annotation files, and recalculate the clusters every 6 months. To do so, we take advantage of the evolutionary techniques included in the proposed algorithm that allows us to update the clusters after running few generations by using the previous clusters as a seed (34). This incremental learning approach (35) accelerates and reduces the computational complexity of the updating process, leaving the full recalculus to extreme and unusual cases (R.R.Z, C.D.V. and I.Z. manuscript in preparation).
Ready2Go version
Input
The input file is a list of accession numbers belonging to one of the organisms for which the GO project provides annotation (2) (Figure 1, panel A). This input file consists of sequence identifiers, one per line, from any of the databases previously mentioned.
Parameters
There are two parameters to be specified: organism and P-value. The organism can be selected from any of the different genomes annotated by the GO Consortium listed in the menu; the menu includes eukaryotes, microorganisms and multispecies (Figure 1, panel A). The second parameter is the P-value (36) and represents the probability of observing by chance a specific intersection between the gene products given by the user and the gene products belonging to the precalculated clusters. The threshold can take values between 0 and 1, where lower values represent greater reliability.
Output
Results are shown as a HTML table containing each of the clusters found in no particular order. The clusters can be ordered by the number of genes or by the cluster P-value using the buttons shown above the table (Figure 1, panel B). The table contains the following fields: cluster identification number (i.e. Cluster ID column), BP subontology GO terms and descriptions belonging to the cluster (i.e. Biological Process and BP Description column, respectively), MF subontology GO terms and descriptions belonging to the cluster (i.e. Molecular Function and MF Description column, respectively), CC subontology GO terms and descriptions belonging to the cluster (i.e. Cellular Component and CC Description column, respectively), the list of accession numbers belonging to the cluster (i.e. ACC column) and the P-value between the set of the given accession numbers and the precalculated EMO-CC clusters for the selected organism (i.e. P-value column). In addition to the HTML version, the output file can be downloaded as a comma separated version (.csv, suitable for MS Excel) and as a tab separated text (.txt).
Advanced version
The Onto-CC advanced version allows obtaining a set of GO descriptions for a list of user input accession numbers by using custom GO annotation information. In this case the substructures (conceptual clusters) will be calculated on the fly based on the annotation files provided by the user. For this protocol two steps are needed: Step 1, creation of custom conceptual clusters and Step 2, creation of a GO description of a list of accession numbers using the previous calculated conceptual clusters.
Step 1
Input
The input file for this step is a custom GO annotation file. This file describes the relationship between a gene product/protein and GO terms. The annotation file contains a description per line, where the identifier of the gene/protein is separated from its GO description by a comma. Each identifier can have multiple GO terms, which are separated by semicolons and can belong to any of the ontologies in the GO project.
Parameters
EMO-CC is a multiobjective EA. An EA uses some mechanisms inspired by biological evolution to optimize the solutions of the problem, such as, reproduction, mutation, recombination, natural selection and survival of the fittest. Several parameters can be modified in an EA, but only two are available to the user: the population size and the number of evaluations. Changes in these parameters modify the algorithm performance and have an effect in the number and quality of clusters found. EAs rely on a population of abstract representations, called chromosomes, of candidate solutions, called individuals, to an optimization problem, and evolve toward better solutions. Bigger population sizes will result in slower performance, but in better results. By increasing the size of the population, more space is made available to save diverse solutions, therefore, promoting the evolution to better areas. As the size of the population increases, the number of evaluations performed must also increase. The population size can be changed by the user in the range [10–1000] with a default value of 200. This value is appropriate for a list of 2000 annotated IDs approximately. Usually, an initial population of randomly generated candidate solutions comprises the first generation. During each successive generation, a proportion of the existing population is selected to breed a new generation. A cost function is used to guide the search and it is applied to the candidate solutions and any subsequent offspring to quantify the optimality of a solution, also termed chromosome, in an EA so that a particular chromosome may be ranked against all the other chromosomes. Each of these cost function evaluations can be used to determine when to stop an EA execution. The user can specify the maximal number of evaluations for the EA, where values range from 100 to 99 999 and the default value is 20 000. As a rule of dumb, the number of evaluations should be a multiple of the population size. This multiple number will be approximately the number of generations to perform.
Output
The HTML table shows each of the clusters found in no particular order. The table is very similar to the output table of Ready2GO but without the PI column and with the addition of specificity and support columns. Specificity values ranging [0–1] with 1 as the best-case scenario meaning that all gene products, described by the cluster, shared the same GO terms. Support is the second objective function used by the optimization algorithm and ranges [0–1] with 1 as the best-case scenario, meaning that the cluster describe all the gene products in the input file. Again, the output file can be downloaded as a comma separated version (.csv, suitable for MS Excel) and as a tab separated text (.txt), in addition to the HTML version.
Step 2
This step needs two inputs. One is the custom-clustering file obtained from Step 1 and the second is the input file containing the IDs that the user wants to analyze. The output is the same as the one for Ready2GO.
DISCUSSION
The GO vocabulary has been extensively explored to analyze the functions of coexpressed genes (4,5). However, despite its extended use, there are still high levels of controversy about its usefulness to validate hypotheses of gene groupings. We proposed Onto-CC as an automatic method specially suited for independent explanation/validation of gene grouping hypotheses (e.g. coexpressed genes) based on GO clusters (i.e. expression versus GO), instead of the widespread use of GO terms to conduct searches of gene expression profiles (i.e. expression + GO) (4) (see Appendix B). The clustering method used in our approach is robust enough for reproducing results independently of the organism annotation specific levels (Supplementary Figures 3–5). Experiments on the algorithm performance over GO databases of different complexities showed similar distribution of solutions (Supplementary Figure 4A and B). The reduced complexity of a database increases the number of highly specific solutions with a low support found, which indicates the presence of overlapped clusters (i.e. fuzzy clusters) caused by more general and condensed GO terms. Although runs over different complexity GO databases of the same organisms achieve small differences in the cluster's specificity evaluations, most of the best-ranked clusters recognized in the full version and the slimmed one, characterize the same genes (Supplementary Figure 4C).
Onto-CC approach reduces the uncertainty of the queries by identifying optimal conceptual clusters that combine terms from different ontologies simultaneously, as well as terms defined at different levels of specificity in the GO hierarchy. Indeed, one gene can belong to more than one cluster (37), thus, providing alternative but still optimal explanations that can generate new insights for a given problem.
The Onto-CC server using the EMO-CC conceptual clustering methodology has been successfully applied to a large inflammatory response study carried out partially at the Cell Injury and Adaptation Laboratory, Washington University School of Medicine. The obtained results have promoted the identification of novel relationships among gene expression profiles that regulate the temporal integration of the complex human inmunoinflammatory response (6,11) (Appendix B). The obtained GO substructures were validated using a high-quality hand-curated database termed Ingenuity Pathways Knowledge Base (http://www.ingenuity.com), which is, at the moment, a gold standard for metabolic pathways. We queried this database with the web-based entry tool developed by Ingenuity Pathways Analysis (IPA) (http://www.ingenuity.com). For example, by using a list of genes sharing a common gene expression behavior, the best description identified by IPA (score 45, focus genes 21) functionally corresponds to an inflammatory network Inflammatory Disease (Appendix B, Figure 5 and Tables 4 and 5). Moreover, the inflammatory disease is the prevalent function of this network with P-values between 1.15 × 10−5 and 8.83 × 103, suggesting that the given genes and the Onto-CC substructures obtained constitute a meaningful biological association.
The methodology highlight is its flexibility to integrate different sources of knowledge based on statistical tests (11), which facilitates the use of Onto-CC in combination with other sources of independent annotation such as IPA. The computational validation of the methodology used by Onto-CC, as well as its performance in comparison with other approaches typically used in GO databases is published in elsewhere (11), and briefly described in the Appendix B. The development of the server presented here has been user driven from the beginning. Its functionality is continually being updated and extended in response to requests and suggestions emerging from our core users.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENT
We would like to thank the referees for their helpful comments. This work was supported in part by the Spanish Ministry of Science and Technology under project TIN-2006-12879 and in part by The Consejeria de Innovacion, Investigacion y Ciencia de la Junta de Andalucia under project TIC-02788. Coral del Val was supported by “Programa de Retorno de Investigadores” from Junta de Andalucia, and Igor Zwir is also a senior research scientist supported by Howard Hughes Medical Institute. Funding to pay open access publication charges for this article were provided by Spanish Ministry of Science and Technology under project TIN-2006-12879.
Conflict of interest statement. None declared.
REFERENCES
- 1.Beissbarth T. Interpreting experimental results using gene ontologies. Methods Enzymol. 2006;411:340–352. doi: 10.1016/S0076-6879(06)11018-6. [DOI] [PubMed] [Google Scholar]
- 2.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khatri P, Drăghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Shahrour F, Minguez P, Tárraga J, Medina I, Alloza E, Montaner D, Dopazo J. FatiGO + : a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 2007;35:W91–W96. doi: 10.1093/nar/gkm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz S, Tainsky MA. Onto-tools, the toolkit of the modern biologist: onto-express, onto-compare, onto-design and onto-translate. Nucleic Acids Res. 2003;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, et al. Inflamm and Host Response to Injury Large Scale Collab. Res. Program. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 7.Zwir I, Huang H, Groisman EA. Analysis of differentially-regulated genes within a regulatory network by GPS genome navigation. Bioinformatics. 2005;21:4073–4083. doi: 10.1093/bioinformatics/bti672. [DOI] [PubMed] [Google Scholar]
- 8.Jonyer I, Cook DJ, Holder LB. Discovery and evaluation of graph-based hierarchical conceptual clusters. J. Mach. Learn. Res. 2001;2:19–43. [Google Scholar]
- 9.Mitchell TM. Machine Learning. 1st edn. McGraw-Hill. ISBN:0070428077; 1997. [Google Scholar]
- 10.Ruspini E, Zwir I. Automated generation fo qualitative representations of complex object by hybrid soft-computing methods. In: Pal S, Pal A, editors. Pattern Recognition: From Classical to Modern Approaches. Singapore: World Scientific Company; 2001. pp. 453–474. [Google Scholar]
- 11.Romero-Zaliz RC, Rubio-Escudero C, Cobb JP, Herrera F, Cordón O, Zwir I. A multi-objective evolutionary conceptual clustering methodology for gene annotation within structural databases: a case of study on the gene ontology database. IEEE T. Evolut. Comput. 2008 (in press). [Google Scholar]
- 12.McCue L, Thompson W, Carmack C, Ryan MP, Liu JS, Derbyshire V, Lawrence CE. Phylogenetic footprinting of transcription factor binding sites in proteobacterial genomes. Nucleic Acids Res. 2001;29:774–782. doi: 10.1093/nar/29.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Antonio A, Collado-Vides A. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Cook D, Holder L, Su S, Maglothin R, Jonyer I. Structural mining of molecular biology data. IEEE Eng. Med. Biol. 2001;4:67–74. doi: 10.1109/51.940050. [DOI] [PubMed] [Google Scholar]
- 15.Kohavi R, John GH. Wrappers for feature subset selection. Artif. Intell. 1997;97:273–324. [Google Scholar]
- 16.Arnaud MB, Costanzo MC, Skrzypek MS, Binkley G, Lane C, Miyasato SR, Sherlock G. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 2005;33:D358–D363. doi: 10.1093/nar/gki003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashurst JL, Chen CK, Gilbert JG, Jekosch K, Keenan S, Meidl P, Searle SM, Stalker J, Storey R, Trevanion S, et al. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2005;33:D459–D465. doi: 10.1093/nar/gki135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chisholm RL, Gaudet P, Just EM, Pilcher KE, Fey P, Merchant SN, Kibbe WA. dictyBase, the model organism database for Dictyostelium discoideum. Nucleic Acids Res. 2006;34:D423–D427. doi: 10.1093/nar/gkj090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM, The FlyBase Consortium FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2005;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson JD, Umayam LA, Dickinson TM, Hickey EK, ,,, , White O. The comprehensive microbial resource. Nucleic Acids Research. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eppig JT, Bult CJ, Kadin JA, Richardson JE, Blake JA, Members of the Mouse Genome Database Group The Mouse Genome Database (MGD): from genes to mice—a community resource for mouse biology. Nucleic Acids Res. 2005;33:D471–D475. doi: 10.1093/nar/gki113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ, the RGD Team The Rat Genome Database, update 2007—easing the path from disease to data and back again. Nucleic Acids Res. 2007;35:D658–D662. doi: 10.1093/nar/gkl988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387(Suppl. 6632):67–73. [PMC free article] [PubMed] [Google Scholar]
- 29.Winsor GL, Lo R, Sui SJ, Ung KS, Huang S, Cheng D, Ching WK, Hancock RE, Brinkman FS. Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res. 2005;33:D338–D343. doi: 10.1093/nar/gki047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bieri T, Antoshechkin I, Bastiani C, Blasiar D, Canaran P, Chan JC, Chen WJ, Davis P, Fiedler TJ, Girard L, et al. WormBase: new content and better access. Nucleic Acids Res. 2007;35:D506–D510. doi: 10.1093/nar/gkl818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sprague J, Clements D, Conlin T, Edwards P, Frazer K, Schaper K, Segerdell E, Song P, Sprunger B, Westerfield M. The Zebrafish information network (ZFIN): the zebrafish model organism database. Nucleic Acids Res. 2003;31:241–243. doi: 10.1093/nar/gkg027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guan SH, Zhu F. An incremental approach to genetic-algorithms-based classification. IEEE Trans. Syst. Man Cybern. B. 2005;35:227–239. doi: 10.1109/tsmcb.2004.842247. [DOI] [PubMed] [Google Scholar]
- 35.Giraud-Carrier C. A note on the utility of incremental learning. AI Commun. 2000;13:215–223. [Google Scholar]
- 36.Tavazoie S, Hughes J, Campbell M, Cho R, Church G. Systematic determination of genetic network architecture. Nat. Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 37.Gasch AP, Eisen MB. Exploring the conditional coregulation of yeast gene expression through fuzzy k-means clustering. Genome Biol. 2002;3:1–22. doi: 10.1186/gb-2002-3-11-research0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.